Abstract
The amino acid sequence requirements in the hinge of human immunoglobulin A1 (IgA1) for cleavage by IgA1 proteases of different species of Streptococcus were investigated. Recombinant IgA1 antibodies were generated with point mutations at proline 227 and threonine 228, the residues lying on either side of the peptide bond at which all streptococcal IgA1 proteases cleave wild-type human IgA1. The amino acid substitutions produced no major effect upon the structure of the mutant IgA1 antibodies or their functional ability to bind to Fcα receptors. However, the substitutions had a substantial effect upon sensitivity to cleavage with some streptococcal IgA1 proteases, with, in some cases, a single point mutation rendering the antibody resistant to a particular IgA1 protease. This effect was least marked with the IgA1 protease from Streptococcus pneumoniae, which showed no absolute requirement for either proline or threonine at residues 227 to 228. By contrast, the IgA1 proteases of Streptococcus oralis, Streptococcus sanguis, and Streptococcus mitis had an absolute requirement for proline at 227 but not for threonine at 228, which could be replaced by valine. There was evidence in S. mitis that proteases from different strains may have different amino acid requirements for cleavage. Remarkably, some streptococcal proteases appeared able to cleave the hinge at a distant alternative site if substitution prevented efficient cleavage of the original site. Hence, this study has identified key residues required for the recognition of the IgA1 hinge as a substrate by streptococcal IgA1 proteases, and it marks a preliminary step towards development of specific enzyme inhibitors.
Immunoglobulin A (IgA) plays a principal role in the defense of the mucosal surfaces of the human body from damage by microorganisms and their products. IgA in its secretory form protects by inhibiting microbial adhesion to mucosae and subsequent colonization. IgA can neutralize the activity of toxins, enzymes, and viruses (39). Necessitated by its role, and as a consequence of its unique structure, secretory IgA is possibly the most resistant of all immunoglobulin types to proteolytic degradation.
However, a small number of bacteria, both important human pathogens and selected commensals at mucosal surfaces, produce proteolytic enzymes termed IgA1 proteases that cleave the heavy chain of IgA1, one of the two human IgA isotypes, thereby destroying its protective properties (reviewed in references 21 and 30). The IgA1 proteases of these pathogens are thought to be important virulence factors because they are produced in vivo (5, 16, 31), because convalescing patients have neutralizing antibodies to them (7, 10, 12), and because the three principal causes of bacterial meningitis, though genetically distinct, all produce an IgA1 protease (21, 30). However, because the substrate of IgA1 proteases is restricted almost exclusively (4, 41) to IgA1 from only humans, gorillas, chimpanzees, and orangutans (37), a convenient animal model is not available, and therefore, it is difficult to assess the contribution of IgA1 protease production to virulence.
Among the streptococci, only Streptococcus pneumoniae, a major cause of lobar pneumonia and meningitis, Streptococcus oralis, Streptococcus sanguis, and certain strains of Streptococcus mitis biovar 1, have been found to produce IgA1 protease (31, 38). The latter three organisms comprise part of the indigenous oral and pharyngeal flora of humans. Although they occasionally cause endocarditis, they are more frequently encountered as the important major colonizers of the tooth surface, where they initiate the formation of dental plaque that may progress to caries and periodontal disease (28). The streptococcal IgA1 proteases are known to be produced in vivo (31) and, by interfering with the action of the major immune defense mechanism of the upper respiratory tract, are thought to promote colonization of mucosal surfaces and invasiveness (18) and may even compromise protection against allergens, leading to atopic sensitization (19).
The IgA1 proteases of streptococci are all metalloproteinases that cleave the Pro227-Thr228 peptide bond in the IgA1 hinge (20) (Fig. 1), but in contrast to the serine-type IgA1 proteases of Haemophilus and Neisseria species, there is no information about the exact amino acid sequence requirements of potential substrates. By creating mutated IgA1 molecules with amino acid substitutions at these residues, this study sought both to examine the site requirements in IgA1 for cleavage by different streptococcal IgA1 proteases and to obtain insight into potential alternative substrates and functions and thereby gain information that might aid in the design of IgA1 protease inhibitors. Such reagents would permit the role of IgA1 proteases as virulence factors to be evaluated and might be of some therapeutic value.
