Abstract
Edwardsiella tarda is an important cause of hemorrhagic septicemia in fish and also of gastro- and extraintestinal infections in humans. Here, we report the identification of 14 virulence genes of pathogenic E. tarda that are essential for disseminated infection, via a genome-wide analysis. We screened 490 alkaline phosphatase fusion mutants from a library of 450,000 TnphoA transconjugants derived from strain PPD130/91, using fish as an infection model. Compared to the wild type, 15 mutants showed significant decreases in virulence. Six mutants had insertions in the known virulence-related genes, namely, fimA, gadB, katB, pstS, pstC, and ssrB. Some mutants corresponded to known genes (astA, isor, and ompS2) that had not been previously shown to be involved in pathogenesis, and three had insertions in two novel genes. In vivo infection kinetics experiments confirmed the inability of these attenuated mutants to proliferate and cause fatal infection in fish. Screening for the presence of the above-described virulence genes in six virulent and seven avirulent strains of E. tarda indicated that seven of the genes were specific to pathogenic E. tarda. The genes identified here may be used to develop vaccines and diagnostic kits as well as for further studying the pathogenesis of E. tarda and other pathogenic bacteria.
Edwardsiella tarda is a facultative aerobic enterobacterium that causes hemorrhagic septicemia in fish. Infections caused by E. tarda have been reported for many commercially important cultured fish, often leading to extensive losses in both freshwater and marine aquaculture (43). E. tarda has a wide host range and is known to cause infections in higher animals, including humans (33). In humans, it causes not only gastrointestinal infections (23) but also extraintestinal infections such as myonecrosis (39), bacteremia (53), septic arthritis (2, 32), and wound infections (3).
Pathogenesis of E. tarda is multifactorial. Several potential virulence properties have been suggested to contribute to pathogenesis of E. tarda, namely, production of dermatotoxins (44) and hemolysins (21) and the ability to resist phagocyte-mediated (40) and serum-mediated (22, 26) killing and to invade epithelial cells (22, 26). Although both virulent and avirulent E. tarda strains were able to invade cultured cells in vitro, only virulent strains could enter the host, multiply, and spread to various organs, causing mortality (25). In general, very little is known about the genes responsible for virulence and their roles in E. tarda pathogenesis.
Identification of virulence genes is essential for understanding the pathogenesis of bacteria. There are several approaches that can be used for discovering infection-related genes. Some of them are in vivo expression technology (28), signature-tagged mutagenesis (19), random insertion mutagenesis (5), and transposon mutagenesis (4). Here, we have used TnphoA transposon mutagenesis, which allows specific targeting of the secreted, periplasmic, and outer membrane proteins of E. tarda. Attenuated mutants and the corresponding mutant genes were identified through a genome-wide analysis with gourami fish as an infection model. We have also carried out a survey to examine the distribution of these virulence genes in pathogenic and nonpathogenic E. tarda strains and in other pathogenic bacterial genomes. The results of this study assist in furthering our understanding of E. tarda pathogenesis and may be used for the development of diagnostic kits and vaccines for E. tarda and other enteric infections.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used in the present study are shown in Table 1. Cultures were routinely grown at 25°C on tryptic soy agar (TSA) (Difco) or in tryptic soy broth (TSB) (Difco). Stock cultures were maintained at −80°C as a suspension in supplemented TSB containing 25% (vol/vol) glycerol. When required, the antibiotics (all from Sigma) ampicillin, neomycin, and colistin were added at final concentrations of 100, 50, and 12.5 μg/ml, respectively.
TABLE 1.
Bacterial strains and vectors used in this study
| Bacterial strain or plasmid | Characteristics (source) | Reference(s) |
|---|---|---|
| E. tarda strains | ||
| PPD130/91 | Wild type, virulent, Colr AVAb, Singapore | 29, 40 |
| 1-490 | Neor, Colr, TnphoA mutants of E. tarda PPD130/91 | 29, 40 |
| Pathogenica | ||
| AL9379 | Wild type (Aubum University, United States) | 26 |
| E381 | Wild type (University of Tokyo, Japan) | Y. P. Tan and K. Y. Leung, unpublished data |
| NE8003 | Wild type (University of Tokyo, Japan) | Y. P. Tan and K. Y. Leung, unpublished data |
| NUF251 | Wild type (University of Tokyo, Japan) | Y. P. Tan and K. Y. Leung, unpublished data |
| SU226 | Wild type (University of Tokyo, Japan) | Y. P. Tan and K. Y. Leung, unpublished data |
| Nonpathogenica | ||
| AL92448 | Wild type (Aubum University, United States) | Y. P. Tan and K. Y. Leung, unpublished |
| ATCC15947T | Wild type (ATCCc, United States) | 26 |
| PPD76/87 | Wild type (AVA, Singapore) | 26 |
| PPD125/87 | Wild type (AVA, Singapore) | 26 |
| PPD129/87 | Wild type (AVA, Singapore) | 26 |
| PPD453/86 | Wild type (AVA, Singapore) | 26 |
| PPD499/84 | Wild type (AVA, Singapore) | 26 |
| E. coli strains | ||
| JM109 | Neos, Cols, Amps | Promega |
| Top10 F′ | Neos, Cols, Amps | Clontech |
| Plasmids | ||
| pGEMT Easy | Blue-white selection, Ampr | Promega |
| pBluescript SK+ | Blue-white selection, Ampr | Stratagene |
Pathogenic bacteria have LD50s of <106.5; non-pathogenic bacteria have LD50s of >106.5.
AVA, Agri-Veterinary Authority of Singapore.
ATCC, American Type Culture Collection.
Screening of mutants for attenuation and LD50 studies.
Healthy blue gourami (Trichogaster trichopterus Pallas) were obtained from a commercial fish farm, maintained in well-aerated dechlorinated water at 25 ± 2°C, and acclimatized to the laboratory conditions for at least 15 days. Fish were approximately 13 g and were about 3 months old. For screening of attenuated mutants, all 490 alkaline phosphatase (PhoA+) fusion mutants were injected individually through the intramuscular route into six naive blue gourami with a dose 1 log unit higher than the 50% lethal dose (LD50) of the wild-type E. tarda PPD130/91 (105.0). Bacterial cultures were prepared as described previously (26). Mortality of fish was recorded over a period of 7 days postinjection.
For the estimation of LD50, three groups of 10 fish each were injected intramuscularly with 0.1 ml of phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4 [pH 7.2]) containing washed bacterial cells adjusted to the required concentrations. LD50s were calculated by the method of Reed and Muench (36).
In vitro characterization of attenuated mutants.
