Abstract
Identification of host responses at the gene transcription level provides a molecular profile of the events that occur following infection. Brucella abortus is a facultative intracellular pathogen of macrophages that induces chronic infection in humans and domestic animals. Using microarray technology, the response of macrophages 4 h following B. abortus infection was analyzed to identify early intracellular infection events that occur in macrophages. Of the >6,000 genes, we identified over 140 genes that were reproducibly differentially transcribed. First, an increase in the transcription of a number of proinflammatory cytokines and chemokines, such as tumor necrosis factor alpha, interleukin-1β (IL-1β), IL-1α, and members of the SCY family of proteins, that may constitute a general host recruitment of antibacterial defenses was evident. Alternatively, Brucella may subvert newly arriving macrophages for additional intracellular infection. Second, transcription of receptors and cytokines associated with antigen presentation, e.g., major histocompatibility complex class II and IL-12p40, were not evident at this 4-h period of infection. Third, Brucella inhibited transcription of various host genes involved in apoptosis, cell cycling, and intracellular vesicular trafficking. Identification of macrophage genes whose transcription was inhibited suggests that Brucella utilizes specific mechanisms to target certain cell pathways. In conclusion, these data suggest that B. abortus can alter macrophage pathways to recruit additional macrophages for future infection while simultaneously inhibiting apoptosis and innate immune mechanisms within the macrophage, permitting intracellular survival of the bacterium. These results provide insights into the pathogenic strategies used by Brucella for long-term survival within a hostile environment.
Bacterial infections require significant interaction between host and pathogen, and the consequences of this interaction determine the outcome of infection. Host defense mechanisms often lead to rapid clearance of the pathogen, and the uptake of bacteria into macrophages is usually fatal for the bacteria. Although many bacteria elude destruction by avoiding macrophages altogether, intracellular bacteria are capable of surviving, and often replicating, inside the macrophage. Thus, for intracellular bacteria, survival and replication within phagocytic cells is the key to pathogenesis.
Brucella abortus, a gram-negative facultative intracellular bacterium and zoonotic pathogen, causes hepatitis, arthritis, and endocarditis in humans and spontaneous abortion in cattle (17). Although, the specific mechanisms of intracellular survival by Brucella are not clearly understood, bacteria often alter normal host function to avoid immune detection. Successful strategies for intracellular survival include the ability to survive in acidified membrane-bound vesicles (25, 26), alteration of macrophage apoptosis (5, 8, 14, 19), prevention of phagosome-lysosome fusion (1), and utilization of detoxification and repair mechanisms. Defining the interaction between a host cell and Brucella is crucial to understanding the infectious process.
The goal of this study was to define the transcript profile of macrophages exposed to B. abortus for 4 h, thus evaluating the early host response to this facultative intracellular bacterium. Microarray technology permits identification of the host response at the gene transcription level and can provide a molecular profile of virulence-associated responses, as well as host defense mechanisms, that occur following infection. As a consequence of infection, we identified 148 macrophage genes that were differentially transcribed in response to 4-h infection by B. abortus. Up-regulation in the transcription of proinflammatory cytokines and chemokines likely represents an antibacterial response by host cells. However, transcription of genes involved in cell cycling, apoptosis, and intracellular trafficking was decreased. The last group of genes may permit bacterial intracellular survival in macrophages. Therefore, these data provide a comprehensive foundation of early host gene expression to further understand the infectious process of B. abortus.
MATERIALS AND METHODS
Bacteria and cell line.
B.abortus strain S2308 (National Animal Disease Center, Ames, Iowa) was grown in 12- by 75-mm tubes on a shaker platform in 4 ml of brucella broth (Difco) or on plates of brucella broth containing 1.5% agar. The cultures were grown at 37°C for 3 days. The mouse macrophage cell line RAW264.7 (ATCC TIB71) was maintained at 37°C with 5% CO2 in supplemented RPMI 1640 (10% fetal bovine serum, 0.2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml) (Sigma, St. Louis, Mo.).
Cell infection.
RAW cells were plated in 75-cm2 flasks in supplemented RPMI 1640 without antibiotics 1 day prior to infection at a concentration of 6 × 106 per flask. The cells were infected for 4 h with 1 ml of 3-day B. abortus culture (multiplicity of infection, ∼100). Following infection, the cells were washed three times with phosphate-buffered saline to remove extracellular bacteria.
Target preparation for microarray analysis.
Target RNA was prepared according to protocols in the Affymetrix Gene Chip Expression Analysis technical manual (Affymetrix, Inc., Santa Clara, Calif.). Total RNA was isolated from RAW cells using the RNeasy Mini Kit (Qiagen, Valencia, Calif.) according to the manufacturer's protocol with the following modifications. Following lysis, the supernatants were centrifuged for 2 min to remove intact bacterial cells. RNA integrity was determined by gel electrophoresis. RNA was converted to double-stranded cDNA using a synthesis kit (GIBCO BRL, Rockville, Md.), except that T-7-(dT)24 oligomer (Genset Corp., La Jolla, Calif.) was used. The cDNA was phenol-chloroform extracted and ethanol precipitated prior to the performance of in vitro transcription and labeling with biotin (Enzo Diagnostics, Inc., Farmingdale, N.Y.). The labeled cRNA was fragmented in 50 mM Tris-acetate, pH 8.1, 100 mM KOAc, and 30 mM MgOAc at 94°C for 35 min.
Probe arrays.
Briefly, 16 μg of fragmented labeled cRNA was hybridized to murine U74A gene chips (Affymetrix). Washing and staining with streptavidin-phycoerythrin was done using a GeneChip Fluidics station 400 (Affymetrix). Scanning was performed with an Affymetrix GeneArray. GeneChip expression analysis software (Affymetrix) was used to scan and analyze data. The output was stored in an Excel spreadsheet. Difference calls were assigned the following values: increased, 2; marginally increased, 1; no change, 0; marginally decreased, −1; and decreased, −2. The sum of the six difference calls from the intergroup comparisons of two uninfected and three Brucella-infected arrays (2 × 3) was calculated. A sum of ≥8 or ≤−8 was the cutoff value for increase and decrease, respectively. Expressed sequence tag sequences were eliminated from the analysis.
Probes for Northern analysis.
