Abstract
Macrophage inflammatory protein 1α (MIP-1α) (CCL3) is an important mediator of leukocyte recruitment and activation in a variety of inflammatory states, including infection. A recombinant human type 5 adenovirus containing the murine MIP-1α cDNA (AdMIP-1α) was constructed to determine the effect of transient intrapulmonary expression of MIP-1α on leukocyte recruitment, activation, and bacterial clearance in a murine model of Klebsiella pneumoniae pneumonia. The intratracheal administration of AdMIP-1α resulted in both time- and dose-dependent expression of MIP-1α mRNA and protein within the lung. Importantly, the intrapulmonary overexpression of MIP-1α resulted in a maximal 35- and 100-fold reduction in lung and blood bacterial burden, respectively, in animals cochallenged with K. pneumoniae, which was associated with a significant increase in neutrophil and activated NK cell accumulation. Furthermore, the transgenic expression of MIP-1α during bacterial pneumonia resulted in enhanced expression of gamma interferon mRNA, compared to that observed in Klebsiella-challenged animals pretreated with control vector. These findings indicate an important role for MIP-1α in the recruitment and activation of selected leukocyte populations in vivo and identify this cytokine as a potential immunoadjuvant to be employed in the setting of localized bacterial infection.
The generation of inflammation in the setting of infectious challenge is a complex and dynamic process, which involves the balanced expression of both pro- and anti-inflammatory cytokines (29, 35). In most instances, the compartmentalized elaboration of activating and/or chemotactic cytokines is required to generate sufficient phagocyte recruitment and activation to eradicate microbial pathogens.
Chemokines are a family of small, structurally related molecules that play a fundamental role in the development, homeostasis, and function of the immune system. Four closely related families of chemokines have now been characterized (24, 37). Of these, members of two families in particular have been shown to participate in pulmonary antimicrobial host responses. The CXC chemokines, which consist of both the ELR+ and ELR− subfamilies, have been most closely linked to the development of protective immunity in bacterial infection. The ELR+ CXC chemokines, which include CXCL1-8 and CXCL15, have predominant neutrophil stimulatory and chemotactic activities, whereas the ELR− CXC chemokines exert predominant chemotactic and/or activating effects on macrophages, specific T-cell populations, and NK cells (37). Selected murine ELR+ CXC chemokines or their common receptor (CXCR2) plays an instrumental role in the recruitment of neutrophils in response to bacterial challenge (14, 31, 33, 34). While a definitive role for ELR− CXC chemokines in antibacterial host defense is unproven, these chemokines are associated with the protective type 1 immune responses and several members of this family exert defensin-like bactericidal effects on Escherichia coli and Listeria monocytogenes when present in high concentrations (7, 16).
The CC chemokines, most notably macrophage inflammatory protein 1α (MIP-1α) (CCL3), have been shown to play an important role in the recruitment and activation of mononuclear cells in both the innate and acquired immune response to microbial pathogens. Specifically, mice deficient in MIP-1α (CCL3) display impaired early mononuclear cell influx and fail to develop protective type 1 cytokine responses after intratracheal (i.t.) challenge with Cryptococcus neoformans (22). Similarly, neutralization of MIP-1α in neutropenic mice challenged with Aspergillus fumigatus conidia resulted in the development of invasive pulmonary infection and increased lethality, which were associated with impaired influx of mononuclear phagocytes (20). Furthermore, mice with a targeted deletion of CCR1 (one of several receptors for MIP-1α) were predisposed to the development of invasive aspergillosis after intravenous administration of A. fumigatus conidia (12). More recent data indicate that MIP-1α may also contribute to innate responses within the lung in bacterial pneumonia, in part by providing necessary signals required for optimal resident macrophage activation (18). Specifically, we have shown that the i.t. administration of Klebsiella pneumoniae to MIP-1α knockout mice resulted in a considerable decrease in the clearance of bacteria from the lung compared to that in wild-type mice and higher mortality than in wild-type mice (18). The accumulation of mononuclear phagocytes and neutrophils post-K. pneumoniae challenge did not differ between the two groups. However, alveolar macrophages obtained from MIP-1α-deficient mice demonstrated reduced phagocytic activity, compared to control alveolar macrophages. Collectively, these studies suggest that MIP-1α can participate in innate and acquired immune responses against both intracellular and extracellular pathogens and that MIP-1α mediates this effect through the recruitment and/or activation of T cells and monocytes/macrophages (11, 24, 37).
MIP-1α has also been shown to exert chemotactic effects on neutrophils and NK cells (8, 19, 32). However, there is limited evidence to support a role for MIP-1α in recruitment of these cells in vivo. In this study, we demonstrate that the intrapulmonary transient transgenic expression of MIP-1α (CCL3) can augment bacterial clearance from the lung, which is associated with augmented neutrophil influx, as well as significant NK cell recruitment and activation.
MATERIALS AND METHODS
Reagents.
Polyclonal rabbit anti-murine MIP-1α antibodies and biotinylated anti-murine MIP-1α antibody used in the enzyme-linked immunosorbent assay (ELISA) were produced from sera of rabbits preimmunized with purified recombinant, murine MIP-1α (PeproTech, Inc., Rocky Hill, N.J.) as previously described (28).
Generation of a recombinant adenovirus encoding murine MIP-1a.
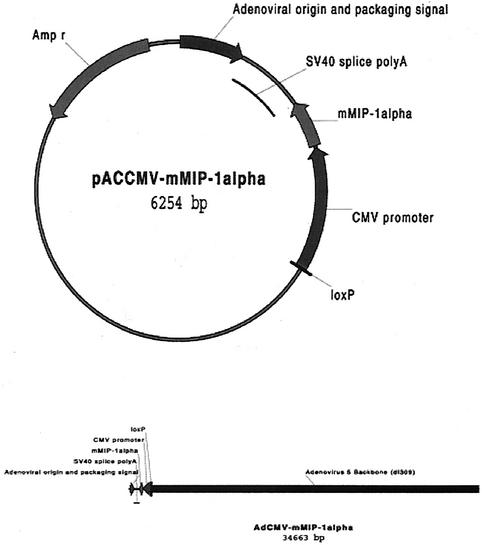
The murine MIP-1α cDNA was cut by restriction enzymes AgeI and NheI from pORF9-mMIP-1α (InvivGen Co., San Diego, Calif.), resulting in a 315-bp mMIP-1α fragment. Plasmid pAcmvplpa(-)Loxp.ssp (5,951 bp) was obtained from the Vector Core, University of Michigan, and was digested by XbaI (931) and XbaI (942) to get a 5,940-bp backbone. The MIP-1α gene and pAcmvplpa(-)Loxp.ssp backbone were ligated, resulting in a 6.2-kp recombinant plasmid, which was linearized by NheI (Fig. 1). The adenoviral shuttle plasmid containing MIP-1α was linearized with NheI and inserted into an adenoviral type 5 backbone with a partial E 3 deletion (dl 309) by using Cre-loxP recombination (1). The product was transfected into 293 helper cells by using calcium phosphate to produce the MIP-1α expressing adenovirus (15). Clones were amplified and tested for MIP-1α protein expression by ELISA. A clone was amplified in the 911 helper cell line and purified as previously described (30). Purified virus was titered by 911 cell plague assay.