PhoA+ fusion mutants were characterized by standard procedures for their ability to grow on TSA and in TSB and phosphate-limiting medium (PLM); production of hemolysin and catalase; and ability to withstand low-pH conditions (pH 5.8) and serum. Briefly, growth of mutants in TSB and on TSA was recorded after 24 h of incubation. Growth under phosphate-limiting conditions was examined by culturing the bacterial cells in a modified defined minimal medium (9) with phosphate salts replaced by 3 μM Na2HPO4 and the pH adjusted to 7.0 with 30 mM HEPES. Hemolysin production was determined by inoculating bacteria on TSA with 5% (vol/vol) heparinized gourami whole blood. A clear zone around a bacterial colony indicated hemolysin production. Catalase production was assayed by adding a drop of H2O2 to a fresh bacterial colony on a TSA plate. Brisk effervescence was associated with the breakdown of H2O2 by endogenous catalase (20). No effervescence was observed for mutants that lacked catalase production. The pH sensitivity of mutants was characterized by estimating their ability to grow in low-pH conditions (16). Overnight bacterial cultures were inoculated into TSB, grown for 3 h to obtain 108 CFU/ml, and inoculated into TSB at pH 5.8 ± 0.1. Subsequently, bacterial growth was monitored over a period of 24 h. Survival of the mutants in serum was determined by incubating the washed bacteria (108 CFU/ml) in 50% (vol/vol) fresh gourami serum for 1 h (48). The serum survival rate was calculated by dividing the viable bacterial population after serum treatment by the initial population. For the serum resistance assay, serum-sensitive (PPD76/87) and serum-resistant (PPD130/91) strains were used as controls. The adhesion assay was carried out in triplicate as described previously (26). All serum data were expressed as means ± standard errors of the means. The data were analyzed by using one-way analysis of variance and a Duncan multiple-range test (SAS software [SAS Institute]). P values of <0.05 were considered significant.
DNA manipulations and Southern hybridization.
Bacterial genomic DNA was extracted as described in the manual of the Genome DNA kit (BIO 101). Plasmid DNA was extracted by using QIAprepmini columns (Qiagen). Restriction endonuclease digestion was accomplished by standard methods (37). Southern blotting was performed to confirm the presence of transposon insertions in mutants by using the BluGene nonradioactive nucleic acid detection system (Invitrogen) as described previously (40). Similarly, the presence of virulence genes in different pathogenic and nonpathogenic strains of E. tarda was detected by carrying out Southern hybridization with the respective virulence genes as the probes by the protocol described above.
Cloning of chromosomal segments flanking TnphoA insertions, genome walking, and DNA sequencing.
BamHI-digested fragments of mutant genomic DNA flanking the transposon were cloned into the pBluescript SK(+) (Ampr) vector and transformed into Escherichia coli Top10F′ competent cells (Clontech). Transformants bearing TnphoA and flanking E. tarda chromosomal sequences were selected by their ability to grow on LB agar containing ampicillin and neomycin. These clones were later sequenced as described below.
To obtain full-length sequences of all of the mutants, a genome walker library of wild type E. tarda PPD130/91digested with EcoRV, PvuII, and StuI was constructed according to the procedure described in the Universal Genome Walker kit (Clontech). PCR amplification was performed with primers that are specific to known upstream nucleotide sequences of mutants and the adapter primer 1. PCR was carried out with Advantage polymerase 2 (Clontech), and the cycling parameters were as follows: 7 cycles of 15 s at 94°C and 3 min at 72°C and 32 cycles of 15 s at 94°C and 3 min at 67°C. The amplified fragments were cloned into the pGEMT Easy vector system (Promega), transformed into E. coli JM109 competent cells, and sequenced with adapter primer 1 and mutant-specific primers.
DNA sequencing was carried out on an ABI PRISM377 automated DNA sequencer by using the ABI PRISMBigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems). Sequence assembly and further editing were carried out with DNASIS DNA analysis software (Hitachi Software). BLASTX and BLASTP sequence homology and protein conserved-domain analyses (CD search) were performed by using the National Center for Biotechnology Information BLAST network service.
In vivo characterization of attenuated mutants.
An intramuscular route of administration was used to study the infection kinetics of E. tarda in vivo. Briefly, fish were injected with 1.0 × 105 CFU of E. tarda PPD130/91 (colistin resistant [Colr]) and mutants 19, 135, and 260 (Colr Neor) separately. A control group of fish were injected with 0.1 ml of PBS. Four fish from each group were sampled on days 1, 3, 5, and 7 postinfection. The gall bladder, spleen, kidney, intestine, liver, and heart were aseptically removed. Blood was aseptically collected from the caudal vein. A piece of body muscle from the site of injection, measuring approximately 1 by 1 cm, was also taken. Samples from each treatment were pooled based on organ type and put into sterile sample bags (Whirl-Pak). One milliliter of PBS was added to all of the sample bags, and the contents were homogenized with a Stomacher Lab-Blender (model 80; Seward Medical). The homogenized samples were serially diluted in PBS, plated in triplicate on appropriate media (either TSA with colistin for PPD134/91 or TSA with colistin and neomycin for mutants), and incubated at 25°C for 48 h.
RESULTS AND DISCUSSION
Screening of attenuated mutants.
Secreted and outer membrane proteins are known to play an important role in causing infection (15). In the present study, we have employed TnphoA transposon mutagenesis to target these proteins, which have been shown to be vital for virulence in other pathogenic bacteria, such as Mycobacterium tuberculosis (7), Vibrio cholerae (1), Salmonella enterica serovar Typhimurium (24), and Pseudomonas aeruginosa (42). We have constructed a library of 490 PhoA+ fusion mutants from a total of 450,000 transconjugants derived from E. tarda PPD130/91 (29, 40). No identical mutants were found when we screened for five virulence factors, namely, the production of siderophores and catalase, motility, serum resistance, and stimulation of reactive oxygen intermediates in phagocytes. We were able to isolate corresponding mutants for any phenotype for which screening systems were available. The results indicated that the library was a good collection of random and nonredundant mutants. In this study, 490 PhoA+ mutants were individually injected intramuscularly into gourami fish at a dose equivalent to 10 times the LD50 of the wild type. The wild type and 402 PhoA+ fusion mutants killed all six injected fish within 3 to 4 days (Table 2). Eleven mutants could not kill any fish, and four killed one fish out of six. The in vivo screening with fish allowed us to identify mutants that may be deficient in invading, surviving, and replicating within the host by overcoming the host defense mechanisms. We have thus succeeded in identifying 15 attenuated mutants involved in the virulence of E. tarda by using fish as an infection model.
TABLE 2.