Probes for β-actin and glyceraldehyde-3-phosphate dehydrogenase (G3PDH) were purchased from Clontech (Palo Alto, Calif.). MIP-2 probe (CTTGGCAGGGTCTTCAGGCATTGACAGCGCAGTTCACTGGCCACAACAGC) was obtained from Oligos, Etc. (Wilsonville, Oreg.). Suppressor of cytokine signaling 3 (SOCS3) and tumor necrosis factor alpha (TNF-α) probes were generated by PCR. The SOCS3 probe was amplified by PCR using the sense primer 5′ ATGGTCACCCACAGCAAGTT and the antisense primer 5′ GCCCCCAGAATAGATGTAGT (Oligos, Etc). Primers for TNF-α were obtained from Maxim Biotech, Inc. (San Francisco, Calif.), and PCR was performed according to the manufacturer's protocol. The 531-bp SOCS3 and 351-bp TNF-α PCR products were gel purified using a Qiaex II agarose gel extraction kit (Qiagen) prior to being labeled. The probes (100 ng) were labeled with horseradish peroxidase (HRP) using a North2South Direct HRP labeling, hybridization, and detection kit (Pierce, Rockford, Ill.).
Northern blot analysis.
Detection was performed using a North2South Direct HRP labeling, hybridization, and detection kit according to the manufacturer's directions. Briefly, total RNA was electrophoresed on a 1% denaturing gel, transferred onto a nitrocellulose membrane, and cross-linked by UV light. The membranes were prehybridized for 30 min, and HRP-labeled probe was added at a final concentration of 6 ng per ml. The prehybridization and hybridization temperatures were as follows: TNF-α and MIP-2, 50°C; G3PDH, 52°C; SOCS3, 54°C; and β-actin, 55°C. After a hybridization time of 90 min, the membranes were washed at hybridization temperature three times with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% sodium dodecyl sulfate for 5 min and one time with 1× SSC containing 0.1% sodium dodecyl sulfate for 5 min, followed by three washes with 2× SSC at room temperature for 5 min. Substrate development was performed according to the manufacturer's protocol (Pierce). The membranes were wrapped in plastic and exposed to film until the signal was obtained. Blots were stripped of bound probe by boiling the membrane in 0.1× SSC followed by addition of new probe to the total RNA bound to the membrane.
RT-PCR.
Total RNA was isolated using the RNeasy Mini Kit according to the manufacturer's protocol, including DNase treatment. RNA (5.0 μg) was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (M-MLV RT) (Promega, Madison, Wis.) for 1 hour at 37°C. The reverse-transcription mix was diluted 1:10 in DNase- and RNase-free water (Life Technologies, Rockville, Md.). The single-stranded cDNA was then subjected to FailSafe PCR from Epicenter (Madison, Wis.) under standard reaction conditions. Each PCR included 1× FailSafe buffer F, 0.5 to 1.0 μM sense and antisense primers for the gene of interest, 0.04 to 0.08 μM control β-actin primers (Clontech), and 5 μl of cDNA template. Sense and antisense primers for the tested genes are shown in Table 1. PCR was performed for 25 cycles at 95°C for 30 s, 62°C for 30 s, and 72°C for 60 s, followed by 72°C for 5 min. Negative controls for PCR were performed with RNA without M-MLV RT and confirmed the lack of DNA in each reaction mixture. The resultant PCR products were electrophoresed on a 1.5% agarose gel. The gel images were saved as BMP files (Foto/Analyst Archiver; Fotodyne, Hartland, Wis.) and analyzed with Un-Scan-IT Gel software (Silk Scientific Corp., Orem, Utah). Negative segment analysis was performed for each band. The correction factor was determined by dividing the digitized image of the actin band from uninfected cells by the digitized image of the actin band from infected cells.
TABLE 1.
Sense and antisense primers for RT-PCR-amplified genes
| Gene name | Forward primer | Reverse primer | Product size (bp) |
|---|---|---|---|
| Ptgs2 | 5′ GAGAGAAGGAAATGGCTGCAGAA | 5′ GGCTTCCAGTATTGAGGAGAACAGA | 193 |
| Scya4 (MIP-1β) | 5′ ATTCCTGACCAAAAGAGGCAGACA | 5′ TGGGAGAGACGCGTCCTATAAACTA | 295 |
| c-myc | 5′ GGAAGAAATTGATGTGGTGTCTGTG | 5′ TCTTGTCGTTTTCCTCCGTGTCT | 290 |
| Cish3 (SOCS3) | 5′ ATGGTCACCCACAGCAAGTT | 5′ AATCCGCTCTCCTGCAGCTT | 137 |
| Traf1 | 5′ GGGAGCCCACAATCCATGCA | 5′ TCGCTTCCACAGCTGCCTGA | 175 |
| Irg1 | 5′ CTCTGTGGTGGGGACTCTGGGAAGT | 5′ ATAGAAGGCACCGAACCCTGACCC | 256 |
| IL-1β | 5′ GTGTGGATCCCAAGCAATACCCA | 5′ CCAGCCCATACTTTAGGAAGACACAGA | 221 |
| Fcgr1 | 5′ TCCCTTTAGAGCTGTTTACCACGC | 5′ CCTCACACCAGTAGAATCCAGCATC | 234 |
| Rab3d | 5′ AAGCGAGATCCCACTGAGATGG | 5′ GTCTTGACCTTGAAGTCGATGCC | 206 |
| Map3k4 | 5′ GAACACACCGAGTCAGTCTCCACA | 5′ GCTACAGCCGAGACTGAGGTAAGG | 232 |
| Rab9 | 5′ AATAATTCTTCTTGGAGATGGTGGAGTTG | 5′ ACTAAATGTAAGCAGGCAACAGTCAGAAC | 238 |
| Mnt | 5′ AGCGGGAACGTGAGAGAGAGCA | 5′ GAGACTGCGGGGAATTGGTCAC | 210 |
| caspase 3 | 5′ CCTGGAGAAATTCAAAGGACGGG | 5′ GCATGGACACAATACACGGGATCT | 192 |
RESULTS
Effect of B. abortus infection on RAW cell gene expression.
Differential gene transcription was determined using RAW264.7 cells infected with B. abortus strain 2308. Three independent experiments were performed, and the RNA transcription levels for over 6,000 genes were determined. Transcripts for ∼40% of the probe sets were detected. Of the RAW cells infected with B. abortus, ∼3% of the expressed transcripts showed an increase in gene transcription. An equal percentage demonstrated a reduction in transcription.
Table 2 lists the genes up-regulated 4 h after Brucella infection, an early time following macrophage infection. Up-regulation of transcription occurred reproducibly in 69 genes. The majority of genes up-regulated during infection were associated with inflammation or apoptosis. The proinflammatory genes, IL-1α, Scya2 (MCP-1), and Scyb2 (MIP-2), were strongly induced following B. abortus infection, with average increases of >100-fold. Additionally, IL-1α,TNF-α, and glucocortoid-regulated inflammatory prostaglandin (Ptgs2) genes were also up-regulated, with increases of 42- and 67-fold, respectively. The up-regulated apoptotic genes included those for apoptosis inhibitors, such as Naf1, Gadd45b, A1-b, and the zinc finger protein A20, as well as the proapoptotic genes Fas, LT, Cash, and Tnfrsf1b. Changes for these genes ranged from 1.9- to >17-fold (Table 2). Other genes that were greatly increased in expression were associated with the cell membrane, e.g., the gene for a macrophage C-type lectin, which was increased >25-fold. Extracellular matrix protein gene transcription was increased, e.g., that for MMP9, a metalloproteinase (17-fold). Regulatory gene transcription was increased, e.g., that of SOCS3 increased 56-fold. Similarly, the TNF receptor-associated factor 1 gene (TRAF1) was increased 36-fold.
