Abstract
Pneumococcal surface protein A (PspA) can elicit protection against Streptococcus pneumoniae in mouse infection models. PspA is classified by serology and amino acid sequence into two major families that are divided by sequence into five clades. The most variable portion of the molecule is the α-helical domain, which comprises the N-terminal half of PspA. Prior studies of a family 1 PspA protein observed that protective antibodies are reactive with epitopes in the α-helical domain and that most cross-protective epitopes mapped to the 108 most C-terminal amino acids of the α-helical region. In these studies, we have used six overlapping recombinant fragments of family 2, clade 3 PspA/EF3296 to map the protection-eliciting regions of its α-helical domain. The three fragments, which included the 104 most C-terminal amino acids of the α-helical domain (314 to 418), could each elicit protection against EF3296. A fragment comprising amino acids 75 to 305 failed to elicit significant protection. A fragment containing amino acids 1 to 115 elicited protection against EF3296 in BALB/c mice but not in CBA/N mice. All three fragments containing amino acids 314 to 418 were able to elicit cross-protection against pneumococci expressing PspA proteins of clades 2, 3, 4, and 5. Cross-protection elicited by these three fragments was easier to demonstrate in CBA/N mice than in BALB/c mice. The 1-to-115 fragment, however, elicited some cross-protection against clades 2 and 4 in BALB/c mice but not in CBA/N mice. These studies provide support for the importance of the C-terminal 104 and N-terminal 115 amino acids of the α-helical region of PspA in the elicitation of cross-protection.
Streptococcus pneumoniae is a common cause of respiratory tract infections, otitis media, sepsis, and meningitis in young children and in the elderly. It is a major cause of mortality in developing parts of the world and the major cause of hospital visits among children in the United States (4, 32, 39).
The 23-valent polysaccharide vaccine and the recently developed 7-valent polysaccharide-protein conjugate vaccine exhibit incomplete protection against carriage of and otitis media and bacteremic disease caused by nonvaccine types; this has made it important to examine other vaccine candidates (21, 22, 37). PspA and pneumolysin have been the most extensively examined pneumococcal proteins used to elicit protective immunity in animal models (11, 34). A number of other protection-eliciting pneumococcal proteins have also been described (6, 7, 17, 24, 27, 28, 36, 40).
PspA is present on all pneumococci (19, 20) and is serologically variable, cross-reactive (25, 29, 33), and cross-protective (14). PspA is made up of three major amino acid sequence domains. The choline-binding domain at the C terminus attaches the protein to the cell surface (43). Upstream of this domain is the proline-rich domain, which is thought to span the cell wall and capsule layer (14, 42). N terminal to the proline-rich domain is the α-helical domain, which is exposed on the bacterial surface and is thought to form an antiparallel coiled-coil structure (23, 26, 42) reminiscent of many other fibrillar surface proteins on gram-positive bacteria.
Most of the epitopes detected by a panel of protective monoclonal antibodies to PspA/Rx1 were mapped to the C-terminal 119 amino acids of the α-helical region of PspA/Rx1. Overlapping fragments that contained the 108 C-terminal amino acids of the PspA/Rx1 α-helical region were found to elicit protection against strains of different capsular types (16, 29, 38). It was also observed that the N-terminal 115 amino acids of PspA/Rx1 could elicit protection, but the ability of the fragment to elicit cross-protection was not examined (13). The ability of fragments from the middle of the α-helical domain of PspA/Rx1 to elicit protection was also not examined.
The role of the approximately 108 C-terminal amino acids of the α-helical region in eliciting cross-protection makes this region important for the characterization of PspA diversity with regard to its use in vaccines. Two additional features of this region further contribute to its importance for characterizing PspA diversity. (i) The relationship between sequences of different PspA proteins in this region was observed to be similar to that of the relationship between the entire α-helical sequences of the same PspA proteins (25). (ii) The dendrograms based on the relationships between differences in sequence for the ∼108 C-terminal amino acids of the α-helical domain of PspA were found to yield dendrograms that were more statistically significant than those based on other regions of the α-helical domain (25). For these reasons, the ∼108 C-terminal amino acids in the α-helical domain have been designated the CDR (clade-defining region) of PspA (25).
On the basis of the amino acid sequence diversity in the CDR, PspA proteins fall into two major sequence families. Family 1 is composed of clades 1 and 2; family 2 is composed of clades 3, 4, and 5. The amino acid sequences of the PspA proteins of families 1 and 2 can differ by as much as 60% in the CDR (25).
The classification of PspA proteins into families and clades on the basis of CDR structure could assist in the formulation of a PspA vaccine containing different PspA proteins if it were known that the CDR of PspA proteins in family 2 was also important in the elicitation of cross-protection. In this study, we have mapped the protection-eliciting regions of a family 2 PspA protein by comparing the protection elicited by overlapping fragments of PspA/EF3296. We chose strain EF3296 because prior studies demonstrated that it is particularly difficult to protect against, thus making it particularly well suited as a challenge strain for resolving differences between the protective capacities of different PspA fragments.
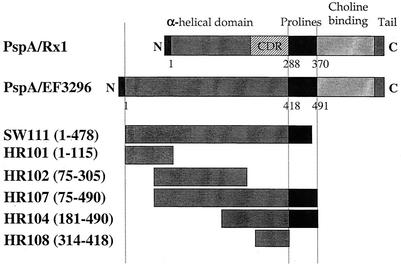
In PspA/EF3296, the α-helical domain extends from amino acid 1 to amino acid 418. On the basis of homology to PspA/Rx1, the CDR of PspA/EF3296 has been considered to extend from amino acid 314 to amino acid 418 (25). Recombinant fragments composed of overlapping segments of the α-helical domain were expressed and used to immunize BALB/c and CBA/N mice, which were then challenged with one of a panel of strains expressing PspA proteins of the five major PspA clades.
MATERIALS AND METHODS
Reagents.
Nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate were from Fisher Scientific (Atlanta, Ga.). Streptavidin-alkaline phosphatase (AP) and AP-conjugated goat anti-mouse and AP-conjugated goat anti-rabbit antibodies were from Southern Biotechnology Associates (Birmingham, Ala.). Bacto Todd-Hewitt medium and yeast extract were from Difco Laboratories (Detroit, Mich.). Protein markers and ready gels were from Bio-Rad (Hercules, Calif.)
Anti-PspA monoclonal antibodies were produced as previously described (29). Anti-PspA family 2 serum was produced as previously described (19, 23). PC3.1 reactive with EF3296 was a gift from Aventis Inc., Toronto, Ontario, Canada (unpublished data).
Bacteria.
The pneumococcal strains were stored at −80°C in 12% glycerol (1), transferred to blood agar plates, and incubated at 37°C in a 5% CO2 atmosphere overnight. Colonies grown on blood agar were used to inoculate liquid growth medium (Todd-Hewitt medium containing 0.5% yeast extract). Upon reaching late log phase, the bacteria were harvested by centrifugation at 1,500 × g for 15 min and suspended in sterile 60 mM phosphate-buffered saline (PBS; pH 7.2). The bacterial concentration was adjusted by measuring absorbance at 600 nm and confirmed by viable counts.
PspA and PspA fragments.