FIG. 1.
Schematic of the adenovirus construction system by Cre-loxP recombination in vitro. The coding region of mouse MIP-1α was excised from pORF-9-mMIP-1α with AgeI and NheI and was then ligated into the XmaI and XbaI sites of the adenovirus shuttle plasmid pAcmvplpa(-)Loxp.ssp (A). SV40, simian virus 40; CMV, cytomegalovirus. The adenoviral shuttle plasmid containing MIP-1α was linearized with PmeI and joined to an adenoviral type 5 backbone with a partial E3 deletion (dl 309) using Cre-loxP recombination (B).
Mice.
Female specific-pathogen-free 6- to 8-week-old C57BL/6J mice were purchased from Jackson Laboratory (Bar Harbor, Maine) and housed in specific-pathogen-free conditions within the animal care facility at University of Michigan until the day of sacrifice.
K. pneumoniae.
K. pneumoniae strain 43816, serotype 2, was obtained from the American Type Culture Collection (Rockville, Md.). For infection, bacteria were grown to stationary phase (18 h) in tryptic soy broth (Soybean-Casein digest; Difco, Detroit, Mich.) in a vented 50-ml conical tube at 37°C, 5% CO2 (6). The concentration of bacteria in broth was determined by measuring the amount of absorbance at 600 nm on a DU-64 Spectrophotometer (Beckman Instruments Inc., Fullerton, Calif.) and was compared to a standard curve of absorbance. The bacterial suspension was diluted with saline to the desired concentration in saline, and bacterial counts were subsequently confirmed by planting out the suspension (14).
i.t. inoculation of K. pneumoniae.
Mice were anesthetized by intraperitoneal injection of a ketamine-xylazine mixture. The trachea was exposed, and 30 μl of inoculum was administered via a sterile 26-gauge needle (2 to 3 × 103 CFU of K. pneumoniae). The skin incision was closed via surgical staples. An aliquot of the K. pneumoniae inoculum was serially diluted onto blood agar plates to determine actual dose of i.t. injected bacteria (14).
Total lung leukocyte preparation.
Lungs were removed from euthanatized animals, and leukocytes were prepared as previously described (21). Briefly, lungs were minced with scissors to a fine slurry in 15 ml of digestion buffer per lung containing RPMI medium, 5% fetal calf serum, 1-mg/ml collagenase (Boehringer Mannheim Biochemical), and 30-μg/ml DNase (Sigma, St. Louis, Mo.). Lung slurries were enzymatically digested for 30 min at 37°C. Any undigested fragments were further dispersed by drawing the solution through the bore of a 10-ml syringe. The total lung cell suspension was pelleted, resuspended, and spun through a 20% Percoll gradient to enrich for leukocytes for flow analysis. Cell counts and viability were determined with trypan blue exclusion counting on a hemacytometer. Cytospin slides were prepared and stained with a modified Wright-Giemsa stain.
Multiparameter flow cytometry analyses.
Cells from infected and uninfected control mice were isolated from lung digests as described above. For analyses of T-cell subsets, isolated leukocytes were stained with biotinylated anti-γδ-T-cell receptor (TCR), anti-αβ-TCR, anti-NK1.1-phycoerythrin (PE), anti-CD8-PE, and anti-CD4-fluorescein isothiocyanate (FITC) plus CD8-FITC, anti-NK1.1-FITC, and anti-CD69−-FITC as previously described (21). TCR expression was detected by the addition of streptavidin-PE (all reagents from PharMingen, San Diego, Calif., unless otherwise noted). In addition, cells were stained with anti-CD45-Tricolor (Caltag Laboratories, South San Francisco, Calif.), allowing discrimination of leukocytes from nonleukocytes and thus eliminating any nonspecific binding of T-cell surface markers on nonleukocytes. T-cell subsets were analyzed by first gating on CD45-positive “lymphocyte-sized” leukocytes and were then examined for FL1 and FL2 fluorescence expression. Cells were collected on a FACScan or FACScalibur cytometer (Becton Dickinson, San Jose, Calif.) with CellQuest software (Becton Dickinson). Analyses of data were performed with the CellQuest software package. Percent positive cells indicated in histogram plots represent the percentage of positive cells back calculated to total leukocytes.
Lung harvesting.
At a designated time point, mice were sacrificed by CO2 asphyxia. Prior to lung removal, the pulmonary vasculature was perfused via the right ventricle with 1 ml of phosphate-buffered saline (PBS) containing 5 mM EDTA. Whole lungs were then harvested for assessment of cytokine protein expression and assessment of bacterial number. After removal, whole lungs were homogenized in 1.0 ml of PBS with protease inhibitor (Boehringer Mannheim, Indianapolis, Ind.) by using a tissue homogenizer (Biospec Products, Inc.) under a vented hood. Portions of homogenates (10 μl) were inoculated on tryptic soy-blood agar after serial 1:5 dilutions with PBS to determine the number of CFU. The remaining homogenates were incubated on ice for 30 min and were then centrifuged at 2,500 rpm for 10 min in a Beckman model CS-6R centrifuge. Supernatants were collected, passed through a 0.45-μm-pore-size filter (Gelman Sciences, Ann Arbor, Mich.), and then stored at −20°C for assessment of cytokine levels.
Isolation and RT-PCR amplification of whole-lung mRNA.
Whole lung was harvested, immediately snap-frozen in liquid nitrogen, and stored at −70°C, and then reverse transcription-PCR (RT-PCR) was performed as previously described (31, 34). Briefly, total cellular RNA from the frozen lungs was isolated, reversed transcribed into cDNA, and then amplified as previously described (4) with specific primers for MIP-1α, with β-actin serving as a control. The primer had the sequences 5′-GCC-CTT-GCT-GTT-CTT-CTC-3′ and 5′-GGC-AAT-CAG-TTC-CAG-GTC-3′ for MIP-1α and 5′-ATG-GAT-GAC-GAT-ATC-GCT-C-3′ and 5′-GAT-TCC-ATA-CCC-AGG-AGG-G-3′ for β-actin. After amplification, the samples (20 μl) were separated on a 2% agarose gel containing ethidium bromide (5 μl/100 ml) (10 mg/ml; Sigma), and bands were visualized and photographed using UV transillumination.
Real-time quantitative PCR.
