Abstract
Expression of the virB operon, encoding the type IV secretion system required for Brucella suis virulence, occurred in the acidic phagocytic vacuoles of macrophages and could be induced in minimal medium at acidic pH values. To analyze the production of VirB proteins, polyclonal antisera against B. suis VirB5 and VirB8 were generated. Western blot analysis revealed that VirB5 and VirB8 were detected after 3 h in acidic minimal medium and that the amounts increased after prolonged incubation. Unlike what occurs in the related organism Agrobacterium tumefaciens, the periplasmic sugar binding protein ChvE did not contribute to VirB protein production, and B. suis from which chvE was deleted was fully virulent in a mouse model. Comparative analyses of various Brucella species revealed that in all of them VirB protein production increased under acidic conditions. However, in rich medium at neutral pH, Brucella canis and B. suis, as well as the Brucella abortus- and Brucella melitensis-derived vaccine strains S19, RB51, and Rev.1, produced no VirB proteins or only small amounts of VirB proteins, whereas the parental B. abortus and B. melitensis strains constitutively produced VirB5 and VirB8. Thus, the vaccine strains were still able to induce virB expression under acidic conditions, but the VirB protein production was markedly different from that in the wild-type strains at pH 7. Taken together, the data indicate that VirB protein production and probably expression of the virB operon are not uniformly regulated in different Brucella species. Since VirB proteins were shown to modulate Brucella phagocytosis and intracellular trafficking, the differential regulation of the production of these proteins reported here may provide a clue to explain their role(s) during the infection process.
Bacteria belonging to the genus Brucella are gram-negative facultative intracellular pathogens of various wild and domestic mammals, and they also cause severe zoonotic infections in humans. Traditionally, three major species are distinguished by their preferences for certain animal hosts; Brucella abortus has a preference for cattle, Brucella melitensis has a preference for caprines, and Brucella suis has a preference for hogs. Whereas B. abortus is the livestock pathogen with the greatest economic impact, B. melitensis and B. suis account for most clinical cases in humans (15, 42).
In an attempt to unravel Brucella virulence factors by transposon mutagenesis, the crucial role of an operon similar to the virB operon of Agrobacterium tumefaciens encoding a type IV secretion system (T4SS) was revealed (35). The importance of the virB operon for Brucella virulence was further confirmed by signature-tagged mutagenesis both in vitro in a human macrophage infection model (24) and in vivo with mice (26). Further studies indicated that a complete Brucella virB operon was required for wild-type virulence in mice (47) or in macrophage-like cells (52, 53). In nonphagocytic HeLa cells, the absence of some functional VirB proteins (B2, B4, and B9) did not affect bacterial entry or prevention of the phagolysosomal fusion (17). However, integrity of the virB operon was required for Brucella to reach the proper niche and to replicate in HeLa cells (13, 47).
Of the gene products deduced from the 12 open reading frames of the B. suis virB operon, the first 11 proteins exhibit significant sequence similarity to the VirB proteins of the A. tumefaciens T4SS and also with Tra proteins required for the transfer of broad-host-range plasmids from the IncP, -N, and -W incompatibility groups (6, 12, 16). A. tumefaciens- and plasmid-encoded systems presumably form a multicomponent pore, which spans both bacterial membranes and allows transport of a single-stranded DNA-protein complex into a recipient plant or bacterial cell. The sequence similarity of the Brucella VirB proteins to the proteins of the T4SS of A. tumefaciens does not necessarily indicate that Brucella transfers DNA through its VirB-like complex, because T4SS from Bordetella pertussis (14) and Helicobacter pylori (36) are known to translocate proteins. It could be speculated that T4SS present in other intracellular pathogens, such as Rickettsia prowazekii (2) and Bartonella henselae (45), may have similar functions in intracellular survival, although effector proteins have not been described yet for these bacteria.
The virulence regulon of A. tumefaciens is induced in response to chemical signals at the plant wound site by a two-component system composed of the sensor VirA and the transcription factor VirG (11). Plant signals, including low pH and phenolic compounds, such as acetosyringone, induce virulence gene expression, which is potentiated by monosaccharides (5, 10). Sugars like galactose and arabinose bind to the periplasmic multiple sugar binding protein ChvE, which is encoded by an operon composed of chvE, gguA, and gguB; the latter two genes encode sugar transporter proteins. Upon sugar binding to ChvE, the complex potentiates the response to phenolic molecules (38, 46). In B. suis, we recently cloned a chvE operon similar to that of A. tumefaciens, which is specifically required for d-(+)-galactose utilization (1). The B. suis operon was dispensable for intracellular survival and multiplication of the bacteria in J774 macrophage-like cells (1).
Despite the high similarity between the virB and chvE operons of A. tumefaciens and B. suis, our previous attempts to identify a Brucella ortholog of the VirA-VirG two-component system by DNA-DNA hybridization failed. These results are in accord with recent analyses of the B. suis (37) and B. melitensis (18) genomes, which failed to detect orthologs of the A. tumefaciens virA or virG genes. However, there is evidence for regulated expression of the B. suis virB operon. Use of promoter virB::gfp reporter gene fusions and analysis of the virB mRNAs showed that the B. suis virB operon is expressed intracellularly in macrophages, and this effect could be mimicked in vitro in minimal medium (MM) at an acidic pH (8). This finding is in agreement with the requirement of an acidified phagosome for survival and multiplication of B. suis in macrophages (3, 39). However, use of lacZ reporter gene fusions showed that the virB operon of B. abortus 2308 is expressed during the stationary phase without a requirement for acidic induction conditions (47).