Full-length PspA was purified from S. pneumoniae EF3296 as previously described (13, 43). Fragment SW111 of PspA/EF3296, constituting amino acids 1 to 478, was kindly provided by Aventis Inc. PspA fragments were produced from cloned, PCR-amplified DNA as follows. DNAs encoding protein fragments HR101 (primer pair ABW23-LSM12), HR102 (primer pair HR10-HR11), HR104 (primer pair HR12-HR14), HR107 (primer pair HR10-HR14), and HR108 (primer pair HR19-HR20) were amplified from S. pneumoniae EF3296. The primers used for PCR have been described earlier (23), except for HR19 (5′-AGCTGCATGCTTAGCAAAAAAACAAACAGA-3′) and HR20 (5′-AGCTCTGCAGAGTTTCTTCTTCATCTCCAT-3′). Amplicons of pspA were cloned into either BglII-HindIII- or SphI-SalI-digested pQE40 or pQE30 (HR108 only) vector (Qiagen Inc., Valencia, Calif.) and transformed into M15(pREP4), a K-12-derived Escherichia coli strain containing a plasmid that encodes a lac repressor allowing control over expression. Clones containing the different pspA inserts were verified by sequencing. Expression of positive clones was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) during growth at room temperature. The overexpressed protein fragments were purified by affinity chromatography with a nickel resin in accordance with the manufacturer's (Qiagen) instructions. The different constructs encoded PspA fragments with predicted molecular masses of 38.6 kDa (HR101), 52 kDa (HR102), 59.3 kDa (HR104), 71.8 kDa (HR107), and 11.4 kDa (HR108), which were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and quantified by using the Bio-Rad DC protein assay (Bio-Rad Laboratories, Hercules, Calif.). Fragments HR101, HR102, HR104, and HR107, which were cloned into pQE40, were all fusion proteins whose N terminus was murine dihydrofolate reductase (DHFR). HR108 consisted only of the cloned PspA sequence.
Western blotting.
PspA and fragments of PspA (0.5 μg) were run on polyacrylamide gels (ready gels; Bio-Rad Laboratories), and the gels were electroblotted to a 0.45-μm-pore-size nitrocellulose membrane (Bio-Rad) in Tris-glycine buffer (20% methanol, 25 mM Tris, 192 mM glycine, pH 8.1 to 8.4) at 100 V for 1 h at 4°C. The blotted membrane was incubated with 1% bovine serum albumin in PBS-T (PBS containing 0.05% Tween 20) for 1 h at room temperature and washed three times (for 5 min each time) with PBS-T. The membranes were exposed to anti-PspA antibodies (polyclonal anti-PspA family 2 antisera or PC3.1 monoclonal antibodies) or antihistidine antibodies (Qiagen) for 30 min at 37°C, washed and incubated with a mixture of biotinylated goat anti-mouse or anti-rabbit antibodies (diluted 1:1,000 in PBS-T) and AP-conjugated streptavidin (1:500 dilution in PBS-T) for 30 min at 37°C. After washing, the membranes were developed with nitroblue tetrazolium at 0.1 mg/ml and 5-bromo-4-chloro-3-indolylphosphate at 0.5 mg/ml in 0.15 M Tris-HCl, pH 8.8.
Mouse immunization and challenge.
Five- to 8-week-old CBA/CAHN/XID (CBA/N) or BALB/cByJ (BALB/c) mice (Jackson Laboratory, Bar Harbor, Maine) were used for protection studies. These strains were selected because they were strains on which we had significant baseline data for infections with these and other strains of pneumococci and because they represented two extremes of resistance to pneumococcal infection. CBA/N mice have an X-linked genetic defect, and BALB/c mice are immunologically normal (8, 9, 12). The group size was generally between 7 and 21 mice, except for two challenge groups of 3 to 5 mice (see Table 3). Whenever the existence of protection was equivocal, the numbers of mice were increased until a clear result was obtained. The mice were immunized subcutaneously with 1 to 5 μg of purified recombinant protein SW111, HR101, HR102, HR104, HR107, or HR108, as indicated, in alum (100 μg of AlOH3/ml) for primary immunization. A booster dose of antigen in alum was administered subcutaneously 2 weeks later. Fourteen days after the boosts, the animals were challenged intravenously through the tail vein with S. pneumoniae strain BG7322, A66.1, EF3296, 3JYP2670, or ATCC 6303 at a dose that was about 2 logs greater than the 50% lethal dose. The challenge doses of these strains for CBA/N mice were 400, 200, 2,000, 700, and 800 CFU, respectively. The challenge doses of these strains for BALB/c mice were 3.6 × 105, 4.8 × 106, 2.0 × 106, 1.0 × 105, and 2.8 × 106 CFU, respectively. The infected animals were monitored daily, and the day of death was recorded. Mice were scored as dead once their bodies reached room temperature, as determined by touch. Mice alive at 21 days postinoculation were considered protected against death. Experience indicates that if deaths do occur in this model, they are generally observed at least 7 to 14 days before this time point (8, 10).
TABLE 3.
Cross-protective immunity elicited by PspA/EF3296 fragments in BALB/c mice
| Immunogena and strain | Capsular type | PspA clade(s)b | Median no. of days to deathc
|
P value | Alive/deadd ratio
|
P value | ||
|---|---|---|---|---|---|---|---|---|
| Nonimmune | Immune | Nonimmune | Immune | |||||
| SW111 | ||||||||
| A66.1 | 3 | 1 + 2e | 2 | 2 | 0.456 | 0:7 | 0:7 | 1.000 |
| BG7322 | 6B | 2 | 7 | 21 | 0.026 | 0:7 | 5:2 | 0.020 |
| 3JYP2670 | 3 | 4 | 4 | 21 | 0.026 | 0:7 | 5:2 | 0.020 |
| ATCC 6303 | 3 | 5 | 3 | 21 | 0.018 | 0:7 | 6:1 | 0.005 |
| HR101 | ||||||||
| A66.1 | 3 | 1 + 2 | 5 | 21 | 0.360 | 3:5 | 6:4 | 0.630 |
| BG7322 | 6B | 2 | 4 | 21 | 0.043 | 0:7 | 5:5 | 0.044 |
| 3JYP2670 | 3 | 4 | 6 | 21 | 0.036 | 0:3 | 5:0 | 0.017 |
| ATCC 6303 | 3 | 5 | 5 | 7.5 | 0.515 | 2:6 | 4:6 | 0.640 |
| HR107 | ||||||||
| A66.1 | 3 | 1 + 2 | 4 | 4 | 0.561 | 1:10 | 4:8 | 0.310 |
| BG7322 | 6B | 2 | 8 | 13.5 | 0.621 | 3:8 | 5:7 | 0.660 |
| 3JYP2670 | 3 | 4 | 2 | 2 | 0.920 | 0:4 | 0:5 | 1.000 |
| ATCC 6303 | 3 | 5 | 3 | 12 | 0.136 | 2:9 | 6:6 | 0.190 |
| HR108 | ||||||||
| A66.1 | 3 | 1 + 2 | 4.5 | 3 | 0.111 | 2:18 | 2:17 | 1.000 |
| BG7322 | 6B | 2 | 9 | 21 | <0.001 | 2:13 | 14:3 | <0.001 |
| 3JYP2670 | 3 | 4 | 2 | 3 | 0.110 | 2:15 | 6:12 | 0.230 |
| ATCC 6303 | 3 | 5 | 3 | 21 | <0.001 | 0:17 | 11:6 | <0.001 |
Mice were immunized and boosted 2 weeks apart with 1 μg of protein and challenged 2 weeks after the boost.
Clades 1 and 2 are in family 1; clades 3, 4, and 5 are in family 2. For protection elicited by these fragments against family 2, clade 3 strain EF3296, see Table 2.