TABLE 2.
Macrophage gene expression induced by B. abortus infectiona
| Probe set | Accession no. | Fold increaseb | Gene symbol | Protein or gene |
|---|---|---|---|---|
| Inflammation and chemokines | ||||
| 103486 | M15131 | 188.58 | Il-1β | IL-1β |
| 94755 | M14639 | 42.97 | Il-1α | IL-1α |
| 104647 | M88242 | 67.35 | Ptgs2 | Glucocorticoid-regulated inflammatory prostaglandin |
| 101160 | X53798 | 102.88 | Scyb2 (MIP-2) | Macrophage inflammatory protein 2 |
| 94142 | M13926 | 20.55 | Csf3 | Granulocyte colony-stimulating factor |
| 102736 | M19681 | 126.23 | Scya2 | Platelet-derived growth factor-inducible protein |
| 94761 | X70058 | 8.07 | Scya7 | Small inducible cytokine A7 |
| 93858 | M33266 | 6.42 | Scyb10 | Macrophage interferon-inducible protein 10 (IP-10) |
| 94146 | X62502 | 7.95 | Scya4 (MIP-1β) | Macrophage inflammatory protein 1β |
| 102629 | D84196 | 57.98 | TNF-α | TNF-α |
| 93871 | L32838 | 8.35 | Il-1rn | IL-1 receptor antagonist IL-1rn |
| 95344 | U65747 | 6.97 | IL-13ra2 | IL-13 receptor alpha 2 |
| 102712 | X03505 | 32.30 | Saa3 | Serum amyloid A protein (SAA) |
| Cell membrane | ||||
| 92217 | U05265 | 3.73 | Gp49b | Glycoprotein 49B |
| 100325 | M65027 | 2.2 | Gp49a | Glycoprotein 49A |
| 104469 | M73748 | 5.83 | Gp38 | Glycoprotein 38 |
| 99434 | AF001036 | 2.72 | Cd83 | CD83 antigen |
| 92415 | L15435 | 3.38 | Tnfsf9 | TNF (ligand) superfamily, member 9 |
| 92962 | M83312 | 5.52 | Tnfrsf5 | TNF receptor superfamily, member 5 |
| 92730 | L07264 | 4.22 | Hegfl | Heparin binding epidermal growth factor-like growth factor |
| 96551 | AB024717 | 25.70 | Clecsf9 | Macrophage C-type lectin |
| Apoptosis | ||||
| 104755 | AJ242778 | 3.65 | Naf1 | A20-binding inhibitor of NF-κB activation |
| 102921 | M83649 | 5.02 | Fas | Fas antigen |
| 102779 | X54149 | 17.15 | Gadd45b | Myeloid differentiation primary-response gene |
| 102940 | U16985 | 6.78 | Ltb | Lymphotoxin beta |
| 103217 | Y14041 | 1.62 | Cash | CASH alpha protein |
| 99392 | U19463 | 5.87 | Tnfaip3 (A20) | TNF-induced protein 3 |
| 94928 | X87128 | 8.05 | Tnfrsf1b | TNF receptor superfamily, member 1b |
| 102914 | U23778 | 1.88 | A1-b | Hematopoietic-specific early-response A1-b protein |
| 93869 | U23781 | 2.12 | A1-d | Hematopoietic-specific early-response A1-d protein |
| Adhesion | ||||
| 103005 | X66084 | 2.95 | Cd44 | CD44 |
| 96752 | M90551 | 3.77 | ICAM1 | Intracellular adhesion molecule 1 |
| 102280 | AB006758 | 2.40 | Pcdh7 | Pcdh7 mRNA for BH-protocadherin-a |
| Cell cycle and proliferation | ||||
| 94246 | J04103 | 3.33 | Ets2 | Avian leukemia oncogene 2 |
| 102292 | U00937 | 16.33 | Gadd45a | GADD45 protein |
| 103349 | M57696 | 1.67 | Lyn | Yamaguchi sarcoma viral (v-yes-1) oncogene homologue |
| 92472 | AF099973 | 4.15 | Slfn2 | Schlafen 2 |
| 96285 | AJ001616 | 2.55 | Myadm | Mycloid-associated differentiation protein |
| Extracellular matrix | ||||
| 94147 | M33960 | 5.55 | Serpine1 | Plasminogen activator protein PAI-1 |
| 92978 | X16490 | 5.53 | Serpine2 | Plasminogen activator protein PAI-2 |
| 101561 | K02236 | 5.38 | Mt2 | Metallothionein II (MT-II) |
| 99957 | X72795 | 17.55 | MMP9 | Matrix metalloproteinase |
| Transcription | ||||
| 98427 | M57999 | 3.68 | Nfkb1 | NF-κB transcription factor |
| 92925 | M61007 | 2.08 | Cebpb | CCAAT/enhancer binding protein (C/EBP) beta |
| 102362 | U20735 | 4.35 | JunB | JunB |
| 94189 | AB011665 | 3.45 | Bazf | BcL6-associated zinc finger protein |
| 104712 | L00039 | 7.88 | c-myc | Myelocytomatosis oncogene |
| 92855 | Z50159 | 1.75 | Suil-rsl | Suppressor of initiator codon mutations-related sequence Sui1 |
| 98007 | AJ131021 | 3.83 | Rsk3 | pp90 ribosomal protein S6 kinase 3 |
| Stress | ||||
| 101995 | U40930 | 3.58 | Sqstml | Oxidative stress-induced protein |
| 96042 | L35528 | 2.43 | MnSOD | Manganese superoxide dismutase (MnSOD) gene |
| 95722 | AB013137 | 3.95 | GLRX | Glutaredoxin |
| Transport | ||||
| 103065 | M73696 | 4.70 | Slc20aI | Solute carrier family 20 |
| 102198 | AF042487 | 1.90 | Kcnn4 | Intermediate conductance potassium channel mIK1/PICK> |
| Signal transduction | ||||
| 101457 | L16956 | 2.13 | Jak2 | Janus kinase 2 |
| 93680 | D89728 | 3.03 | Stk10 | Serine-threonine kinase 10 |
| 94378 | U94828 | 3.83 | Rgs16 | Retinally abundant regulator of G-protein signaling |
| Regulatory | ||||
| 99109 | M59821 | 2.65 | ler2 | Growth factor-inducible protein (pip92) |
| 102957 | U20159 | 2.37 | SLP-76 | 76-kDa tyrosine phosphoprotein SLP-76 |
| 92232 | U88328 | 55.72 | Cish3/SOCS3 | Suppressor of cytokine signaling 3 |
| 94186 | L35302 | 36.30 | Traf1 | TNF receptor-associated factor 1 |
| Other | ||||
| 100515 | X54056 | 1.93 | PC3 | Proprotein convertase subtilisin-kexin type 3 |
| 102313 | L09737 | 2.30 | Gch | GTP cyclohydrolase 1 |
| 94085 | M34603 | 2.30 | Prg | Proteoglycan core protein |
| 102663 | X62700 | 9.70 | Plaur | Urokinase plasminogen activator |
| 98774 | L38281 | 54.33 | Irg1 | Immunoresponsive gene 1 |
| 94384 | X67644 | 9.70 | ler3 | Growth factor-inducible immediate-early gene gly96 |
| 101554 | U57524 | 6.12 | Nfkbia | I κBα |
| 100981 | U43084 | 10.97 | lfit1 | Interferon-induced protein with tetratricopeptide repeats 1 |
Difference calls were assigned the following values: increased, 2; marginally increased, 1; no change, 0; marginally decreased, −1; and decreased, −2. The sum of the six difference calls from the intergroup comparisons of two uninfected and three Brucella-infected arrays (2 × 3) was calculated. A sum of ≥8 was the cutoff value for increase determination.