In order to investigate the regulation of virB operon expression in various Brucella wild-type and attenuated strains, we raised specific antisera against the VirB5 and VirB8 proteins. These new tools allowed determination of VirB protein production for the first time. Analysis of different Brucella species revealed marked differences in VirB protein production under various growth conditions, suggesting that the VirB proteins play different roles in infection of the different hosts.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The characteristics of the bacterial strains and plasmids used are described in Table 1. Escherichia coli strains were routinely grown at 37°C in Luria-Bertani medium, whereas B. suis and other Brucella strains were grown in tryptic soy (TS) broth. TS broth was supplemented with kanamycin (50 μg ml−1) to grow Brucella mutants. Brucella strains were grown for 20 h at 37°C in TS broth with or without antibiotics to the stationary phase (optical density at 600 nm, 1.5 to 1.8). Four volumes of phosphate-buffered saline (PBS) was added before centrifugation at 2,500 × g for 25 min, and the pellets were resuspended in 4 volumes of PBS, resulting in optical density values around 0.5. For interspecies comparisons, the optical density at 600 nm was adjusted at this step to 0.5. Two-milliliter aliquots of the various bacterial suspensions were sedimented by centrifugation for 2 min at 13,000 × g, and the pellets were then resuspended in 0.5 ml of MM at pH 4.5 or 7 as indicated below. The Eppendorf tubes were incubated horizontally with shaking at 37°C for various times. MM, derived from MMA (4), was composed of 7.56 mM (NH4)2SO4, 33 mM KH2PO4, 60.3 mM K2HPO4, 1.7 mM sodium citrate supplemented with 1 mM MgSO4, 10 mM glucose (or other carbon sources as indicated below), 0.1% yeast extract, 2 μg of vitamin B6 per ml, 2 μg of vitamin B1 per ml, and 0.0005 μg of biotin per ml, as previously described (1). The pH was adjusted with 2 M citric acid. The eukaryotic cell culture medium, RPMI 1640, was obtained from Gibco (Life Technologies, Cergy Pontoise, France).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains or plasmid | Relevant genotype and/or description | Reference or sourcea |
|---|---|---|
| E. coli BL21 (λDE3) | F−hsdS gal1 λDE3 | 49 |
| B. suis strains | ||
| 1330 | Biotype 1; ATCC 23444T | ATCC |
| 1330 ΔchvE | B. suis 1330 ΔchvE::kan | 1 |
| 1330 omp25 | B. suis 1330 omp25::kan | 28 |
| 1330 nikA | B. suis 1330 nikA::kan | 28 |
| 1330 virB5 | B. suis virB5::kan | 8 |
| 1330 virB12 | B. suis virB12::kan | 35 |
| B. melitensis strains | ||
| 16M | Biotype 1; ATCC 23456T | ATCC |
| Rev.1 | Vaccine strain | Jiménez de Bagüés |
| B. abortus strains | ||
| 544 | Biotype 1; ATCC 23448T | ATCC |
| 2308 | Biotype 1; wild type, smooth, virulent | Jiménez de Bagüés |
| S19 | Vaccine strain, smooth | Jiménez de Bagüés |
| RB51 | Vaccine strain, rough | Jiménez de Bagüés |
| B. canis | ATCC 23365T | ATCC |
| B. ovis | Reo 198 | Jiménez de Bagüés |
| Plasmid pT7-H6-TrxFus | Ampr, trxA fusion plasmid for T7 promotor-controlled overproduction | 30 |
ATCC, American Type Culture Collection; Jiménez de Bagüés, M. P. Jiménez de Bagüés.
Production of the VirB5 and VirB8 recombinant proteins and antiserum preparation.
DNA fragments encoding B. suis VirB5 and VirB8 without signal peptides were amplified by PCR with the following primers derived from the B. suis 1330 sequence (GenBank accession no. AF 141604): the oligonucleotides used for amplification of virB5 were T7B5suis5′ (CAGGGTACCCGCGCACGCGCAGCTCC) and T7B5suis3′ (GAGCTGCAGCTAATAGGCGGCTTCCAGTGC), and the oligonucleotides used for amplification of virB8 were T7B8suis5′ (CAGGGTACCCCGCGTCAACGCACAGAC) and T7B8suis3′ (GAGCTGCAGCTATTGCACCACTCCCATTTCTGG). PCR fragments were cleaved with Acc65I/PstI and ligated into a similarly cleaved pT7-H6-TrxFus vector (30). Hexahistidyl-thioredoxin fusion proteins were overproduced in BL21 by isopropyl-β-d-thiogalactopyranoside (IPTG) induction of the T7 promoter and were purified by immobilized metal affinity chromatography performed as recommended by the manufacturer (Amersham Bioscience, Saclay, France). Polyclonal antisera were raised in New Zealand White rabbits, and VirB5 antisera were further purified by affinity chromatography as described previously (41)
Western blot analysis.
After cell cultivation for various times, bacteria were sedimented, and the pellets were resuspended in Laemmli sample buffer and heated to 100°C for 5 min. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed in 15% (wt/vol) acrylamide separating gels (31). The proteins were transferred onto Immobilon polyvinylidene difluoride membranes (Millipore, Saint Quentin-Yvelines, France) by using a semidry transfer procedure, and the membranes were stained with Coomassie blue for detection of reference proteins. Immunodetection of proteins in total cell lysates was performed with polyclonal VirB8 antiserum (1/5,000), VirB5 antiserum (1/3,000), or affinity-purified anti-VirB5 serum (1/5,000). Horseradish peroxidase-conjugated goat anti-rabbit antibodies (Jackson Immunoresearch Laboratories Inc.; obtained from Immunotech, Marseilles, France) were used in combination with the ECL system (Amersham Bioscience Saclay, France) to develop the chemiluminescence for visualization on Kodak X-AR film (Sigma-Aldrich, Saint-Quentin Falavier, France). The molecular weight markers were obtained from Sigma or Amersham Bioscience.
Infection of mice, preparation of spleens, and bacterial counts.