Median numbers of days to death were compared between immune and nonimmune mice with the two-tailed Mann-Whitney U test (nonparametric, two-sample rank test). Boldface P values indicate statistically significant differences.
Fisher's exact test was used to compare alive/dead ratios of immune and nonimmune mice in survival experiments. Boldface P values indicate significant differences.
A66.1 expresses both clade 1 and clade 2 PspA proteins (10).
Determination of anti-PspA antibody levels in serum.
Mice were bled retroorbitally 24 h before challenge. Titers were evaluated by enzyme-linked immunosorbent assay (ELISA) by using the homologous recombinant fragment used for immunization and the recombinant full-length α-helical fragment (SW111) to coat plates. The coated plates were incubated overnight at 4°C and then blocked by incubation for 2 h at room temperature with 1% bovine serum albumin in PBS-T. After 3 h of incubation at 37°C with different dilutions of mouse sera, the plates were washed with PBS-T and biotinylated goat anti-mouse antibodies (1:1,000 dilution in PBS-T) were used to detect the mouse serum antibodies. After further incubation with AP-conjugated streptavidin (1:1,000 dilution in PBS-T), p-nitrophenyl phosphate (1 mg/ml; Sigma) was used for color development. Absorbance was read at 405 nm. The concentration of antibody reactive with PspA was calculated as micrograms per milliliter by using a serum standard of pooled immune sera from groups of mice immunized with HR101 or HR107.
Binding of anti-PspA serum antibodies to the bacterial surface.
Bacteria were grown on blood agar plates or in Todd-Hewitt medium containing 0.5% yeast extract and were suspended in PBS at a concentration of approximately 108/ml. The bacterial suspension (80 μl) was mixed with 20 μl of mouse preimmune or immune sera (final dilution, 1:40) or with anti-PspA PC3.1 monoclonal antibodies (20 μg/ml) for 30 min at room temperature and washed by centrifugation at 1,500 × g for 5 min in PBS. Fluorescein isothiocyanate-conjugated goat anti-mouse antibodies (1:100 dilution in PBS) were added for an additional 30 min at room temperature, and after a final wash in PBS, the cells were inspected by epifluorescence microscopy with a Leitz upright microscope (Leitz, Wetzlar, Germany). Binding was quantitated by flow cytometry with a FACScalibur flow cytometer (Becton-Dickinson Biosciences) as the fold increase compared to the fluorescence obtained with nonimmune mouse sera from the same strain of mice.
Statistical analyses.
Antibody levels and numbers of days to death were compared with the two-tailed Mann-Whitney U test (nonparametric, two-sample rank). Comparisons of live-versus-dead numbers in survival experiments were done by using two-by-two contingency tables and Fisher's exact test.
RESULTS
Production of recombinant fragments of PspA/EF3296.
The α-helical domain of PspA is known to be exposed on the bacterial cell surface (23, 29, 30). Studies of PspA/Rx1 (family 1) have shown that a major cross-protection-eliciting region is located in the CDR of the α-helix located in the C-terminal part of this domain (29). To investigate the protection-eliciting regions of PspA/EF3296 (family 2/clade 3), overlapping fragments of PspA/EF3296 spanning its α-helical domain were produced (Fig. 1). PCR products were cloned in frame into pQE40, an expression vector in which the cloned fragments are located C terminal to DHFR, which has a polyhistidine tag at the N-terminal end. The DHFR used (187 amino acids, 26 kDa) is from the mouse and is expected to be nonimmunogenic in mice (5). For HR108, pQE30, an expression vector similar to pQE40 but without DHFR, was used. Recombinant plasmids containing inserts were selected on the basis of antibiotic resistance, the insert was verified by sequencing, and the recombinant protein fragments were expressed and purified.
FIG. 1.
PspA/EF3296 fragments used in this study. Protection-eliciting epitopes of Rx1 were mapped previously (29) to the α-helical region. The CDR has been shown to contain major cross-protection-eliciting epitopes (16, 29). This region has subsequently been used in sequence analysis as the CDR for PspA proteins (25).
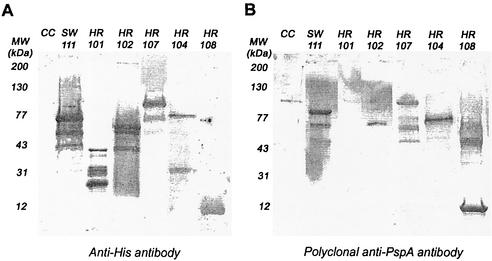
All recombinant PspA (rPspA) protein fragments were of the expected sizes in a Western blot developed with anti-His antibodies (Fig. 2A). Western blots visualized with anti-family 2 PspA polyclonal antibodies (Fig. 2B) or the PC3.1 anti-PspA/EF3296 monoclonal antibody (not shown) showed the same recognition pattern as did the anti-His antibodies, except for HR101 (amino acids 1 to 115), which was not recognized by either the PC3.1 monoclonal antibody or the polyclonal antibodies (Fig. 2B). This nonreactivity of polyclonal serum against the N-terminal domain of PspA in Western analyses has been observed in earlier studies using antibodies to both PspA/Rx1 and PspA/EF3296 (23).
FIG. 2.
Western blotting of fragments of PspA/EF3296. Fragments of PspA/EF3296 were run on 12% gels and subjected to Western blotting with anti-His antibodies (A) or polyclonal anti-PspA antibody (B). Both blots display a choline eluate from strain EF3296 (lane CC), as well as fragments SW111 (lane 2), HR101 (lane 3), HR102 (lane 4), HR107 (lane 5), HR104 (lane 6), and HR108 (lane 6). MW, molecular mass.
The fact that PC3.1 bound both HR102 and HR108, which are nonoverlapping fragments, suggests that this antibody may recognize a part of the repeated sequence motif that is a feature of family 2 proteins (20). Within the EF3296 (and TIGR4) sequence, there is a very strong repeat of 30 amino acids that extends from amino acid 255 to amino acid 285 (residing within HR102) and from amino acid 336 to amino acid 366 (residing within HR108). Within these amino acid regions, the sequences are identical except for the change of a Q at position 257 to a K at position 338. The repeated region extends with a little less fidelity about 20 amino acids in both directions from these 30-amino-acid blocks.
Antibody responses to PspA/EF3296 fragments as measured by ELISA and surface staining.
To assess the relative protection-eliciting capacities of the recombinant fragments, groups of BALB/c and CBA/N mice (5 to 10 mice per group) were immunized and boosted 2 weeks apart with 1 μg of specific fragments (SW111, HR101, HR102, HR104, HR107, or HR108) or with purified DHFR adjuvanted with alum (Table 1). Two weeks after the boost, the mice were bled and the serum was collected for analysis by ELISA. The immune sera to the fragments showed no reactivity to microtiter plates coated with recombinant DHFR (data not shown), demonstrating that detectable levels of antibody were not elicited by this sequence, which was fused to most of the rPspA fragments. Thus, all antibody reactive with the immunizing proteins by ELISA is assumed to be reactive with rPspA.
TABLE 1.
