Abstract
As part of a search for homologous members of the Plasmodium falciparum Pf60 multigene family in the intraerythrocytic protozoan parasite Babesia canis, we report here the characterization of a cDNA of 1,115 bp, which was designated Bcvir for its potential viral origin. The Bcvir cDNA contained two overlapping open reading frames (ORFs) (ORF1 from nucleotide [nt] 61 to 486 and ORF2 from nt 417 to 919), where Bcvir15, the deduced ORF1 peptide (M1 to I141), is the main expressed product. The Bcvir cDNA was derived from an extrachromosomal dsRNA element of 1.2 kbp that was always found associated with a double-stranded RNA (dsRNA) of 2.8 kbp by hybridization, and no copy of this cDNA sequence was found in B. canis genomic DNA. Biochemical characterization of Bcvir15, by using polyclonal rabbit sera directed against recombinant proteins, indicated that it is a soluble protein which remained associated with the cytoplasm of the B. canis merozoite. Interestingly, purified immunoglobulins from the anti-glutathione S-transferase-Bcvir15 (at a concentration of 160 μg/ml) induced 50% inhibition of the in vitro growth of B. canis, and the inhibitory effect was associated with morphological damage of the parasite. Our data suggest that the extrachromosomal dsRNA-encoded Bcvir15 protein might interfere with the intracellular growth of the parasite rather than with the process of invasion of the host cell by the merozoite. Epitope mapping of Bcvir15 identified three epitopes that might be essential for the function of the protein.
Extrachromosomal double-stranded RNA (dsRNA) genetic elements are widespread in pathogenic protozoa (26, 46) and fungi (for reviews see references 31 and 47). In protozoa, dsRNAs have been described for parasites from the genera Leishmania, Trichomonas, Giardia, Eimeria, Babesia, and Cryptosporidium. As has been demonstrated for most dsRNAs from fungi, dsRNAs from protozoa appear to be closely associated with viral particles and to be encapsidated (27, 46). Most of the protozoan dsRNA viruses are organized similarly to members of the fungal virus family Totiviridae (i.e., with an unsegmented dsRNA viral genome), whose well-characterized members corresponded to the dsRNA viruses from the yeast Saccharomyces cerevisiae (47). They contain a large nonsegmented genomic dsRNA (L-dsRNA), whose size ranges from 4.3 to 7.5 kbp (46), which encodes structural proteins necessary for the encapsidation and replication of viruses, i.e., the capsid and an RNA-dependent RNA polymerase (RDRP). Smaller dsRNAs of less than 1 kbp were also described for the viruses of Trichomonas vaginalis and Eimeria nieschulzi, and those from T. vaginalis were characterized as dsRNA satellites (24, 25, 36). Another common feature of most of the pathogenic protozoa and fungal dsRNA viruses is their lack of infectivity and their maintenance in the cytoplasm environment of their host (31, 46). For this reason, it was initially assumed that these viruses play no role in host biology. However, data from fungal dsRNA viruses analysis now clearly demonstrate that dsRNA-encoded products may play a critical role in host virulence. The best-known example is the toxin responsible for the killer phenomenon in yeast (47), in which a yeast strain that produces the toxin is able to kill similar nonproducing strains. This toxin is encoded by the smaller satellite M-dsRNA from yeast viruses. Other examples concern virus-infected plant pathogenic fungi, where the presence of dsRNA elements has been reported to reduce or increase fungal virulence, thus acting on the pathogenicity induced in the infected plants (1, 31). In protozoa, although numerous data are available on the genomes of the dsRNA viruses, the precise roles of these dsRNA viruses and their encoded products in host-parasite relationships remain largely unknown. Viral density was reported to affect the growth of the Giardia parasite, and in the case of Trichomonas, the presence of dsRNA was associated with phenotypic modifications of the infected cells (23, 35, 45, 46). In the Leishmania virus, an endoribonuclease activity was demonstrated to be associated with the capsid protein (29, 30). These data strongly suggested that extrachromosomal dsRNAs might interfere with the infected cells and that their encoded products might modulate the transcription level of the infected cells. However, to date, encoded products from either larger or smaller dsRNAs from protozoan parasites have never been directly linked to a change in the host virulence or host-induced pathogenicity.
Here, in a search for a homologous sequence of the Plasmodium falciparum Pf60 multigene family in the Apicomplexan intraerythrocytic protozoan parasite Babesia canis, which is responsible for the widespread tick-borne canine babesiosis in Europe, we have identified and characterized a 1.2-kbp cDNA that was called Bcvir. This 1.2-kbp cDNA encoded a major protein of 15 kDa (Bcvir15), and we demonstrate that antibodies directed against Bcvir15 are able to inhibit the in vitro growth of B. canis. Hybridization experiments indicate that Bcvir cDNA was derived from an extrachromosomal dsRNA element of 1.2 kbp that was always associated with a 2.8-kbp dsRNA, suggesting a viral origin for Bcvir cDNA. Furthermore, our data suggest that the 1.2-kbp dsRNA might correspond to a smaller dsRNA satellite rather than an L-dsRNA from a potential B. canis virus. The putative function of the dsRNA-encoded protein Bcvir15 in the intracellular growth of B. canis and the relationship of this extrachromosomal dsRNA to the P. falciparum Pf60 multigene family are discussed.
MATERIALS AND METHODS
Parasites and in vitro culture of B. canis.
Isolates A and B from B. canis and the isolate of B. canis subsp. rossi were derived from dogs that contracted babesiosis in France and in South Africa, respectively (44). The two other isolates from B. canis (designated G and R) were collected from dogs that contracted babesiosis in southern France, like for A and B, but from other regions. These isolates, from the European and South African species of large Babesia parasites that infect dogs, were maintained in in vitro culture by using dog erythrocytes at a hematocrit of 2% in culture medium supplemented with 10% (vol/vol) normal dog serum (38).
Isolation of the Bcvir cDNA clone and DNA analysis.
A B. canis cDNA library was constructed with the ZAP Express cDNA Gigapack II Gold cloning kit (Stratagene) as previously described (12), using purified mRNA from European isolate A of B. canis. The cDNA library was immunologically screened with a rabbit polyclonal serum directed against the Pf60.1 clone from P. falciparum (11) at a 1:100 dilution. The pBK-CMV plasmids from positive clones were in vitro excised and purified with JetQuick plasmid miniprep spin columns (Genomed) (12). Double-stranded DNA was sequenced (Genome Express S.A, Grenoble, France) by using the dideoxy chain termination method (37) with T3 and T7 universal primers initially and with primers derived from the established sequence of each strand thereafter. Hybridization and dehybridization procedures for Southern and pulsed-field gel electrophoresis (PFGE) analyses with Bcvir-derived probes were performed as previously described (13). A cDNA probe derived from the single-copy gene Bc12D3 from B. canis (C. Carret et al., unpublished data) was used as a control.
RT-PCR.
Total RNA from isolate A of B. canis was extracted with the Trizol reagent (Gibco BRL) and used as starting material for reverse transcription-PCR (RT-PCR) by the two-step protocol of the enhanced avian RT-PCR kit (Sigma). Briefly, the retrotranscription step was performed during 50 min at 42°C in a final volume of 20 μl containing 250 pg of total RNA, 20 U of both enhanced avian myeloblastosis virus retrotranscriptase and RNase inhibitor, specific Bcvir cDNA primer at a final concentration of 1 μM, and the deoxyribonucleotide mix for a final concentration of 500 μM for each deoxyribonucleotide. Specific Bcvir cDNA primers used in the retrotranscription step were reverse primer P15.2 (5′-AATGACATACTCACAGGAAGC-3′) and forward primers P1 (5′-GACGTTTGATGTGATGAGGGAAGC-3′) and P5 (5′-AGGGAGCTGTCACGGAAGATT-3′). As control for DNA contamination, the enhanced avian reverse transcriptase was also omitted from the first-step reaction for each primer tested. Subsequently, one-fifth of the first-step reaction product was used as a template for the PCR, which was performed at an annealing temperature of 55°C and with the AccuTaq DNA polymerase (Sigma). The primers used to analyze the amplified products were P1-P2 (reverse primer P2, 5′-ATGAGTCTATTGACTCCTTG-3′) and P5-P15.2. As a control, an RT-PCR experiment was also performed with reverse and forward specific primers that were derived from the single-copy gene Bc12D3 from B. canis.
Northern blot analysis.