Eight-week-old female BALB/c mice obtained from IFFA Credo were challenged intraperitoneally with 5 × 104 CFU of either wild-type B. suis 1330 or the chvE null mutant as described previously (22). Brucellae were grown on TS agar (Life Technologies) supplemented with 0.1% (wt/vol) yeast extract (Difco, Detroit, Mich.) (TSA-YE). Viable counts were determined retrospectively by enumeration on TSA-YE plates. Five infected mice for each B. suis strain were sacrificed on days 1, 7, 14, 21, 35, and 56 by cervical dislocation. Spleens were harvested, weighed, and frozen at −20°C. After homogenization in a buffered saline solution, bacterial counts were determined on TSA-YE plates. Normalization and statistical analysis of the results were carried out as previously reported (9, 22).
RESULTS
Both acidic conditions and MM are required for in vitro production of B. suis VirB8.
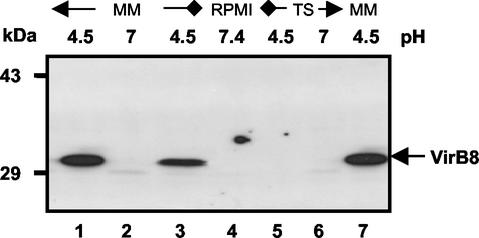
Polyclonal rabbit antisera were raised against B. suis VirB5 and VirB8 hexahistidyl-thioredoxin fusion proteins. The sera were tested by Western blotting by using lysates of B. suis cultivated in MM at either pH 7.0 or pH 4.5, because previous studies had shown that there was increased virB mRNA in acidic MM (8). After 5 h of cultivation in acidic MM containing 10 mM glucose (Fig. 1, lanes 1 and 7) or galactose (lane 3), the VirB8 antiserum detected a protein with an apparent molecular mass that was consistent with the predicted molecular mass, 31.5 kDa. In contrast, when bacteria were grown at neutral pH in MM, RPMI 1640, or TS medium (Fig. 1, lanes 2, 4, and 6), the VirB8 antiserum did not detect any protein. Furthermore, acidification of TS medium (lane 5) or of RPMI 1640 (data not shown) to pH 4.5 was not sufficient to trigger VirB8 production in this time. To determine the optimal pH for in vitro VirB8 production by B. suis, the bacteria were incubated in MM adjusted to pH values ranging from 4.0 to 7.0. The maximal amount of VirB8 detected after 6 h of incubation was obtained at pH values of 4.5 to 5.5 (data not shown). At pH 4.0, VirB8 was not detected, although the presence of virB mRNA was reported previously (8). This suggests that biosynthesis and the stability of the protein were impaired in MM at pH values below 4.5, while transcription was still operative at pH 4.0.
FIG. 1.
In vitro induction of B. suis VirB8 in different media. B. suis wild-type strain 1330 was grown in TS broth to the stationary phase and washed with PBS by centrifugation, and the bacteria were resuspended in different media as follows. Further cultivation was carried out at 37°C for 5 h in MM at pH 7.0 (lane 2) or at pH 4.5 (lanes 1, 3, and 7), in TS medium, or in RPMI 1640. Lane 3 contained lysates from a B. suis culture grown in MM containing d-(+)-galactose instead of d-(+)-glucose. VirB8 production in cells was evaluated by SDS-15% PAGE of cell lysates, followed by Western blotting with VirB8-specific antiserum and chemoluminescent detection.
B. suis virB5 and virB8 genes are expressed as part of the same operon.
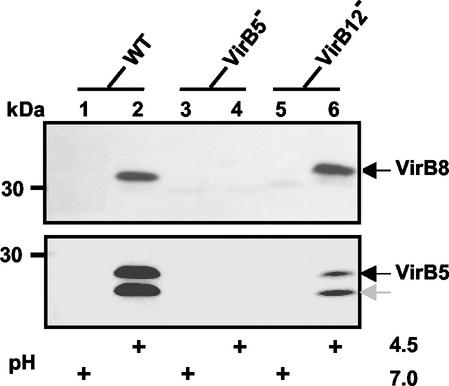
In contrast to the easily detectable amount of VirB8 in the B. suis wild type at pH 4.5 (Fig. 2, lane 2), no VirB8-specific immunostaining was detected in lysates from the virB5::kan mutant (Fig. 2, lane 4). Insertion of the kanamycin cassette into virB5 obviously exerted a polar effect towards the downstream genes, thus confirming that virB8 was part of the virB operon. Accordingly, knockout of the 12th gene of the virB operon should not have affected expression of the downstream virB8 gene, and analysis of the virB12::kan insertion mutant confirmed this assumption (Fig. 2, lane 6). Parallel observations were made with the affinity-purified VirB5 antiserum (Fig. 2, lower panel). However, in this case, two proteins were detected, and they had apparent molecular masses of 28 and 26 kDa, which are slightly greater than the molecular mass expected for the full-length protein or the mature form after removal of the signal peptide (35). The lower-molecular-mass form of VirB5 may be a degradation product, as suggested for the A. tumefaciens VirB5 ortholog (44). Together, use of VirB8- and VirB5-specific antisera demonstrated for the first time that B. suis VirB proteins are produced in acidic MM, and the results further supported the operon structure predicted previously.
FIG. 2.
Comparison of production of VirB8 in acidic MM by the B. suis wild type (WT) and by virB5- and virB12-disrupted mutants. Bacteria grown in TS broth to the stationary phase were washed with PBS and resuspended in MM at pH 7.0 or 4.5. Further incubation was carried out at 37°C for 6 h prior to evaluation of VirB8 (upper panel) and VirB5 (lower panel) production by Western blotting.
Kinetics of B. suis VirB5 and VirB8 protein production in acidic MM.
Whereas the experiments described above provided information about the VirB5 and VirB8 levels after 5 to 6 h of incubation in the virB-inducing medium, we sought to determine the in vitro onset of VirB protein synthesis. B. suis was incubated in MM at pH 4.5 for periods ranging from 1 to 24 h. Synthesis of the VirB5 protein started after 2 h of cultivation in acidic MM (data not shown) and was clearly apparent after 3 h (Fig. 3, lower panel). In contrast, the VirB8 protein was first detected after 4 h, and the amount increased steadily after that (Fig. 3, upper panel). Thus, we concluded that under inducing conditions B. suis required about 3 h to produce detectable amounts of VirB5 and VirB8. Since the VirB8- and VirB5-specific antisera gave similar results and the former exhibited lower background reactivity, it was used in most subsequent experiments, and a standard incubation time of 6 h was considered suitable for obtaining virB-induced cells. Next, we evaluated whether B. suis VirB8 production was affected either by ChvE, as it is in A. tumefaciens, or by an omp25 gene defect, since the absence of Omp25 resulted in attenuated strains.