Screening for attenutated PhoA+ mutants of E. tarda PPD130/91 with fish as a model
| Strain(s) | No. of fish deada (% mortality) | Description |
|---|---|---|
| E. tarda PPD130/91 | 6 (100) | Virulent |
| E. tarda PPD125/87 | 0 (0) | Avirulent |
| 402 PhoA+ mutants | 6 (100) | |
| 40 PhoA+ mutants | 5 (83) | |
| 14 PhoA+ mutants | 4 (67) | |
| 13 PhoA+ mutants | 3 (50) | |
| 6 PhoA+ mutants | 2 (33) | |
| 4 PhoA+ mutants | 1 (17) | Slightly attenuated |
| 11 PhoA+ mutants | 0 (0) | Moderately and highly attenuated |
All of the strains were individually injected intramuscularly into six gourami fish at 106 CFU, and the mortality was recorded at 7 days postinjection.
LD50 determination and Southern hybridization with attenuated mutants.
The 15 mutants had LD50s ranging from 1.2 to more than 3 log units higher than that for the wild-type PPD130/91. They were divided into three groups, i.e., highly, moderately, and slightly attenuated mutants, depending on the level of attenuation (Table 3).
TABLE 3.
Characterization of PhoA+ fusion mutants derived from E. tarda PPD130/91
| Strain | Growth in: |
Acid tolerancec | Motilityd | Catalasee | Serum resistancef (mean ± SD) | LD50 | ||
|---|---|---|---|---|---|---|---|---|
| TSBa | TSAb | PLMa | ||||||
| PPD130/91 (wild type) | ++ | ++ | + | R | ++ | + | 1.5 ± 0.1 a | 105.0 |
| Highly attenuated mutants | ||||||||
| 135 | ++ | + | − | R | ++ | + | 1.0 ± 0.2 b,c | >108.0 |
| 227 | ++ | + | − | R | ++ | + | 1.1 ± 0.1 b,c | >108.0 |
| 280 | ++ | + | − | R | ++ | + | 1.1 ± 0.2 b,c | >108.0 |
| Moderately attenuated mutants | ||||||||
| 247 | ++ | ++ | + | R | ++ | + | 1.2 ± 0.1 b | 108.0 |
| 249 | ++ | ++ | + | R | ++ | + | 1.3 ± 0.2 b | 107.2 |
| 257g | ++ | ++ | + | R | ++ | + | 1.1 ± 0.1 b,c | 107.9 |
| 260 | ++ | ++ | + | S | ++ | + | 1.2 ± 0.2 b | 107.5 |
| 305 | ++ | ++ | + | R | ++ | + | 1.2 ± 0.1 b | 107.7 |
| 271h | ++ | ++ | + | R | ++ | + | 0.9 ± 0.1c | 107.6 |
| 309h | ++ | ++ | + | R | ++ | − | 1.0 ± 0.1 b,c | 107.9 |
| 364 | ++ | ++ | + | R | ++ | + | 1.2 ± 0.2 b | 108.0 |
| Slightly attenuated mutants | ||||||||
| 2A | + | ± | + | R | ++ | + | 1.5 ± 0.1 a | 106.2 |
| 19 | + | ± | + | R | + | + | 1.5 ± 0.2 a | 106.3 |
| 34 | + | ± | + | R | − | − | 1.7 ± 0.1 a | 106.6 |
| 337 | ++ | ++ | + | R | ++ | + | 1.1 ± 0.4 b,c | 106.6 |
+, little growth; ++, significant growth.
±, pinpoint growth; +, little growth; ++, significant growth.
R, resistant; S, sensitive.
−, nonmotile; +, slightly motile; ++, motile.
−, no effervescence; +, more effervescence.
Values with the same letters do not differ significantly (P > 0.05).
Mutant 257 has a triple transposon insertion.
Mutants 271 and 309 have double transposon insertions.
Southern hybridization was performed with the transposon as the probe for all of the mutants except 2A and 34, since they have been previously reported to have single transposon insertions (29). All of the mutants except mutants 257, 271, and 309 had single transposon insertions in their genomic DNA (data not shown). Mutants 271 and 309 had double transposon insertions, and mutant 257 had triple transposon insertions. The hybridized fragments of all mutants were larger than 8.0 kb, which is the size of the transposon. No band was found for the PPD130/91 genomic DNA.
Cloning and sequence analyses of disrupted genes.
The genes interrupted by transposon insertions in each of the 15 mutants were cloned and sequenced. Sequence analyses showed that 12 mutants had insertions in different gene loci with significant homology to known genes (Table 4). Mutants 249, 257, and 305 had the transposon inserted in unknown genes. All these mutants were grouped based on the level of attenuation and the gene products encoded by mutated genes, as described below.
TABLE 4.
Genes disrupted by transposon insertion in PhoA+ fusion mutants of E. tarda
| Mutant | Accession no. | Sequence, bp (aa)c | Identity |
Putative or known function | |||
|---|---|---|---|---|---|---|---|
| Gene | Bacterium | % (spand) | Accession no. | ||||
| Highly attenuated | |||||||
| 135 | AY090560 | 969 (323) | pstC | E. cloacae | 86 (271) | BAA22862 | Peripheral membrane protein C |
| 227 | AY090560 | 777 (259) | pstB | E. tarda | 100 (259) | AF324340 | ATP binding protein B |
| 280 | AF491965 | 1,041 (346) | pstS | E. tarda | 100 (313) | AF324342 | Phosphate binding protein |
| Moderately attenuated | |||||||
| 247 | AY78508 | 1,750 (446) | isor | S. enterica serovar Typhimurium | 84 (439) | NP_459829 | Iron-sulfur oxidoreductase |
| 249 | AF326582 | 1,000 (97) | orfA | NAe | NA | NA | No homolog |
| 257a | AY078507 | 1,589 (357) | orf20 | Y. enterocolitica | 27 (348) | CAB46587 | Hypothetical protein |
| AF492456 | 571+ (171+) | ssrB | S. enterica serovar Typhimurium | 39 (147) | CAB09344 | Secretory system regulator | |
| AF491963 | 562+ (187+) | citC | S. enterica serovar Typhimurium | 65 (187) | AAL19575 | Citrate lyase ligase | |
| 260 | AY078505 | 1,800 (464) | gadB | E. coli | 78 (463) | NP_310125 | Glutamate decarboxylase isozyme |
| 271b | AY078509 | 1,563 (427) | ompS2 | S. enterica serovar Typhimurium | 66 (372) | Q56111 | Outer membrane protein |
| AF492456 | 571+ (171+) | ssrB | S. enterica serovar Typhimurium | 39 (147) | CAB09344 | Secretory system regulator | |
| 305 | AF326582 | 1,000 (97) | orfA | NA | NA | NA | No homolog |
| 309b | AY078506 | 1,900 (552) | katB | P. aeruginosa | 77 (496) | Q59635 | Catalase precursor |
| AF492456 | 571+ (171+) | ssrB | S. enterica serovar Typhimurium | 39 (147) | CAB09344 | Secretory system regulator | |
| 364 | AF492456 | 361+ (171+) | ssrB | S. enterica serovar Typhimurium | 39 (147) | CAB09344 | Secretory system regulator |
| Slightly attenuated | |||||||
| 2A | AF324343 | 619+ (206+) | astA | E. amnigenus | 74 (206) | AF012826 | Arylsulfate transferase |
| 19 | AF491964 | 600 (154) | fimA | S. marcescens | 65 (180) | P22595 | Fimbrial protein precursor |
| 34 | AY078506 | 1,900 (552) | katB | P. aeruginosa | 77 (496) | Q59635 | Catalase precursor |
| 337 | AY078510 | 547+ (245+) | mukF | E. coli | 80 (180) | P36567 | Killing factor |
Mutant 257 has a triple transposon insertion.