Average of six (2 × 3) increase values.
Genes down-regulated during infection are shown in Table 3. The majority of genes down-regulated during infection were associated with cell cycle and proliferation or intracellular trafficking. The transcription of 22 genes involved in cell cycle proliferation or differentiation was down-regulated in the presence of B. abortus. For example, Cdc6 and Lyl1 were decreased >4-fold, and BRCA1 was decreased 6-fold. Additionally, a decrease in the transcription of nine genes involved in intracellular trafficking was observed. Members of the Rab family (Rab9 and Rab3d) and kinesin motor proteins (Kif1 and Kif4) were down-regulated, as well as genes (Gsn and Pip5k). Six genes involved in apoptosis also showed decreased transcription; one of these genes, caspase 3, had a >5-fold reduction in expression. The transcription of the other apoptotic genes, Nix, Bip3, Bad, Birc5, and Siva, was decreased ∼2-fold (Table 3). MEK kinase 4b was decreased fivefold, while transcription of other genes, e.g., that for a nuclear protein (acidic nuclear phosphoprotein 32), was decreased 9-fold.
TABLE 3.
Macrophage gene expression decreased by B. abortus infectiona
| Probe set | Accession no. | Fold decreaseb | Gene symbol | Protein or gene |
|---|---|---|---|---|
| Inflammatory cytokines and chemokines | ||||
| 102794 | Z80112 | 5.0 | Cmkar4 | lcr-1 gene |
| Cell membrane | ||||
| 101793 | X70980 | 10.2 | Fcgr1 | Fc receptor; immunoglobulin G; high-affinity I |
| 94425 | AB007599 | 1.6 | Ly86 | MD-1 |
| Apoptosis | ||||
| 93836 | AF041054 | 2.4 | Bnip3 | E1B 19K/Bcl-2-binding protein homologue (Nip3) |
| 99670 | L37296 | 2.6 | Bad | BAD protein |
| 97828 | AF033115 | 2.7 | Siva | Proapoptotic protein (Siva) gene |
| 96255 | AF067395 | 2.4 | Nix | NIX (Nix) mRNA; nuclear-gene-encoding mitochondrial protein |
| 101521 | AB013819 | 2.1 | Birc5 | TIAP mouse homologue of inhibitor of apoptosis |
| 98436 | U54803 | 5.4 | caspase 3 | Caspase 3; apoptosis-related cysteine protease |
| Intracellular trafficking | ||||
| 97415 | M89777 | 4.3 | Rab3d | GTP-binding protein (Rab3D) mRNA |
| 95516 | AB027290 | 3.0 | Rab9 | SID 99 mRNA for small GTP-binding protein |
| 102221 | AJ002306 | 2.7 | Syngr1 | Synaptogyrin 1b |
| 93750 | J04953 | 1.7 | Gsn | Gelsolin |
| 102318 | X86000 | 2.9 | Siat8d | N-Glycan alpha 2,8-sialyltransferase |
| 104644 | D12646 | 1.8 | Kif4 | Kinesin heavy-chain, member 4 |
| 99541 | AJ223293 | 2.2 | Kif11 | Kinesin-related mitotic motor protein |
| 101109 | U43512 | 3.3 | Dag1 | Dystroglycan 1 |
| 98428 | AJ246002 | 2.0 | Spg4 | Spastin protein orthologue (Spast gene) |
| 101865 | AB009615 | 2.5 | Pip5k2a | Type II phosphatidylinositolphosphate kinase alpha |
| Adhesion | ||||
| 99577 | M57647 | 4.1 | KitI | Mouse mast cell growth factor (MGF) |
| 95016 | D50086 | 1.9 | Nrp | Neuropilin |
| Cell cycle, differentiation, proliferation | ||||
| 92481 | AF086905 | 2.9 | Chk2 | Protein kinase Chk2 (Chk2) |
| 103821 | AJ223087 | 4.4 | Cdc6 | Cdc6-related protein |
| 93666 | M64360 | 2.5 | Lmo2 | LIM only |
| 100467 | X57687 | 4.7 | Lyl1 | LYL gene |
| 103001 | U43836 | 1.9 | Vegfb | VEGF-related factor mvrf186 precursor mRNA |
| 93319 | U20238 | 3.8 | Rasa3 | GTPase-activating protein GAPIII |
| 101027 | AF069051 | 1.7 | PTTG | Pituitary tumor transforming gene protein |
| 101484 | U73039 | 2.0 | Nbr1 | Next to Brca 1 |
| 102976 | U32446 | 6.1 | BRCA1 | Breast cancer 1 |
| 104476 | U27177 | 3.4 | Rb11 | Retinoblastoma-like 1 (p107) |
| 99076 | U09504 | 2.9 | Thra | Thyroid hormone receptor alpha |
| 93356 | D26091 | 1.6 | Mcmd7 | mCDC47 |
| 92262 | AF012923 | 2.3 | Wig1 | p53-inducible zinc finger protein (Wig-1) |
| 99564 | D87908 | 1.9 | Np95 | Nuclear protein np95 |
| 97963 | D11374 | 3.2 | Spa1 | Signal-induced proliferation-associated gene 1 |
| 99632 | U83902 | 1.8 | Mad211 | Mitotic checkpoint component Mad2 |
| 93099 | U01063 | 2.4 | Plk | Polo-like kinase homologue |
| 99532 | D78382 | 2.1 | Tob1 | Tob family |
| 97468 | AB025409 | 1.7 | CksI | sid1334p |
| 92210 | AF004326 | 2.5 | Agpt2 | Angiopoietin 2 |
| 100427 | U37465 | 3.0 | Ptpro | Protein tyrosine phosphatase; receptor type; O |