FIG. 3.
Kinetics of B. suis VirB5 and VirB8 production in MM. B. suis grown in TS broth to the stationary phase was washed, resuspended in MM at pH 4.5, and incubated at 37°C. After 3 h bacteria were divided into aliquots and centrifuged. The pellets were resuspended in MM at pH 4.5. After various incubation times at 37°C (3 to 24 h, as indicated), VirB protein production was evaluated by using purified VirB5 antiserum (lower panel) or VirB8 antiserum (upper panel). For direct comparison, lane 24* was loaded with bacteria incubated continuously for 24 h at pH 7.0. The data are data from a single representative experiment that was repeated three times.
B. suis chvE operon is not required for induction of the virB operon.
Recent animal trials indicated that Δomp25 Brucella mutants were attenuated in cattle (19), goats (20), and mice (21). Furthermore, these omp25 mutant strains raised a protective immune response against a later challenge with wild-type strains, and use of these strains as vaccines is under investigation (20). Moreover, attenuated B. abortus bvrR-bvrS mutants were recently reported to be unable to produce the outer membrane protein Omp25 (25). To establish whether the attenuation of omp25 mutants is caused by their inability to synthesize VirB proteins, we included the B. suis omp25 kanamycin insertion mutant in our analysis. As a control, another B. suis mutant was included (nikA::kan), which was generated by the same method. The B. suis mutants impaired in Omp25 production and nickel uptake (nikA) produced VirB8 like the wild type (Fig. 4). In contrast, VirB8 was not detected in lysate from the chvE mutant (Fig. 4, lane 5). This finding prompted us to evaluate whether alterations of the chvE operon could lead to an attenuated behavior in mice. A comparison of the infection kinetics of the B. suis wild-type strain with the infection kinetics of the ΔchvE variant indicated that the bacterial charges of both strains decreased similarly 1 week after infection (Fig. 5A). After this, the mutated strain persisted in mouse spleens even longer (8 weeks) than the parental strain (3 weeks). Slower elimination of a mutant than of wild-type B. suis was reported previously in the case of the ΔclpA mutant (22). Although the reason for the difference is not clear, our results suggested that the ΔchvE strain was still able to produce VirB proteins required for Brucella virulence. In an attempt to reconcile these conflicting in vitro and in vivo data, the effects of various carbon sources on VirB8 production were investigated. It was previously shown that galactose uptake, glucose uptake, and erythritol uptake were totally, partially, and not dependent on expression of the chvE operon, respectively (1). Growth in the presence of each of these sugars allowed production of VirB8 by the wild type (Fig. 5B). In contrast, the VirB8 protein was produced by the ΔchvE mutant in the presence of erythritol (lane 8) and to some extent also in the presence of glucose, but no VirB protein was ever detected in the presence of galactose. These experiments demonstrated that the ChvE protein was not required for in vivo or in vitro VirB8 production, as long as carbon sources other than galactose were available to the bacteria. The obvious specificity of the ChvE protein for uptake of glucose and galactose suggests that this protein likely does not play a general role in the regulation of VirB protein production in the natural infection process.
FIG. 4.
Analysis of VirB8 induction in various B. suis mutants. Wild-type B. suis (WT) grown in TS medium to the stationary phase was washed and resuspended in MM at either pH 7.0 (lane 1) or pH 4.5 (lanes 2 and 4). B. suis mutants ΔchvE (lane 5), nikA::kan (NikA−) (lane 6), and omp25::kan (Omp25−) (lane 7) were grown in TS medium with kanamycin (50 μg/ml) and similarly resuspended in MM at pH 4.5. Cell cultivation was carried out for 6 h at 37°C. Lane 3 contained molecular weight standards (Markers).
FIG. 5.
Comparison of mouse infection and VirB8 production for wild-type B. suis and the ΔchvE mutant. (A) Mice were infected with B. suis wild-type strain 1330 (•) and the ΔchvE mutant (○). The infections were assessed by determining the number of bacteria present in the spleen. The data are means ± standard deviations for five animals per condition. The results obtained with the transporter ΔgguA mutant were not significantly different from the results obtained with the ΔchvE mutant. (B) Wild-type B. suis (WT) (lanes 1 to 4) and the isogenic ΔchvE mutant (lanes 5 to 8) were cultivated for 6 h in MM containing 10 mM d-(+)-glucose (glu) at pH 7.0 (lanes 1 and 5) or at pH 4.5 (lanes 2 and 6) or in pH 4.5 medium in which glucose was replaced by either d-(+)-galactose (gal) (lanes 3 and 7) or meso-erythritol (ery) (lanes 4 and 8). The data are representative of the results of the four experiments performed. The VirB8 levels obtained under acidic conditions in the presence of glucose and galactose were sometimes higher than those shown in lanes 6 and 8, while VirB8 was never detected in the presence of galactose.
B. melitensis, B. abortus, and Brucella ovis produce VirB5 and VirB8 in rich TS medium.
Due to the high levels of similarity among orthologous genes in brucellae, it was anticipated that our antisera would detect VirB5 and VirB8 in all Brucella strains. Since the B. abortus virB promoter was activated in rich culture medium during the stationary phase (47), we analyzed the presence of the VirB components in various Brucella strains grown in rich TS medium (Fig. 6). B. suis did not produce detectable amounts of either VirB protein, and only small amounts were detected in Brucella canis (Fig. 6, lanes 1 and 2). In contrast, B. melitensis, B. abortus 544, and B. ovis produced easily detectable quantities of VirB5 and VirB8 (Fig. 6, lanes 3 to 5). Interestingly, the VirB5 form detected under these growth conditions corresponded almost exclusively to the higher-molecular-mass form detected in B. suis in acidic MM conditions (Fig. 2 and 3).