One microgram of total RNA purified with Trizol reagent (Gibco BRL) or 100 ng of mRNA purified by using the polyATtract mRNA isolation system (Promega) was separated on a 0.8% Seakem GTG agarose (FMC) gel under denaturing conditions with the NorthernMax Kit (Ambion). Subsequently, the RNA was transferred onto a Nytran N nylon membrane (Schleicher & Schuell) by using a vacuum blotter (Bio-Rad) with 10× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1mM EDTA [pH 7.7]) for 2 h under 5 × 105 Pa of pressure. The membranes were then hybridized with sense (mRNA sequence) or antisense (complementary mRNA sequence) digoxigenin (DIG)-11-UTP-labeled riboprobes from ORF1 or ORF2 sequences derived from the Bcvir cDNA in presence of the ULTRAHyb reagent (Ambion). The hybridization was developed by incubation of the membrane with an anti-DIG alkaline phosphatase-conjugated antibody followed by CPD-Star Reagent as described in the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche).
Nuclease S1 digestion assays.
Total RNA purified with Trizol reagent (Gibco BRL) was digested (50 U/μg) with nuclease S1 enzyme (Promega) for 1, 2, or 3 h at 37°C in the appropriate reaction buffer. Treated RNA was then extracted with phenol-chloroform (pH 4.7) (Sigma) and treated for Northern blotting as described above. Membranes were hybridized with the sense ORF1 riboprobe. Untreated total RNAs were used as controls. As a control for the correct digestion of single-stranded RNA, nuclease S1-digested RNA was also hybridized with an antisense riboprobe derived from the encoding region of cDNA Bc28 from B. canis (P. Cibrelus et al., unpublished data).
Recombinant protein purification.
Both glutathione S-transferase (GST)- and His6-tagged recombinant proteins, including the predicted ORF1 protein (i.e., Bcvir15), were produced. For the production of GSTBcvir15 (polypeptide A12 to I141), a 5′-incomplete Bcvir cDNA clone was excised from the pBK-CMV vector by EcoRI-XhoI enzymatic digestion, purified, and subcloned in EcoRI-XhoI-digested pGEX-4T3 vector (Pharmacia Biotech). The GSTBcvir15 protein was purified by affinity chromatography on glutathione-agarose beads (Sigma) as described elsewhere (42). For the production of (His)6Bcvir15 (polypeptide M25 to I141), the ORF1 sequence of the Bcvir cDNA was amplified by PCR with specific internal modified primers. The forward primer contained an in-frame 5′ SphI restriction site (5′-ATGAGGGAAGCATGCCTTCGTGTA-3′), and the reverse primer contained a 3′ KpnI restriction site (5′-TACCTACGTGGTACCTTCTTA-3′) in frame with the vector stop codon. The PCR product was purified, digested, and subcloned in an SphI-KpnI-digested pQE-30 vector (Qiagen). The (His)6Bcvir15 protein was purified by affinity chromatography on Ni-nitrilotriacetic acid beads (Qiagen) under denaturing conditions according to the manufacturer's instructions.
Immunization.
Two polyclonal antisera were produced against the Bcvir15 protein, using either the GSTBcvir15 or the (His)6Bcvir15 recombinant protein for immunization of rabbits. For the first immunization, New Zealand White rabbits were subcutaneously injected with 100 μg of GST- or His6-tagged recombinant Bcvir15 (emulsified in Freund's complete adjuvant or saponin adjuvant, respectively). The two subsequent injections were performed intramuscularly, with 100 μg of purified recombinant proteins, emulsified in incomplete Freund's adjuvant or saponin adjuvant, respectively. Injections of animals were performed at 4-week intervals, and serum samples were collected 8 days before each immunization.
Immunoblot analysis.
Parasitized red blood cells from in vitro cultures of B. canis were lysed in 10 ml of RPMI supplemented with 40 U of α-hemolysin from Staphylococcus aureus (Sigma) per ml in a water bath at 37°C for 30 min. The sample was centrifuged (3,500 × g, 30 min), and the pellet of free merozoites and red blood cell membranes was washed extensively in Tris-buffered saline (TBS). The merozoites were separated from contaminating erythrocyte membranes by centrifugation (3,500 × g, 30 min) through a discontinuous Percoll gradient (lower layer density of 1.09 and upper layer density of 1.02). The merozoites were retrieved at the interface of the gradient. They were centrifuged (15,000 × g, 15 min), washed, and boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer for 5 min at 100°C. The proteins were separated by electrophoresis in an SDS-15% (wt/vol) polyacrylamide gel and transferred onto nitrocellulose membranes. The membranes were saturated by incubation for 1 h at room temperature with 5% (wt/vol) skim milk powder and incubated for 1 h with the sera diluted 1:100 in TBS. The membranes were then washed extensively with TBS-0.3% (vol/vol) Tween 20 and incubated for 1 h with goat anti-rabbit immunoglobulin G (IgG) peroxidase-conjugated antibodies (Sigma) diluted 1:500 in TBS. The detection procedure was performed with 4-chloro-1-naphthol as a chromogen.
Indirect immunofluorescence assay.
Blood smears of RPMI-washed B. canis in vitro cultures were fixed with a cold methanol-acetone (1:4) mixture for 20 min at −80°C. Fixed infected erythrocytes were incubated for 1 h at room temperature with sera diluted 1:100. Specific antibodies were detected with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (Sigma) diluted 1:250. Controls were blood smears incubated with preimmune sera or with secondary conjugated antibodies. Parasite nuclei were stained with 10 μg of 4,6-diamidino-2-phenylindole per ml. Slides were mounted with Citifluor solution and analyzed with a fluorescence microscope (Axioscope; Zeiss).
Immunoprecipitation.
[35S]methionine in vitro radiolabeling of B. canis cultures was performed with 10% starting parasitemia. Prior to labeling, infected erythrocytes were washed three times with methionine-deficient medium (Gibco BRL). Labeling was performed by adding 50 μCi of [35S]methionine (>1,000 Ci/mmol; Amersham Pharmacia Biotech) per ml during 12 h at 37°C. Radiolabeled parasitized erythrocytes and culture supernatants (exoantigen fraction) from B. canis in vitro cultures were then separated by centrifugation (1,800 × g, 5 min). Volumes of 200 μl (106 cpm) from radiolabeled culture supernatant were precleared with protein A-Sepharose CL4B beads (Amersham Pharmacia Biotech) for 30 min at 4°C before immunoprecipitation experiments. The erythrocyte pellet was washed extensively and was either dissociated into 9 volumes of radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (2% Triton X-100, 600 mM KCl, 150 mM NaCl, 5 mM EDTA, 10 mM Tris-HCl [pH 7.8], 3 mM phenylmethylsulfonyl fluoride, 1% aprotinin, 2.5% iodoacetamide) or lysed with α-hemolysin as described above. The radiolabeled lysate resulting from RIPA buffer treatment was maintained for 1 h on ice and centrifuged at 15,000 × g for 15 min, and the supernatant was collected (total antigen fraction). Radiolabeled parasitized red blood cells lysed with α-hemolysin were centrifuged (3,500 × g, 30 min), and the supernatant (infected-erythrocyte stroma fraction) was collected. The pellet was processed through a Percoll gradient as described above to collect the purified radiolabeled merozoites free of contaminating erythrocyte membranes. One part of the purified merozoite fraction was used for phase separation of the proteins in Triton X-114 (Sigma) according the method of Bordier (10) as modified by Precigout et al. (34). Immunoprecipitation experiments were performed with total, supernatant, infected-erythrocyte stroma, purified merozoite, Triton X-114 aqueous (soluble antigens), and Triton X-114 detergent (insoluble antigens) fractions as the source of radiolabeled antigens of B. canis. They were performed by adding 5 μl of each tested antiserum to volumes of radiolabeled exoantigens or antigens corresponding to 106 cpm. Incubation was performed overnight at 4°C with constant stirring. The antigen-antibody complexes were precipitated with 75 μl of protein A-Sepharose CL4B beads (Amersham Pharmacia Biotech) for 1 h at room temperature. The complexes were then washed four times with RIPA buffer and two times with TBS. Subsequently, immune complexes and beads were dissociated in 50 μl of SDS-PAGE sample buffer. Radiolabeled proteins were separated on by SDS-15% PAGE. After electrophoresis, gels were fixed, dried, and autoradiographed on Biomax MR films (Eastman Kodak Co).
In vitro inhibition assays.