Measurement of gene expression was performed with the ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, Calif.). Primers and probes for β-actin and gamma interferon (IFN-γ) were designed using Shortcat for Primer Express software (Applied Biosystems). The primers, placed in different exons, were confirmed not to amplify genomic DNA. Primer and probe nucleotide sequences for murine IFN-γ (GenBank accession number K00083) were as follows: forward primer, 5′-CTG CGG CCT AGC TCT GAG A-3′; reverse primer, 5′-CAG CCA GAA ACA GCC ATG AG-3′; and TaqMan probe, 5′(FAM)-CAC ACT GCA TCT TGG CTT TGC AGC TC-(TAMR) 3′. Primer and probe nucleotide sequences for murine β-actin (GenBank accession number M12481) were as follows: forward primer, 5′-CCG TGA AAA GAT GAC CCA GAT C-3′, reverse primer, 5′-CAC AGC CTG GAT GGC TAC GT-3′; and TaqMan probe, 5′(FAM)-TTT GAG ACC TTC AAC ACC CCA GCC A-TAMRA-3′. They were purchased from Applied Biosystems Custom Oligo Synthesis Service. Thermal cycling parameters for use with the TaqMan One-Step RT-PCR Master Mix Reagents Kit included 30 min at 48°C, 10 min at 95°C, and 40 cycles involving denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min. Total RNA was extracted from a frozen lung specimen by using a commercial Trizol Reagent (Life Technologies, Invitrogen). These experiments were performed in duplicate. To normalize the amount of total RNA present in each reaction, the housekeeping gene β-actin was amplified, which is assumed to be constant in both AdMIP-1α and Adcmvplpa samples. The amount of IFN-γ, normalized to β-actin and relative to a calibrator, is given as follows: target amount = 2−ΔΔCt, where ΔΔCt = {[CtIFN-γ sample − Ctβ-actin sample] − [CtIFN-γ calibrator − Ctβ-actin calibrator]}. This method is based on the assumption that the target IFN-γ and β-actin display equal amplification efficiencies. To verify this, ΔCt (CtIFN-γ − Ctβ-actin) variations were assessed according to template dilution. To this end, a standard curve was generated composed of five different dilutions of total RNA, corresponding to 50, 25, 12.5, 6.25, and 3.125 ng. The slope of this curve was 0.086. To assure the appropriate amplification efficiency, the slope of the standard curve should be <0.1.
Murine cytokine ELISA.
Murine MIP-1α was quantitated with a modification of a double ligand method as previously described (28). Standards were one-half log dilution of murine recombinant cytokine from 1 pg/ml to 100 ng/ml. This ELISA method consistently detected murine MIP-1α concentrations above 50 pg/ml. The ELISA did not cross-react with other cytokines, such as interleukin 1 (IL-1), IL-2, IL-4, and IL-6. In addition, the ELISA did not cross-react with other members of the murine chemokine family, including murine MIP-2, KC, or RANTES.
Lung histological evaluation.
Mice were sacrificed with carbon dioxide inhalation on day 2 and day 3 after inoculation with K. pneumoniae. The trachea was exposed, intubated with a 1.7-mm-outside-diameter polyethylene catheter, and lung was perfused with 4% paraformaldehyde in PBS. The trachea was tied with 18-inch silk black braided thread. Lung was removed and fixed for up to 48 h in 4% paraformaldehyde in PBS (pH 7.4) at room temperature. The lungs were then embedded in paraffin, and sections were cut and stained with hematoxylin and eosin under standard techniques.
Statistical analyses.
Statistical significance was determined with one-way analysis of variance with Bonferroni's posttest for three or more groups or the Mann-Whitney test for two groups. Calculations were performed with Prism 3.0 for Windows 95 and NT (GraphPad Software).
RESULTS
Time-dependent expression of MIP-1α mRNA and protein within the lung after i.t. administration of Adcmvplpa or AdMIP-1α.
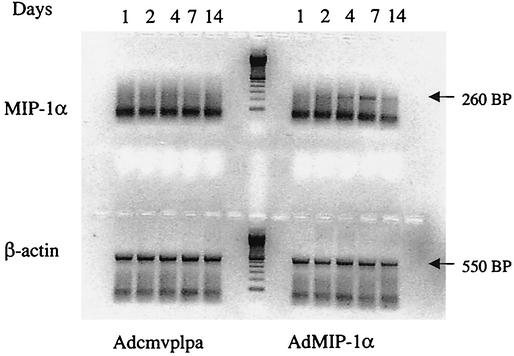
To assess the efficacy of i.t. delivery of Adcmv-mMIP-1α on subsequent MIP-1α production, C57BL/6J mice were administered either 5 × 108 PFU of control adenovirus (Adcmvplpa[-]Lox.ssp) or recombinant MIP-1α adenovirus (AdMIP-1α) and then lungs were harvested at various time points thereafter. As shown in Fig. 2, MIP-1α mRNA was expressed within the lung by day 2 after i.t. administration of AdMIP-1α, with maximal expression of MIP-1α observed at day 7. In contrast, no MIP-1α mRNA expression was detected in the lungs at any time point after i.t. administration of Adcmvplpa (control vector).
FIG. 2.
Time-dependent expression of MIP-1α mRNA after i.t. AdMIP-1α administration. Lungs were harvested on days 1, 2, 4, 7, and 14 after i.t. administration of Adcmvplpa (5 × 108 PFU) and AdMIP-1α (5 × 108 PFU), and then MIP-1α and β-actin mRNA expression was determined by RT-PCR (30 and 25 cycles for MIP-1α and β-actin, respectively). Molecular weight markers are shown to the right. Each lane represents two animals per time point; experimental n = 4.
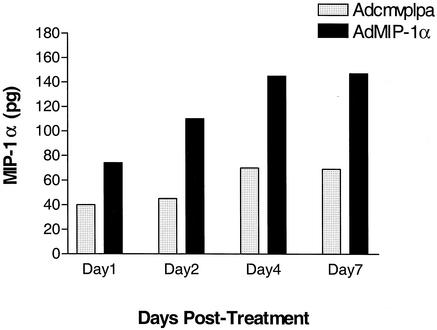
The expression of MIP-1α protein within the lung after AdMIP-1α or Adcmvplpa was also determined. As shown in Fig. 3, no significant increase in MIP-1α was seen in lung at day 1 and day 2, while a modest increase in lung MIP-1α protein levels was detected at days 4 and day 7 after i.t. administration of Adcmvplpa. However, a significant increase in lung MIP-1α levels was noted as early as day 1 after AdMIP-1α administration, with maximal levels observed at days 4 and 7 post-AdMIP-1α administration. Maximal expression of MIP-1α mRNA and protein occurred at a dose of 5 × 108 PFU, as no additional increase was observed at higher concentrations of vector (data not shown).
FIG. 3.
Time-dependent production of MIP-1α in lung homogenates. Mice were i.t. administered AdMIP-1α (5 × 108 PFU) or Adcmvplpa (5 × 108 PFU), and then MIP-1α levels were determined by specific ELISA. P < 0.01 compared with animals receiving Adcmvplpa at each point in time. Experimental n = 4 per group.
Effect of intrapulmonary MIP-1α transient transgenic expression on bacterial clearance in murine Klebsiella pneumonia.