FIG. 6.
Comparison of the VirB5 and VirB8 contents in various Brucella wild-type strains growth in rich TS medium. The wild-type bacteria B. suis (lane 1), B. canis (lane 2), B. melitensis (lane 3), B. abortus 544 (lane 4), and B. ovis (lane 5) were grown to the stationary phase in TS medium, washed once with PBS, and centrifuged. VirB8 (upper panel) and VirB5 (lower panel) protein contents were evaluated after SDS-PAGE and electrotransfer by Western blotting by using VirB8 antiserum or affinity-purified VirB5 antiserum. The data are the data from one experiment that was replicated once with similar results.
Acidic MM enhances VirB8 production in different Brucella species.
Because some Brucella species produced VirB proteins in rich TS medium, we determined whether the B. suis virB-inducing conditions were also relevant for the other strains. To do this, Brucella strains grown to the stationary phase were incubated for 6 h in MM at pH 7.0 or 4.5 (Fig. 7). There were no significant changes in the VirB8 levels at pH 7.0 in the various strains compared to the levels reached in TS medium (Fig. 7, lanes 1, 3, 5, 7, 9, 11, 13, and 15), but in all strains the VirB8 content increased under the B. suis virB-inducing conditions (lanes 2, 4, 6, 8, 10, 12, 14, and 16). In the case of B. ovis, however, a decreased VirB8 content was observed, which probably resulted from poor adaptation of this strain to the acidic conditions, as indicated by the dramatic protein degradation seen after SDS-PAGE (data not shown).
FIG. 7.
Constitutive and inducible VirB8 production in the various Brucella strains. The wild-type bacteria B. suis, B. canis, B. melitensis, B. abortus 544, and B. abortus A2308 and the vaccine strains S19, RB51, and Rev.1 were grown to the stationary phase in TS medium and washed in PBS. After 6 h of incubation in MM at either pH 7.0 (lanes 1, 3, 5, 7, 9, 11, 13, and 15) or pH 4.5 (lanes 2, 4, 6, 8, 10, 12, 14, and 16), VirB8 protein production was evaluated by Western blotting by using the VirB8 antiserum. The data are the data from one experiment that was representative of the three experiments performed.
Because of the importance of the virB operon for the pathogenicity of Brucella, we considered the possibility that attenuated Brucella strains used as vaccine strains were affected in the ability to produce T4SS. To examine this, we determined the VirB8 contents in B. abortus S19 (also referred to as B19) and in the rough B. abortus RB51 strain, as well as in B. melitensis Rev.1 (23). Compared to the levels in the wild-type strains, only low levels of VirB8 were detected at neutral pH (Fig. 7, lanes 11, 13, and 15), but all vaccine strains produced substantial amounts of VirB8 protein in MM at pH 4.5 (Fig. 7, lanes 12, 14, and 16). This observation demonstrated that the vaccine strains were not impaired in terms of the ability to express the virB genes in an acidic environment, which is believed to be required for intracellular multiplication in the natural infection process. Interestingly, the in vitro regulation of VirB8 production in these vaccine strains was similar to that in B. suis and B. canis and notably different from that in the parental B. abortus and B. melitensis strains.
DISCUSSION
Different reports have highlighted the importance of virB operon integrity for the virulence of Brucella species. In addition, the operon-encoded VirB proteins exhibit significant sequence similarity to the proteins encoded by the well-studied T4SS of A. tumefaciens. However, so far there is no evidence concerning the proper assembly of the VirB proteins into a secretory apparatus, and substrates of the Brucella T4SS were not identified. Nevertheless, recent studies have indicated that the absence of some VirB proteins leads to alterations in the early steps of infection, cell entry (52), or intracellular trafficking (13), thus providing clues about the possible role(s) of VirB proteins. Similarly, analysis of the environmental conditions in which Brucella expresses its virB operon certainly should contribute to a better understanding of the functions of these gene products in the various steps of Brucella infection. With this in mind, induction and regulation of the virB operon, previously studied by using gene reporters (8, 47, 50), were investigated further. VirB5- and VirB8-specific antisera were raised to determine the presence of these proteins in various Brucella wild-type strains and mutants.
The first part of this study, which was devoted to analysis of induction of VirB proteins specifically in B. suis, demonstrated that VirB5 and VirB8 were either absent or only weakly produced at neutral pH either in MM or in rich TS medium. In accord with the analysis of mRNA levels (8), we showed for the first time that growth in acidic MM triggers the synthesis of both VirB5 and VirB8 proteins, although VirB5 was detected before VirB8. A comparison of the VirB8 and VirB5 contents after induction of the B. suis wild type and of virB5 and virB12 mutants further confirmed that the virB5 and virB12 genes belong to the same operon. Knockout of the 12th open reading frame of the virB operon does not prevent production of VirB8, indicating that VirB12 is not essential for expression of the virB operon. The absence of Omp25 in putative vaccine strains of B. suis did not affect their ability to produce VirB8 in MM at pH 4.5. Similarly, B. suis chvE deletion strains produced VirB proteins provided that suitable sugar sources were available. Whereas the conditions for induction of the virB operon of this organism share suggestive features with the conditions for induction of the virB operon of A. tumefaciens, the B. suis chvE strain survived well in macrophages (1), as well as in infected mice. Therefore, exogenous sugars may not play a role in the intracellular induction of virB expression. In addition to the requirement for acidic conditions, growth in MM was apparently another prerequisite for virB expression in B. suis. The intracellular vacuole is believed to be a nutrient-poor environment, and this is consistent with the recent finding that many attenuated transposon insertion mutants were affected in their basic metabolic functions (29). Thus, intracellularly, B. suis is in a nutrient-poor environment, and the bacteria may link the starvation response to induction of their virulence functions.