B. canis parasites were cultured at 2% (vol/vol) hematocrit and a starting parasitemia of 1% (day 0) as previously described. All experiments were performed in triplicate, and the growth-inhibitory effect of anti-Bcvir15 antibodies (as crude sera or purified IgG) was analyzed by either microscopic determination of parasitemia or [3H]hypoxanthine incorporation. In a first series of experiments, inhibition assays were performed with 8% (vol/vol) crude sera in 1-ml cultures in vitro performed in 24-well plates (Nunclon Δ surface; Nunc). After 24 h of incubation at 37°C (day 1), the cultures were centrifuged at 1,800 × g for 5 min, and the medium was completely removed and replaced by prewarmed fresh medium containing the same amount of serum that was added at day 0. A supplementary 24-h incubation of the cultures was performed at 37°C, and the infected erythrocytes were collected (day 2). Reversion of inhibition was performed by addition of 20 or 40 μg of purified GSTBcvir15 recombinant protein (that was dialyzed overnight against RPMI) to wells containing a culture supplemented with 8% (vol/vol) crude anti-GSTBcvir15 serum at day 0 of the experiment. As a control, the same amount of GSTBcvir15 protein was added to wells of normal cultures. Blood smears of each well were made at days 0, 1, and 2, and the inhibitory effect was determined by microscopic assessment of parasitemia. In a second series of experiments, inhibition assays were performed with total purified IgG that was added (at a final concentration of 20 to 200 μg of total IgG/ml of culture) to 100 μl of in vitro cultures maintained in 96-well plates (Nunclon Δ surface; Nunc) under the conditions described above. Anti-GSTBcvir15 preimmune, irrelevant anti-GSTBc12D3 and anti-GST antibodies were purified by affinity chromatography on a HITrap protein G column (Amersham Pharmacia Biotech). Purified IgG was then dialyzed overnight at 4°C against RPMI, and IgG titers were determined with the Coomassie blue protein reagent kit (Pierce). Each sample was then resuspended to a final concentration of 2 mg of IgG per ml in RPMI, and the ratio of IgG specifically directed against the GST part of GST-tagged recombinant proteins (GSTBcvir15 or GSTBc12D3) that were used to immunize rabbits was analyzed by enzyme-linked immunosorbent assay; this was evaluated to be half of the total amount of purified IgG for the two sera. Thus, purified IgGs were diluted in RPMI to 2.0, 1.6, 0.8, 0.4, and 0.2 mg/ml, and 10 μl per well was added to 90-μl cultures containing 50 μCi of [3H]hypoxanthine (in aqueous solution; ICN) per ml. Cultures were incubated for 24 h at 37°C. Subsequently, the infected erythrocytes were collected with a Cell Collector apparatus (Bioblock) onto a filter. Incorporation of radiolabel was determined by liquid scintillation counting in the presence of 2 ml of Emulsifier Safe Scintillant (Packard).
Epitope mapping.
Sixty-five overlapping dodecapeptides covering the entire Bcvir15 amino acid sequence were synthesized on a polyvinylidene difluoride membrane by the method of Franck (16) as modified by Molina et al. (32). Epitope mapping of Bcvir15 was performed with anti-GSTBcvir15 and anti-(His)6Bcvir15 sera diluted 1:1,000 and 1:300, respectively. Corresponding preimmune rabbit sera were used at the same dilutions.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper are available in the GenBank, EMBL, and DDBJ databases under accession number AJ494862.
RESULTS
Isolation and sequence analysis of Bcvir cDNA.
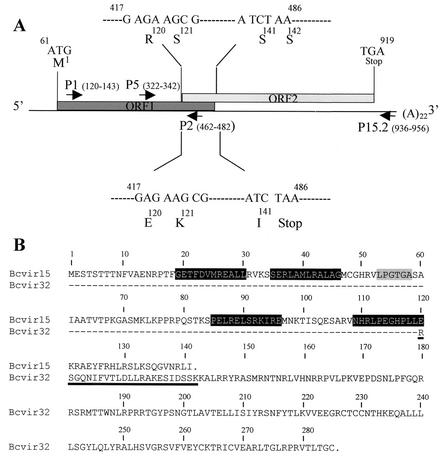
As part of a search for a homologous sequence from the P. falciparum Pf60 multigene family (8, 9, 11) in the agent of canine babesiosis in Europe, the anti-Pf60.1 antiserum (11) was used to perform an immunological screening of a B. canis (isolate A) cDNA library. The Bcvir cDNA (accession number AJ494862) was 1,115 nucleotides (nt) long, with a poly(A)22 tail and with the translation initiation codon at position 61 (Fig. 1A). An interesting feature of this cDNA was that it contained two open reading frames (ORFs) which were +1 frameshifted and overlapped by 69 nt (nt 417 to 486) (Fig. 1A). ORF1 (nt 61 to 486) was predicted to encode a polypeptide of 141 amino acids (aa) (M1 to I141, designated Bcvir15) starting with a methionine (Fig. 1B), with an expected molecular mass of 15.7 kDa and a pI of 10.9. ORF2 (nt 417 to 919) started with an arginine residue (Fig. 1B). Sequence analysis of the overlapping region of the Bcvir cDNA suggested that a +1 ribosomal frameshift would produce an ORF1-ORF2 fusion protein of 285 residues (M1 to C285, designated Bcvir32) (Fig. 1B), with a predicted molecular mass of 32.3 kDa and a pI of 10.5. Due to the +1 shift between ORF1 and ORF2, the predicted Bcvir15 and Bcvir32 polypeptides would show different amino acid sequences in the overlapping region (Fig. 1B).
FIG. 1.
Schematic representation of the Bcvir cDNA. (A) The predicted ORF1 (nt 61 to 486) and ORF2 (nt 417 to 919) sequences and the ORF1-ORF2 overlapping region (nt 417 to 486) of Bcvir cDNA are indicated by boxes. Reverse (P2 and P15.2) and forward (P1 and P5) primers derived from the Bcvir cDNA sequence are also positioned at the cDNA. Above and below the ORF1-ORF2 overlapping region are indicated the beginning and the end of the nucleotide and deduced amino acid sequences for the translation of ORF2 or ORF1 in that area. Because of +1 frameshifted translation of ORF2 (upper nucleotide sequence) in comparison to that of ORF1 (lower nucleotide sequence), their deduced amino acid sequences in that overlapping region are different. (B) Full amino acid sequences of Bcvir15 and putative Bcvir32, respectively, deduced from the translation of ORF1 or from the translation of ORF1-ORF2 through a +1 frameshift mechanism within the overlapping region of the Bcvir cDNA. Within the Bcvir32 sequence, the dashed line corresponds to the Bcvir15 sequence that is common between the two products upstream from the overlapping area of the cDNA. The underlined area represents the specific amino acid sequence of Bcvir15 and Bcvir32 within the overlapping area because of the +1 frameshift translation mechanism between ORF1 and ORF2 in that area. The four epitopes recognized by the anti-Bcvir15 antisera [either anti-GSTBcvir15 or anti-(His)6Bcvir15] are indicated in black boxes. The gram-positive surface protein-anchoring consensus hexapeptide (PROSITE PS50847) present in Bcvir15 is indicated in a grey box.
Sequence database screening with the Bcvir cDNA sequence as a query was performed with both the BLAST (2) and PSIBLAST (3) programs. It failed to indicate any significant distant homology. However, two specific motifs were found within the Bcvir15 amino acid sequence in the PRINTS (5) and PROSITE (6) databases: the PRINT PR00349 virion infectivity factor Vif of HIV1 retrovirus and the PROSITE PS50847 gram-positive anchoring motif present in coccus surface proteins. This last motif, corresponding to the consensus hexapeptide sequence L-P-X-T-G-[STGAVDE], was found as an LPGTGA sequence (aa 53 to 58 of Bcvir15) (Fig. 1B). Interestingly, when the FUGUE program was used for recognizing distant homologues by sequence-structure comparison (40), the Bcvir15 protein was found to be homologous (>95%) to the b chain protein core from the paramyxovirus simian virus 5, and a coiled-coil structure might be predicted with the last amino acid sequence from Bcvir15. Analysis of the hydrophobic profile (33) from the Bcvir15 amino acid sequence did not predict a putative peptide signal sequence at the N-terminal end, suggesting that Bcvir15 is not a secreted protein.
Bcvir cDNA has no copy in the genome of B. canis.