Mutants 271 and 309 have double transposon insertions.
Length of the nucleotide sequence obtained. +, partial sequence.
Span, percent identity over total amino acids.
NA, not applicable.
Highly attenuated mutants with mutations in the PST operon.
All three mutants (mutants 135, 227, and 280) in the highly attenuated group had LD50s more than 3 log units higher than that of PPD130/91 (Table 3). They had insertions in the pstSCAB-phoU operon, which is required for phosphate transport and is a part of a phosphate (Pho) regulon. This is a high-affinity phosphate-specific transport (PST) operon belonging to the family of ATP binding cassette (ABC) transporters and includes five genes encoding PstS, a phosphate binding protein; PstC and PstA, transmembrane proteins; PstB, an ATP binding protein; and PhoU, a protein required for repression of the Pho regulon (49). The PST operon transports inorganic phosphate (Pi) from the external environment into the bacteria and is induced when the availability of Pi is limited (<4 μM). Low Pi concentrations are found in intracellular spaces of phagocytic and epithelial cells, and this may stimulate a bacterial response to switch on virulence genes so as to survive and replicate within the host. Valdivia and Falkow (45) also identified the phoS (pstS) gene as a macrophage-inducible gene in S. enterica serovar Typhimurium.
Initial characterization of these mutants showed that they formed smaller colonies on TSA than the wild type and were growth deficient in PLM, indicating that these mutants are deficient in the transport of phosphate (Table 3). All three mutants were serum resistant; however, they showed significantly lower replication rates during the 1-h serum treatment than the wild type and were highly attenuated. Daigle and coworkers (10) reported that mutation in pstC gene of an E. coli strain reduced the serum resistance and also the pathogenicity, making it unable to cause septicemia in pigs. Since Pi is an essential element required for most of the biochemical and physiological processes in bacteria, mutation in this operon may affect the uptake of Pi, thereby leading to growth inhibition or the death of the bacteria.
Disruption of genes upstream of phoU may affect the function of phoU due to the polar effect. PhoU is known to act as a repressor of the Pho regulon (35), which includes a PhoB-PhoR two-component regulatory system. Mutation of phoB in V. cholerae has been shown to affect intestinal colonization and therefore pathogenesis (47). Analysis of the pstS mutant of S. enterica serovar Typhimurium indicated that hilA and invasion genes were repressed by the response regulator PhoB in the absence of the PST high-affinity inorganic phosphate uptake system (27). Thus, mutations in PST operon genes may either directly affect the uptake of Pi or indirectly affect the expression of virulence-related genes, leading to attenuation.
Moderately attenuated mutants. (i) Regulatory mutants.
Out of eight moderately attenuated mutants, four (mutants 257, 271, 309, and 364) had transposon insertions in different positions of a gene with 45% identity to the secretory system regulatory gene (ssrB) of S. enterica serovar Typhimurium. This gene is known to regulate the type III secretion system (TTSS), a contact-dependent secretory system of Salmonella pathogenicity island 2. Salmonella pathogenicity island 2 plays a central role in systemic infection and in intracellular pathogenesis. The TTSS is required for replication inside macrophages and for systemic infection (18). Mutations in the ssrB gene in S. enterica serovar Typhimurium lead to a loss of virulence (38), as also observed for our mutants. Worley and coworkers (52) have also shown that SsrB activates the global regulon of horizontally acquired genes. Growth of these mutants on TSA and in TSB and PLM was not affected and all were serum resistant, but all had significantly lower replication rates in serum than the wild type (Table 3). None of the ssrB mutants caused severe infection in fish; they all had LD50s well above 100 times that of the wild type. As in the case of Salmonella, SsrB may play an important role in pathogenesis of E. tarda. Further research is required to elucidate whether E. tarda has a pathogenicity island required for the TTSS. Since E. tarda replicates inside phagocytes (40) and is also biochemically similar to Salmonella (23), it might have a similar cluster of genes to facilitate systemic infection.
(ii) Secretion mutants.
Three of the mutants (mutants 260, 247, and 309) had insertions in the secreted enzymes GadB, Isor, and KatB, respectively. These enzymes are known to play an important role in providing resistance to bacteria towards phagocyte-mediated killing inside the host. Mutant 260 had single insertion in the gadB gene, which is involved in acid resistance (AR) in E. coli (8). Acidic conditions are common inside phagosomes and the gastrointestinal tract. In order to cause infection, it is necessary for the bacteria to survive in the harsh acidic environment inside phagocytes. Mutant 260 was also sensitive to acidic conditions in vitro (Table 3). E. coli has three different systems for AR, namely, oxidative or glucose-repressed oxidative AR, arginine-dependent AR, and glutamate-dependent AR systems (8). The glutamate-dependent AR system requires the glutamate decarboxylase gene for protection under acidic conditions. The Gad system neutralizes acidity and enhances survival under extreme acid conditions. Transposon insertion in the gadB gene of E. tarda resulted in attenuation of the mutant in vivo and acid sensitivity in vitro, indicating that the mutant was unable to survive and cause infection inside the host.
In order to survive inside the host, microbes have developed complex strategies to avoid or overcome the damaging effects of reactive oxygen species (ROS). We have found an attenuated mutant (mutant 247) that has an insertion in a gene homologous to a putative Fe-S oxidoreductase (isor). Sequence analysis of the isor gene indicated the presence of a CXXXCXXC domain, which is known to bind iron in the 4Fe-4S form (41). Since 4Fe-4S enzymes act as “circuit breakers” halting the production of toxic ROS by temporarily stopping cellular oxidative metabolism, they help the bacteria to survive the microbicidal action of ROS (30). This is the first report that disruption of the isor gene leads to attenuation of pathogenic bacteria.