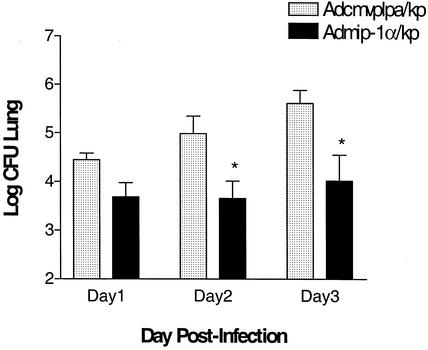
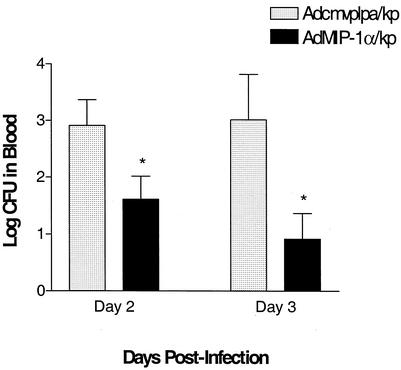
Experiments were next performed to assess the effect of transient transgenic expression of MIP-1α on bacterial clearance in a murine Klebsiella pneumonia model. In these studies, control adenovirus or AdMIP-1α (5 × 108 PFU) was administered i.t., followed 4 days later by the i.t. administration of K. pneumoniae (2 × 103 to 3 × 103 CFU). Then lungs and blood were harvested on days 1, 2, and 3 post-bacterial challenge for assessment of K. pneumoniae CFU. As shown in Fig. 4, there was a time-dependent increase in the number of bacterial CFU in Adcmvplpa-pretreated mice, maximal at day 3 post-K. pneumoniae administration. In contrast, no significant increases in lung bacterial burden were observed over 3 days in animals pretreated with AdMIP-1α. Importantly, lung bacterial burden was approximately 16- and 35-fold greater, respectively, in the Adcmvplpa-treated mice at days 2 and 3 than in animals pretreated with AdMIP-1α. No difference in K. pneumoniae CFU was observed between animals pretreated with control vector, compared to animals receiving no vector prior to bacterial challenge (data not shown). Similarly, treatment with AdMIP-1α resulted in a significant reduction in bacterial burden in blood, with a maximal 100-fold reduction in the number of K. pneumoniae CFU observed at 3 days post-bacterial administration, compared to that found in infected animals treated with control vector (Fig. 5, P < 0.05).
FIG. 4.
K. pneumoniae CFU in lung digests of Klebsiella-challenged mice pretreated with AdMIP-1α or Adcmvplpa. Mice were i.t. administered AdMIP-1α or Acmvplpa (5 × 108 PFU), followed 4 days later by the i.t. administration of K. pneumoniae (3 × 103 CFU). On days 1, 2, and 3 post-K. pneumoniae administration, lung digests were prepared and the numbers of CFU were determined. P < 0.05 compared to control. n = 5 mice per group.
FIG. 5.
K. pneumoniae CFU in blood of Klebsiella-challenged mice pretreated with AdMIP-1α or Adcmvplpa. Mice were i.t. administered AdMIP-1α or Acmvplpa (5 × 108 PFU), followed 4 days later by the i.t. administration of K. pneumoniae (3 × 103 CFU). On days 2 and 3 post-K. pneumoniae administration, the numbers of CFU in blood were determined. P < 0.05 compared to control. n = 10 to 15 mice per group. No K. pneumoniae CFU were isolated at day 1 post-bacterial challenge; thus, this time point was excluded.
Effect of intrapulmonary MIP-1α transgene expression on lung histopathology after challenge with K. pneumoniae.
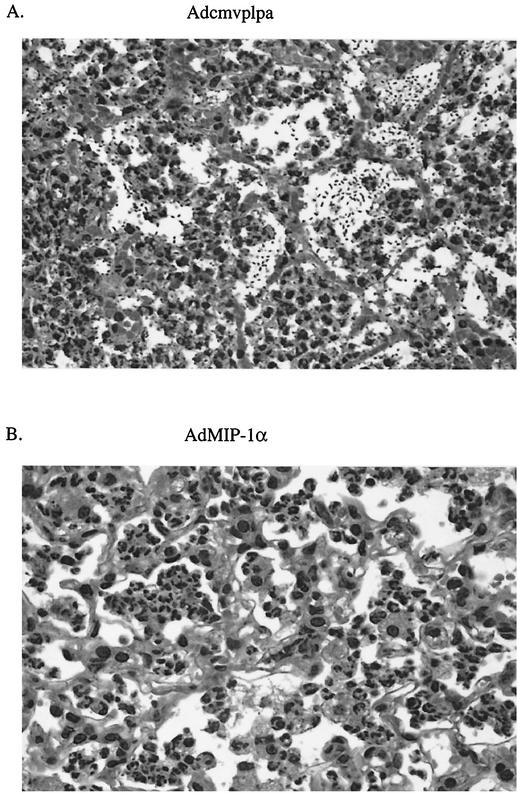
The next series of studies was performed to determine the effect of MIP-1α transgene expression on lung histopathology after bacterial challenge. Mice were administered either Adcmvplpa or AdMIP-1α i.t. (5 × 108 PFU), followed by K. pneumoniae administration (2 × 103 to 3 × 103 CFU) 4 days later. Histology performed 3 days post-bacterial administration revealed a prominent infiltration of inflammatory cells, predominantly neutrophils, in the lungs of AdMIP-1α-treated mice (Fig. 6B). By comparison, animals pretreated with Adcmvplpa displayed a less prominent influx of leukocytes, while showing clear evidence of extracellular bacteria present within alveolar spaces of involved lungs (Fig. 6A).
FIG. 6.
Effect of intrapulmonary MIP-1α transgene expression on lung histopathology after K. pneumoniae challenge. Representative lung histology in Adcmvplpa-pretreated mice (A) or AdMIP-1α (5 × 108 PFU)-pretreated mice (B) 3 days post-i.t. administration of K. pneumoniae (2 × 103 CFU). Experimental n = 2 per group.
Effect of intrapulmonary transient transgenic expression of MIP-1α on inflammatory cell influx in murine Klebsiella pneumonia.
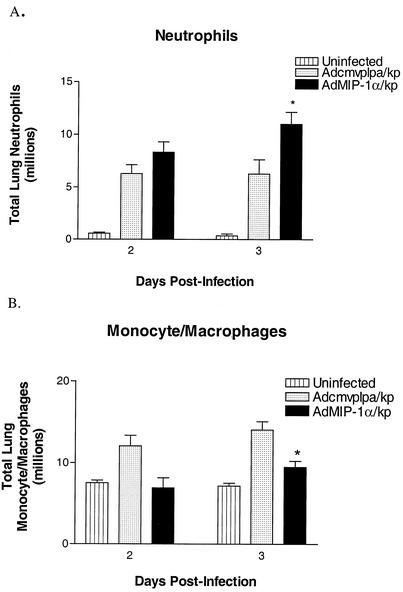
To determine if the beneficial effect of AdMIP-1α administration on bacterial clearance was attributable to augmented inflammatory cell recruitment, lungs from Adcmvplpa- or AdMIP-1α-treated animals were harvested 2 or 3 days following administration of K. pneumoniae administration. These time points were chosen, as maximal influx of inflammatory cells occurs at these times. As shown in Table 1, treatment with AdMIP-1α (5 × 108 PFU) induced modest but statistically significant increases in total lung neutrophils, lymphocytes and activated NK cells (NK1.1+ CD69+) and a trend toward increases in NK1.1+ CD69+ cells (P = 0.08) at 2 days posttreatment. Compared to uninfected controls, both Adcmvplpa- and AdMIP-1α-pretreated mice experienced a marked increase in lung neutrophils in response to bacterial administration (Fig. 7). Interestingly, the numbers of neutrophils in AdMIP-1α-treated mice was approximately 1.7-fold greater 3 days after bacterial challenge than in Klebsiella-infected animals pretreated with control virus (P < 0.05). Conversely, animals pretreated with AdMIP-1α were found to have fewer recruited monocytes/macrophages in lungs at 3 days post-K. pneumoniae administration than did similarly treated control animals (P < 0.05). K. pneumoniae administration resulted in no significant change in overall numbers of T or B lymphocytes over those seen in uninfected animals (data not shown).