| 103057 | AF024570 | 2.8 | Pold1 | DNA polymerase delta 1; catalytic domain |
| Transcription | ||||
| 94698 | X59421 | 2.4 | FliI | Friend leukemia integration 1 |
| 94296 | AF043220 | 2.5 | Gtf2i | TFII-I protein short-form mRNA; alternatively spliced |
| 98122 | AF074600 | 2.5 | Lmo4 | LIM domain transcription factor LMO4 |
| 92300 | Y07609 | 5.7 | Mnt | Max-binding protein |
| 104591 | L13171 | 3.1 | Mefc2 | Myocyte enhancer factor 2C |
| 99602 | AF064088 | 2.1 | Tieg | Transcription factor GIF mRNA |
| 102963 | L21973 | 3.7 | E2F-1 | E2F transcription factor 1 |
| Stress | ||||
| 94897 | D87896 | 1.6 | Gpx4 | Glutathione peroxidase 4 |
| 102792 | U55040 | 1.8 | Ung | Uracil-DNA glycosylase (ung) gene; exon 1 |
| 98398 | U22262 | 1.8 | Apobec1 | Apolipoprotein B editing complex 1 |
| 98071 | X77731 | 2.5 | Dck | Deoxycytidine kinase |
| 97327 | L26320 | 1.9 | Fen1 | Flap structure-specific endonuclease 1/PICK> |
| Transport | ||||
| 102892 | U65592 | 2.3 | Kcnab2 | K+ channel beta 2 subunit mRNA |
| Signal transduction | ||||
| 97411 | L11316 | 2.0 | Ect2 | Ect2 oncogene |
| 103070 | AB018194 | 3.1 | Ptpns1 | BIT |
| 104272 | U85608 | 5.4 | Map3k4 | MEK kinase 4b (MEKK4b) |
| Regulatory | ||||
| 101966 | AF037206 | 1.9 | Rnf13 | RING zinc finger protein (Rzf) |
| 94061 | M13018 | 1.8 | Crip | Cysteine-rich intestinal protein |
| 92975 | L14543 | 2.4 | Sh3bp2 | SH3 binding protein 3BP2 |
| Other | ||||
| 93320 | AF017175 | 8.4 | Cpt1a | Carnitine palmitoyltransferase 1; liver |
| 92608 | D88793 | 1.8 | Csrp | Cysteine-rich protein |
| 93582 | AF080580 | 1.7 | Coq7 | CLK-1 (clk-1) |
| 94324 | U49878 | 1.8 | Hmgcl | hydroxy-3-methylglutaryl-coenzyme A lyase |
| 96310 | L07508 | 1.7 | Mbp | Golli-mpb |
| 96887 | Y08702 | 1.7 | Np15.6 | Neuronal protein 15.6 |
| 98989 | AF057368 | 2.5 | Dhcr7 | 7-Dehydrocholesterol reductase |
| 96110 | U3196 | 3.7 | Cbr1 | 6:Carbonyl reductase |
| 93908 | X16670 | 2.1 | Ccr4 | Type IIB intracisternal A-particle (IAP) element-encoding integrase |
| 93372 | U73478 | 9.1 | Anp32 | Acidic nuclear phosphoprotein 32 |
| 100323 | Z23077 | 1.9 | Samdc | S-Adenosylmethionine decarboxylase 3 |
| 100596 | M32032 | 4.7 | Selenbp1 | Selenium binding protein 1 |
| 101104 | AB001990 | 2.2 | dcra | Dcra |
| 100978 | U62105 | 1.9 | FSHD | FSHD region gene 1 |
| 103032 | AF038008 | 1.5 | TPST-1 | Tyrosylprotein sulfotransferase-1 |
| 94815 | X13586 | 3.3 | Bpgm | 2,3-Bisphosphoglycerate mutase |
| 96081 | X60980 | 6.1 | TK | TK gene encoding thymidine kinase |
Difference calls were assigned the following values: increased, 2; marginally increased, 1; no change, 0; marginally decreased, −1; and decreased, −2. The sum of the six difference calls from the intergroup comparisons of two uninfected and three Brucella-infected arrays (2 × 3) was calculated. A sum of ≤−8 was the cutoff value for decrease determination.
Average of six (2 × 3) decrease values.
Confirmation of array data.
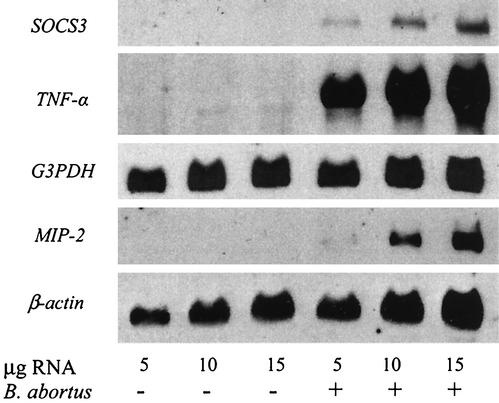
To further validate the gene array results, Northern blot analysis or RT-PCR was performed on selected genes. For Northern blot analysis, total RNA from RAW cells infected for 4 h with B. abortus was isolated, and 5, 10, and 15 μg were loaded onto a 1% formaldehyde gel. Transcription of the MIP-2, TNF-α, and SOCS3 genes was assessed. With uninfected RAW cells, no MIP-2, TNF-α, or SOCS3 transcription was detected. However, in the presence of B. abortus, all three transcripts were present (Fig. 1). The eukaryotic housekeeping genes, the β-actin gene and G3PDH, were used as references for comparison of gel loadings.
FIG. 1.
Northern blot analysis of SOCS3, TNF-α, and MIP-2 transcription in RAW264.7 macrophages infected with B. abortus or uninfected. Total RNA was isolated from RAW cells infected for 4 h with B. abortus and compared to RNA from uninfected cells. RNA (5, 10, or 15 μg) was separated by denaturing gel electrophoresis and transferred to a nitrocellulose membrane. The blots were hybridized with probes to SOCS3, TNF-α, and MIP-2. Probes to the housekeeping genes G3PDH and β-actin were used as controls.