Antibody responses detected by ELISA and flow cytometry
| Mouse strain and immunogen | No. of serum samples tested | Avg antibody concn (μg/ml ± SD) detected by ELISAa
|
Surface binding to EF3296
|
||
|---|---|---|---|---|---|
| SW111 | Immunizing fragment | No. of serum samples tested | Times control (avg ± SD)b | ||
| BALB/c | |||||
| None | 1c | 0.00 ± 0.00 | NDd | 5 | 1.00 |
| DHFR | 5 | 0.00 ± 0.00 | 0.00 ± 0.00 | 5 | ND |
| SW111 | 10 | 507 ± 565 | 507 ± 565 | 16 | 9.25 ± 1.71 |
| HR101 | 8 | 0.003 ± 0.001 | 0.22 ± 0.13 | 16 | 1.51 ± 0.09 |
| HR102 | 8 | 37 ± 19 | 88 ± 90 | 16 | 6.36 ± 2.44 |
| HR104 | 8 | 0.09 ± 0.05 | 50 ± 25 | 16 | 0.94 ± 0.03 |
| HR107 | 10 | 48 ± 17 | 74 ± 76 | 16 | 6.64 ± 0.80 |
| HR108 | 8 | 23 ± 13 | 225 ± 139 | 16 | 2.24 ± 0.28 |
| CBA/N | |||||
| None | 1c | 0.00 ± 0.00 | ND | 5 | 1.00 |
| DHFR | 5 | 0.00 ± 0.00 | 0.00 ± 0.00 | 5 | ND |
| SW111 | 8 | 43 ± 14 | 43 ± 14 | 8 | 5.47 ± 1.79 |
| HR101 | 8 | 0.01 ± 0.003 | 3.73 ± 5.00 | 8 | 1.06 ± 0.06 |
| HR102 | 12 | ND | 2.79 ± 5.07 | ND | |
| HR107 | 8 | 172 ± 118 | 187 ± 119 | 8 | 6.61 ± 1.15 |
| HR108 | 8 | 11.3 ± 3.9 | 189 ± 38 | 8 | 6.81 ± 2.29 |
Immune serum samples from BALB/c or CBA/N mice were tested for their anti-PspA antibody concentrations either against full-length α-helical PspA/EF3296 (SW111) or against the fragment used for immunization. Antibody concentrations and standard deviations were determined as described in Materials and Methods.
Binding to the EF3296 bacterial surface was determined for each serum sample as the fluorescent signal of the sample divided by the fluorescent signal of a preimmune serum control.
Pooled serum samples from five individual mice were used.
ND, not done.
When evaluated by ELISA for reactivity to SW111, the largest of the α-helical fragments, the levels of antibody elicited by the different fragments varied greatly. With BALB/c mice, the highest ELISA reactivity was seen with sera obtained by immunization with SW111, with an average concentration of 507 μg of PspA antibodies per ml. Fragments HR102, HR107, and HR108 consistently resulted in moderate levels of antibody (20 to 50 μg/ml) reactive with SW111, while HR101 and HR104 only elicited nanogram levels of antibody reactive with SW111 (Table 1). All of the PspA fragments, however, elicited higher levels of antibody reactive with the immunizing fragment than antibody reactive with SW111. Especially high discrepancies were seen for fragments HR101 and HR104, which elicited negligible levels of antibodies capable of binding SW111. HR101 elicited 70-fold more antibody reactive with HR101 than antibody reactive with SW111. In the case of the antibody elicited by HR104, 500-fold higher reactivity was observed against the homologous fragment, HR104, than against SW111. For PspA/EF3296 fragments SW111, HR101, HR102, HR107, and HR108, the same general observations about cross-reactivity with SR111 were made for the antibodies elicited in CBA/N mice as were made for the antibodies elicited in BALB/c mice. HR104 was not used to immunize CBA/N mice.
The poorer reactivity of the antibodies elicited to each fragment with SW111 (the largest fragment of PspA/EF3296 examined) demonstrate that the conformation of some of the fragments is probably different in many places from that of the same sequence in the complete α-helical domain. For both the BALB/c and CBA/N mice, we observed that the antibody elicited by HR107 showed higher relative reactivity with SW111 than did the antibody elicited by any of the other fragments of the α-helical domain. This finding probably indicates that of all of the fragments, HR107, which was also the largest, had the most native conformation. The smallest fragment, HR108, which is the only one to lack DHFR, elicited antibody 9 to 17 times as reactive with itself as with SW111. By these criteria, it appears that the size and composition of the fragments, rather than the attached DHFR, were the factors primarily responsible for their lack of a completely native conformation.
Surface staining by antibody to PspA fragments.
In an attempt to estimate the levels of antibody elicited by each fragment that could react with intact pneumococci, we determined the relative abilities of the antibodies elicited by the different fragments to mediate fluorescent surface staining of intact EF3296 pneumococci (Table 1). The antisera from BALB/c and CBA/N mice immunized with fragments SW111, HR107, and HR108 showed levels of binding to the bacterial cells 2 to 10 times higher than that of the preimmune serum control (Table 1). Sera from BALB/c mice immunized with HR102 showed a different pattern, characterized by highly variable binding capacities of individual sera. Eight out of 16 serum samples tested had a binding ability less than 1.5 times that of the control, 5 showed binding between 1.5 and 5 times that of the control, and 3 showed considerably higher binding (11, 29, and 31 times that of the control) to the bacterial surface. None of the sera from BALB/c mice immunized with fragment HR101 or HR104 showed significant surface binding in the fluorescence assay. This result was entirely consistent with the fact that these two immunogens elicited very little antibody reactive by ELISA against the full α-helical domain.
There was, in general, a good correlation between surface binding and antibody reactivity to SW111, which expresses the complete α-helical domain. The only exception was immunity to HR108. These antibodies exhibited the second highest binding to EF3296 but showed the seventh highest reactivity with SW111 by ELISA. This is consistent with our view that this region of PspA is probably the most surface-exposed portion of the molecule (26, 29).
Protection of BALB/c and CBA/N mice against S. pneumoniae EF3296 by immunization with PspA/EF3296 fragments.
To evaluate the protection induced by immunization with family 2 PspA, BALB/c mice were immunized with full-length α-helical fragment SW111 and challenged with S. pneumoniae EF3296 at a dose 2 logs higher than the 50% lethal dose (2 × 106 CFU). Immunization with SW111 resulted in an increase in the median time to death of the mice from 5 days in the control group to 14.5 days in the immunized group (P < 0.05) (Table 2). Five of 10 immunized mice survived the challenge, compared with none in the nonimmunized group (P < 0.05). The lack of complete protection is consistent with earlier data suggesting that type 4 encapsulated S. pneumoniae strains are more difficult to protect against by immunization with PspA than are capsular type 3, 6A, and 6B strains (35, 38).
TABLE 2.
Protection of mice challenged with S. pneumoniae EF3296 by prior immunization with recombinant PspA/EF3296 fragments
| Mice strain immunized and immunogena (amino acids) | Median no. of days to deathb
|
P value | Alive/deadc ratio
|
P value | ||
|---|---|---|---|---|---|---|
| Nonimmuned | Immune | Nonimmuned | Immune | |||
| BALB/c | ||||||
| SW111 (1-478) | 5 | 14.5 | 0.036 | 0:10 | 5:5 | 0.032 |
| HR101 (1-115) | 3 | 21 | <0.001 | 0:15 | 14:7 | <0.001 |
| HR102 (75-305) | 5 | 5 | 0.570 | 0:23 | 3:20 | 0.230 |
| HR104 (181-490) | 5 | 21 | 0.011 | 3:13 | 11:5 | 0.011 |
| HR107 (75-490) | 5 | 9 | 0.008 | 1:22 | 9:13 | 0.004 |
| HR108 (314-418) | 2.5 | 21 | 0.008 | 3:7 | 9:1 | 0.020 |
| CBA/N | ||||||
| SW111 (1-478) | 2 | 21 | 0.001 | 0:7 | 5:2 | 0.020 |
| HR101 (1-115) | 3 | 4 | 1.000 | 0:7 | 0:7 | 1.000 |
| HR102 (75-305) | 5 | 9.5 | 0.044 | 1:11 | 5:7 | 0.155 |
| HR107 (75-490) | 4 | 21 | <0.001 | 1:9 | 10:0 | <0.001 |
| HR108 (314-418) | 2 | 21 | 0.015 | 0:8 | 5:3 | 0.026 |
Mice were immunized and boosted 2 weeks apart with 1 μg of protein and challenged 2 weeks after the boost with 2 × 106 EF3296 bacteria.