TABLE 1.
Effect of i.t. adMIP-1α or Adcmvplpa on leukocyte influx on days 2 and 4 postadministrationa
| Day | Drug given | No. (105) of cells
|
|||||
|---|---|---|---|---|---|---|---|
| Total cells | Monocytes/macrophages | Polymorphonuclear leukocytes | Lymphocytes | NK1.1+ CD69+ cells | NK1.1+ CD69+ cells | ||
| 2 | Saline | 109.2 ± 5.2 | 98.9 ± 5.1 | 5.0 ± 1.1 | 4.6 ± 0.8 | 1.0 ± 0.1 | 0.07 ± 0.01 |
| AdMIP-1α | 157.6 ± 12* | 137.5 ± 10.6 | 11.7 ± 0.9* | 7.0 ± 0.6* | 1.48 ± 0.2 | 0.18 ± 0.02* | |
| Adcmvplpa | 131.1 ± 9.0 | 119.5 ± 9.6 | 7.7 ± 0.7 | 4.6 ± 0.3 | 1.1 ± 0.2 | 0.10 ± 0.01 | |
| 4 | Saline | 131.3 ± 10.0 | 121.4 ± 9.0 | 5.9 ± 1.1 | 3.6 ± 0.6 | ND | ND |
| AdMIP-1α | 123.7 ± 17 | 116.4 ± 10.7 | 3.9 ± 0.5 | 3.5 ± 0.5 | ND | ND | |
| Adcmvplpa | 129.7 ± 9.3 | 126.8 ± 4.6 | 2.6 ± 0.4 | 3.1 ± 0.2 | ND | ND | |
*, P < 0.05 compared to saline. ND = not done. Experimental n = three to six per group.
FIG. 7.
Effect of pretreatment with Adcmvplpa or AdMIP-1α on accumulation of neutrophils and monocytes/macrophages in murine Klebsiella pneumonia. Lung neutrophils and monocytes/macrophages were quantitated from enzymatically digested lung tissue at days 2 and 3 post-K. pneumoniae administration. *, P < 0.01 compared to untreated mice and P < 0.05 compared to Adcmvplpa-treated mice. Experimental n = 3 for uninfected mice and 8 for Klebsiella-infected mice.
Effect of AdMIP-1α administration on numbers and activational state of NK cells following i.t. challenge with K. pneumoniae.
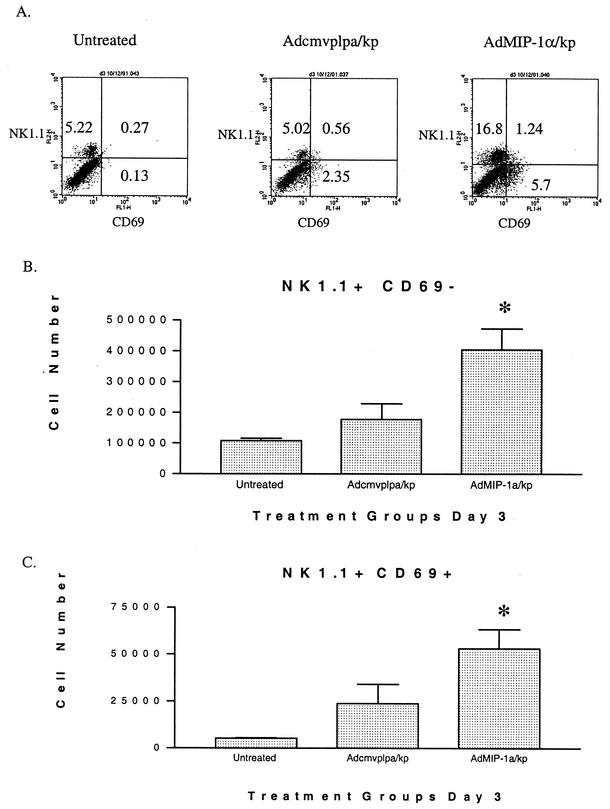
NK cells have previously been shown to be an important source of activating cytokines, including chemokines and IFN-γ (2, 23). In fact, these cells are the predominant intrapulmonary source of IFN-γ in certain bacterial infections of the lung, most notably pneumonia due to Legionella pneumophila (9). Given that MIP-1α has been demonstrated to induce NK cell activation and migration in vitro, studies were performed to assess the effect of intrapulmonary MIP-1α expression on the accumulation and activation of NK cells during bacterial pneumonia. The cell surface expression of CD69 was used as a measure of cellular activation (4), while NK cells were identified with the NK-specific marker NK1.1. As shown, in Fig. 8A (dot plot) and 8B, no significant change in the numbers of inactivated NK cells (NK1.1+ CD69−) were noted in Klebsiella-infected animals pretreated with Adcmvplpa, compared to those found in uninfected control mice. However, pretreatment with AdMIP-1α resulted in a significant increase in the number of NK1.1+ CD69− cells by 3 days following K. pneumoniae administration (P < 0.05). Furthermore, while K. pneumoniae-infected mice control mice had a small increase (P = 0.10) in NK1.1+ CD69+ cells recovered from the lung, compared to uninfected animals, pretreatment with AdMIP-1α resulted in a 10-fold increase in the number of NK1.1+ CD69+ cells after K. pneumoniae challenge in comparison to no treatment (Fig. 7C, P < 0.01). Notably, we observed no changes in the numbers of either CD69− or CD69+ NK-T cells (NK1.1+ TCR+) in either control or AdMIP-1α-treated mice during the course of Klebsiella infection, indicating that the changes in NK1.1+ cells reflect true changes in the NK cell population instead of changes in NK-T cells (data not shown).
FIG. 8.
Effect of pretreatment with Adcmvplpa or AdMIP-1α on accumulation of activated and unactivated NK cells post-K. pneumoniae challenge. Cells were isolated from lungs at 3 days post-K. pneumoniae administration, stained for surface expression of NK1.1 and CD69, and then analyzed by flow cytometry with gating for lymphocytes by size and complexity characteristics. Panel A is a representative dot plot; panels B and C give total numbers of NK1.1+ CD69− and NK1.1+ CD69+ cells, respectively. *, P < 0.01 compared to untreated and P < 0.05 compared to Adcmvplpa-treated mice. Experimental n = 3 for uninfected mice and 6 for Klebsiella-infected mice.