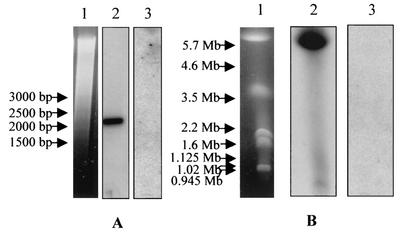
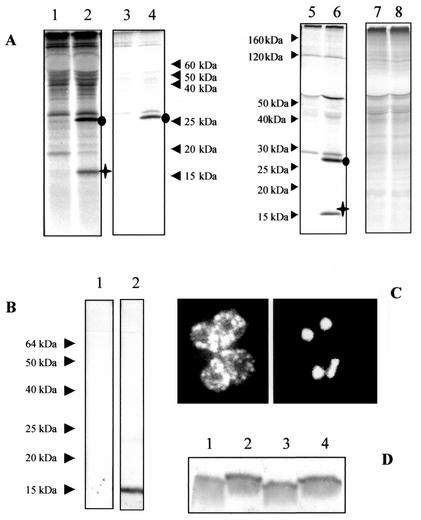
The ORF1-derived cDNA sequence was used to probe either B. canis genomic DNA digested with restriction enzyme or chromosomes of B. canis separated by PFGE. Surprisingly, the Bcvir cDNA probe failed to hybridize both to Southern blots of genomic DNA (Fig. 2A, lane 3) or to PFGE-separated chromosomes of B. canis (Fig. 2B, lane 3). In contrast, the cDNA probe that was derived from the single-copy gene Bc12D3 from B. canis gave strong signals with the same blots (Fig. 2, lanes 2). In controls, the ORF1-derived probe was reactive on the plasmid that carried the Bcvir cDNA, and there was no hybridization of this probe to canine lymphocyte DNA (data not shown). PCR experiments using genomic DNA as the template and Bcvir cDNA sequences as primers were unable to amplify regions from this cDNA in the genome of B. canis. The same primers tested on the recombinant plasmid carrying the Bcvir cDNA and primers derived from the single-copy gene Bc12D3 from B. canis tested on B. canis genomic DNA amplified DNA fragments with expected sizes (data not shown).
FIG. 2.
Analysis of the Bcvir sequence within B. canis genomic DNA. Analysis was performed by hybridization on a Southern blot of genomic DNA digested with the restriction enzyme XbaI (A) and on PFGE-separated chromosomes of B. canis (B). Ethidium bromide staining of digested DNA and chromosomes are indicated in lanes 1. The same membrane was consecutively hybridized with a cDNA probe derived either from the single-copy gene Bc12D3 of B. canis (lanes 2) or from the entire Bcvir sequence (lanes 3).
Bcvir cDNA is carried by dsRNA elements of B. canis.
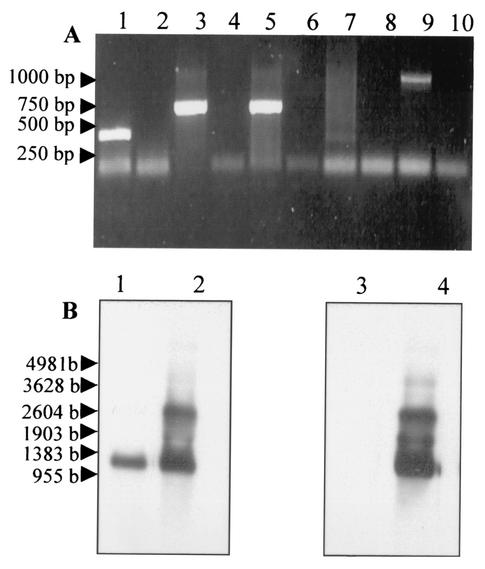
As there appeared to be no DNA copy of the Bcvir cDNA in the genome of B. canis, a series of experiments were performed at the RNA level to search for the derived genetic element corresponding to the Bcvir cDNA sequence. An RT-PCR experiment was first performed with either Bcvir cDNA sequence-derived forward or reverse primers for the first retrotranscription step (Fig. 3A, lanes 1 to 6). As a control for this experiment, a similar approach was used with primers derived from the cDNA that was derived from the single-copy gene Bc12D3 from B. canis (Fig. 3A, lanes 7 to 10). As expected, with the control a 1-kbp product was amplified when the reverse primer was used for the first retrotranscription step (Fig. 3A, lane 9), and it was negative when this step was performed with the forward primer (Fig. 3A, lane 7). No genomic DNA contamination was observed, since no products were amplified when reverse transcriptase was omitted before RT-PCR (Fig. 3A, lanes 8 and 10). Such experiments with a reverse primer derived from the Bcvir cDNA used as retrotranscription step primer revealed an amplified product with the expected size (Fig. 3A, lane 5). Unexpectedly however, the same experiments performed with forward primers as retrotranscription step primers were also able to amplify products with the expected sizes (Fig. 3A, lanes 1 and 3).
FIG. 3.
RT-PCR and Northern blot analysis of the Bcvir sequence. (A) RT-PCR was performed on total RNA from B. canis with primers derived either from the Bcvir cDNA (lanes 1 to 6) or from a cDNA corresponding to a single-copy gene of B. canis (lanes 7 to 10). For both sequences analyzed, the first retrotranscription step was performed either with (lanes 1, 3, 5, 7, and 9) or without (lanes 2, 4, 6, 8, and 10) enhanced avian myeloblastosis virus retrotranscriptase enzyme. Reverse primer P15.2 (lanes 5 and 6) and forward primers P1 (lanes 1 and 2) and P5 (lanes 3 and 4) from the Bcvir cDNA sequence were used for this first step. PCRs were performed with the primer pair P1-P2 (lanes 1 and 2) or P5-P15.2 (lanes 3 to 6) in the second step. Likewise, the first step was performed with reverse (lanes 9 and 10) and forward (lanes 6 and 7) primers derived from the single-copy gene Bc12D3 of B. canis, and PCRs were performed with both primers (lanes 7 to 10). (B) Northern blot analysis of the Bcvir sequence was performed either on purified mRNA (lanes 1 and 3) or on total RNA (lanes 2 and 4) from B. canis. RNA were hybridized with antisense (lanes 1 and 2) or sense (lanes 3 and 4) riboprobes derived from the ORF1 sequence of the Bcvir cDNA.
As this suggested that two opposite cRNA strands might carry the Bcvir cDNA sequence, the precise nature of RNA element bearing the Bcvir cDNA sequence was analyzed by Northern blotting (Fig. 3B). Hybridizations were performed with riboprobes corresponding either to the ORF1 mRNA sequence (sense riboprobe) (Fig. 3B, lanes 3 and 4) or to its complementary nucleotide sequence (antisense riboprobe) (Fig. 3B, lanes 1 and 2). Both sense and antisense ORF1-derived riboprobes were able to detect two RNA products of 1.2 and 2.8 kb when tested on total RNA extract from B. canis (Fig. 3B, lanes 2 and 4), suggesting that the Bcvir cDNA sequence was carried by two dsRNAs. In contrast, a single 1.2-kb mRNA corresponding to the size of Bcvir cDNA was detected with the antisense riboprobe when hybridization was performed with purified mRNA (Fig. 3B, lane 1). No signal was detected with the sense probe on this extract (Fig. 3B, lane 3). Similar results were obtained when hybridizations were performed with ORF2-derived riboprobes (data not shown).
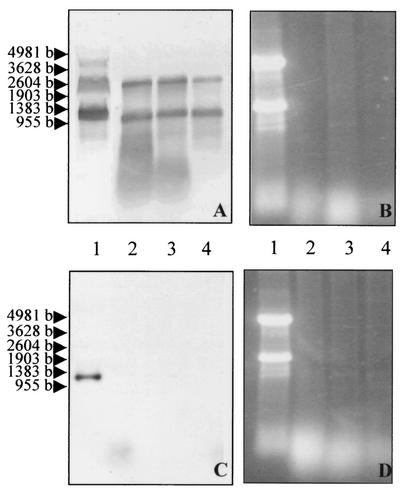
The dsRNA status of genetic elements that carried the Bcvir cDNA sequence was confirmed by treating the total RNA extract with nuclease S1 prior to Northern blot hybridization with the ORF1-derived riboprobe (Fig. 4A and B). As a control for proper activity of nuclease S1, a similar approach was used with an antisense riboprobe derived from the encoding region of the Bc28 cDNA from B. canis (Fig. 4C and D). As expected, with the control a 1-kb mRNA was detected when the riboprobe was hybridized on untreated total RNA (Fig. 4C, lane 1), but no signal was obtained with the nuclease S1-treated extract (Fig. 4C, lanes 2 to 4), confirming that no single-stranded RNA was present in the extract. In contrast, hybridization with the sense ORF1-derived riboprobe gave a strong signal both with the nuclease S1-treated extract (Fig. 4A, lanes 2 to 4) and with the untreated extract (Fig. 4A, lane 1), indicating that the Bcvir cDNA sequence was carried by two extrachromosomal dsRNAs of 1.2 and 2.8 kbp within B. canis.
FIG. 4.
Northern blot analysis of the Bcvir sequence on dsRNA from B. canis. Undigested (lanes 1) and nuclease S1-digested (lanes 2 to 4) total RNA were ethidium bromide stained for 1 to 3 h (B and D) and hybridized either with a sense ORF1-derived riboprobe (A) or with an antisense riboprobe derived from the encoding region of the cDNA Bc28 from B. canis (C).
Biochemical characterization and location of Bcvir15, the main expressed product from Bcvir cDNA-derived dsRNA.