Median numbers of days to death were compared with the two-tailed Mann-Whitney U test (nonparametric, two-sample rank). Boldface P values indicate significant differences from the control value.
Fisher's Exact test was used to compare alive/dead ratios in survival experiments. Boldface P values indicate significant differences obtained from the control value.
There is no reason to expect that the results obtained with the nonimmune mice would not have been the same for each mouse strain. The slight differences in frequency of survivors for the different control groups are not significant. Even so, these groups were not pooled prior to analysis.
To investigate the location of protection-eliciting epitopes of PspA/EF3296, different groups of BALB/c mice were immunized with each of the overlapping set of PspA/EF3296 fragments (HR101, HR102, HR104, HR107, and HR108). Two weeks after the second immunization, the mice were challenged intravenously with S. pneumoniae EF3296. Immunization with HR101 showed the highest degree of protection against EF3296, with 14 of 21 mice surviving the challenge (P < 0.001; Table 2). The strong protection elicited with this fragment was somewhat surprising, in view of the low antibody response to HR101 detected by both ELISA and surface staining. This finding indicates that the antibody that was elicited was highly efficacious against EF3296 infection.
Immunization with HR104, HR107, and HR108 also resulted in statistically significant protection of BALB/c mice against EF3296, as measured by both time to death and fraction alive (Table 2). There was no direct association between the recognition of the antigen in ELISA or on the bacterial surface and the ability to protect. For example, HR104 elicited low concentrations of antibodies against SW111 when probed by ELISA and the antibodies did not show detectable binding to the EF3296 bacterial surface (Table 1) but were protective against EF3296 infection (Table 2).
In contrast, HR102 did not significantly protect mice from death, even though it elicited substantial antibody titers, as detected by ELISA and surface binding (Table 1). To see if a higher dose of HR102 might result in protection, an additional group of five BALB/c mice (with five alum-only controls) were immunized with 5 μg of HR102. The larger dose resulted in a somewhat longer time to death than did immunization with 1 μg, but the effect was still not statistically significant (data not shown).
Fragments SW111, HR101, HR102, HR107, and H108 were further studied for protection against EF3296 infection in CBA/N mice (Table 2). These mice lack a functional response to polysaccharides. As a result they are devoid of natural antibodies to the phosphocholine epitope of pneumococcal teichoic acids. This absence contributes to their higher susceptibility to pneumococcal disease than is the case for mice of most other strains (3, 15, 31). As a result of their greater susceptibility than BALB/c mice, proportionately smaller challenge doses were used.
Fragments HR107 and HR108 elicited significantly better protection in CBA/N mice than in BALB/c mice (P < 0.01 and P < 0.01 for HR107 and P < 0.05 and P < 0.05 for HR108 when median days to death and alive/dead ratios were compared, respectively). SW111 also appeared to elicit better protection in CBA/N mice than in BALB/c mice, even though the difference in protection was not statistically significant. HR102, which had not elicited protection in BALB/c mice, also failed to protect CBA/N mice against death, even though it did result in a statistically significant (P < 0.05) increase in the median number of days to death from 5 to 9.5 days. One might be tempted to explain these results by speculating that the challenge dose, relative to the resistance of the mice, was larger for BALB/c mice than for CBA/N mice. However, the results obtained by immunization with fragment HR101 make it clear that the true explanation may be a little more complex. In contrast to its ability to protect BALB/c mice, HR101 did not protect CBA/N mice against a challenge with EF3296. The difference in the efficacy of HR101 immunization in BALB/c versus CBA/N mice was statistically significant (P < 0.05 and P < 0.01 when comparing median numbers of days to death and alive/dead ratios, respectively).
These studies with BALB/c and CBA/N mice make it clear that PspA can elicit protection against this type 4 encapsulated strain and demonstrate that EF3296 fragments that include the C-terminal and α-helical ends of the α-helical region could both elicit protection. The fact that the C-terminal fragments elicited protection in both mouse strains makes this region of somewhat greater interest with regard to vaccine epitopes.
Ability of immunity to PspA/EF3296 fragments to elicit protection against S. pneumoniae expressing different clades of PspA.
To further evaluate the protective responses of PspA/EF3296, BALB/c and CBA/N mice were immunized and challenged with strains of capsular types 3 and 6 expressing different clades of PspA (Tables 3 and 4).
TABLE 4.
Cross-protective immunity of PspA/EF3296 immunization in CBA/N mice
| Immunogena and strain (capsular type) | PspA clade(s)b | Median no. of days to deathc
|
P value | Alive/dead ratiod
|
P value | ||
|---|---|---|---|---|---|---|---|
| Nonimmune | Immune | Nonimmune | Immune | ||||
| SW111 | |||||||
| A66.1 (3) | 1 + 2e | 2 | 4 | 0.622 | 0:7 | 3:4 | 0.191 |
| BG7322 (6B) | 2 | 6 | 21 | 0.018 | 0:7 | 6:1 | 0.005 |
| 3JYP2670 (3) | 4 | 1 | 21 | 0.002 | 0:7 | 5:2 | 0.020 |
| ATCC 6303 (3) | 5 | 2 | 21 | <0.001 | 0:7 | 7:0 | <0.001 |
| HR101 | |||||||
| A66.1 (3) | 1 + 2 | 2 | 2 | 1.000 | 0:7 | 0:7 | 1.000 |
| BG7322 (6B) | 2 | 5 | 6 | 0.379 | 0:7 | 0:7 | 1.000 |
| 3JYP2670 (3) | 4 | 1 | 1 | 0.945 | 0:7 | 0:7 | 1.000 |
| ATCC 6303 (3) | 5 | 2 | 3 | 0.002 | 0:7 | 0:7 | 1.000 |
| HR107 | |||||||
| A66.1 (3) | 1 + 2 | 2 | 3 | 0.692 | 0:5 | 0:5 | 1.000 |
| BG7322 (6B) | 2 | 6 | 21 | 0.016 | 0:5 | 3:2 | 0.167 |
| 3JYP2670 (3) | 4 | 2 | 21 | <0.001 | 0:10 | 9:1 | <0.001 |
| ATCC 6303 (3) | 5 | 2 | 21 | 0.016 | 0:5 | 3:2 | 0.167 |
| HR108 | |||||||
| A66.1 (3) | 1 + 2 | 2 | 2 | 0.72 | 0:8 | 0:8 | 1.000 |
| BG7322 (6B) | 2 | 3.5 | 21 | 0.015 | 0:8 | 6:2 | 0.007 |
| 3JYP2670 (3) | 4 | 1 | 21 | <0.001 | 0:8 | 6:2 | 0.007 |
| ATCC 6303 (3) | 5 | 1 | 21 | <0.001 | 0:7 | 6:1 | 0.026 |
Mice were immunized and boosted 2 weeks apart with 1 μg of protein and challenged 2 weeks after the boost.
Clades 1 and 2 are in family 1; clades 3, 4, and 5 are in family 2. For protection against family 2, clade 3 strain EF3296, see Table 2.