Mutants 34 and 309 had insertions in the catalase (katB) gene at different positions. Catalase breaks down toxic H2O2 into water and oxygen. H2O2 is an ROS, which can damage cellular constituents such as cell membranes, enzymes, and DNA. H2O2 is produced by phagocytes for microbicidal action. Mutant 34 had a single insertion and was slightly attenuated, having an LD50 1.5 log units higher than that of the wild type (29), while mutant 309 had double transposon insertions, one in the katB gene and another in the ssrB gene, and had an LD50 2 to 3 log units higher than that of the wild type. Neither of these two mutants produced effervescence upon contact with H2O2, indicating that they had impaired catalase production (Table 3). Catalase has been shown to be a virulence factor in bacterial pathogens such as Haemophilus influenzae (6) and Mycobacterium bovis (50). Although there were some variations in the two catalase mutants (mutants 34 and 309) of E. tarda, both of them were attenuated in fish, indicating the role of these mutations in virulence.
(iii) Outer membrane protein mutant.
Outer membrane proteins (Omps) are known to play a protective role for pathogenic bacteria by causing resistance to serum- and complement-mediated killing. DsrA, an outer membrane protein of Haemophilus ducreyi, conferred resistance to serum-mediated killing (12). Mutant 271 had an insertion in a gene having 75% identity to ompS2 of S. enterica serovar Typhimurium and was also slightly serum sensitive (Table 3). Serum resistance has been shown to be critical for survival and establishment of disease in the host for several bacteria. The disruption of the omp gene might have led to the serum sensitivity, thereby making the mutant avirulent. Mutant 271 also had another insertion in the ssrB gene. Further studies with isogenic mutations in these genes would help in understanding their individual role in the pathogenesis of E. tarda.
(iv) Other virulence gene mutants.
Mutant 257 had insertions in a gene having homology to a hypothetical protein of Y. enterocolitica (orf20) and the citrate lyase ligase gene (citC) of E. coli. orf20 is one of several genes present in the high-pathogenicity island of Y. enterocolitica (34). The high-pathogenicity island is known to mediate biosynthesis and uptake of the siderophore yersinibactin and to contribute to the mouse-lethal phenotype. The citrate lyase ligase gene (citC) is one of the five genes in the citrate lyase operon. This operon helps in cleaving citrate to oxaloacetate and acetate. Since mutant 257 had triple insertions, association of citC with virulence of bacteria could be validated only by testing for attenuation of isogenic mutants.
Two of the mutants (249 and 305) had single transposon insertions at different positions in a novel gene (orfA). This gene may encode for an important secreted or membrane protein, since these mutants were PhoA+. Further studies are required to elucidate the function of this gene.
Slightly attenuated mutants. (i) Adherence mutants.
Fimbriae are known to help in adhesion of bacteria to host cells (11). Mutant 19 had an insertion in the fimA gene, the first gene in fimABCDE operon. Compared to the wild-type E. tarda PPD130/91, the adherence to epithelioma papillosum of carp cells by mutant 19 was 5 times lower (adhesion to epithelioma papillosum of carp cells by PPD130/91 and mutant 19 was 3.9 ± 0.7 and 0.8 ± 0.2, respectively [n = 3]). Adherence is the primary step for invading the host and causing infection. Hence, any mutation in genes involved in attachment would affect the infective ability of the bacteria, which would have led to attenuation of this mutant. Vander Velden and coworkers (46) also have shown that multiple fimbrial adhesins are required for full virulence of S. enterica serovar Typhimurium in mice. They reported that mutations in fimbrial operons reduced the virulence of the bacteria.
(ii) Other mutants.
Mutant 337 had a transposon insertion in a gene having homology to mukF (kicB) of E. coli (Table 4). In E. coli, a null mutation of the mukF gene led to temperature-dependent colony formation, anucleate cell production, chromosome cutting by septum closure, and hypersensitivity to novobiocin (31). MukF is present in a complex of MukF (KicB)-MukE (KicA) and MukB proteins. Feng and coworkers (14) suggested that the kicB (mukF) gene encodes a killing factor which kills cells and that the kicA gene codes for a protein that suppresses the killing function of kicB gene product. They also showed that a kicA null mutant is nonviable but that kicB mutants are able to grow. In the present study we found that a mutation in mukF gene led to a slight attenuation of E. tarda, indicating its possible role in affecting virulence. Mutants 2A, with an insertion in astA, and 34, with an insertion in katB, were deficient in siderophore and catalase production, respectively, rendering them slightly attenuated, as discussed elsewhere (29).
In vivo infection kinetics of attenuated mutants.
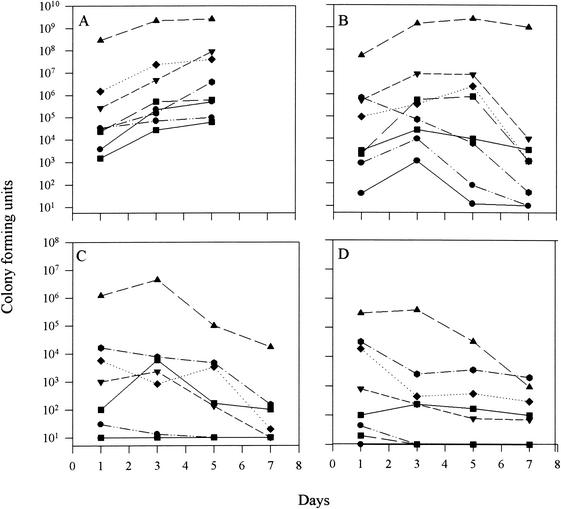
Infection kinetics experiments were carried out to investigate the ability of the mutants to proliferate and cause infection in vivo. This measures the potential of the bacteria to invade, survive, colonize, and replicate in various organs of the host. An intramuscular infection model was used to simulate infection by physical injury under normal conditions. Hence, one representative mutant from each of the three groups described above and the wild type were injected separately into fish, and the infection kinetics was studied over a period of 7 days. Fish injected with PPD130/91 showed high numbers of bacteria in all of the different organs, followed by mutants 19 (slightly attenuated), 260 (moderately attenuated), and 135 (highly attenuated) (Fig. 1). By day 1, wild-type PPD130/91 could survive, colonize, and proliferate to reach very high numbers in all of the organs sampled, thereby causing mortality of fish within 3 to 5 days (Fig. 1A). In the case of mutant 19, the bacterial numbers were slightly lower than for the wild type but higher than for mutants 260 and 135 (Fig. 1B). Mutant 19 could survive and colonize in the muscle and reached levels comparable to that of wild type, but it still could not cause fish mortality. The bacterial numbers of this mutant were reduced to lower levels in all of the organs except muscle by 3 days postinfection. For both mutants 260 and 135, bacteria could not survive and colonize in all of the organs and were possibly killed by the host defense mechanisms (Fig. 1C and D). None of the fish succumbed to infection when injected with mutants 260 and 135. Fish injected with the wild type and mutant 19 showed hemorrhages around the site of injection, which persisted and progressed in the case of the wild type but decreased and healed in the fish injected with mutant 19. For the fish injected with mutants 260 and 135, no hemorrhages were observed at any time during the experiment. This clearly indicated that the wild type and mutant 19 could survive and colonize in muscle and cause an extensive inflammatory response compared to mutants 260 and 135. Infection kinetics experiments also demonstrated the inability of mutants with defective fimA, gadB, and pstC genes to establish fatal infection in fish.