To identify and characterize the main predicted expressed protein from the Bcvir cDNA-derived dsRNA sequence within B. canis, i.e., Bcvir15, specific sera were raised against GST- and His6-tagged recombinant Bcvir15 proteins (polypeptides A12 to I141 and M25 to I141, respectively). In immunoprecipitation assays, both polyclonal antisera identified Bcvir15 in the [35S]methionine-radiolabeled infected red blood cell extract (Fig. 5A, lanes 2 and 3) and a 27-kDa protein was coprecipitated with Bcvir15 by anti-(His)6Bcvir15 (Fig. 5A, lane 2). The preimmune serum, corresponding to the rabbit immunized with the recombinant protein (His)6-Bcvir15, was unreactive on these proteins (Fig. 5A, lane 1), and the anti-GST serum was not reactive on Bcvir15 (Fig. 5A, lane 4). Bcvir15 was not immunoprecipitated from the supernatant fraction of B. canis (Fig. 5B, lane 2), suggesting that it is not a protein that might be secreted or released from lysed infected erythrocytes.
FIG. 5.
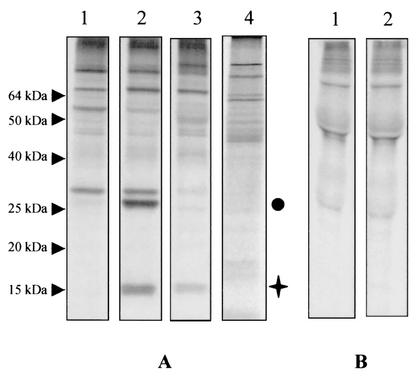
Analysis of Bcvir15 in the total or exoantigen fractions of [35S]methionine-radiolabeled antigens from B. canis. (A) Reactivities of the anti-(His)6Bcvir15 (lane 2) and its corresponding preimmune (lane 1), anti-GSTBcvir15 (lane 3), and anti-GST (lane 4) sera in immunoprecipitation experiments with the total fraction. (B) Reactivity of the anti-(His)6Bcvir15 (lane 2) and of its corresponding preimmune (lane 1) serum in immunoprecipitation experiments with the exoantigen fraction. The Bcvir15 protein is indicated by a cross, and the coprecipitated 27-kDa protein is indicated by a circle.
This was confirmed by immunoprecipitation experiments performed with the anti-(His)6Bcvir15 serum on the infected-erythrocyte stroma and purified merozoite fractions collected after α-hemolysin treatment of the [35S]methionine-radiolabeled infected red blood cells (Fig. 6A). The results showed that Bcvir15 was present in purified merozoites, whereas it was not detected in stroma of infected erythrocytes (Fig. 6A, lanes 2 and 4, respectively). Moreover, Triton X-114 partitioning of [35S]methionine-radiolabeled antigens from purified merozoites revealed that Bcvir15 was immunoprecipitated from the hydrophilic fraction but not from the hydrophobic one (Fig. 6A, lanes 6 and 8, respectively), suggesting that Bcvir15 was a soluble antigen within merozoites. Like Bcvir15, the coprecipitated 27-kDa antigen was detected in the purified merozoite fraction and in its hydrophilic fraction (Fig. 6A, lanes 2 and 6). In contrast to the case for Bcvir15, it was detected in the infected-erythrocyte stroma (Fig. 6A, lane 4). The preimmune serum did not recognize these antigens either in immunoprecipitation experiments (Fig. 6A, lanes 1, 3, 5, and 7) or in Western blotting (Fig. 6B, lane 1). On immunoblots of purified merozoites from isolate A of B. canis, the anti-(His)6Bcvir15 serum reacted with Bcvir15 but not with the 27-kDa antigen (Fig. 6B, lane 2). The relationship between Bcvir15 and the coprecipitated 27-kDa antigen remains unclear, but the fact that the 27-kDa antigen was detected in extracts of purified merozoites by immunoprecipitation and not by immunoblot experiments suggests that it shared a conformational epitope with Bcvir15.
FIG. 6.
Biochemical characterization and location of Bcvir15. (A) Immunoprecipitation of [35S]methionine-radiolabeled antigens from purified merozoites (lanes 1 and 2), infected-erythrocyte stroma (lanes 3 and 4), and soluble (lanes 5 and 6) and insoluble (lanes 7 and 8) Triton X-114-extracted antigens from purified merozoites. Analyses were performed with the anti-(His)6Bcvir15 (lanes 2, 4, 6, and 8) and corresponding preimmune (lanes 1, 3, 5, and 7) sera. The Bcvir15 protein is indicated by a cross, and the coprecipitated 27-kDa protein is indicated by a circle. (B) Western blot of purified B. canis merozoites with the anti-(His)6Bcvir15 (lane 2) or corresponding preimmune (lane 1) serum. (C) Location of Bcvir15 by indirect immunofluorescence assay with the anti-(His)6Bcvir15 serum on fixed B. canis-infected erythrocytes. Bcvir15-associated fluorescence on merozoites from a typical Babesia tetrad form (left panel) and corresponding 4′,6′-diamidino-2-phenylindole nucleus staining (right panel) are shown. (D) Analysis of Bcvir15 polymorphism with the anti-(His)6Bcvir15 serum by Western blotting. The four different isolates of B. canis analyzed were isolate A (lane 1), isolate B (lane 2), isolate G (lane 3), and isolate R (lane 4).
The location of the Bcvir15 antigen was then investigated by indirect immunofluorescence assay with both anti-(His)6Bcvir15and anti-GSTBcvir15 sera. The two antisera gave the same image; i.e., a strong fluorescence signal was obtained as dots within the merozoites, as shown on the four merozoites of B. canis from the Babesia tetrad form presented in Fig. 6C (left panel). These dots were distributed all over the cytoplasm of the merozoite, and they were not distributed within the infected-erythrocyte host cells. Moreover, nuclear staining with 4,6-diamidino-2-phenylindole suggested that they did not colocalize with the nucleus of the merozoite (Fig. 6C, right panel). Neither the preimmune sera, the anti-GST serum, nor the secondary conjugated antibody was reactive in indirect immunofluorescence assay (data not shown).
Finally, immunoblotting of purified merozoites from four isolates of B. canis, originating from different regions in France, with the anti-(His)6Bcvir15 serum revealed a slight size polymorphism of Bcvir15 (Fig. 6D). Two profiles were observed among the four tested isolates, and the size polymorphism between two reference isolates of B. canis for vaccination experiments, i.e., isolates A and B, was clear (Fig. 6D, lanes 1 and 2, respectively).
In vitro inhibition assays.
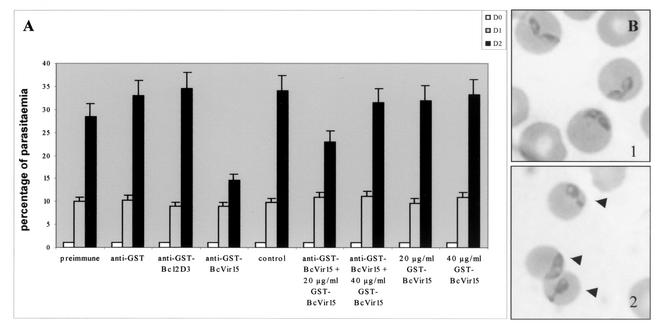
As Bcvir15 was found to share similarity with viral and bacterial infectivity factors, we further studied such a function for Bcvir15 in the parasite B. canis by performing a series of in vitro inhibition tests. Moreover, as biochemical analysis of Bcvir15 with the anti-(His)6Bcvir15 serum described above indicated that a 27-kDa antigen shared a conformational epitope, inhibition tests of the in vitro culture of B. canis were performed with an anti-GSTBcvir15 serum that did not cross-react with this antigen (Fig. 5A, lane 3). A pilot experiment showed that 8% (vol/vol) anti-GSTBcvir15 serum in the culture medium inhibited the in vitro growth of B. canis to 50% after 2 days of culture (data not shown). Thus, further inhibition assays were performed at this defined concentration of anti-GSTBcvir15 serum, using either crude serum (Fig. 7A) or purified immunoglobulins (IgG) (Fig. 8). In the first series of experiments, the inhibitory effect of 8% (vol/vol) anti-GSTBcvir15 serum was estimated by parasitemia counting (performed in triplicate). It confirmed the 50% inhibitory effect of this concentration on the in vitro growth of B. canis after 2 days of culture (Fig. 7A), as was observed with a similar concentration of the anti-(His)6Bcvir15 serum (data not shown). As controls, similar concentrations of either preimmune sera, anti-GST serum, or irrelevant anti-GSTBc12D3 serum had no inhibitory effect (Fig. 7A). Moreover, a competitive inhibition assay demonstrated that addition of GSTBcvir15 protein to the medium of a culture treated with 8% (vol/vol) anti-GSTBcvir15 serum showed 50 and 100% reversion of the inhibition at concentrations of 20 and 40 μg/ml, respectively (Fig. 7A). The recombinant protein itself had no inhibitory effect on B. canis cultures (Fig. 7A).