Median numbers of days to death were compared between immune and nonimmune mice with the two-tailed Mann-Whitney U test (nonparametric, two-sample rank test). Boldface P values indicate significant differences.
Fisher's exact test was used to compare alive/dead ratios of immune and nonimmune mice in survival experiments. Boldface P values indicate significant differences.
A66.1 expresses both clade 1 and clade 2 PspA proteins (10).
When median numbers of days to death and alive/dead ratios were compared, the most broadly cross-protective immunogen in BALB/c mice was SW111, which contained all of the α-helical sequence. Immunization of BALB/c mice with SW111 protected them against BG7322 (clade 2, family 1), 3JYP2670 (clade 4, family 2), and ATCC 6303 (clade 5, family 2). In the studies whose results are depicted in Table 2, we observed that this fragment also elicited protection against clade 3 strain EF3296. No protection, however, was elicited against A66.1 (clades 1 and 2, family 1). A66.1 is a virulent capsular type 3 strain that is known to express both clade 1 and clade 2 PspA proteins (25).
When HR101 was used to immunize BALB/c mice, the mice were protected against death induced by BG7322 and 3JYP2670 but not against death induced by ATCC 6303 or A66.1 (Table 3). HR108 showed protection against BG7322 and ATCC 6303, with a trend toward protection against 3JYP2670. HR107 immunization did not result in statistically significant protection against any of the strains, although increased survival was seen in infections with ATCC 6303.
Immunization of CBA/N mice with SW111, HR107, and HR108 gave results similar to those obtained with BALB/c mice, except that this time HR108 was as cross-protective as the much larger fragment SW111. Immunization with HR107 showed almost as much protection against 3JYP2670, ATCC 6303, and BG7322 as did SW111 and HR108. In contrast to the results obtained with BALB/c mice, immunization of CBA/N mice with HR101 failed to elicit meaningful protection against any of the challenge strains (Table 4).
Evidence that the protective efficacies of antibodies elicited by different regions of PspA/EF3296 differ.
Comparisons of the protective capacities of immunization with different fragments with the titers of antibodies reactive with the full α-helical region (SW111) has revealed very large differences in the protective efficacies of the antibodies elicited. On the basis of reactivity to SW111, this fragment (Table 1) was 100,000 times as immunogenic as HR101 in BALB/c mice but the antibodies elicited by HR101 protected a slightly higher percentage of immunized mice from EF3296 infection (Table 2). HR102 was 10,000 times as immunogenic as HR101, but the response to HR102 was not protective against death. These comparisons indicate that the antibody elicited by HR101 in BALB/c mice is probably much more effective, per microgram, than that elicited by the other two fragments. Similar results were obtained in the cross-protection experiments. HR108 elicited half as much antibody in BALB/c mice as did HR107 (Table 1) and resulted in good cross-protection against the clade 2 and clade 5 strains (Table 3). In contrast, the response to HR107 did not elicit any significant cross-protection in BALB/c mice.
DISCUSSION
From the data gathered in this study, it is apparent that although the approximately 104 C-terminal amino acids of the α-helical domain of PspA/EF3296 are particularly important for elicitation of cross-protection, epitopes in the N-terminal 115 amino acids of the α-helical domain of PspA can also elicit protection against pneumococcal infection. Knowing which portions of PspA do and do not elicit protective responses may, in time, help us to determine how a serologically variable molecule like PspA can elicit antibodies that are able to cross-react and cross-protect. This information may also lead to the construction of a PspA vaccine that maximizes the fraction of the elicited antibodies that are protective.
Potential use of epitope-mapping techniques to develop a superior PspA-containing vaccine.
These studies have shown not only that different PspA fragments are more protection eliciting than others, but they have also shown that, even among the protection-eliciting fragments, the amount of protection per ELISA titer unit indicates that not all antibodies to PspA are protective and implies that not all PspA epitopes are protection eliciting. By careful construction of rPspA fragments, one might avoid eliciting nonprotective antibody and therefore further improve the efficacy of a PspA-containing vaccine beyond that obtained by immunizing with the entire α-helical domain.
The N-terminal region and CDR elicit protection and cross-protection.
As had been observed for clade 2 PspA/Rx1 (13, 29), fragments containing the N-terminal 115 amino acids or the CDR of PspA/EF3296 were able to elicit protection against the challenge strains of the same PspA clade. The fact that both ends of the α-helical domain of family 2 PspA proteins have been found to elicit protection suggests that this may be true of most PspA proteins.
HR102 (amino acids 75 to 305), the fragment lacking either end of the PspA/EF3296 α-helical domain, did not elicit protection in immunocompetent BALB/c mice. Although HR102 also failed to elicit protection against death in immunocompromised CBA/N mice, it did provide a significant increase in time to death. These results suggest that there is a paucity of protection-eliciting epitopes in this region. At the very least, the results indicate that, in the absence of the flanking PspA sequences, this central region does not express important protection-eliciting epitopes. When the original mapping studies were conducted with PspA/Rx1, a similar central fragment was not used as an immunogen but it was observed that none of the epitopes detected by protective monoclonal antibodies mapped to this region (13, 29).
When cross-protection of BALB/c mice was examined, the N-terminal fragment, HR101, and the CDR fragment, HR108, both elicited some cross-protection. The two fragments were, however, somewhat complementary with regard to the pneumococcal strains they protected against. PspA/EF3296 fragment SW111, which contained the amino acids comprising the sequences of both HR101 and HR108, protected against all of the strains that were protected against by either HR101 or HR108. This suggests that SW111 probably was more broadly cross-protective because it expressed more cross-protective epitopes and therefore was able to share individual epitopes with more strains. In BALB/c mice, there was no clear dominance of the CDR over the N-terminal 115 amino acids in elicitation of cross-protection.
In contrast to the CDR-containing fragments, the central α-helical fragment, HR102, had a poor ability to elicit even homologous protection. These findings suggest that rPspA immunogens designed to lack the epitopes encoded by the central region might be better vaccine antigens by being able to elicit a higher proportion of the antibody that might be protective. This might be achieved by immunizing with a mixture of N-terminal and CDR fragments or with a fusion protein containing only the N-terminal 115 amino acids and the CDR.
Differences between the protection-eliciting responses in BALB/c and CBA/N mice.
The fact that some differences in the results obtained with the two strains of mice were observed emphasizes the importance of examining the protective properties of potential vaccine proteins in more than one strain of mice, as well as the importance of the eventual human antibodies to establish the applicability of the findings to Homo sapiens.
In the original studies to identify protective regions of PspA/Rx1, all passive and active immunizations were conducted with CBA/N mice (29). In the present study, we observed that with some PspA fragments, very different results were obtained with BALB/c versus CBA/N mice. In BALB/c mice, HR101, which contains the first 115 amino acids of PspA/EF3296, was strongly protection eliciting. In CBA/N mice, this fragment elicited no less antibody but failed to elicit protection against EF3296. In contrast, fragment HR107, which contained all but the most N-terminal 74 amino acids of the mature α-helical region, was very cross-protective in CBA/N mice but was not cross-protective at all in BALB/c mice. When cross-protection of BALB/c mice was examined, SW111 elicited better cross-protection than the other fragments tested. In CBA/N mice, however, SW111, HR107, and HR108 all showed very similar levels of cross-protection.