Effect of AdMIP-1α administration on lung IFN-γ mRNA expression following i.t. challenge with K. pneumoniae.
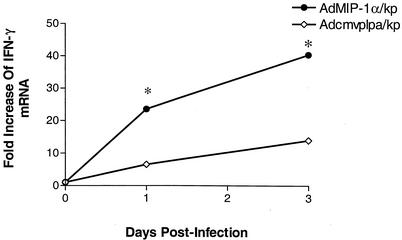
Given that activated NK cells are an important cellular source of IFN-γ and that MIP-1α has been associated with type 1 immune responses in other infection or inflammation models, we next assessed the effect of transgenic expression of MIP-1α on lung IFN-γ mRNA expression in bacterial pneumonia. As shown in Fig. 9, the i.t. administration of K. pneumoniae in mice pretreated with Adcmvplpa resulted in a time-dependent induction of IFN-γ mRNA expression, as determined by quantitative real-time PCR, with a maximal 14-fold induction observed at 72 h post-bacterial challenge. Pretreatment with AdMIP-1α, however, resulted in a substantially greater induction of IFN-γ mRNA at both 24 and 72 h post-K. pneumoniae administration (P < 0.05 at both time points) than in infected animals pretreated with control virus.
FIG. 9.
Effect of pretreatment with Adcmvplpa or AdMIP-1α on lung IFN-γ mRNA accumulation post-K. pneumoniae challenge. Lungs were harvested at days 1 and 3 post-K. pneumoniae administration, and then IFN-γ mRNA levels were determined by quantitative real-time PCR. Each value represents the mean of five animals per group. *, P < 0.05 compared to the infected, Adcmvplpa-pretreated mice.
DISCUSSION
Innate immunity against bacterial pathogens of the lung requires the activation and recruitment of phagocytic cells. The alveolar macrophage is the primary resident phagocyte within the alveolus and has been shown to be instrumental in the clearance of bacteria that reach the distal airspaces. When the number or virulence of invading bacteria overwhelms the ability to contain them, neutrophils are vigorously recruited and are necessary for successful eradication of extracellular bacterial organisms (5, 29, 35). It has been previously shown that MIP-1α is produced in response to intrapulmonary challenge with K. pneumoniae and that animals deficient in MIP-1α display an impaired clearance of bacteria and increased mortality, compared to similarly challenged wild-type animals (18). In this study, we have constructed a recombinant adenovirus incorporating the murine MIP-1α cDNA, which results in the time- and dose-dependent overexpression of MIP-1α mRNA and protein in lung after i.t. administration. Importantly, the forced transgenic expression of MIP-1α during murine Klebsiella pneumonia enhanced bacterial clearance, which was associated with increased recruitment of neutrophils, accumulation of CD69+ and CD69− NK cells, and enhanced expression of IFN-γ mRNA during the evolution of pneumonia. We and other investigators have previously used adenoviral gene therapy approaches to enhance the expression of inflammatory genes, which have generally resulted in improved innate responses in bacterial pneumonia (9, 13, 17, 30). The reason(s) why the endogenous expression of MIP-1α and other stimulatory cytokines can often be suboptimal during innate responses to infectious challenge is unclear but may be attributable to the local presence of anti-inflammatory cytokines (e.g., IL-10), which are released by the host in an attempt to limit leukocyte-mediated tissue injury. Collectively, these studies support the notion that means to augment immunity are usually beneficial when faced with localized infection with encapsulated, highly invasive bacterial organisms.
The cellular mechanism(s) by which transgenic expression of MIP-1α enhances clearance in pneumonia has not been completely defined. However, we observed a modest but significant increase in the influx of neutrophils in the lung after MIP-1α expression in uninfected mice, compared to results found in animals receiving control virus alone. Furthermore, the sequestration of neutrophils was significantly enhanced in the lungs of Klebsiella-infected mice overexpressing MIP-1α and represented an approximately 1.7-fold increase in neutrophil influx over that seen in animals pretreated with control adenovirus. While the biologic properties of MIP-1α have been most consistently directed toward mononuclear cells (20, 22, 24, 37), some evidence exists to indicate that, under certain circumstances, this chemokine can stimulate neutrophil chemotaxis and activation. For instance, murine neutrophils isolated from inflammatory exudates display chemotactic responses and calcium flux in response to MIP-1α (8). Interestingly, resting human neutrophils express low levels of CCR1 at baseline and fail to migrate in response to MIP-1α (3, 36). However, treatment with IFN-γ or granulocyte-macrophage colony-stimulating factor can dramatically increase CCR1 expression, as well as calcium flux and chemotactic responses to MIP-1α (3, 6). In vivo, the inhibition of MIP-1α or deficiency of CCR1 has been shown to attenuate the recruitment of neutrophils to the lung in rodents in response to i.t. or systemic endotoxin administration (27, 28) or during pulmonary paramyxovirus infection (10). Thus, the neutrophil chemotactic and activating effects of MIP-1α may be of some functional relevance, particularly in the setting of pyogenic bacterial infection. Importantly, the recruitment of neutrophils in response to transgenic MIP-1α expression was not dependent on NK cell recruitment or activation, as depletion of NK cells (using anti-NK1.1 antibody) did not block the effects of MIP-1α gene therapy on neutrophil influx during Klebsiella infection (data not shown). Our findings are somewhat at odds with the fact that no decrease in neutrophil influx was observed in MIP-1α-deficient mice challenged with K. pneumoniae (18). However, it should be noted that bacterial burden was considerably increased in knockout animals, likely driving the expression of other neutrophil chemoattractants to compensate for a deficiency in MIP-1α.
In addition to changes in neutrophil numbers, we also observed MIP-1α-induced changes in the numbers of CD69+ and CD69− NK cells. Specifically, adenoviral infection alone induced a modest increase in the number of activated NK cells, whereas the overexpression of MIP-1α concomitant with bacterial infection resulted in an increase in the number of both CD69+ and CD69− NK cells. MIP-1α, along with several members of the CC and ELR− CXC chemokine families, has been shown to induce NK cell chemotaxis, degranulation, and tumoricidal activity in vitro (19, 32). Furthermore, in vivo NK chemoattractant effects of MIP-1α have been suggested by the finding of reduced hepatic NK cell infiltration during murine cytomegalovirus infection in animals treated with MIP-1 neutralizing antibody (26). However, the present study provides the most direct evidence linking MIP-1α to localized NK cell accumulation and activation in vivo. Based on the fact that neither MIP-1α, nor other chemokines for that matter, has been shown to induce NK cell proliferation, we have presumed that the accumulation of NK cells in animals overexpressing MIP-1α occurs as a result of recruitment rather than proliferation. Although our studies indicate a significant effect of forced MIP-1α expression on both NK cell accumulation and activation, the functional significance of this effect remains unclear, as the depletion of NK cells during Klebsiella infection failed to appreciably alter either bacterial clearance or survival in this model. Furthermore, depletion of NK cells resulted in only a modest and statistically insignificant attenuation of MIP-1α protective effects on bacterial clearance in pneumonia (data not shown). These data would suggest that the beneficial effects of MIP-1α gene transfer in pneumonia are more directly attributable to effects on neutrophil recruitment/activation (and possibly macrophage activation) rather than changes in NK cell trafficking and activation.