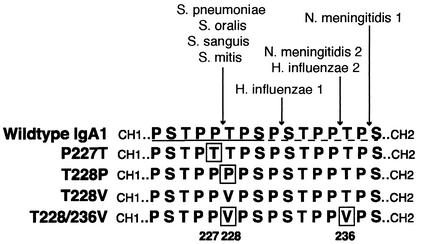
FIG. 1.
Sequence of amino acids in the hinge of the α chain of human IgA1 and the four IgA1 mutants. The wild-type IgA1 hinge contains two identical duplicated halves, one underlined by a solid line and the other underlined by a dashed line. The sites of cleavage of some bacterial IgA1 proteases in the wild-type IgA1 hinge are indicated above. The residues mutated in this study are boxed and numbered at the bottom.
MATERIALS AND METHODS
Generation of mutant IgA1 expression vectors.
Recombinant IgA1 vectors with mutations in the hinge region were prepared by PCR overlap extension (15) by using the plasmid pMB2 containing the wild-type human α1 heavy chain sequence as template DNA, as described previously (1, 22). The 5′ flankingprimer (5′-GCGCGCGCGGATCCGGTCCAACTGCAGGC-3′) annealedaround 140 bp 5′ of the start of the Cα1 domain sequence and incorporated a BamHI restriction site (designated in italics) to facilitate cloning of the PCR product. The 3′ flanking primer (5′-TTCTGAACCTAAGAGCAGGTCC-3′) annealed 3′ of a unique XhoI site in the α1 sequence. Each set of paired mismatch primers annealed within the region encoding the IgA1 hinge. In each case, the mutated PCR products were ligated into unique BamHI and XhoI restriction sites in the expression vectors, replacing the wild-type sequence in that region.
In mutant T228P (using mismatch primer 5′-CTCAACTCCACCTCCCCCATCTCCCTC-3′ and its complement) an ACC-to-CCC substitution changed Thr228 to Pro. In mutant T228V, mismatch sense primer 5′-AACTCCACCTGTCCCATCTCCCT-3′ and antisense primer 5′-GAGGGAGATGGGACAGGTGGAGTTGAG-3′ were used to encode valine (GTC) at residue 228 instead of wild-type threonine (ACC). For mutant P227T, mismatch primer 5′-CCCTCAACTCCAACTACCCCATCT-3′ and its complement replaced wild-type CCT with ACT, thereby encoding threonine instead of wild-type proline at residue 227. The double mutant T228/236V was generated by using mismatch primer 5′-TCAACTCCACCGGTACCATCTCCCTCA-3′ and its complement. As a result of fortuitous primer slippage in the duplicated sequence of the IgA1 hinge (Fig. 1), overlap PCR generated a mutant in which wild-type threonines at residues 228 and 236 were both replaced with valine. Each resultant plasmid was sequenced by the dideoxy chain termination method (9) to confirm both the presence of each mutation and that no additional base changes had arisen during the PCR process.
Expression and purification of mutated IgA1 antibodies.
CHO-K1 cells stably transfected with an appropriate mouse λ light chain (26) were transfected with the plasmid vectors for each of the mutated IgA1 antibodies by using calcium phosphate as described previously (26). Positive transfectants were selected by growth in medium supplemented with hypoxanthine and thymidine (HT supplement; Life Technologies, Paisley, United Kingdom), xanthine (0.25 mg/ml), and mycophenolic acid (10 μg/ml) (27). Clones secreting high levels of mutated recombinant antibody were identified by an enzyme-linked immunosorbent assay measuring binding to the antigen NIP (3-nitro-4-hydroxy-5-iodophenylacetate) as described previously (26) before they were expanded into large cultures. The recombinant mutated antibodies were purified from supernatants of CHO-K1 transfectants by affinity chromatography on NIP-Sepharose as described previously (26). The purified antibodies were supplemented with 0.04% sodium azide and stored in small aliquots at −20°C.