These findings suggested that the antibodies produced by the two strains of mice in response to the same molecule may have very different specificities and/or biologic activities. Other factors may be involved, however. BALB/c mice are known to have levels of antibody to phosphocholine in their serum that can protect against small challenge inocula of S. pneumoniae. These antibodies are not in the serum of CBA/N mice, which express the Xid immunodeficiency trait (3, 15, 31). It is possible that the antiphosphocholine antibodies are needed to synergize with antibodies to the N-terminal 115 amino acids for protection to be observed. This could explain why HR101 elicited protection in BALB/c but not CBA/N mice.
In the case of the broad cross-protection of CBA/N mice due to immunization with fragment HR107, a different mechanism may be at work. It is possible that the antibody elicited is protective against the low concentrations of bacteria needed to kill CBA/N mice but not against the much larger challenge doses needed to kill BALB/c mice. It is known that systemic infection with high concentrations of pneumococci can decrease complement levels and that PspA acts by interfering with complement deposition on pneumococci (2, 41). As is the case for antibodies to capsule (18), protection by antibody to PspA is probably optimal in a host that still has circulating complement (16). Such antibodies would be expected to be less effective once the host became septic.
Effect of protein context on the ability of epitopes to elicit protective responses.
The expression of immunogenic protection-eliciting epitopes of PspA/EF3296 could be affected by the context in which the sequence is presented. For example, fragment HR107 (amino acids 75 to 490) contains fragment HR108 (amino acids 314 to 418) but HR107 failed to elicit cross-protection against pneumococci expressing clades 2 and 5 in BALB/c mice whereas the smaller fragment, HR108, elicited good cross-protection against the same strains. An explanation might be that the cross-protective epitopes of HR108 are not expressed in the larger fragment because of conformational problems in HR107. Alternatively, HR107 may contain epitopes that are highly immunogenic in BALB/c mice but not protection eliciting. If these epitopes are missing in HR108, their absence might permit the anti-HR108 response to be more focused on its protection-eliciting epitopes.
Successful protection against a difficult challenge strain.
Previous studies had demonstrated that certain capsular type 2, 4, and 5 strains were difficult to protect against by immunization with non-family 2 PspA proteins (35, 38). In the present studies, highly statistically significant protection was obtained against EF3296 (capsular type 4) and the HR101 and HR108 fragments appeared to confer on BALB/c mice even higher levels of protection against EF3296 than did the full-length α-helical fragment, SW111. These findings suggest that the best challenge strains for use in the future to identify cross-protective epitopes may be those that are the most difficult to protect against.
Potential surrogate assays of protection.
Efforts to develop successful vaccines are greatly augmented by the use of effective surrogate assays of protection. The only useful surrogate assay for protective antibodies to PspA is passive protection in mice. Since the different fragments of EF3296 elicited very different levels of protection, it is possible to use the antisera elicited by these fragments to test the validity of potential surrogate assays. In this paper, we have demonstrated conclusively that levels of antibody detected by ELISA do not correlate with protection and thus do not provide a surrogate of protection. Similarly, surface staining by antibodies to PspA also failed to provide a useful correlate with protection. Surface staining, but not good protection, was observed, for example, when immune sera from mice that were immunized with HR102 were used. The failure to obtain surface staining with some of the immune sera to protection-eliciting fragments may not have been because the antibodies do not bind the surface but because surface fluorescence is not sensitive enough to detect them at the low concentrations present.
Acknowledgments
We acknowledge Flora Gathof, whose handling of administrative details greatly facilitated this study, and Alex Smith for help in producing fragment HR108. We are also grateful to our collaborators at Aventis Inc. for providing monoclonal antibody PC3.1 and PspA/EF3296 construct SW111.
This study was supported by the Swedish Cancer Society (A.H.), National Institutes of Health grants AI21548 and HL54818, and the Carsten Cole Buckley Pediatric Meningitis Research Fund (D.B.).
Editor: D. L. Burns
REFERENCES
- 1.Aaberge, I. S., J. Eng, G. Lermark, and M. Løvik. 1995. Virulence of Streptococcus pneumoniae in mice: a standardized method for preparation and frozen storage of the experimental bacterial inoculum. Microb. Pathog. 18:141-152. [DOI] [PubMed] [Google Scholar]
- 2.Abeyta, M. 1999. Pneumococcal surface protein A and capsular polysaccharide in virulence of Streptococcus pneumoniae. Ph.D. thesis. University of Alabama at Birmingham.
- 3.Amsbaugh, D. F., C. T. Hansen, B. Prescott, P. W. Stashak, D. R. Barthold, and P. J. Baker. 1972. Genetic control of the antibody response to type III pneumococcal polysaccharide in mice. I. Evidence that an X-linked gene plays a decisive role in determining responsiveness. J. Exp. Med. 136:931-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. 1997. Prevention of pneumococcal disease: recommendations of the Advisory Committee on Immunization Practices. Morb. Mortal. Wkly. Rep. 46:1-24. [PubMed] [Google Scholar]
- 5.Anonymous. 2001. The QIAexpressionist handbook, 5th ed. Qiagen, Valencia, Calif.
- 6.Berry, A. M., and J. C. Paton. 2000. Additive attenuation of virulence of Streptococcus pneumoniae by mutation of the genes encoding pneumolysin and other putative pneumococcal virulence proteins. Infect. Immun. 68:133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briles, D. E., E. Ades, J. C. Paton, J. S. Sampson, G. M. Carlone, R. C. Huebner, A. Virolainen, E. Swiatlo, and S. K. Hollingshead. 2000. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect. Immun. 68:796-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briles, D. E., C. Forman, and M. Crain. 1992. Mouse antibody to phosphocholine can protect mice from infection with mouse-virulent human isolates of Streptococcus pneumoniae. Infect. Immun. 60:1957-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briles, D. E., C. Forman, J. C. Horowitz, J. E. Volanakis, W. H. Benjamin, Jr., L. S. McDaniel, J. Eldridge, and J. Brooks. 1989. Anti-pneumococcal effects of C-reactive protein and monoclonal antibodies to pneumococcal cell wall and capsular antigens. Infect. Immun. 57:1457-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briles, D. E., S. K. Hollingshead, J. King, A. Swift, P. A. Braun, M. K. Park, L. M. Ferguson, M. H. Nahm, and G. S. Nabors. 2000. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J. Infect. Dis. 182:1694-1701. [DOI] [PubMed] [Google Scholar]
- 11.Briles, D. E., S. K. Hollingshead, E. Swiatlo, A. Brooks-Walter, A. Szalai, A. Virolainen, L. S. McDaniel, K. A. Benton, P. C. Aerts, H. van Dijk, and M. J. Crain. 2000. Pneumococcal proteins PspA and PspC: their potential for use as vaccines, p. 253-260. In A. Tomasz and R. Austrian (ed.), Molecular biology of pneumococci and pneumococcal diseases. Mary Ann Liebert Inc., Larchmont, N.Y.