Members of the CC chemokine family, including MIP-1α, are known to induce the recruitment of mononuclear cells, particularly monocytes. Therefore, it is somewhat surprising that the transgenic expression of MIP-1α did not increase the influx of mononuclear phagocytes. In fact, a decrease in monocytes was observed in infected animals overexpressing MIP-1α. A possible explanation for this finding is that the transgenic expression of MIP-1α resulted in an early and persistent decrease in bacterial burden, which might serve to modify the magnitude and timing of monocyte recruitment in response to bacterial challenge. We have demonstrated diminished phagocytic responses in alveolar macrophages recovered from MIP-1α-deficient mice, indicating important macrophage-activating effects of this chemokine (18). Thus, it is quite possible that resident lung macrophage antimicrobial properties are augmented in the setting of MIP-1α transgenic expression.
Previous studies have found that neutralization of MIP-1α during pulmonary inflammation can alter that expression of other inflammatory cytokines, most notably tumor necrosis factor alpha (22, 27). In our model, the overexpression of MIP-1α in pneumonia was not associated with appreciable alterations in lung tumor necrosis factor alpha levels. Furthermore, we observed no changes in the expression of relevant chemokines, including the ELR+ CXC chemokines MIP-2 and KC and the CC chemokine MCP-1 (data not shown). However, we did observe substantial induction of IFN-γ in infected animals pretreated with AdMIP-1α. IFN-γ has previously been shown to play a crucial role in lung host defense against both extracellular and intracellular bacterial pathogens (9, 25), in part through enhancement of both neutrophil and macrophage antimicrobial properties. Thus, it is likely that augmented expression of IFN-γ contributes to the improved innate immunity observed in the setting of transgenic expression of MIP-1α by facilitating polymorphonuclear leukocyte influx (via upregulation of CCR1 [3]) and by directly enhancing leukocyte effector cell function. A predominant source of IFN-γ in lung bacterial infection is activated NK cells (9; unpublished observations). However, other cell populations, including inflammatory macrophages and γδ-T cells, may also be relevant cellular sources of enhanced IFN-γ expression. The importance of γδ-T cells in the initial expression of IFN-γ during Klebsiella pneumonia has been demonstrated previously (21).
In conclusion, our findings illustrate an important role for MIP-1α in the recruitment and/or activation of neutrophils and NK cells in bacterial infection of the respiratory tract. Furthermore, these studies identify MIP-1α as a potential therapeutic target in the compartmentalized treatment of patients with life-threatening bacterial infection of the lung. While adenoviral approaches represent an attractive and efficient means to deliver genes experimentally, better vehicles are needed to limit the potential toxicities associated with this form of gene delivery.
Acknowledgments
This research was supported by National Institutes of Health grants HL57243 and P50HL60289 (T.J.S.) and AI49448 (T.A.M.).
Editor: D. L. Burns
REFERENCES
- 1.Aoki, K., C. Barker, X. Danthinne, M. J. Imperiale, and G. J. Nabel. 1999. Efficient generation of recombinant adenoviral vectors by Cre-lox recombination in vitro. Mol. Med. 5:224-231. [PMC free article] [PubMed] [Google Scholar]
- 2.Biron, C. A. 1997. Activation and function of natural killer cell responses during viral infections. Curr. Opin. Immunol. 9:24-34. [DOI] [PubMed] [Google Scholar]
- 3.Bonecchi, R., N. Palentarutti, W. Luini, A. Borsatti, S. Bernasconi, M. Locati, C. Power, A. Proudfoot, T. N. C. Wells, C. McKay, A. Mantovani, and S. Sozzani. 1999. Upregulation of CCR1 and CCR3 and induction of chemotaxis to CC chemokines by IFN-γ in human neutrophils. J. Immunol. 162:474-479. [PubMed] [Google Scholar]
- 4.Borrego, F., M. J. Robertson, J. Ritz, J. Pena, and R. Solana. 1999. CD69 is a stimulatory receptor for natural killer cells and its cytotoxic effect is blocked by CD94 inhibitory receptor. Immunology 97:159-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broug-Holub, E., G. B. Toews, J. F. Van Iwaarden, R. M. Strieter, S. L. Kunkel, R. Paine III, and T. J. Standiford. 1997. Alveolar macrophages are required for protective pulmonary defenses in murine Klebsiella pneumonia: elimination of alveolar macrophages increases neutrophil recruitment but decreases bacterial clearance and survival. Infect. Immun. 65:1139-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, S. S., J. J. Lai, N. W. Lukacs, and S. L. Kunkel. 2001. Granulocyte-macrophage colony stimulating factor up-regulates CCR1 in human neutrophils. J. Immunol. 166:1178-1184. [DOI] [PubMed] [Google Scholar]
- 7.Cole, A. M., T. Ganz, A. M. Liese, M. D. Burdick, L. Liu, and R. M. Strieter. 2001. Cutting edge: IFN-inducible ELR− CXC chemokines display defensin-like antimicrobial activity. J. Immunol. 167:623-627. [DOI] [PubMed] [Google Scholar]
- 8.Davatelis, G., T. Tekamp-Olsen, S. D. Wolpe, K. Hermsen, C. Luedke, C. Gallegos, D. Coit, J. Merryweather, and A. Cerami. 1988. Cloning and characterization of a cDNA for murine macrophage inflammatory protein (MIP), a novel heparin-binding protein with inflammatory and chemokinetic properties. J. Exp. Med. 167:1939-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng, J. C., K. Tateda, X. Zeng, and T. J. Standiford. 2001. Transient transgenic expression of gamma interferon promotes Legionella pneumophila clearance in immunocompetent hosts. Infect. Immun. 69:6382-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domachowske, J. B., C. A. Bonville, J. L. Gao, P. M. Murphy, A. J. Easton, and H. F. Rosenberg. 2000. The chemokine macrophage inflammatory protein-1 alpha and receptor CCR1 control pulmonary inflammation and antiviral host defense in paramyxovirus infection. J. Immunol. 155:2677-2682. [DOI] [PubMed] [Google Scholar]
- 11.Fahey, T. J., III, K. J. Tracey, P. Tekamp, S. D. Wolpe, L. S. Cousens, W. G. Jones, G. T. Squires, A. Cerami, and B. Sherry. 1992. Macrophage inflammatory protein-1 modulates macrophage function. J. Immunol. 148:2764-2769. [PubMed] [Google Scholar]