Rosette assays.
Rosette assays to assess binding to FcαR on isolated neutrophils (33) were performed as described previously (32, 43).
Bacterial IgA1 proteases.
The IgA1 proteases used were from S. pneumoniae strain SK690 (PK81); S. sanguis strains SK1 (ATCC 10556) (biovar 1), SK4 (biovar 2), and SK49 (biovar 4); S. oralis strain SK10; and S. mitis biovar 1 strains SK564, SK597, and SK599. They were grown in 2TY broth (1.6% tryptone, 1% yeast extract, and 0.5% sodium chloride in distilled water [pH 7]) at 37°C in air containing 5% CO2. The culture supernatants were precipitated with ammonium sulfate at 60% saturation, and the precipitates were dissolved in a small volume of phosphate-buffered saline (PBS) buffer (pH 7.2) containing 0.1% sodium azide. After dialysis against this buffer, the resultant protease preparations were stored in small aliquots at −20°C. Purified IgA1 proteases from Neisseria meningitidis (type 1 enzyme) strain HF 48, N. meningitidis (type 2 enzyme) strain HF13, Haemophilus influenzae (type 1 enzyme) strain HK368 and H. influenzae (type 2 enzyme) strain HK224 were also used.
S. pneumoniae glycosidases.
S. pneumoniae strain SK1015, an IgA1 protease-negative deletion mutant developed by Knud Poulsen, Department of Medical Microbiology and Immunology, University of Aarhus, Denmark, was used as source of pneumococcal glycosidases. These were prepared from 2TY broth culture supernatants in the same way as the streptococcal IgA1 proteases.
Digestion of IgA1 by microbial IgA1 proteases and analysis of the products.
Before incubation with the mutant antibodies, all of the streptococcal IgA1 proteases were adjusted to equivalent potency after assaying the activity of a range of dilutions of the protease preparations on wild-type IgA1 after 16 h of incubation. Thereafter, appropriate amounts of IgA1 and IgA1 proteases in PBS (pH 7.2) containing 0.1% sodium azide in a total volume of 20 to 30 μl were incubated at 37°C for 16 h. The reaction mixture was then electrophoresed in sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis gels under reducing and nonreducing conditions. Wild-type recombinant human IgA1 and IgA2m(1) (26) served as positive and negative controls, respectively, of enzyme activity. The electrophoresed proteins were transferred to nitrocellulose membranes which were then blocked by agitation for 30 min in 5% nonfat dried milk powder in PBS. After washing in PBS, the membranes were immersed in an appropriate antibody, usually at a dilution of 1 in 1,000, in PBS containing 0.1% Tween 20 and agitated for 2 h at room temperature.
Detection of Fab or Fc fragments of IgA1 was difficult with some IgA-specific antibodies because many seemed to combine with epitopes in the hinge region of the uncleaved antibody, and their affinity was considerably reduced when the antibody was cleaved and in the reduced form. Among the antibodies used were alkaline phosphatase-labeled mouse antibodies specific for either the Fc part of the human α1 or the human α2 chain (Southern Biotechnology Associates, Inc., Birmingham, Ala.), horseradish peroxidase-labeled antibodies specific either for the Fab part of the human α chain (Kirkegaard and Perry, Gaithersburg, Md.) or to the mouse λ light chain (Nordic Immunological Laboratories, Tilburg, The Netherlands), and mouse monoclonal antibody HP6116 to the Fc part of human IgA (25) used in conjunction with, as a secondary antibody, a horseradish peroxidase-labeled antibody to the Fc part of mouse IgG (Sigma).