- 12.Briles, D. E., J. Horowitz, L. S. McDaniel, W. H. Benjamin, Jr., J. L. Claflin, C. L. Booker, G. Scott, and C. Forman. 1986. Genetic control of the susceptibility to pneumococcal infection. Curr. Top. Microbiol. Immunol. 124:103-120. [DOI] [PubMed] [Google Scholar]
- 13.Briles, D. E., J. D. King, M. A. Gray, L. S. McDaniel, E. Swiatlo, and K. A. Benton. 1996. PspA, a protection-eliciting pneumococcal protein: immunogenicity of isolated native PspA in mice. Vaccine 14:858-867. [DOI] [PubMed] [Google Scholar]
- 14.Briles, D. E., G. S. Nabors, A. Brooks-Walter, J. Paton, C., and S. Hollingshead. 2001. The potential for using protein vaccines to protect against otitis media caused by Streptococcus pneumoniae. Vaccine 19:S87-S95. [DOI] [PubMed] [Google Scholar]
- 15.Briles, D. E., M. Nahm, K. Schroer, J. Davie, P. Baker, J. Kearney, and R. Barletta. 1981. Antiphosphocholine antibodies found in normal mouse serum are protective against intravenous infection with type 3 Streptococcus pneumoniae. J. Exp. Med. 153:694-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briles, D. E., R. C. Tart, H.-Y. Wu, B. A. Ralph, M. W. Russell, and L. S. McDaniel. 1996. Systemic and mucosal protective immunity to pneumococcal surface protein. Ann. N. Y. Acad. Sci. 797:118-126. [DOI] [PubMed] [Google Scholar]
- 17.Brooks-Walter, A., D. E. Briles, and S. K. Hollingshead. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect. Immun. 67:6533-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown, E. J., S. W. Hosea, and M. M. Frank. 1983. The role of antibody and complement in the reticuloendothelial clearance of pneumococci from the bloodstream. Rev. Infect. Dis. 5 Suppl. 4:S797-S805. [DOI] [PubMed] [Google Scholar]
- 19.Coral, M. C. V., N. Fonseca, E. Castaneda, J. L. Di Fabio, S. K. Hollingshead, and D. E. Briles. 2001. Families of pneumococcal surface protein A (PspA) of Streptococcus pneumoniae invasive isolates recovered from Colombian children. Emerg. Infect. Dis. 7:832-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crain, M. J., W. D. Waltman II, J. S. Turner, J. Yother, D. F. Talkington, L. S. McDaniel, B. M. Gray, and D. E. Briles. 1990. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect. Immun. 58:3293-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dagan, R. 2000. Treatment of acute otitis media—challenges in the era of antibiotic resistance. Vaccine 19(Suppl. 1):S9-S16. [DOI] [PubMed] [Google Scholar]
- 22.Dagan, R., R. Melamed, M. Muallem, L. Piglansky, D. Greenberg, O. Abramson, P. M. Mendelman, N. Bohidar, and P. Yagupsky. 1996. Reduction of nasopharyngeal carriage of pneumococci during the second year of life by a heptavalent conjugate pneumococcal vaccine. J. Infect. Dis. 174:1271-1278. [DOI] [PubMed] [Google Scholar]
- 23.Håkansson, A., H. Roche, S. Mirza, L. S. McDaniel, A. Brooks-Walter, and D. E. Briles. 2001. Characterization of binding of human lactoferrin to pneumococcal surface protein A. Infect. Immun. 69:3372-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammerschmidt, S., S. R. Talay, P. Brandtzaeg, and G. S. Chhatwal. 1997. SpsA, a novel pneumococcal surface protein with specific binding to secretory immunoglobulin A and secretory component. Mol. Microbiol. 25:1113-1124. [DOI] [PubMed] [Google Scholar]
- 25.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jedrzejas, M. J., S. K. Hollingshead, J. Lebowitz, L. Chantalat, D. E. Briles, and E. Lamani. 2000. Production and characterization of the functional fragment of pneumococcal surface protein A. Arch. Biochem. Biophys. 373:116-125. [DOI] [PubMed] [Google Scholar]
- 27.Lock, R. A., J. C. Paton, and D. Hansman. 1988. Comparative efficacy of pneumococcal neuraminidase and pneumolysin as immunogens protective against Streptococcus pneumoniae. Microb. Pathog. 5:461-467. [DOI] [PubMed] [Google Scholar]
- 28.Lopez, R., E. Garcia, P. Garcia, and J. L. Garcia. 1997. The pneumococcal cell wall degrading enzymes: a modular design to create new lysins? Microb. Drug Resist. 3:199-211. [DOI] [PubMed] [Google Scholar]
- 29.McDaniel, L. S., B. A. Ralph, D. O. McDaniel, and D. E. Briles. 1994. Localization of protection-eliciting epitopes on PspA of Streptococcus pneumoniae between amino acid residues 192 and 260. Microb. Pathog. 17:323-337. [DOI] [PubMed] [Google Scholar]
- 30.McDaniel, L. S., G. Scott, K. Widenhofer, J. Carroll, and D. E. Briles. 1986. Analysis of a surface protein of Streptococcus pneumoniae recognized by protective monoclonal antibodies. Microb. Pathog. 1:519-531. [DOI] [PubMed] [Google Scholar]
- 31.Mosier, D. E., I. M. Zitron, J. Mond, A. Aftab, I. Scher, and W. E. Paul. 1977. Surface immunoglobulin D as a functional receptor for a subclass of B lymphocytes. Immunol. Rev. 37:89-104. [DOI] [PubMed] [Google Scholar]
- 32.Mulholland, E. K. 1997. A report prepared for the scientific advisory group of experts, global programme for vaccines and immunization. World Health Organization, Geneva, Switzerland.
- 33.Nabors, G. S., P. A. Braun, D. J. Herrmann, M. L. Heise, D. J. Pyle, S. Gravenstein, M. Schilling, L. M. Ferguson, S. K. Hollingshead, D. E. Briles, and R. S. Becker. 2000. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) variant stimulates broadly cross-reactive antibodies to heterologous PspA molecules. Vaccine 18:1743-1754. [DOI] [PubMed] [Google Scholar]
- 34.Ogunniyi, A. D., R. L. Folland, D. B. Briles, S. K. Hollingshead, and J. C. Paton. 2000. Immunization of mice with combinations of pneumococcal virulence proteins elicits enhanced protection against challenge with Streptococcus pneumoniae. Infect. Immun. 68:3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ralph, B. A., D. E. Briles, and L. S. McDaniel. 1994. Cross-reactive protection-eliciting epitopes of pneumococcal surface protein A. Ann. N. Y. Acad. Sci. 730:361-363. [DOI] [PubMed] [Google Scholar]
- 36.Rosenow, C., P. Ryan, J. N. Weiser, S. Johnson, P. Fontan, A. Ortqvist, and H. R. Masure. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25:819-829. [DOI] [PubMed] [Google Scholar]
- 37.Shinefield, H. R., and S. Black. 2000. Efficacy of pneumococcal conjugate vaccines in large scale field trials. Pediatr. Infect. Dis. J. 19:394-397. [DOI] [PubMed] [Google Scholar]
- 38.Tart, R. C., L. S. McDaniel, B. A. Ralph, and D. E. Briles. 1996. Truncated Streptococcus pneumoniae PspA molecules elicit cross-protective immunity against pneumococcal challenge in mice. J. Infect. Dis. 173:380-386. [DOI] [PubMed] [Google Scholar]
- 39.Tomasz, A. 1997. Antibiotic resistance in Streptococcus pneumoniae. Clin. Infect. Dis. 24:S85-S88. [DOI] [PubMed] [Google Scholar]
- 40.Tong, H. H., L. E. Blue, M. A. James, and T. F. DeMaria. 2000. Evaluation of the virulence of a Streptococcus pneumoniae neuraminidase-deficient mutant in nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect. Immun. 68:921-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tu, A. H., R. L. Fulgham, M. A. McCrory, D. E. Briles, and A. J. Szalai. 1999. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect. Immun. 67:4720-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yother, J., and D. E. Briles. 1992. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J. Bacteriol. 174:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yother, J., and J. M. White. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J. Bacteriol. 176:2976-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]