Incubation of B. suis in MM at pH 4.5 for 3 to 4 h was required for accumulation of detectable amounts of VirB5 and VirB8. This relatively long lag time might prevent the bacteria from inadequate induction of the virB operon under transiently changing environmental conditions. It is also possible that virB operon expression requires the synthesis of another sensing system, and this may account for the delay, similar to the delay observed for the pmrA and pmrB genes induced by the PhoP/PhoQ sensor in Salmonella (54). If present, such regulatory Brucella gene products would then induce production of VirB proteins leading to T4SS assembly. The observed in vitro lag time is in accord with the finding that intracellular survival was inhibited after neutralization of the acidic Brucella-containing phagosome with NH4Cl 1 h postinfection (39). However, when the phagosome was neutralized 7 h postinfection, bacterial multiplication was not affected. This suggests that all Brucella signals were transmitted to the eukaryotic cells within the first 7 h and ensured inhibition of the phagolysosome fusion, allowing its survival and its replication.
In the second part of this study, we demonstrated the different abilities of various Brucella strains to produce both VirB5 and VirB8 at neutral pH in nutrient-rich TS medium. First, B. canis and B. suis biovar 1, whose genomic physical maps are identical (33), produced only marginal amounts of VirB8 at neutral pH. After cultivation in acidic MM, both organisms produced easily detectable amounts of VirB8. Thus, these two organisms can be considered virB operon-inducible bacteria. Second, the other Brucella wild-type strains produced substantial amounts VirB8 in nutrient-rich TS medium or in MM at neutral pH and can be considered virB constitutive. In acidic induction medium, however, B. abortus biovar 1 (strains 544 and 2308) and B. melitensis increased their VirB8 contents further.
With regard to these two VirB expression patterns, the results obtained after analysis of the B. abortus and B. melitensis vaccine strains more closely resembled the results obtained with the inducible organism B. suis than the results obtained with their parental strains. The vaccine strains exhibited reduced T4SS production at neutral pH in a medium presumably encountered in the body fluids. The similarity between the vaccine strains and B. suis, however, is not limited to the low VirB content at neutral pH. Similarly, the kinetics of mouse infection by B. suis, which are characterized by a rapid decrease in the bacterial content after only 1 week (Fig. 5A), are very similar to the kinetics of the attenuated vaccine strains B. abortus S19 (43) and B. melitensis Rev.1 (27; data not shown). This finding differs from the results of previous studies of the mouse colonization patterns of virulent B. abortus 2308 (7, 43) and B. melitensis 16M (27). In both cases, a plateau phase with very high numbers of brucellae per spleen was observed, which lasted 8 weeks before there was a gradual reduction in bacterial content. The constitutive production of VirB proteins may therefore be a marker of wild-type virulence of B. abortus and B. melitensis. In the future, it will be very interesting to assess whether the rapid elimination of B. suis and the vaccine strains in the mouse infection model is solely due to the low production of VirB proteins under the neutral and nutrient-rich conditions of body fluids.
The concomitant presence of VirB5 and VirB8 in strains which constitutively expressed virB may be taken as an indication of proper assembly of all the VirB proteins into the T4SS. If this occurs, the T4SS apparatus should protrude outside the bacterial cell surface, and, besides its putative intracellular function, it may play a role in the early events of infection. Indeed, it was recently shown that a functional virB operon determined the mode of entry of the wild-type B. abortus strain in mouse bone marrow-derived macrophages (53). This mode of internalization required the integrity of cell surface lipid rafts and the presence of an intact virB operon in B. abortus. The presence of intact lipid rafts was also required for entry and short-term survival of B. suis (34), but the role of the virB operon in the initial infection steps of this species remains to be elucidated.
In summary, development of specific VirB5 and VirB8 antisera permitted for the first time monitoring of the production of the corresponding VirB proteins in the wild type, in vaccine strains, and in mutants of different Brucella strains. Analysis of the presence of VirB5 and VirB8 indicated that these two virB products are produced simultaneously and revealed that there is differential in vitro regulation of the virB operon in the Brucella strains. This may reflect in vivo differences in the requirement for the T4SS function(s) during the infection process. The distinction between virB-constitutive and virB-inducible strains underlines the fact that the limited differences between Brucella genomes (51) can influence bacterial virulence by modulation of the expression of essential genes. In the future, these antisera could be used to assess whether other interesting attenuated mutants have altered VirB protein levels. Among the candidates for such an analysis are strains that are not able to replicate in macrophages or epithelial cells, such as the B. abortus mutants with mutations in the hfq gene (40) and in the BvrR/BvrS system (48). Interestingly, the A. tumefaciens BvrR/BvrS orthologs are involved in acid sensing (32), suggesting that this two-component system may confer the acid inducibility of the virB operon.
Acknowledgments
We thank Maria Pilar Jiménez de Bagüés Picazo (University of Cantabria, Santander, Spain) and Véronique Maurin for providing various wild-type and mutant Brucella strains. We also thank A. Böck for support and discussions and J. Armand for technical assistance. We thank the Institute of Molecular Biology and Medicine (University of Scranton, Scranton, Pa.) for providing access to preliminary B. melitensis sequence data.
M.T.A.M. was supported by the FRM (Fondation pour la Recherche Médicale) and by the Languedoc-Roussillon region. This work was supported by INSERM, by the European Union Frame Programme 5 under contract QLK2-CT-2001-01200, by an EGIDE/DAAD French-German exchange program (PROCOPE no. 00356UJ), and by the French Cancer Research Association (ARC grant 5566).