After thorough washing of the membranes in PBS, the alkaline phosphatase-labeled antibodies were developed in the dark at 37°C in 10 ml of developing buffer (100 mM Tris-HCl [pH 9.5] containing 100 mM NaCl and 10 mM MgCl2) to which had been added 30 μl of bromo-chloro-indolyl phosphate solution (50 mg/ml in dimethyl formamide) and 30 μl of nitroblue tetrazolium solution (100 mg/ml in 70% dimethyl formamide in water). The peroxidase-labeled antibodies were developed in 10 ml of 50 mM Tris-HCl (pH 7.6) buffer containing 0.3 mg of nickel chloride/ml, 10 mg of diaminobenzidine, and 60 μl of 30% hydrogen peroxide.
RESULTS
Analysis of mutated IgA1 antibodies.
Dideoxy sequencing of the plasmids encoding the different α1 mutants confirmed the presence of the desired mutations and that the nucleotide sequence of the remainder was the same as that of wild-type human α1. Analysis of the four mutated IgA1 antibodies after electrophoresis and immunoblotting under reducing conditions revealed the expected heavy and light chain bands. For antibodies P228T, T228V, and T228/236V, under nonreducing conditions the expected band corresponding to intact antibody (H2L2) was seen in each case. However, under nonreducing conditions, antibody T228P gave the expected H2L2 band and another band of ca. 85 kDa. The latter was reactive with both anti-α and anti-λ antibodies and thus most likely represents an IgA half molecule comprising one α and one λ chain (HL). Such half molecules are assumed to associate noncovalently in free solution to give H2L2 IgA monomers. Studies investigating the maturation of IgA have shown half molecules to be the major intermediary molecule in folding to the mature antibody structure (17). Thus, the altered residue in antibody T228P may in some way prevent efficient formation of inter-heavy chain disulfide bridges during antibody assembly, resulting in the secretion of a percentage of IgA molecules lacking such bridges.
Binding to Fcα receptors.
The ability of the four mutant antibodies to bind to Fcα receptors expressed on neutrophils was determined by rosette assays. The amount of each antibody giving half of the maximum binding to Fcα receptors lay in the range from 40 to 100 μg/ml (data not shown), values broadly comparable to that for wild-type IgA1 (32). Thus, the introduced mutations did not appear to affect the ability of the antibodies to bind to the Fcα receptor. These results are consistent with the view that the mutations had caused no major changes in the conformation of the mutant antibodies and are in keeping with the localization of the FcαR binding site to the region between the CH2 and CH3 domains of IgA Fc, some distance from the hinge (32, 8).
Susceptibility of the mutant IgA1 antibodies to cleavage with bacterial IgA1 proteases.
All of the protease preparations of the different bacteria cleaved recombinant wild-type IgA1 to Fab and Fc fragments within 16 h of incubation, whereas recombinant wild-type IgA2 was resistant to cleavage even after 72 h of incubation (results not shown). For all mutant antibodies, prolonging incubation to 72 h did not alter the extent of cleavage seen at 16 h. The cleavage of IgA1 by the protease preparations from all of the streptococcal strains was inhibited by the presence of 25 mM EDTA. These results confirmed that all of the preparations had IgA1 protease activity and that for the streptococcal preparations this was a metalloprotease activity. The mutant IgA1 antibodies P227T, T228P, and T228V were also cleaved to Fab and Fc fragments by the type 1 and type 2 IgA1 proteases of N. meningitidis and H. influenzae. This result was expected because all of these enzymes cleave wild-type IgA1 at sites distant to the peptide bond between residues 227 and 228 (Fig. 1). However, mutant antibody T228/236V was also cleaved by these enzymes despite the fact that the type 2 IgA1 proteases of N. meningitidis and H. influenzae cleave wild-type IgA1 at Pro235-Thr236 (Fig. 1). Thus, we conclude that there is no requirement for IgA1 to have threonine at 236 for it to be cleaved by the type 2 proteases of N. meningitidis and H. influenzae, in keeping with the results of an earlier study (34).