FIG. 1.
Infection kinetics of E. tarda strains in blue gourami. Fish were injected with 1.0 × 105 CFU of PPD130/91 (A), mutant 19 (B), mutant 260 (C), and mutant 135 (D). Four fish each were sampled per datum point, and the mean numbers of E. tarda per sample in triplicates are shown. For PPD130/91, sampling was done until day 5, when all of the fish died due to severe infection. Fish from the other groups were sampled until day 7. Blood (—•—), body muscle (▴), liver (▾), kidney (⧫), gall bladder (- -▪- -), spleen ( ), heart (· · - - · ·), and intestine (—▪—) were dissected and homogenized, and bacterial enumeration was done by plating onto TSA supplemented with appropriate antibiotics.
Distribution of virulence genes in E. tarda and other pathogens.
Pathogenic bacteria may have virulence genes that are absent in nonpathogenic bacteria, making them virulent. Virulence genes may also be present in both pathogenic and nonpathogenic bacteria but may be functional only in pathogenic ones. Hence, we determined whether the 14 genes discussed above are specific to pathogenic bacteria. Southern hybridization was carried out to survey the presence of the above-described genes in six different virulent and seven avirulent strains of E. tarda (Table 5). Our results indicated that seven of the genes, namely, orfA, citC, fimA, gadB, katB, mukF, and ssrB, were present only in virulent and not in avirulent strains, indicating that they are specific to pathogenic E. tarda (Table 5). These genes can therefore be used as biomarkers to perform diagnosis of pathogenic E. tarda. Grant and coworkers (17) have used gadAB genes as a prescreening marker for detection of pathogenic E. coli groups.
TABLE 5.
Distribution of virulence genes in various virulent and avirulent strains of E. tarda
| Strain | Presence of virulence gene |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| orf20 | orfA | citC | fimA | gadB | katB | mukF | ssrB | astA | isor | ompS2 | pstB | pstC | pstS | |
| Virulent E. tarda | ||||||||||||||
| PPD130/91 | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| AL9379 | − | + | + | + | + | + | + | + | + | + | + | + | + | + |
| E381 | − | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NE8003 | − | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NUF251 | − | + | + | + | + | + | + | + | + | + | + | + | + | + |
| SU226 | − | + | + | + | + | + | + | + | + | + | + | + | + | + |
| No. positive/total | 1/6 | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 |
| Avirulent E. tarda | ||||||||||||||
| AL92448 | − | − | − | − | − | − | − | − | + | + | + | + | + | + |
| ATCC 15947T | − | − | − | − | − | − | − | − | + | + | + | + | + | + |
| PPD76/87 | − | − | − | − | − | − | − | − | + | + | + | + | + | + |
| PPD125/87 | − | − | − | − | − | − | − | − | + | + | + | + | + | + |
| PPD129/87 | − | − | − | − | − | − | − | − | + | + | + | + | + | + |
| PPD453/86 | − | − | − | − | − | − | − | − | + | + | + | + | + | + |
| PPD499/84 | − | − | − | − | − | − | − | − | + | + | + | + | + | + |
| No. positive/total | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 7/7a | 7/7a | 7/7a | 7/7a | 7/7a | 7/7a |
A hybridization band pattern different from that of pathogenic strains was seen in all of the nonpathogenic strains.
A gene having homology to a Y. enterocolitica hypothetical gene (orf20) was present only in E. tarda PPD130/91 and not in any of the other strains, and it thus may be specific to this strain. Other genes, such as astA, isor, ompS2, and pstSCB, hybridized to the DNAs of most of the strains, but with a different profile (data not shown). This may be due to polymorphism in the genetic organization in the various E. tarda strains. The Omp may have certain motifs that are present in many Omps that could result in cross hybridization.
Biocomputational analysis was carried out to determine the distribution of the these genes in other pathogenic bacteria. Most of the 14 virulence genes required for E. tarda infection have related sequences in other common human pathogens, such as enterohemorrhagic E. coli, S. enterica serovar Typhimurium, V. cholerae, and others (Table 6). Genes such as fimA, pstS, and ssrB play an important role in pathogenesis of S. enterica serovar Typhimurium (18, 27, 51). Some other genes are known to contribute to virulence in other bacterial pathogens whose complete genomes are not available. This is not unexpected given the genetic relatedness of some of these bacteria and their similarity as enteric and systemic pathogens. These common virulence genes will be useful in determining how pathogenic bacteria interact with the host and cause systemic infections. They may also form the basis for the design of novel therapeutics and common antigens in vaccine development to protect hosts against systemic diseases.
TABLE 6.
Distribution of virulence genes in other pathogenic bacteria and their E values
| Strain | E valuea |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| orf20 | orfA | citC | fimA | gadB | katB | mukF | ssrB | astA | isor | ompS2 | pstB | pstC | pstS | |
| E. coli O157:H7 | 2e-64 | 4e-11 | 0.0 | 8e-90 | 1e-78 | 0.0 | e-139 | e-126 | e-132 | e-163 | ||||
| H. influenzae Rd | c-100c | 9e-64 | 4e-37 | e-105 | 3e-92 | e-100 | ||||||||
| M. leprae | 1e-42 | 3e-67 | 2e-38 | 2e-32 | ||||||||||
| M. tuberculosis H37Rv | e-112 | 4e-05 | 2e-47 | 4e-69 | 6e-41 | 2e-40 | ||||||||
| P. aeruginosa PA01 | 0.0*d | 3e-07 | 1e-13 | 0.0 | 1e-83 | 4e-12 | 1e-09 | |||||||
| P. multocida pm70 | 6e-92 | 2e-67 | 3e-05 | 0.0 | e-105 | 9e-92 | e-108 | |||||||
| S. enterica serovar Typhimurium LT2 | 4e-66 | 3e-13*b | 9e-91 | 2e-78 | 3e-17*e | 6e-52 | 0.0 | e-137 | e-125 | e-132 | e-167*f | |||
| S. enterica subsp. enterica serovar Typhi CT18 | 1e-64 | 3e-09 | 9e-91 | 2e-78 | 3e-17 | 6e-52 | 0.0 | e-142 | e-125 | e-132 | e-168 | |||
| V. cholerae El Tor N16961 | 9e-40 | e-169 | 5e-54 | 0.0 | 5e-24 | 6e-75 | 5e-31 | 2e-12 | ||||||
| Y. pestis CO92 | 1e-05 | 1e-98 | 3e-81 | 9e-19 | 5e-40 | e-129 | e-127 | e-141 | e-166 | |||||
Conclusions.