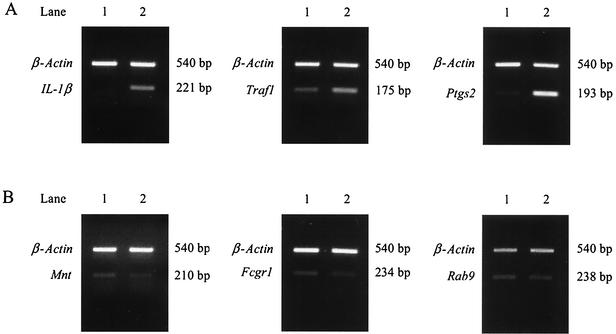
RT-PCR was performed for 13 genes. All reactions were done with and without RT (data not shown) and with actin primers. No bands were observed in reactions without RT, confirming the lack of DNA in RNA samples. Transcription of β-actin gene was used as a positive control and to ensure equal amounts of cDNA in each reaction and that the PCR products were equally loaded onto the gel. RT-PCRs were performed at least twice for each gene. Representative gels are shown in Fig. 2. Increased transcription of IL-1α, Traf1, and Ptgs2 (Fig. 2A, lane 2) and decreased transcription of Mnt, Fcgr1, and Rab9 (Fig. 2B, lane 2) was observed, as well as equal concentrations of the β-actin product. Gel images were digitized, and the percent change in transcription between infected and uninfected macrophage RNA was determined. As shown in Table 4, Traf, SOCS3, Ptgs2, Irg, MIP-1α, c-myc, and IL-1α were up-regulated during infection. The percent change ranged from 17 to >200%. Six genes, Map3k4, Rab3d, Fcgr1, Rab9, Mnt, and caspase 3, exhibited decreased transcription with B. abortus infection. These results confirm the gene array data.
FIG. 2.
RT-PCR analysis of selected gene transcription in RAW264.7 macrophages infected with B. abortus or uninfected. Total RAW cell RNA from uninfected (lanes 1) cells or 4 h postinfection with B. abortus (lanes 2) was reverse transcribed with M-MLV RT, and PCR was performed. Transcription of up-regulated (A) and down-regulated (B) genes is shown. β-Actin (540 bp) was included in all reactions to verify equal cDNA concentrations in the PCR and on the gel.
TABLE 4.
Changes in gene expression detected by RT-PCR
| Gene | Digitized counts
|
% Changeb | |
|---|---|---|---|
| Without B. abortus | With B. abortusa | ||
| IL-1β | 25,787 | 59,434 | 130 |
| Ptgs2 | 51,368 | 155,445 | 203 |
| MIP-1β/Scya4 | 222,869 | 308,561 | 38 |
| c-myc | 115,055 | 215,706 | 88 |
| Cish3/SOCS3 | 75,038 | 168,048 | 124 |
| Traf1 | 448,160 | 522,370 | 17 |
| Irg1 | 135,260 | 239,045 | 77 |
| Fcgr1 | 51,011 | 41,805 | −18 |
| caspase 3 | 32,502 | 29,505 | −9 |
| Rab3d | 16,429 | 11,814 | −28 |
| Rab9 | 42,724 | 37,378 | −13 |
| Mnt | 59,939 | 51,396 | −14 |
| Map3k4 | 6,092 | 4,772 | −22 |
Digitized image of band from infected macrophages was normalized to uninfected band by calculating the correction factor. Correction factors ranged from 0.95 to 1.03.
Percent change was calculated using the formula percent change = (value for B. abortus treated − value for not treated/value for not treated) × 100.
DISCUSSION
Although the entry of pathogens into macrophages is usually fatal, Brucella cells not only survive but replicate within these hostile cells (16). Macrophages respond to pathogens by producing cytokines, eliciting an inflammatory response, and inducing the death of the bacteria (4, 8, 28). Pathogens like Brucella have developed sophisticated evasion strategies, often utilizing normal host cell functions to avoid destruction (6, 20, 25). To better understand the complex interaction between the host cell and B. abortus, we have analyzed the differential transcription of >6,000 murine genes following a 4-h B. abortus infection of RAW264.7 macrophages. At 4 h, transcription of host genes would likely be activated as an early response to the intracellular infection, and likewise, the bacteria might influence the transcription of host genes at this early period of infection to help ensure their intracellular survival.
Inflammation is a powerful protective mechanism coordinated and controlled by cytokines and chemokines. Increases in macrophage cytokines have been detected in gene array experiments using other gram-negative intracellular bacteria, such as Salmonella enterica serovar Typhimurium (27) and Listeria monocytogenes (2). Similar up-regulation of inflammation-associated genes was observed during B. abortus infection. Up-regulation of transcription in B. abortus-infected cells was verified by RT-PCR with five genes, Ptgs2, MIP2, IL-1β, MIP-1α, and TNF-α. Additionally, our results confirm reports that B. abortus up-regulates TNF-α in mouse macrophages (10). Thus, B. abortus-infected macrophages mount a powerful inflammatory response in an effort to clear this pathogen. Alternatively, the influx of inflammatory cells to the site of infection may provide additional host cells for B. abortus to infect.
Sustained or excessive production of inflammatory cytokines can have damaging consequences. A strong inflammatory response can enhance the invasiveness of some bacteria by increasing tissue destruction, permitting bacterial dissemination (29). To counterbalance inflammatory cytokines, anti-inflammatory cytokines and/or inhibitors of signal transduction are produced. The balance between pro- and anti-inflammatory signals may influence the outcome of disease (34). Anti-inflammatory cytokines include interleukin 10 (IL-10), transforming growth factor β, and IL-1 receptor antagonist (IL-1ra) (12, 23). Transcription of IL-1ra was increased in the presence of B. abortus. During Brucella infection, anti-inflammatory signals may decrease the potentially damaging effects of proinflammatory cytokines on host tissue.
Alterations in cell surface and adhesion molecules may facilitate bacterial clearance or macrophage infection. For example, up-regulation of the metalloproteinase MMP9 may facilitate macrophage-bacterium contact through extracellular matrix digestion, while expression of macrophage C-type lectin on the macrophage surface may serve to enhance the uptake of bacteria (31).
Cell function depends on multiple signaling pathways that control the decision to proliferate, differentiate, or initiate apoptosis (33). Disruption of these pathways by pathogens leads to alterations in both proliferation and cell death. Central to the control of cell proliferation are the retinoblastoma (Rb) and p53 genes. p53 inhibits the proliferation of damaged cells, thus inhibiting cell cycle progression and inducing apoptosis. Also involved in the p53 signaling pathway are Chk2 kinase, which triggers the p53 pathway, and Wig-1, which is induced in human cells following DNA damage (13). Transcription factor E2F1 activates the transcription of genes involved in cell cycle progression, as well as DNA synthesis. However, the association of E2F1 with Rb prevents cell proliferation. Furthermore, this E2F/Rb pathway interacts with pathways that control apoptosis. As with the E2F/Rb and p53 pathways, c-myc influences both proliferation and apoptosis pathways. Down-regulation of cell cycle genes during B. abortus infection may permit cell cycle progression and inhibition of apoptosis.