Antibodies P227T and T228P were the most resistant of the mutant antibodies to the streptococcal proteases, and in the main, they showed similar susceptibilities. P227T was resistant to cleavage by the proteases of all of the S. sanguis and S. mitis strains and was at least partially resistant to that of S. oralis but was readily cleaved by the protease of S. pneumoniae. Results for representative strains are shown in Fig. 2, and the results for all strains are summarized in Table 1. EDTA inhibited this proteolytic action (Fig. 2, lane 8). Among the streptococci, S. pneumoniae and S. oralis strains, in addition to producing IgA1 proteases, secrete glycosidases which copurify with their IgA1 proteases. The slight decrease in the mass of undegraded antibody when treated by the protease preparations from these bacteria is due to the action of their glycosidases (Fig. 2, lanes 4 and 8) (40). The mass of the Fab fragment arising from the cleavage of P227T by the pneumococcal protease was consistent with cleavage at or close to the peptide bond between residues 227 and 228 and was quite different from the mass of the Fab fragment arising through cleavage of the mutant antibody and recombinant wild-type human IgA1 by N. meningitidis type 2 protease which cleaves at the Pro235-Thr236 peptide bond in the latter (Fig. 1 and 2, lanes 6 and 7).
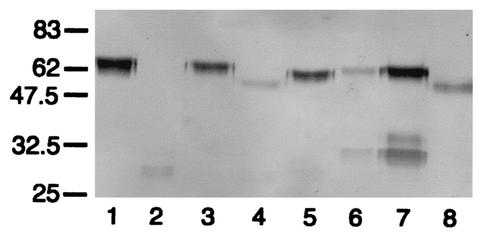
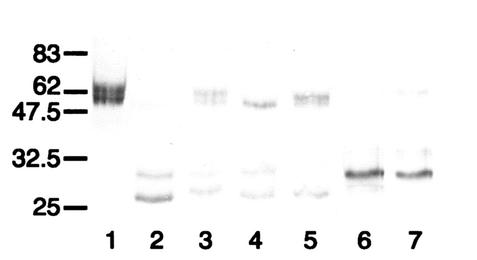
FIG. 2.
Western blot analysis of the action of streptococcal IgA1 proteases on mutant antibody P227T. Digests run under reducing conditions were probed with an anti-human IgA α chain-specific peroxidase conjugate that bound to epitopes in the Fab region. P227T was untreated (lane 1) or digested with IgA1 proteases of S. pneumoniae SK690 (lane 2) (and in the presence of 25 mM EDTA [lane 8]), S. sanguis SK4 (lane 3), S. oralis SK10 (lane 4), S. mitis SK564 (lane 5), and the type 2 IgA1 protease of N. meningitidis HF13 (lane 6). Wild-type recombinant IgA1 treated with the latter enzyme is shown in lane 7. Positions of molecular mass markers (in kilodaltons) are indicated on the left. Antibody P227T was resistant to cleavage by the IgA1 proteases of S. sanguis SK4 andS. mitis SK564 but was cleaved by the EDTA-sensitive IgA1 protease of S. pneumoniae and partially cleaved by the protease of S. oralis SK10 to give Fab fragments with masses consistent with cleavage at or near the peptide bond between residues 227 and 228 and different in mass from that resulting from cleavage with N. meningitidis type 2 IgA1 protease.
TABLE 1.
Summary of effects of hinge mutations in IgA1 on susceptibility to cleavage by IgA1 proteases from strains of different species of streptococci
| IgA1 antibody | Resulta for strain (source of IgA1 protease):
|
|||||||
|---|---|---|---|---|---|---|---|---|
| S. pneumoniae SK690 | S. oralis SK10 | S. mitis SK564 | S. mitis SK597 | S. mitis SK599 | S. sanguis SK4 | S. sanguis SK1 | S. sanguis SK49 | |
| Wild-type IgA1 | + | + | + | + | + | + | + | + |
| P227T | + | +/− | − | − | − | − | − | − |
| T228P | + | +/− | − | + | − | − | − | − |
| T228V | + | +/− | +/− | + | + | +/− | − | +/− |
| T228/236V | + | +/− | +/− | + | + | +/− | − | +/− |
Antibody susceptibility to cleavage by protease: +, sensitive; +/−, partially sensitive; −, resistant.