FIG. 7.
Inhibitory effect of crude anti-GSTBcvir15 serum on the in vitro growth of B. canis. (A) The in vitro inhibition effect of 8% (vol/vol) anti-GSTBcvir15 serum was evaluated by parasitemia counting on Giemsa-stained slides and monitored during a 48-h period (day 0 [D0] to day 2). Controls were preimmune, anti-GST, and anti-GSTBc12D3 sera used at the same concentration and an untreated culture. Reversion of the inhibitory effect of the recombinant GSTBcvir15 was evaluated by addition of 20 to 40 μg of the protein per ml to an untreated or treated culture with 8% (vol/vol) crude anti-GSTBcvir15 serum. Error bars indicate standard deviations. (B) Cytological observations of infected erythrocytes untreated (panel 1) or treated with 8% (vol/vol) crude anti-GSTBcvir15 serum (panel 2). Arrowheads indicate intraerythrocytic abnormal parasite forms.
FIG. 8.
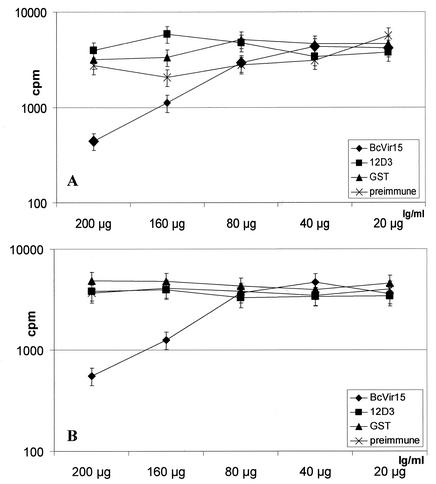
Inhibitory effect of purified anti-GSTBcvir15 IgG on the in vitro growth of B. canis. The in vitro inhibition effect of 20 to 200 μg of purified anti-GSTBcvir15 IgG per ml was evaluated by [3H]hypoxanthine incorporation. Controls were purified IgG from preimmune, anti-GST, and anti-GSTBc12D3 sera used at the same concentrations and an untreated culture. The experiment was performed with isolates A (A) and B (B) of B. canis. Error bars indicate standard deviations.
Cytological examination of blood smears revealed many abnormal parasite forms, such as forms with huge cytoplasm or cytoplasmic extrusion within the host erythrocytes (Fig. 7B, panel 2), compared to parasites from untreated cultures (Fig. 7B, panel 1). Free merozoites were not found in the medium. These observations suggested that anti-GSTBcvir15 inhibited parasite growth and development within the erythrocyte after invasion rather than inhibiting the invasion process per se. This is in agreement with the fact that the inhibitory effect of anti-GSTBcvir15 sera was evident after 2 days of culture.
This was confirmed by a second series of inhibition assays that were performed in triplicate with [3H]hypoxanthine incorporation and purified IgG from GSTBcvir15, irrelevant GSTBc12D3, GST, and preimmune sera (Fig. 8). Twenty to 200 micrograms of purified IgG from each serum was added per milliliter of culture, and the test was performed on the two European isolates A and B of B. canis (i.e., B. canis subsp. canis) (Fig. 8A and B, respectively). The 50% inhibition effect of purified IgG from the anti-GSTBcvir15 on the in vitro cultures was obtained at a concentration of 160 μg/ml, for both isolates A and B of B. canis (Fig. 8A and B, respectively), and no inhibitory effect was observed with purified IgG from control sera. Half of the purified IgG from anti-GSTBcvir15 was estimated to be directed against the GST part of the recombinant protein by enzyme-linked immunosorbent assay. This indicated that 80 μg of IgG per ml, specifically directed against the Bcvir15 part of the recombinant protein, was responsible for the 50% in vitro growth inhibition of B. canis. Interestingly, the polymorphism of Bcvir15 observed between isolates A and B (Fig. 6D, lanes 1 and 2) seems to have no effect on the function of the protein, since the inhibitory effect of antibodies was found to be equivalent for the two isolates. In contrast, when the test with purified IgG from GSTBcvir15 was performed with a South African isolate from B. canis (i.e., B. canis subsp. rossi), no inhibitory effect was observed (data not shown). In this last experiment the growth-inhibitory effect of anti-GSTBcvir15 was evident after 1 day of culture. Combined with the cytological observations described above (absence of free merozoites in the medium and abnormal parasitic forms), this suggested that the inhibitory effect of IgG specifically directed against Bcvir15 on the growth of the parasite occurred after erythrocyte invasion.
Epitope mapping of Bcvir15.
As in vitro inhibition assays indicated that Bcvir15 might interfere with the intracellular growth of B. canis, we further analyzed which epitopes might be implied in that process by synthesizing overlapping dodecapeptides that covered the Bcvir15 amino acid sequence on a polyvinylidene difluoride membrane. Of the 65 overlapping dodecapeptides that covered the Bcvir15 amino acid sequence, four were recognized by the anti-GSTBcvir15 serum and three were recognized by the anti-(His)6Bcvir15 serum (Fig. 1B). Interestingly, the three epitopes, SERLAMLRALAG (aa 35 to 46), PELRELSRKIRE (aa 85 to 96), and NHRLPEGHPPLE (aa 109 to 120), that were recognized by the anti-(His)6Bcvir15 serum were also recognized by the anti-GSTBcvir15 serum (data not shown), suggesting that they might be critical for the function of Bcvir15.
DISCUSSION
The data reported here describe the characterization of a 1.2-kbp cDNA and of its main encoded product, Bcvir15, which might be of particular importance for the intracellular growth of the Apicomplexan parasite B. canis. Our data support a potential viral origin for this cDNA, which was designated Bcvir. This is suggested by its particular structure with the presence of two overlapping ORFs, by the fact that its main encoded product (Bcvir15) might share homology with viral proteins, and especially by the fact that it is not derived from the DNA genome of B. canis. Indeed, the Bcvir cDNA was found to be derived from two extrachromosomal dsRNA genomes of 1.2 and 2.8 kbp. dsRNA elements have already been described as viral genomes present in a large number of parasitic protozoa (46), including another Babesia species, B. bovis (20). Protozoan viruses described to date have been separated into two families, depending on whether their L-dsRNA, i.e., the genome which encodes structural proteins necessary for their encapsidation and replication, is monopartite or fragmented. Most of these viruses harbor a monopartite linear L-dsRNA genome with a size range of 4.3 to 7.5 kbp, characteristic of the Totiviridae virus family (46). An exception is the viral genome of the Apicomplexan parasite Cryptosporidium parvum, which is fragmented into two separated dsRNA molecules of 1.8 and 1.4 kbp and, as such, is classified as a member of the Partitiviridae virus family (26, 27). Because of the dsRNA status of these viruses, an RDRP activity is needed for their replication, and for all protozoan viruses described to date, it was evidenced or predicted to be encoded by the L-dsRNA genome. Attempts to find an RDRP activity and to analyze all of the dsRNA elements that might constitute the complete genome of the potential B. canis virus are under way using [33P]UTP radiolabeling experiments. An RDRP activity was detected within purified merozoites of B. canis (B. Carcy, unpublished data), in agreement with a dsRNA origin for the genome(s) of the potential B. canis virus. However, our data strongly suggested that the Bcvir cDNA does not derive from the L-dsRNA genome of the potential B. canis virus. Indeed, neither Bcvir15, the main encoded product from the 1.2-kbp Bcvir cDNA, nor Bcvir32, the minor product that might be expressed in vitro from this cDNA through a +1 translational recoding mechanism (P. Drakulovski et al., unpublished data) predicts such an RDRP motif. Moreover, probes that derived from the 1.2-kbp Bcvir cDNA always hybridized to the 1.2- and 2.8-kbp dsRNAs (this work), but they do not hybridize to a larger, 6.5-kbp dsRNA detected following nuclease S1 digestion of [33P]UTP-labeled RNA extracted from in vitro cultures (Carcy, unpublished). The size of this supplementary dsRNA is in agreement with the size of an L-dsRNA genome for the potential B. canis dsRNA virus and is also in agreement with the L-dsRNA genome that was identified in the dsRNA virus from the bovine Babesia species B. bovis (20). Thus, the inability of the 1.2-kbp Bcvir cDNA-derived probes to hybridize with this 6.5-kbp dsRNA strongly suggested that the Bcvir cDNA does not derive from the L-dsRNA genome of the potential B. canis virus but derives from smaller dsRNA genomes of 1.2 and 2.8 kbp. Indeed, smaller dsRNA genomes have already been described for the viruses of T. vaginalis and E. nieschulzi, and those from T. vaginalis were characterized as dsRNA satellites (24, 25, 36). Interesting features of these dsRNA satellites are their lack of cross-hybridization with the L-dsRNA (25) and their ability to dimerize, as was demonstrated in the Cryphonectria parasitica virus (19). Our data indicated a close association between the dsRNAs of 1.2 and 2.8 kbp, since they were always detected together by hybridization experiments. However, no mRNA longer than 1.2 kb, i.e., that might derive from the 2.8-kbp dsRNA, was detected by hybridization, and attempts to sequence this 2.8-kbp dsRNA by SMART (switch mechanism at the 5′ end of the RNA transcript; Clontech) or SPAT (single primer amplification technique) methodologies (4) were unsuccessful. Although it remains to be demonstrated, this suggests that the extrachromosomal 2.8-kbp dsRNA might correspond to a dimeric form of the 1.2-kbp dsRNA. Thus, together these results indicated that the Bcvir cDNA and its expressed products, Bcvir15 and Bcvir32, would be derived from a smaller dsRNA genome of 1.2 kbp and that this extrachromosomal element might be a dsRNA satellite.