- 12.Gao, J. L., T. A. Wynn, Y. Chang, E. J. Lee, H. E. Broxmeyer, S. Cooper, H. L. Tiffany, H. Westphal, J. Kwon-Chung, and P. M. Murphy. 1997. Impaired host defense, hematopoiesis, granulomatous inflammation and type 1-type 2 cytokine balance in mice lacking CC chemokine receptor 1. J. Exp. Med. 185:1959-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenberger, M. J., S. L. Kunkel, R. M. Strieter, N. W. Lukacs, J. Bramson, J. Gauldie, F. L. Graham, M. Hitt, J. M. Danforth, and T. J. Standiford. 1996. IL-12 gene therapy protects mice in lethal Klebsiella pneumonia. J. Immunol. 157:3006-3012. [PubMed] [Google Scholar]
- 14.Greenberger, M. J., R. M. Strieter, S. L. Kunkel, J. M. Danforth, R. E. Goodman, and T. J. Standiford. 1996. Neutralization of MIP-2 decreases neutrophil influx in a murine model of Klebsiella pneumonia. J. Infect. Dis. 173:159-165. [DOI] [PubMed] [Google Scholar]
- 15.Hames, B. D., and D. Glover (ed.). 1995. DNA cloning: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 16.Kahn, I. A., J. A. MacLean, F. S. Lee, L. Casciotti, E. DeHaan, J. D. Schwartzman, and A. D. Luster. 2000. IP-10 is critical for effective T cell trafficking and host survival in Toxoplasma gondii infection. Immunity 12:483-494. [DOI] [PubMed] [Google Scholar]
- 17.Kolls, J. K., P. Ye, and J. E. Shellito. 2001. Gene therapy to modify pulmonary host defenses. Semin. Respir. Infect. 16:18-26. [DOI] [PubMed] [Google Scholar]
- 18.Lindell, D. M., T. J. Standiford, P. Mancuso, Z. J. Leshen, and G. B. Huffnagle. 2001. Macrophage inflammatory protein 1α/CCL3 is required for clearance of an acute Klebsiella pneumoniae pulmonary infection. Infect. Immun. 69:6364-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loetscher, P., M. Seitz, I. Clark-Lewis, M. Baggiolini, and B. Moser. 1996. Activation of NK cells by CC chemokines: chemotaxis, Ca2+ mobilization, and enzyme release. J. Immunol. 156:322-327. [PubMed] [Google Scholar]
- 20.Mehrad, B., T. A. Moore, and T. J. Standiford. 2000. Macrophage inflammatory protein-1α is a critical mediator of host defense against invasive pulmonary aspergillosis in neutropenic hosts. J. Immunol. 165:962-968. [DOI] [PubMed] [Google Scholar]
- 21.Moore, T. A., B. B. Moore, M. W. Newstead, and T. J. Standiford. 2000. γδ-T cells are critical for survival and early proinflammatory cytokine gene expression during murine Klebsiella pneumonia. J. Immunol. 165:2643-2650. [DOI] [PubMed] [Google Scholar]
- 22.Olszewski, M. A., G. B. Huffnagle, R. A. McDonald, D. M. Lindell, B. B. Moore, D. N. Cook, and G. B. Toews. 2000. The role of macrophage inflammatory protein-1α/CCL3 in regulation of T cell-mediated immunity to Cryptococcus neoformans infection. J. Immunol. 165:6429-6436. [DOI] [PubMed] [Google Scholar]
- 23.Orange, J. S., B. Wang, C. Terhorst, and C. A. Biron. 1995. Requirement for natural killer cell-produced interferon γ in defense against murine cytomegalovirus infection and enhancement of this pathway by interleukin 12 adminstration. J. Exp. Med. 182:1045-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rollins, B. J. 1997. Chemokines. Blood 90:909-928. [PubMed] [Google Scholar]
- 25.Rubins, J. B., and C. Pomeroy. 1997. Role of gamma interferon in the pathogenesis of bacteremic pneumococcal pneumonia. Infect. Immun. 65:2975-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salazar-Mather, T. P., J. S. Orange, and C. A. Biron. 1998. Early murine cytomegalovirus (MCMV) infection induces liver natural killer (NK) cell inflammation and protection through macrophage inflammatory protein 1α (MIP-1α)-dependent pathways. J. Exp. Med. 187:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shanley, T. P., H. Schmal, H. P. Friedl, M. L. Jones, and P. A. Ward. 1995. Role of macrophage inflammatory protein 1-α (MIP-1α) in acute lung injury in rats. J. Immunol. 154:4793-47802. [PubMed] [Google Scholar]
- 28.Standiford, T. J., S. L. Kunkel, N. W. Lukacs, M. J. Greenberger, J. M. Danforth, R. G. Kunkel, and R. M. Strieter. 1995. Macrophage inflammatory protein-1α mediates lung leukocyte recruitment, lung capillary leak, and early mortality in murine endotoxemia. J. Immunol. 155:1515-1524.7636213 [Google Scholar]
- 29.Standiford, T. J., and G. B. Huffnagle. 1997. Cytokines in host defense against pneumonia. J. Investig. Med. 45:335-345. [PubMed] [Google Scholar]
- 30.Standiford, T. J., J. M. Wilkowski, T. H. Sisson, N. Hattori, B. Mehrad, K. A. Bucknell, and T. A. Moore. 1999. Intrapulmonary tumor necrosis factor gene therapy increases bacterial clearance and survival in murine gram-negative pneumonia. Hum. Gene Ther. 10:899-909. [DOI] [PubMed] [Google Scholar]
- 31.Tateda, K., T. A. Moore, M. W. Newstead, W. C. Tsai, X. Zeng, J. C. Deng, G. Chen, R. Reddy, K. Yamaguchi, and T. J. Standiford. 2001. Chemokine-dependent neutrophil recruitment in a murine model of Legionella pneumonia: potential role of neutrophils as immunoregulatory cells. Infect. Immun. 69:2017-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taub, D. D., T. J. Sayers, C. D. Carter, and J. R. Ortaldo. 1995. α and β chemokines induce NK cell migration and enhance NK-mediated cytolysis. J. Immunol. 155:3877-3888. [PubMed] [Google Scholar]
- 33.Tsai, T. C., R. M. Strieter, J. M. Wilkowski, K. A. Bucknell, M. D. Burdick, S. A. Lira, and T. J. Standiford. 1998. Lung-specific transgenic expression of KC enhances resistance to Klebsiella pneumoniae in mice. J. Immunol. 161:2435-2440. [PubMed] [Google Scholar]
- 34.Tsai, W. C., R. M. Strieter, B. Mehrad, M. W. Newstead, X. Zeng, and T. J. Standiford. 2000. CXC chemokine receptor CXCR2 is essential for protective innate host response in murine Pseudomonas aeruginosa pneumonia. Infect. Immun. 68:4289-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, P., W. R. Summer, G. J. Bagby, and S. Nelson. 2000. Innate immunity and pulmonary host defense. Immunol. Rev. 173:39-53. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, S., B. S. Youn, J. L. Gao, P. M. Murphy, and B. S. Kwon. 1999. Differential effects of leukotactin-1 and macrophage inflammatory protein-1α on neutrophils mediated by CCR1. J. Immunol. 162:4938-4942. [PubMed] [Google Scholar]
- 37.Zlotnik, A., and O. Yoshie. 2000. Chemokines: a new classification system and their role in immunity. Immunity 12:121-127. [DOI] [PubMed] [Google Scholar]