Stress also affects cell cycle progression. Specifically, Lyn and GADD45a are induced in response to DNA damage and function to prevent proliferation of damaged cells (9, 30). JunB represses cyclin D transcription (24), thereby arresting cell cycle progression. An increase in mRNA levels by stress-associated genes may block macrophage proliferation yet allow intracellular Brucella organisms to multiply. Findings with these genes appear contradictory to those with Rb, Wig, Chk2, and c-myc. However, one set of genes may be the result of the host response rather than the influence of the pathogen. Additional experiments will be necessary to identify the contributions of host and pathogen.
Apoptosis plays a significant role in regulating the pathogenesis of infection. To survive, intracellular pathogens may induce and/or block apoptosis. The main components of the apoptotic process include the surface receptors (death domain receptors, FAS, and TNF receptors), cysteine proteases (caspases), and the Bcl-2-like family of proteins. The family of death receptors includes Fas cell surface receptor, FADD, TNFR1, and TRAIL. The binding of Fas ligand to Fas antigen induces apoptosis. Although an increase in expression of Fas was seen in our experiments, no change in transcription was observed with the Fas-associated (FADD-like) protein or TRAIL gene. Furthermore, although the transcription of TNFR2, a death domain gene that can trigger apoptosis, was increased, no increase was observed in its ligand, TRADD.
Several pathogenic organisms alter the caspase pathway, which is important in apoptosis. Shigella (15) and Salmonella (14) utilize similar mechanisms to induce apoptosis via the caspase-1 pathway. Legionella-induced apoptosis of macrophages utilizes caspase 3 (7), as does Mycobacterium (3). Chlamydia protects infected cells against apoptosis early in infection but induces apoptosis during late stages of infection (5, 22). We determined that caspase-3 was down-regulated in B. abortus infection at 4 h. Prevention of apoptosis during infection would permit the survival and replication of intracellular Brucella organisms. Studies are under way to examine the roles of caspase proteins during Brucella infection in greater detail.
The Bcl-2 family is composed of prosurvival (Bcl-2 and A1) and proapoptotic (Bad, Bax, Bak, and Bid) proteins. Our results indicate that the A1 gene is up-regulated in B. abortus infection, similar to a report about Brucella suis in which up-regulation of the A1 gene blocked apoptosis (11). Additionally, genes encoding two proapoptotic Bcl-2 proteins, Bnip-3 and Bnip-31, were down-regulated, as well as Bad. Thus, many of the genes that function in proapoptotic pathways are down-regulated in B. abortus infection of macrophages, while the prosurvival gene A1 is up-regulated. This alteration of apoptosis may result in the increased survival of Brucella within the macrophage.
A key defense mechanism of the host cell during infection is the production of reactive oxygen. The gene encoding the oxidative stress-induced protein, Sqstml, was up-regulated, suggesting the generation of reactive oxygen species. Manganese superoxide dismutase, a reactive-oxygen scavenger, was up-regulated, as well as the metallothionein gene (MT2), which plays a role in the detoxification of heavy metals and scavenging of free radicals. Metallothionein may alter the induction of apoptosis due to oxidative stress (18).
Transcription was down-regulated in nine genes whose proteins are involved in intracellular trafficking. With Salmonella, interference in intracellular membrane trafficking leads to the inhibition of phagosome-lysosome fusion (32). The down-regulation of two intracellular-trafficking genes, those for Rab3d and Rab9, was verified by RT-PCR. The Rab proteins are key regulators of membrane trafficking and function in tethering and vesicle movement. Specifically, Rab9 appears to mediate vesicle transport from endosomes to the trans-Golgi. The modulation of genes involved in intracellular trafficking may serve to increase survival of B. abortus within macrophages by redirecting vesicular movement and preventing phagosome-lysosome fusion.
In summary, determining the macrophage genes that are transcribed during the early stages of Brucella infection can establish new hypotheses regarding the molecular pathogenesis of brucellosis. Using the RAW264.7 macrophage cell line, common host defense mechanisms, as well as pathogen-specific manipulations of those defenses, were identified. First, the increase in a number of proinflammatory cytokines was evident, similar to findings with human macrophages activated by several gram-negative and -positive bacteria (21). These proinflammatory components may constitute a general host recruitment of antibacterial defenses. However, Brucella may subvert newly arriving macrophages for additional intracellular infection. Second, mRNA levels of receptors and cytokines associated with antigen presentation, e.g., IL-12p40 and major histocompatibility complex class II, were not evident at this 4-h period of infection. Third, Brucella infection inhibited the mRNA levels of a number of host genes involved in apoptosis, cell-cycling, and intracellular-trafficking mechanisms among cytoplasmic compartments. Decreased mRNAs of specific genes suggests that Brucella utilizes specific mechanisms to alter the expression of certain genes. Our results provide a unique opportunity to select particular host pathways to determine how intracellular Brucella organisms can alter such pathways to ensure a bacterial advantage for intracellular survival. Pathogen-specific manipulations of host pathways have practical applications in designing vaccines and therapies that engage the innate immune system in a targeted fashion. The DNA microarray expression data in the present study provide a foundation for further understanding the long-term survival of Brucella in an immunocompetent host.
Acknowledgments
This work was supported by grants RO1 AI48490 from the National Institutes of Health and US.-2968-98C from the Binational Agricultural Research and Development (BARD) Fund.
We thank Bruce Jarvis and Gireesh Rajashekara for valuable discussions and critical review of the manuscript. Appreciation is expressed to Sandra Splinter for probe array hybridization and scanning.