Until this analysis, no data were available on the products that might be expressed from smaller dsRNAs for protozoan dsRNA viruses or on their function. The best-known example of an encoded product from a smaller dsRNA genome of viral origin that was demonstrated to play a critical role in the infected host is the toxin responsible for the killer phenomenon in yeast (47). Sequence analysis of Bcvir15 indicated the presence of the consensus sequence L-P-x-T-G-(STGAVDE) on Bcvir15, and this motif is found in numerous surface proteins that play an important role in the virulence of the bacteria (15, 39, 41). Moreover, Bcvir15 might share homology with the retroviral Vif protein, which plays an important role in regulating virus infectivity, since the lack of functional Vif protein was demonstrated to result in the production of virions with reduced or abolished infectivity (14, 28, 43). As it suggested a function of Bcvir15 in the infectivity of the parasite, this was examined by testing the effect of antibodies directed against Bcvir15 on the in vitro growth of B. canis. We found that addition of 8% of crude serum or of 160 μg of purified IgG directed against Bcvir15 per ml led to a 50% inhibition of the in vitro growth of the parasite and that the inhibitory activity of antibodies was associated with morphological damage of the parasite. However, the mechanism for the inhibitory effect of antibodies remains unknown. One possible explanation is that the antibodies block the process of parasite invasion of the erythrocyte. However, the absence of free merozoites in the medium and the presence of abnormal parasitic forms within erythrocytes suggested that the inhibitory effect of anti-Bcvir15 IgG on the growth of B. canis occurred after erythrocyte invasion. Moreover, biochemical characterization of Bcvir15 indicated that it is not a merozoite surface antigen that might be easily accessible to antibodies, thus blocking an interaction of Bcvir15 with a receptor from the host erythrocyte. Bcvir15 is a soluble antigen that remains in the merozoite cytoplasm. This suggested that antibodies have to directly reach the protein Bcvir15 within the cytoplasm of B. canis. Babesia parasites are directly in contact with the cytoplasm of the erythrocyte during their erythrocytic life cycle, and in B. equi and B. caballi, a tubular structure connecting the erythrocyte membrane to parasite was observed and its ultrastructure was described (17, 21, 22). Although the function of these tubular structures remains unknown, it has been speculated that intraerythrocytic Babesia parasites come into direct contact with culture medium or host plasma and that these structures might play a role in the uptake of nutrients. Direct contacts between intraerythrocytic B. canis parasites and the red blood cell surface membrane have been also observed, either through the apex of the merozoites or through a tubular structure (18). Thus, another and most plausible explanation for the inhibitory effect of the anti-Bcvir15 IgG is that it might reach the parasite through these direct contacts and neutralize the intracytoplasmic Bcvir15. Thus, rather than playing a function in the infectivity of the parasite, Bcvir15 might interfere with the metabolic pathway of the parasite that is necessary for its intracellular growth. As a consequence of neutralization of Bcvir15, the parasite metabolism would be modified, and this would be responsible for morphological changes. How and with which parasitic molecules implicated in the metabolic pathway necessary for the intracellular growth of B. canis Bcvir15 might interact remains unknown and will be further studied. The Bcvir15 protein was immunologically identified in all of the European isolates of B. canis (i.e., B. canis subsp. canis) that were tested, whatever their geographic origin. For the four isolates analyzed in this study, we have observed that Bcvir15 presented two different sizes. Interestingly, we observed that the inhibitory effect of antibodies was conserved against an isolate expressing the polymorphic form of Bcvir15, suggesting that, in spite of this polymorphism, functional domains might be conserved. To date, epitope mapping of Bcvir15 suggested that epitopes SERLAMLRALAG (aa 35 to 46), PELRELSRKIRE (aa 85 to 96), and NHRLPEGHPPLE (aa 109 to 120) might be essential for its function, since these three epitopes were commonly recognized by anti-GSTBcvir15 and anti-(His)6Bcvir15 sera, both of which inhibited the in vitro growth of B. canis. The production of a monoclonal antibody directed against Bcvir15 is under investigation, to analyze which epitope(s) might play a crucial role for the function of Bcvir15.
The results presented here on the Apicomplexan parasite B. canis provide an example of an encoded product from a potential dsRNA virus that might play an important role in protozoan parasites. We found that this function would be specific for the European subspecies of B. canis, since the dsRNA was not identified in the South African subspecies of B. canis (i.e., B. canis subsp. rossi) and no inhibitory effect of anti-Bcvir15 IgG was observed with such isolates. Finally, the Bcvir15 product of the Bcvir dsRNA was identified by using a polyclonal antibody directed against a member of the multigene family Pf60 of P. falciparum (8, 9, 11), another Apicomplexan parasite. Surprisingly, whereas no homology was found between Bcvir15 and members of this large family, Bcvir15 shares some features with two members of the Pf60 family. Like the Bcvir cDNA (Drakulovski et al., unpublished), these two members used recoding translational mechanisms for the expression of their products, which function as interacting proteins (7, 8). However, in contrast to Bcvir dsRNA, these Pf60 genes are integrated within the genome of P. falciparum (7, 8). This suggested that both Bcvir dsRNA from B. canis and these genes from the Pf60 family of P. falciparum might originate from a common viral ancestor but that a rerouting of the viral information has occurred in P. falciparum, i.e., that the recoding translational mechanisms have been integrated in the genome to the benefit of the parasite.
Acknowledgments
We thank V. Arnal for efficient technical assistance and M. Guillotte for providing anti-Pf60.1 and anti-GST sera. We are also grateful to O. Mercereau-Puijalon, S. Bonnefoy, E. Précigout, A. Vermeulen, R. Keus, and M. I. Gonzatti for helpful discussions.
Pascal Drakulovski was supported by a fellowship from the Ministère de l'Education Nationale de la Recherche et de la Technologie (France) (contract 98-5-11861). This work was supported by a grant from Intervet International (The Netherlands).
Editor: J. M. Mansfield
REFERENCES
- 1.Ahn, I. P., and Y. H. Lee. 2001. A viral double-stranded RNA up regulates the fungal virulence of Nectria radicicola. Mol. Plant-Microbe Interact. 14:496-507. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attoui, H., F. Billoir, J. F. Cantaloube, P. Biagini, P. de Micco, and X. de Lamballerie. 2000. Strategies for the sequence determination of viral dsRNA genomes. J. Virol. Methods 89:147-158. [DOI] [PubMed] [Google Scholar]
- 5.Attwood, T. K., M. E. Beck, A. J. Bleasby, and D. J. Parry-Smith. 1994. PRINTS—a database of protein motif fingerprints. Nucleic Acids Res. 22:3590-3596. [PMC free article] [PubMed] [Google Scholar]
- 6.Bairoch, A. 1991. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 19(Suppl.):2241-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bischoff, E. 2001. Etude de deux membres de la superfamille multigénique Pf60/var de Plasmodium falciparum, exprimés par recodage traductionnel programmé. Ph.D. thesis. University Paris 7, Paris, France.