Understanding the mechanisms of pathogenesis in bacteria involves the identification and characterization of genes that are specifically required for establishment and maintenance of persistent infection in the host. In the present study, we have successfully used TnphoA transposon mutagenesis to identify 14 genes that are involved in the pathogenesis of E. tarda via a genome-wide analysis. Fifteen of the attenuated mutants had insertions in 14 different gene loci, homologous to transporter, regulator, enzyme secretion, outer membrane protein, adhesion, and unknown functions. The identification of most of the mutations in genes homologous to known virulence factors (fimA, gadB, katB, pstC, pstS, and ssrB) validates the approach used in this study. Some of the mutations (pstS, -C, and -B) may have had polar effects on genes located downstream of the interrupted genes. We also had mutants that had multiple transposon insertions. Complementation or construction of defined deletion or knockout mutations will be necessary to determine which of the disrupted genes is responsible for attenuation.
For any bacterium to cause an infection, it has to adhere to (fimA) and invade the host. Once inside the host, bacteria have to survive and overcome defensive barriers such as serum and phagocyte-mediated killing. These are the major host defenses against septicemia infection. Hence, it is not surprising to find the genes (gadB, isor, katB, ompS2, and ssrB) which neutralize these effects in the present study. Later, during infections, bacteria need to acquire nutrients to proliferate within the host. pst genes and astA help in acquiring nutrients such as phosphate and iron within the host. Some regulatory elements (phoU and ssrB) are also probably required to enable E. tarda to switch on the necessary virulence genes to establish itself inside fish, causing severe systemic infection and eventually death. Here we have made an attempt to connect all of the genes and postulate their combined roles in the systemic infection.
This study has allowed us to identify E. tarda genes required to cause infection in fish. Further study of these genes will improve our understanding of the mechanisms of E. tarda pathogenesis. The genes that were present only in pathogenic bacteria can now be used for developing diagnostic reagents to identify the pathogenic E. tarda strains. Some of the genes identified here may also be future vaccine candidates.
Acknowledgments
We are grateful to the National University of Singapore for providing a research grant for this work.
We are grateful to John Grizzle (Auburn University), H. Wakabayashi (University of Tokyo), and T. Ngiam and H. Loh (Agri-food and Veterinary Authority of Singapore) for providing us with E. tarda isolates from the United States, Japan, and Singapore, respectively. We also acknowledge J. A. Mathew, who carried out the mutagenesis of E. tarda PPD130/91. We thank Peter Howard and Shashikant Joshi for their constructive criticism.
Editor: B. B. Finlay
REFERENCES
- 1.Ali, A., J. A. Johnson, A. A. Franco, D. J. Metzier, T. D. Connell, J. G. Morris, Jr., and S. Sozhamannan. 2000. Mutations in the extracellular protein secretion pathway genes (eps) interfere with rugose polysaccharide production in and motility of Vibrio cholerae. Infect. Immun. 68:1967-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashford, R. U., P. D. Sargeant, and G. D. Lum. 1998. Septic arthritis of the knee caused by Edwardsiella tarda after a catfish puncture wound. Med. J. Aust. 168:443-444. [DOI] [PubMed] [Google Scholar]
- 3.Banks, A. S. 1992. A puncture wound complicated by infection with Edwardsiella tarda. J. Am. Pediatr. Med. Assoc. 82:529-531. [DOI] [PubMed] [Google Scholar]
- 4.Berg, C. M., D. E. Berg, and E. A. Groisman. 1994. Transposable elements and the genetic engineering of bacteria, p. 880-925. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 5.Bijlsma, J. J. E., C. M. J. E. Vandenbroucke-Grauls, S. H. Phandis, and J. G. Kusters. 1999. Identification of virulence genes of Helicobacter pylori by random transposon mutagenesis. Infect. Immun. 67:2433-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishai, W. R., N. S. Howard, J. A. Winkelstein, and H. O. Smith. 1994. Characterization and virulence analysis of catalase mutants of Haemophilus influenzae. Infect. Immun. 62:4855-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braunstein, M., T. J. Griffin, I. V., J. I. Kriakov, S. T. Freidman, N. D. F. Grindley, and W. R. Jacobs, Jr. 2000. Identification of genes encoding exported Mycobacterium tuberculosis proteins using a Tn552phoA in vitro transposition system. J. Bacteriol. 182:2732-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casterine-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, L., and R. L. Thune. 1996. Development of a defined minimal medium for the growth of Edwardsiella ictaluri. Appl. Environ. Microbiol. 62:848-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daigle, F., J. M. Fairbrother, and J. Harel. 1995. Identification of a mutation in the pst-phoU operon that reduces pathogenicity of an Escherichia coli strain causing septicemia in pigs. Infect. Immun. 63:4924-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duguid, J. P., and D. C. Old. 1980. Adhesive properties of Enterobacteriaceae, p. 185-217. In E. H. Beachey (ed.) Bacterial adherence receptors and recognition, vol. 6. Chapman and Hall, London, United Kingdom.