Editor: B. B. Finlay
REFERENCES
- 1.Buchmeier, N. A., and F. Heffron. 1991. Inhibition of macrophage phagosome-lysosome fusion by Salmonella typhimurium. Infect. Immun. 59:2232-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen, P., M. Bouaboula, M. Bellis, V. Baron, O. Jbilo, C. Poinot-Chazel, S. Galiegue, E. H. Hadibi, and P. Casellas. 2000. Monitoring cellular responses to Listeria monocytogenes with oligonucleotide arrays. J. Biol. Chem. 275:11181-11190. [DOI] [PubMed] [Google Scholar]
- 3.Duan, L., H. Gan, J. Arm, and H. G. Remold. 2001. Cytosolic phospholipase A2 participates with TNF-alpha in the induction of apoptosis of human macrophages infected with Mycobacterium tuberculosis H37Ra. J. Immunol. 166:7469-7476. [DOI] [PubMed] [Google Scholar]
- 4.Eckmann, L., and M. F. Kagnoff. 2001. Cytokines in host defense against Salmonella. Microb. Infect. 3:1191-1200. [DOI] [PubMed] [Google Scholar]
- 5.Fan, T., H. Lu, H. Hu, L. Shi, G. A. McClarty, D. M. Nance, A. H. Greenberg, and G. Zhong. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187:487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frenchick, P. J., R. J. Markham, and A. H. Cochrane. 1985. Inhibition of phagosome-lysosome fusion in macrophages by soluble extracts of virulent Brucella abortus. Am. J. Vet. Res. 46:332-335. [PubMed] [Google Scholar]
- 7.Gao, L. Y., and Y. Abu Kwaik. 1999. Activation of caspase 3 during Legionella pneumophila-induced apoptosis. Infect. Immun. 67:4886-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao, L. Y., and Y. A. Kwaik. 2000. The modulation of host cell apoptosis by intracellular bacterial pathogens. Trends Microbiol. 8:306-313. [DOI] [PubMed] [Google Scholar]
- 9.Grishin, A. V., O. Azhipa, I. Semenov, and S. J. Corey. 2001. Interaction between growth arrest-DNA damage protein 34 and Src kinase Lyn negatively regulates genotoxic apoptosis. Proc. Natl. Acad. Sci. USA 98:10172-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross, A., S. Spiesser, A. Terraza, B. Rouot, E. Caron, and J. Dornand. 1998. Expression and bactericidal activity of nitric oxide synthase in Brucella suis-infected murine macrophages. Infect. Immun. 66:1309-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross, A., A. Terraza, S. Ouahrani-Bettache, J. P. Liautard, and J. Dornand. 2000. In vitro Brucella suis infection prevents the programmed cell death of human monocytic cells. Infect. Immun. 68:342-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton, T. A., Y. Ohmori, J. M. Tebo, and R. Kishore. 1999. Regulation of macrophage gene expression by pro- and anti-inflammatory cytokines. Pathobiology 67:241-244. [DOI] [PubMed] [Google Scholar]
- 13.Hellborg, F., W. Qian, C. Mendez-Vidal, C. Asker, M. Kost-Alimova, M. Wilhelm, S. Imreh, and K. G. Wiman. 2001. Human wig-1, a p53 target gene that encodes a growth inhibitory zinc finger protein. Oncogene 20:5466-5474. [DOI] [PubMed] [Google Scholar]
- 14.Hersh, D., D. M. Monack, M. R. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA 96:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilbi, H., J. E. Moss, D. Hersh, Y. Chen, J. Arondel, S. Banerjee, R. A. Flavell, J. Yuan, P. J. Sansonetti, and A. Zychlinsky. 1998. Shigella-induced apoptosis is dependent on caspase-1 which binds to IpaB. J. Biol. Chem. 273:32895-32900. [DOI] [PubMed] [Google Scholar]
- 16.Jones, S. M., and A. J. Winter. 1992. Survival of virulent and attenuated strains of Brucella abortus in normal and gamma interferon-activated murine peritoneal macrophages. Infect. Immun. 60:3011-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madkour, M. M. 2001. Madkour's brucellosis. Springer, New York, N.Y.
- 18.McAleer, M. F., and R. S. Tuan. 2001. Metallothionein protects against severe oxidative stress-induced apoptosis of human trophoblastic cells. In Vitr. Mol. Toxicol. 14:219-231. [DOI] [PubMed] [Google Scholar]
- 19.Monack, D. M., B. Raupach, A. E. Hromockyj, and S. Falkow. 1996. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. USA 93:9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naroeni, A., N. Jouy, S. Ouahrani-Bettache, J. P. Liautard, and F. Porte. 2001. Brucella suis-impaired specific recognition of phagosomes by lysosomes due to phagosomal membrane modifications. Infect. Immun. 69:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nau, G. J., J. F. Richmond, A. Schlesinger, E. G. Jennings, E. S. Lander, and R. A. Young. 2002. Human macrophage activation programs induced by bacterial pathogens. Proc. Natl. Acad. Sci. USA 99:1503-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ojcius, D. M., P. Souque, J. L. Perfettini, and A. Dautry-Varsat. 1998. Apoptosis of epithelial cells and macrophages due to infection with the obligate intracellular pathogen Chlamydia psittaci. J. Immunol. 161:4220-4226. [PubMed] [Google Scholar]
- 23.Opal, S. M., and V. A. DePalo. 2000. Anti-inflammatory cytokines. Chest 117:1162-1172. [DOI] [PubMed] [Google Scholar]
- 24.Passegue, E., and E. F. Wagner. 2000. JunB suppresses cell proliferation by transcriptional activation of p16(INK4a) expression. EMBO J. 19:2969-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porte, F., J. P. Liautard, and S. Kohler. 1999. Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in murine macrophages. Infect. Immun. 67:4041-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rathman, M., M. D. Sjaastad, and S. Falkow. 1996. Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect. Immun. 64:2765-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenberger, C. M., M. G. Scott, M. R. Gold, R. E. Hancock, and B. B. Finlay. 2000. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J. Immunol. 164:5894-5904. [DOI] [PubMed] [Google Scholar]
- 28.Russell, D. G. 1995. Of microbes and macrophages: entry, survival and persistence. Curr. Opin. Immunol. 7:479-484. [DOI] [PubMed] [Google Scholar]
- 29.Sansonetti, P. J., J. Arondel, J. M. Cavaillon, and M. Huerre. 1995. Role of interleukin-1 in the pathogenesis of experimental shigellosis. J. Clin. Investig. 96:884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tong, T., W. Fan, H. Zhao, S. Jin, F. Fan, P. Blanck, I. Alomo, B. Rajasekaran, Y. Liu, N. J. Holbrook, and Q. Zhan. 2001. Involvement of the MAP kinase pathways in induction of GADD45 following UV radiation. Exp. Cell Res. 269:64-72. [DOI] [PubMed] [Google Scholar]
- 31.Tsuiji, M., M. Fujimori, Y. Ohashi, N. Higashi, T. M. Onami, S. M. Hedrick, and T. Irimura. 2002. Molecular cloning and characterization of a novel mouse macrophage C-type lectin, mMGL2, which has a distinct carbohydrate specificity from mMGL1. J. Biol. Chem. 277:28892-28901. [DOI] [PubMed] [Google Scholar]
- 32.Uchiya, K., M. A. Barbieri, K. Funato, A. H. Shah, P. D. Stahl, and E. A. Groisman. 1999. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 18:3924-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Underhill, D. M., and A. Ozinsky. 2002. Phagocytosis of microbes: complexity in action. Annu. Rev. Immunol. 20:825-852. [DOI] [PubMed] [Google Scholar]
- 34.van Dissel, J. T., P. van Langevelde, R. G. Westendorp, K. Kwappenberg, and M. Frolich. 1998. Anti-inflammatory cytokine profile and mortality in febrile patients. Lancet 351:950-953. [DOI] [PubMed] [Google Scholar]