We found that the resistance of P227T to cleavage by the IgA1 proteases of S. sanguis could be overcome if the antibody received prior treatment with pneumococcal glycosidases (Fig. 3). This finding suggests that the P227T mutation may affect either the structure, three-dimensional conformation, or the attachment sites of the sugars attached to the hinge in such a way as to influence the access and/or cleavage efficiency of the protease and may be indicative of a general influence of hinge glycosylation on the efficiency of IgA1 protease cleavage.
FIG. 3.
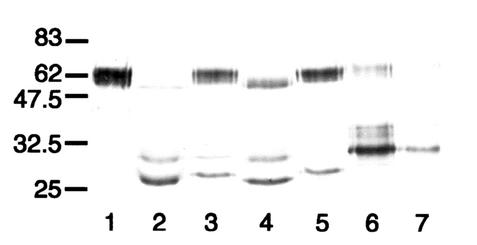
Western blot analysis of the effect of pneumococcal glycosidases on the sensitivity of mutant antibody P227T to cleavage by S. sanguis SK4 IgA1 protease. After separation under reducing conditions, the digests were probed with an anti-human α chain-specific peroxidase conjugate that bound to epitopes in the Fab region. Antibody P227T was untreated (lane 1) or treated singly or in combination with the IgA1 protease of S. pneumoniae SK690 or S. sanguis SK4 or the glycosidases of the IgA1 protease-negative strain SK1015 of S. pneumoniae as follows: lane 2, glycosidases only; lane 3, S. pneumoniae SK690 protease only; lane 4, S. sanguis SK4 protease only; lane 5, glycosidases and S. sanguis SK4 protease. Wild-type IgA1 treated with S. sanguis SK4 protease is shown in lane 6. The positions of molecular mass markers (in kilodaltons) are shown on the left. The IgA1 proteases were used at lower concentrations than those in Fig. 2. S. sanguis SK4 protease was unable to cleave antibody P227 unless it had been deglycosylated by pneumococcal glycosidases.
Antibody T228P reacted in a similar way to antibody P227T when treated with the proteases, except for the protease of S. mitis strain SK597, which cleaved antibody T228P but not antibody P227T (Fig. 4).
FIG. 4.
Western blot analysis of the action of the IgA1 proteases of S. mitis SK597 and S. mitis SK599 on mutant antibodies P227T and T228P. After separation under reducing conditions, digests were probed with an anti-human IgA α chain-specific alkaline phosphatase-conjugated antibody specific for epitopes in the Fc region of uncleaved IgA (the Fc of cleaved IgA1 is not detected). Lanes 1 to 5 and lanes 6 to 7 derive from two identical and simultaneously run gels. Coalignment of molecular mass markers allowed the blots to be realigned before figure preparation. IgA1 antibodies were untreated (wild-type IgA1, lane 1) or treated with IgA1 proteases of S. mitis SK597 (wild-type IgA1, lane 2; P227T, lane 3; T228P, lane 4) or of S. mitis SK599 (wild-type IgA1, lane 5; P227T, lane 6; T228P, lane 7). The positions of molecular mass markers (in kilodaltons) are shown on the left. The IgA1 proteases of different strains of S. mitis differed in their abilities to cleave antibodies P227T and T228P. The protease of S. mitis strain SK599 cleaved neither of the mutated antibody molecules, whereas that of S. mitis strain SK597 was able to cleave T228P but not P227T.
Antibody T228V was somewhat more sensitive to streptococcal IgA1 proteases than P227T and T228P, for it was cleaved not only by the protease of S. pneumoniae but also by that of S. oralis and those of the S. mitis strains and most of the S. sanguis strains (Fig. 5). However, in most cases, cleavage produced Fab fragments of two sizes. The smaller ones were of a size consistent with cleavage at or near the peptide bond between residues 227 and 228, as in wild-type IgA1, whereas the larger Fab fragments were of similar masses to those formed by the action of N. meningitidis type 2 protease on both the mutant and wild-type IgA1 (Fig. 5, lanes 2 to 4, 6, and 7). This suggested that some streptococcal proteases cleaved antibody T228V not only at or close to the peptide bond between residues 227 and 228 bond but also at or close to the bond between residues 235 and 236.