- 12.Elkins, C., K. H. Morrow, Jr., and B. Olsen. 2000. Serum resistance in Haemophilus ducreyi requires outer membrane protein DsrA. Infect. Immun. 68:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkins, J. G., D. J. Hassett, P. S. Stewart, H. P. Schweizer, and T. R. McDermott. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl. Environ. Microbiol. 65:4594-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng, J., K. Yamanaka, H. Niki, T. Ogura, and S. Hiraga. 1994. New killing system controlled by two genes located immediately upstream of the mukB gene in Escherichia coli. Mol. Gen. Genet. 43:136-147. [DOI] [PubMed] [Google Scholar]
- 15.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster, J. W., and H. K. Hall. 1990. Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 172:771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant, M. A., S. D. Weagant, and P. Feng. 2001. Glutamate decarboxylase genes as a prescreening marker for detection of pathogenic Escherichia coli groups. Appl. Environ. Microbiol. 67:3110-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hensel, M. 2000. Salmonella pathogenicity island 2. Mol. Microbiol. 36:1015-1023. [DOI] [PubMed] [Google Scholar]
- 19.Hensel, M., J. E. Shea, M. D. Gleeson, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 20.Hertel, C., G. Schmidt, M. Fischer, K. Oellers, and W. P. Hammes. 1998. Oxygen-dependent regulation of the expression of the catalase gene katA of Lactobacillus sakei LTH677. Appl. Environ. Microbiol. 64:1359-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirono, I., N. Tange, and T. Aoki. 1997. Iron regulated hemolysin gene from Edwardsiella tarda. Mol. Microbiol. 24:851-856. [DOI] [PubMed] [Google Scholar]
- 22.Janda, J. M., S. L. Abott, S. Kroske-Bystrom, W. K. W. Cheung, C. Powers, R. P. Kokka, and K. Tamura. 1991. Pathogenic properties of Edwardsiella species. J. Clin. Microbiol. 29:1997-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janda, J. M., and S. L. Abott. 1993. Infections associated with the genus Edwardsiella: the role of Edwardsiella tarda in human disease. Clin. Infect. Dis. 17:742-748. [DOI] [PubMed] [Google Scholar]
- 24.Kwan, L. Y., and R. E. Isaacson. 1998. Identification and characterization of a phase-variable nonfimbrial Salmonella typhimurium gene that alters O-antigen production. Infect. Immun. 66:5725-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling, S. H. M., X. H. Wang, T. M. Lim, and K. Y. Leung. 2001. Green fluorescent protein-tagged Edwardsiella tarda reveals portal of entry in fish. FEMS Microbiol. Lett. 194:239-243. [DOI] [PubMed] [Google Scholar]
- 26.Ling, S. H. M., X. H. Wang, L. Xie, T. M. Lim, and K. Y. Leung. 2000. Use of green fluorescent protein (GFP) to track the invasive pathways of Edwardsiella tarda in the in vivo and in vitro fish models. Microbiology 146:7-19. [DOI] [PubMed] [Google Scholar]
- 27.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 29.Mathew, J. A., Y. P. Tan, P. S. Srinivasa Rao, T. M. Lim, and K. Y. Leung. 2001. Edwardsiella tarda mutants defective in siderophore production, motility, serum resistance and catalase activity. Microbiology 147:449-457. [DOI] [PubMed] [Google Scholar]
- 30.Miller, R. A., and B. E. Britigan. 1997. Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 10:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onogi, T., M. Yamazoe, C. Ichinose, H. Niki, and S. Hiraga. 2000. Null mutation of the dam or seqA gene suppresses temperature-sensitive lethality but not hypersensitivity to novobiocin of muk null mutants. J. Bacteriol. 182:5898-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osiri, M., T. Tantawichien, and U. Deesomchock. 1997. Edwardsiella tarda bacteremia and septic arthritis in a patient with diabetes mellitus. Southeast Asian J. Trop. Med. Public Health 28:669-672. [PubMed] [Google Scholar]
- 33.Plumb, J. A. 1993. Edwardsiella septicaemia, p. 61-79. In V. Inglis, R. J. Roberts, and N. R. Bromage (ed.), Bacterial diseases of fish. Cambridge University Press, Cambridge, United Kingdom.
- 34.Rakin, A., C. Noelting, S. Schubert, and J. Heesemann. 1999. Common and specific characteristics of the high-pathogenicity island of Yersinia enterocolitica. Infect. Immun. 67:5265-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao, N. N., and A. Torriani. 1990. Molecular aspects of phosphate transport in Escherichia coli. Mol. Microbiol. 4:1083-1090. [DOI] [PubMed] [Google Scholar]
- 36.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent end points. Am. J. Hyg. 27:493-497. [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slaven, E. M., F. A. Lopez, S. M. Hart, and C. V. Sanders. 2001. Myonecrosis caused by Edwardsiella tarda: a case report and case series of extraintestinal E. tarda infections. Clin. Infect. Dis. 32:1430-1433. [DOI] [PubMed] [Google Scholar]
- 40.Srinivasa Rao, P. S., T. M. Lim, and K. Y. Leung. 2001. Opsonized virulent Edwardsiella tarda strains are able to adhere to and survive and replicate within fish phagocytes but fail to stimulate reactive oxygen intermediates. Infect. Immun. 69:5689-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamarit, J., C. Gerez, C. Meier, E. Mulliez, A. Trautwein, and M. Fontecave. 2000. The activating component of the anaerobic ribonucleotide reductase from Escherichia coli. An iron-sulfur center with only three cysteines. J. Biol. Chem. 275:15669-15675. [DOI] [PubMed] [Google Scholar]
- 42.Tan, M. W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 96:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thune, R. L., L. A. Stanley, and R. K. Cooper. 1993. Pathogenesis of gram-negative bacterial infections in warm water fish. Annu. Rev. Fish Dis. 3:37-68. [Google Scholar]
- 44.Ullah, M. A., and T. Arai. 1983. Pathological activities of the naturally occurring strains of Edwardsiella tarda. Fish Pathol. 18:65-70. [Google Scholar]
- 45.Valdivia, R. H., and S. Falkow. 1997. Fluorescence based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 46.Vander Velden, A. W., A. J. Baumler, R. M. Tsolis, and F. Heffron. 1998. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect. Immun. 66:2803-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Von Kruger, W. M., S. Humphreys, and J. M. Ketley. 1999. A role for the PhoBR regulatory system homologue in the Vibrio cholerae phosphate-limitation response and intestinal colonization. Microbiology 145:2463-2475. [DOI] [PubMed] [Google Scholar]
- 48.Wang, X. H., H. L. Oon, G. W. P. Ho, W. S. F. Wong, T. M. Lim, and K. Y. Leung. 1998. Internalization and cytotoxicity are important virulence mechanisms in Vibrio fish epithelial cell interactions. Microbiology 144:2987-3002. [DOI] [PubMed] [Google Scholar]
- 49.Webb, D. C., H. Rosenberg, and G. B. Cox. 1992. Mutational analysis of Escherichia coli phosphate-specific transport system, a member of traffic ATPase (or ABC) family of membrane transporters. J. Biol. Chem. 267:24661-24668. [PubMed] [Google Scholar]
- 50.Wilson, T. M., G. W. de Lisle, and D. M. Collins. 1995. Effect of inhA and katG on isoniazid resistance and virulence of Mycobacterium bovis. Mol. Microbiol. 15:64-68. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, R. L., J. Elthon, S. Clegg, and B. D. Jones. 2000. Salmonella enterica serovars Gallinarum and Pullorum expressing Salmonella enterica serovar Typhimurium type 1 fimbriae exhibit increased invasiveness for mammalian cells. Infect. Immun. 68:4782-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Worley, M. J., K. H. L. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749-761. [DOI] [PubMed] [Google Scholar]
- 53.Yang, C. H., and C. K. Wang. 1999. Edwardsiella tarda bacteraemia complicated by acute pancreatitis and pyomyoma. J. Infect. 38:124-126. [DOI] [PubMed] [Google Scholar]