- 8.Bischoff, E., M. Guillotte, O. Mercereau-Puijalon, and S. Bonnefoy. 2000. A member of the Plasmodium falciparum Pf60 multigene family codes for a nuclear protein expressed by readthrough of an internal stop codon. Mol. Microbiol. 35:1005-1016. [DOI] [PubMed] [Google Scholar]
- 9.Bonnefoy, S., E. Bischoff, M. Guillotte, and O. Mercereau-Puijalon. 1997. Evidence for distinct prototype sequences within the Plasmodium falciparum Pf60 multigene family. Mol. Biochem. Parasitol. 87:1-11. [DOI] [PubMed] [Google Scholar]
- 10.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 11.Carcy, B., S. Bonnefoy, M. Guillotte, C. Le Scanf, P. Grellier, J. Schrevel, T. Fandeur, and O. Mercereau-Puijalon. 1994. A large multigene family expressed during the erythrocytic schizogony of Plasmodium falciparum. Mol. Biochem. Parasitol. 68:221-233. [DOI] [PubMed] [Google Scholar]
- 12.Carret, C., S. Delbecq, G. Labesse, B. Carcy, E. Precigout, K. Moubri, T. P. Schetters, and A. Gorenflot. 1999. Characterization and molecular cloning of an adenosine kinase from Babesia canis rossi. Eur. J. Biochem. 265:1015-1021. [DOI] [PubMed] [Google Scholar]
- 13.Depoix, D., B. Carcy, E. Jumas-Bilak, M. Pages, E. Precigout, T. P. M. Schetters, C. Ravel, and A. Gorenflot. 2002. Chromosome number, genome size and polymorphism of European and South-African isolates of large Babesia parasites that infect dogs. Parasitology 125:313-321. [DOI] [PubMed] [Google Scholar]
- 14.Fisher, A. G., B. Ensoli, L. Ivanoff, M. Chamberlain, S. Petteway, L. Ratner, R. C. Gallo, and F. Wong-Stahl. 1987. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science 237:888-893. [DOI] [PubMed] [Google Scholar]
- 15.Fischetti, V. A., V. Pancholi, and O. Schneewind. 1990. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 4:1603-1605. [DOI] [PubMed] [Google Scholar]
- 16.Franck, R. 1992. Spot-synthesis: an easy technique for positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48:9217-9232. [Google Scholar]
- 17.Frerichs, W. M., and A. A. Holbrook. 1974. Feeding mechanisms of Babesia equi. J. Protozool. 21:707-709. [DOI] [PubMed] [Google Scholar]
- 18.Gorenflot, A., A. Marchand, M. Gorenflot, C. Piette, and M. Piette. 1979. Morphologie des hématies parasitées lors de babésiose bovine (B. divergens) et canine (B. canis). Ann. Pharm. Fr. 37:493-500. [PubMed] [Google Scholar]
- 19.Hillman, B. I., R. Foglia, and W. Yuan. 2000. Satellite and defective RNAs of Cryphonectria hypovirus 3-grand haven 2, a virus species in the family Hypoviridae with a single open reading frame. Virology 276:181-189. [DOI] [PubMed] [Google Scholar]
- 20.Johnston, R. C., N. A. Farias, J. C. Gonzales, H. Dewes, A. Masuda, C. Termignoni, K. Amako, and L. S. Ozaki. 1991. A putative RNA virus in Babesia bovis. Mol. Biochem. Parasitol. 45:155-158. [DOI] [PubMed] [Google Scholar]
- 21.Kawai, S., I. Igarashi, A. Abgaadorjiin, H. Ikadai, Y. Omata, A. Saito, H. Nagasawa, Y. Toyoda, N. Suzuki, and H. Matsuda. 1999. Tubular structures associated with Babesia caballi in equine erythrocytes in vitro. Parasitol. Res. 85:171-175. [DOI] [PubMed] [Google Scholar]
- 22.Kawai, S., I. Igarashi, A. Abgaadorjiin, K. Miyazawa, H. Ikadai, K. Fujisaki, T. Mikami, N. Suzuki, and H. Matsuda. 1999. Ultrastructural characteristics of Babesia caballi in equine erythrocytes in vitro. Parasitol. Res. 85:794-799. [DOI] [PubMed] [Google Scholar]
- 23.Khoshnan, A., and J. F. Alderete. 1994. Trichomonas vaginalis with a double-stranded RNA virus has upregulated levels of phenotypically variable immunogen mRNA. J. Virol. 68:4035-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khoshnan, A., and J. F. Alderete. 1995. Characterization of double-stranded RNA satellites associated with the Trichomonas vaginalis virus. J. Virol. 69:6892-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khoshnan, A., D. Provenzano, and J. F. Alderete. 1994. Unique double-stranded RNAs associated with the Trichomonas vaginalis virus are synthesized by viral RNA-dependent RNA polymerase. J. Virol. 68:7108-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khramtsov, N. V., and S. J. Upton. 2000. Association of RNA polymerase complexes of the parasitic protozoan Cryptosporidium parvum with virus-like particles: heterogeneous system. J. Virol. 74:5788-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khramtsov, N. V., K. M. Woods, M. V. Nesterenko, C. C. Dykstra, and S. J. Upton. 1997. Virus-like, double-stranded RNAs in the parasitic protozoan Cryptosporidium parvum. Mol. Microbiol. 26:289-300. [DOI] [PubMed] [Google Scholar]
- 28.Kishi, M., Y. Nishino, M. Sumiya, K. Ohki, T. Kimura, T. Goto, M. Nakai, M. Kakinuma, and K. Ikuta. 1992. Cells surviving infection by human immunodeficiency virus type 1: vif or vpu mutants produce non-infectious or markedly less cytopathic viruses. J. Gen. Virol. 73:77-87. [DOI] [PubMed] [Google Scholar]
- 29.MacBeth, K. J., and J. L. Patterson. 1998. Overview of the Leishmaniavirus endoribonuclease and functions of other endoribonucleases affecting viral gene expression. J. Exp. Zool. 282:254-260. [PubMed] [Google Scholar]
- 30.MacBeth, K. J., and J. L. Patterson. 1995. Single-site cleavage in the 5′-untranslated region of Leishmaniavirus RNA is mediated by the viral capsid protein. Proc. Natl. Acad. Sci. USA 92:8994-8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCabe, P. M., P. Pfeiffer, and N. K. Van Alfen. 1999. The influence of dsRNA viruses on the biology of plant pathogenic fungi. Trends Microbiol. 7:377-381. [DOI] [PubMed] [Google Scholar]
- 32.Molina, F., D. Laune, C. Gougat, B. Pau, and C. Granier. 1996. Improved performances of spot multiple peptide synthesis. Pept. Res. 9:151-155. [PubMed] [Google Scholar]
- 33.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8:581-599. [DOI] [PubMed] [Google Scholar]
- 34.Precigout, E., A. Gorenflot, A. Valentin, G. Bissuel, B. Carcy, P. Brasseur, Y. Moreau, and J. Schrevel. 1991. Analysis of immune responses of different hosts to Babesia divergens isolates from different geographic areas and capacity of culture-derived exoantigens to induce efficient cross-protection. Infect. Immun. 59:2799-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Provenzano, D., A. Khoshnan, and J. F. Alderete. 1997. Involvement of dsRNA virus in the protein composition and growth kinetics of host Trichomonas vaginalis. Arch. Virol. 142:939-952. [DOI] [PubMed] [Google Scholar]
- 36.Roditi, I., T. Wyler, N. Smith, and R. Braun. 1994. Virus-like particles in Eimeria nieschulzi are associated with multiple RNA segments. Mol. Biochem. Parasitol. 63:275-282. [DOI] [PubMed] [Google Scholar]
- 37.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schetters, T. P., K. Moubri, E. Precigout, J. Kleuskens, N. C. Scholtes, and A. Gorenflot. 1997. Different Babesia canis isolates, different diseases. Parasitology 115:485-493. [DOI] [PubMed] [Google Scholar]
- 39.Schneewind, O., D. Mihaylova-Petkov, and P. Model. 1993. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 12:4803-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi, J., T. L. Blundell, and K. Mizuguchi. 2001. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310:243-257. [DOI] [PubMed] [Google Scholar]
- 41.Singh, B. R. 1996. Critical aspects of bacterial protein toxins. Adv. Exp. Med. Biol. 391:63-84. [DOI] [PubMed] [Google Scholar]
- 42.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 43.Strebel, K., D. Daugherty, K. Clouse, D. Cohen, T. Folks, and M. A. Martin. 1987. The HIV'A'(sor) gene product is essential for virus infectivity. Nature 328:728-730. [DOI] [PubMed] [Google Scholar]
- 44.Uilenberg, G., F. F. Franssen, N. M. Perie, and A. A. Spanjer. 1989. Three groups of Babesia canis distinguished and a proposal for nomenclature. Vet. Q. 11:33-40. [DOI] [PubMed] [Google Scholar]
- 45.Wang, A., C. C. Wang, and J. F. Alderete. 1987. Trichomonas vaginalis phenotypic variation occurs only among trichomonads infected with the double-stranded RNA virus. J. Exp. Med. 166:142-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, A. L., and C. C. Wang. 1991. Viruses of parasitic protozoa. Parasitol. Today 7:76-80. [DOI] [PubMed] [Google Scholar]
- 47.Wickner, R. B. 1996. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol. Rev. 60:250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]