FIG. 5.
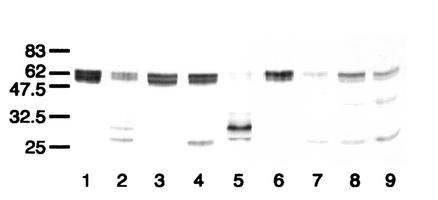
Western blot analysis of the action of streptococcal IgA1 proteases on mutant antibody T228V. After separation under reducing conditions, digests were probed with an anti-human IgA α chain-specific peroxidase conjugate that bound to epitopes in the Fab region. T228V was untreated (lane 1) or digested with the IgA1 proteases of S. pneumoniae SK690 (lane 2), S. sanguis SK4 (lane 3), S. oralis SK10 (lane 4), S. mitis SK564 (lane 5), and the type 2 IgA1 protease of N. meningitidis HF 13 (lane 6). Wild-type recombinant IgA1 treated with the latter enzyme is shown in lane 7. The positions of the molecular mass markers (in kilodaltons) are indicated on the left. Antibody T228V treated with streptococcal IgA1 proteases was cleaved totally (S. pneumoniae) or partially (S. oralis, S. sanguis, and S. mitis SK564) to give Fab fragments. With the exception of the latter enzyme, these were of two sizes. The larger Fab fragment was of a mass similar to that resulting from cleavage with N. meningiditis type 2 protease, whereas the smaller fragment was of a mass consistent with cleavage at or near the peptide bond between residues 227 and 228.
The IgA1 proteases of different strains of S. sanguis reproducibly reacted with the T228V antibody in different ways. That of strain SK49 behaved differently from that of the strain SK4 (Fig. 5, lane 3, and 6, lane 2) in that, like that of S. mitis strain SK564 (Fig. 5, lane 5), it cleaved T228V to only the smaller-sized Fab fragment (Fig. 6, lane 4), whereas that of strain SK1 was unable to cleave the antibody (Fig. 6, lane 3).
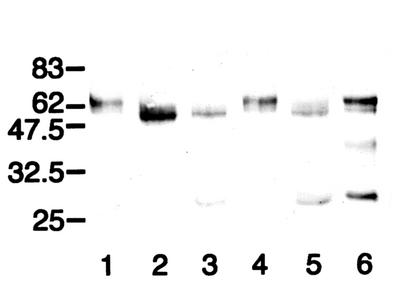
FIG. 6.
Western blot analysis of the action of IgA1 proteases from different S. sanguis strains on mutant antibody T228V. After separation under reducing conditions, digests were probed with an anti-human IgA α chain-specific peroxidase conjugate that bound to epitopes in the Fab region. T228V (lanes 1 to 5) or wild-type IgA1 (lanes 6 to 9) was untreated (lanes 1 and 6) or digested with the IgA1 proteases of S. sanguis SK4 (lanes 2 and 7), S. sanguis SK1 (lanes 3 and 8), S. sanguis SK49 (lanes 4 and 9), or N. meningitidis type 2 HF13 (lane 5). Although the IgA1 proteases of the different strains of S. sanguis cleaved wild-type IgA1, they differed in their abilities to cleave antibody T228V. The enzyme of strain SK1 was unable to cleave the antibody, that of SK49 cleaved it at a site consistent with cleavage at or near the peptide bond between residues 227 and 228, as in wild-type IgA1, and that of SK4 cleaved it at an additional site at or near the peptide bond between residues 235 and 236.
Antibody T228/236V displayed a susceptibility to the streptococcal proteases similar to that of mutant antibody T228V (Fig. 7). Although antibody T228/236V had no Pro-Thr peptide bonds in the hinge, all of the streptococcal proteases, except that from S. mitis SK1, and the type 2 proteases of N. meningitidis and H. influenzae, which cleave wild-type IgA1 at such sites, were able to cleave it. The masses of the cleavage fragments were consistent with cleavage at or close to Pro227-Val228 for all of the streptococcal enzymes and, for most of them, also at the bond between residues 235 and 236, the cleavage site in wild-type IgA1 for the type 2 IgA1 proteases of N. meningitidis and H. influenzae. A summary of the activities of the different streptococcal IgA1 proteases on the mutant antibodies is given in Table 1.