Editor: D. L. Burns
REFERENCES
- 1.Alvarez-Martinez, M.-T., J. Machold, C. Weise, H. Schmidt-Eisenlohr, C. Baron, and B. Rouot. 2001. The Brucella suis homologue of the Agrobacterium tumefaciens chromosomal virulence operon chvE is essential for sugar utilization but not for survival in macrophages. J. Bacteriol. 183:5343-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, S. G. E., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Pontén, U. C. M. Alsmark, R. M. Podowski, A. K. Näslund, A.-S. Eriksson, H. H. Winckler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 3.Arenas, G., A. Staskevich, A. Aballay, and L. Mayorga. 2000. Intracellular trafficking of Brucella abortus in J774 macrophages. Infect. Immun. 68:4255-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, S. F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seichman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 5.Baker, B., P. Zambryski, B. Staskawicz, and S. Dinesh-Kumar. 1997. Signaling in plant-microbe interactions. Science 276:726-733. [DOI] [PubMed] [Google Scholar]
- 6.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: Euroconference on the Biology of the Type IV Secretion Processes. Mol. Microbiol. 43:1359-1365. [DOI] [PubMed] [Google Scholar]
- 7.Bellaire, B., P. Elzer, C. Baldwin, and R. N. Roop. 1999. The siderophore 2,3-dihydroxybenzoic acid is not required for virulence of Brucella abortus in BALB/c mice. Infect. Immun. 67:2615-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J.-P. Liautard, R. M., and D. O'Callaghan. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. USA 99:1544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosseray, N., and M. Plommet. 1976. Transformation normalisant la distribution du nombre de Brucella dans la rate de souris inoculees par voie intraperitoneale. J. Biol. Stand 4:341-351. [DOI] [PubMed] [Google Scholar]
- 10.Burns, D. L. 1999. Biochemistry of type IV secretion. Curr Opin. Microbiol. 2:25-29. [DOI] [PubMed] [Google Scholar]
- 11.Cangelosi, G. A., R. G. Ankenbauer, and E. W. Nester. 1990. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc. Natl. Acad. Sci. USA 87:6708-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christie, P. J. 2001. Type IV secretion: intracellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comerci, D. J., M. J. Martinez-Lorenzo, R. Sieira, J.-P. Gorvel, and R. A. Ugalde. 2001. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell. Microbiol. 3:159-168. [DOI] [PubMed] [Google Scholar]
- 14.Cook, D. M., K. M. Farzio, and D. L. Burns. 1999. Identification and characterization of PtlC, an essential component of the pertussis toxin secretion system. Infect. Immun. 67:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbel, M. 1997. Brucellosis: an overview. Emerg. Infect. Dis. 3:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 17.Delrue, R., M. Martinez-Lorenzo, P. Lestrate, I. Danese, V. Bielarz, P. Mertens, X. De Bolle, A. Tibor, J. Gorvel, and J. Letesson. 2001. Identification of Brucella spp. genes involved in intracellular trafficking. Cell. Microbiol. 7:487-497. [DOI] [PubMed] [Google Scholar]
- 18.DelVecchio, V., V. Kapatral, R. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. Elzer, S. Hagius, D. O'Callaghan, J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edmonds, M. D., A. Cloeckaert, N. Booth, W. T. Fulton, S. Hagius, J. V. Walker, and P. H. Elzer. 2001. Attenuation of a Brucella abortus mutant lacking a major 25 kDa outer membrane protein in cattle. Am. J. Vet. Res. 62:1461-1466. [DOI] [PubMed] [Google Scholar]
- 20.Edmonds, M. D., A. Cloeckaert, S. D. Hagius, L. E. Samartino, W. T. Fulton, J. V. Walker, F. M. Enright, N. J. Booth, and P. H. Elzer. 2002. Pathogenicity and protective activity in pregnant goats of a Brucella melitensis omp25 deletion mutant. Res. Vet. Sci. 72:235-239. [DOI] [PubMed] [Google Scholar]
- 21.Edmonds, M. D., A. Cloekaert, and P. H. Elzer. 2002. Brucella species lacking the major outer membrane protein Omp25 are attenuated in mice and protect against Brucella melitensis and Brucella ovis. Vet. Microbiol. 2373:1-17. [DOI] [PubMed] [Google Scholar]
- 22.Ekaza, E., L. Guilloteau, J. Teyssier, J. P. Liautard, and S. Köhler. 2000. The Clp ATPase ClpA of Brucella suis: cloning and characterization of the gene, and analysis of a knockout mutant in macrophages and in BALB/c mice. Microbiology 146:1605-1616. [DOI] [PubMed] [Google Scholar]
- 23.el Idrissi, A., A. Benkirane, M. el Maadoudi, M. Bouslikhane, J. Berrada, and A. Zerouali. 2001. Comparison of the efficacy of Brucella abortus strain RB51 and Brucella melitensis Rev.1 live vaccines against experimental infection with Brucella melitensis in pregnant ewes. Rev. Sci. Technol. 20:741-747. [DOI] [PubMed] [Google Scholar]
- 24.Foulongne, V., G. Bourg, C. Cazevieille, S. Michaux-Charachon, and D. O'Callaghan. 2000. Identification of Brucella suis genes affecting intracellular survival in an in vitro human macrophage infection model by signature-tagged mutagenesis. Infect. Immun. 68:1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guzman-Verri, C., L. Manterola, A. Sola-Landa, A. Parra, A. Cloeckaert, J. Garin, J. Gorvel, I. Moriyon, E. Moreno, and I. Lopez-Goni. 2002. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. USA 99:12375-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong, P. C., R. M. Tsolis, and T. A. Ficht. 2000. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect. Immun. 68:4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoover, D., R. Crawford, L. Van De Verg, M. Izadjoo, A. Bhattacharjee, C. Paranavitana, R. Warren, M. Nikolich, and T. Hadfield. 1999. Protection of mice against brucellosis by vaccination with Brucella melitensis WR201(16MDeltapurEK). Infect. Immun. 67:5877-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jubier-Maurin, V., A. Rodrigue, S. Ouahrani-Bettache, M. Layssac, M.-A. Mandrand-Berthelot, S. Köhler, and J.-P. Liautard. 2001. Identification of the nik gene cluster of Brucella suis: regulation and contribution to urease activity. J. Bacteriol. 183:426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Köhler, S., V. Foulongne, S. Ouahrani-Bettache, G. Bourg, J. Teyssier, M. Ramuz, and J.-P. Liautard. 2002. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. USA 99:15711-15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kromayer, M., R. Wilting, P. Tormay, and A. Bock. 1996. Domain structure of the prokaryotic selenocysteine-specific elongation factor SelB. J. Mol. Biol. 262:413-420. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Li, L., Y. Jia, Q. Hou, T. C. Charles, E. W. Nester, and S. Q. Pan. 2002. A global pH sensor: Agrobacterium sensor protein ChvG regulates acid-inducible genes on its two chromosomes and Ti plasmid. Proc. Natl. Acad. Sci. USA 99:12369-12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michaux-Charachon, S., G. Bourg, E. Jumas-Bilak, P. Guigue-Talet, A. Allardet-Servent, D. O'Callaghan, and M. Ramuz. 1997. Genome structure and phylogeny in the genus Brucella. J. Bacteriol. 179:3244-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naroeni, A., and F. Porte. 2002. Role of cholesterol and the ganglioside GM(1) in entry and short-term survival of Brucella suis in murine macrophages. Infect. Immun 70:1640-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210-1220. [DOI] [PubMed] [Google Scholar]
- 36.Odenbreit, S., J. Püls, B. Sedlmaier, E. Gerland, W. Fischer, and R. Haas. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497-1499. [DOI] [PubMed] [Google Scholar]
- 37.Paulsen, I., R. Seshadri, K. Nelson, J. Eisen, J. Heidelberg, T. Read, R. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. Deboy, A. Durkin, J. Kolonay, R. Madupu, W. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. Van Aken, S. Riedmuller, H. Tettelin, S. Gill, O. White, S. Salzberg, D. Hoover, L. Lindler, S. Halling, S. Boyle, and C. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 99:13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng, W., Y. Lee, and E. Nester. 1998. The phenolic recognition profiles of Agrobacterium tumefaciens VirA protein are broadened by a high level of the sugar binding protein ChvE. J. Bacteriol. 180:5632-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porte, F., J. Liautard, and S. Köhler. 1999. Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in macrophages. Infect. Immun. 67:4041-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson, G., and R. J. Roop. 1999. The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 34:690-700. [DOI] [PubMed] [Google Scholar]
- 41.Ruckdeschel, K., J. Machold, A. Roggenkamp, S. Schubert, J. Pierre, R. Zumbihl, J. Liautard, J. Heesemann, and B. Rouot. 1997. Yersinia enterocolitica promotes deactivation of macrophage mitogen-activated protein kinases extracellular signal-regulated kinase-1/2, p38, and c-Jun NH2-terminal kinase. J. Biol. Chem. 272:15920-15927. [DOI] [PubMed] [Google Scholar]
- 42.Sangari, F., and J. Aguero. 1996. Molecular basis of Brucella pathogenicity: an update. Microbiol. Semin. 12:207-218. [PubMed] [Google Scholar]
- 43.Sangari, F. J., M. J. Grillo, M. P. Jimenez De Bagues, M. I. Gonzalez-Carrero, J. M. Garcia-Lobo, J. M. Blasco, and J. Aguero. 1998. The defect in the metabolism of erythritol of the Brucella abortus B19 vaccine strain is unrelated with its attenuated virulence in mice. Vaccine 16:1640-1645. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt-Eisenlohr, H., N. Domke, C. Angerer, G. Wanner, P. Zambryski, and C. Baron. 1999. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181:7485-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmiederer, M., and B. Anderson. 2000. Cloning, sequencing, and expression of three Bartonella henselae genes homologous to Agrobacterium tumefaciens VirB region. DNA Cell Biol. 19:141-147. [DOI] [PubMed] [Google Scholar]
- 46.Shimoda, N., A. Toyoda-Yamamoto, S. Aoki, and Y. Machida. 1993. Genetic evidence for an interaction between the VirA sensor protein and the ChvE sugar-binding protein of Agrobacterium tumefaciens. J. Biol. Chem. 268:26552-26558. [PubMed] [Google Scholar]
- 47.Sieira, R., D. J. Comerci, D. O. Sanchez, and R. A. Ugalde. 2000. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J. Bacteriol. 182:4849-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sola-Landa, A., J. Pizarro-Cerda, M.-J. Grillo, E. Moreno, I. Moriyon, J.-M. Blasco, J.-P. Gorvel, and I. Lopez-Goni. 1998. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 29:125-138. [DOI] [PubMed] [Google Scholar]
- 49.Studier, F., and B. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 50.Taminiau, B., D. Mavis, S. Swift, M.-L. Bochiroli, A. Tibor, P. Lestrate, X. De Bolle, D. O'Callaghan, P. Williams, and J.-J. Letesson. 2002. Identification of a quorum sensing signal molecule in the facultative intracellular pathogen Brucella melitensis. Infect. Immun. 70:3004-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsolis, R. 2002. Comparative genome analysis of the alpha-proteobacteria: relationships between plant and animal pathogens and host specificity. Proc. Natl. Acad. Sci. USA 99:12503-12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watarai, M., S. Makino, Y. Fujii, K. Okamoto, and T. Shirahata. 2002. Modulation of Brucella-induced macropinocytosis by lipid rafts mediates intracellular replication. Cell. Microbiol. 4:341-355. [DOI] [PubMed] [Google Scholar]
- 53.Watarai, M., S. Makino, and T. Shirahata. 2002. An essential virulence protein of Brucella abortus, VirB4, requires an intact nucleoside-triphosphate-binding domain. Microbiology 148:1439-1446. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, Z., A. Ribeiro, S. Lin, R. Cotter, S. Miller, and C. Raetz. 2001. Lipid A modifications in polymyxin-resistant Salmonella typhimurium: PMRA-dependent 4-amino-4-deoxy-l-arabinose, and phosphoethanolamine incorporation. J. Biol. Chem. 276:43111-43121. [DOI] [PubMed] [Google Scholar]