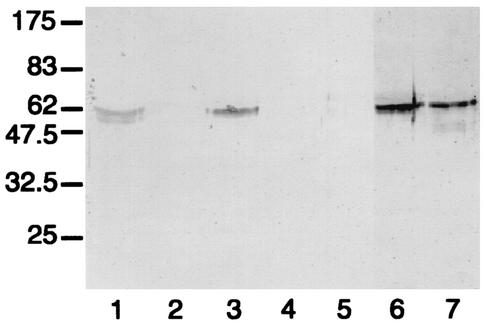
FIG. 7.
Western blot analysis of the action of streptococcal IgA1 proteases on mutant antibody T228/236V. After separation under reducing conditions, digests were probed with an anti-human IgA α chain-specific peroxidase conjugate that bound to epitopes in the Fab region. T228/236V was untreated (lane 1) or treated with the IgA1 proteases of S. pneumoniae SK690 (lane 2), S. sanguis SK4 (lane 3), S. oralis SK10 (lane 4), S. mitis SK564 (lane 5), and the type 2 IgA1 protease of N. meningitidis HF13 (lane 6). Wild-type recombinant IgA1 treated with the latter enzyme is shown in lane 7, and the positions of molecular mass markers (in kilodaltons) are indicated on the left. Mutant antibody T228/236V treated with streptococcal IgA1 proteases was cleaved to give Fab fragments with masses consistent with cleavage at or near the peptide bond between residues 227 and 228, as in wild-type IgA1, and also, for most of them, at or near the bond between residues 235 and 236.
DISCUSSION
As a prelude to ultimately understanding the full biological role of IgA1 protease activity in streptococci, this study sought, by the construction and preparation of a number of mutant forms of IgA1, to elucidate the structural features required in the IgA1 hinge for its recognition and cleavage by different streptococcal IgA1 proteases. Analysis of the IgA1 protease (iga) genes of different Streptococcus spp. has shown that they all code for proteins of ca. 200 kDa which contain sequence motifs characteristic of zinc metalloproteases (13, 35, 36, 44). All of the enzymes cleave the same Pro-Thr peptide bond between residues 227 and 228 in the hinge of the human IgA1 α chain (20, 29, 38).
The four amino acid substitutions made were designed to generate information about IgA1 protease cleavage of the peptide bond between residues 227 and 228. All IgA1 proteases cleave at postproline bonds, and a strong preference for such a bond has been shown for the serine type IgA1 proteases (2). By replacing the proline at residue 227 with threonine, as in the mutant antibody P227T, the preferences of the streptococcal metalloproteases could be examined. In order to examine the importance of the postproline amino acid and the conformation of the site upon cleavage, antibodies T228V and T228P were prepared. The former represents a relatively conservative substitution anticipated to cause minor perturbation to the hinge conformation. However, the latter substitution of proline for threonine may disrupt local conformation around the preferred cleavage site by introducing an extra bend into the hinge polypeptide and would create a stretch of four consecutive proline residues, mirroring the hinge sequence in protease-resistant IgA2. The flexibility of the hinge region in the immediate vicinity would also probably be reduced in this mutant antibody, possibly rendering it less accessible to IgA1 proteases. Antibody T228/236V, carrying the double substitutions of valine for threonine at positions 228 and 236, arose fortuitously as a consequence of primer slippage in the repetitive nucleotide sequence encoding the two duplicated halves of the hinge. Hence, the mismatch primers, designed to anneal around the sequence encoding residue 236, also annealed around the duplicate sequence encoding residue 228. Nevertheless, this mutant was useful in providing information regarding the effect of replacement of threonine at residue 236 with valine and about whether the double mutation might force protease cleavage to take place at a different peptide bond.
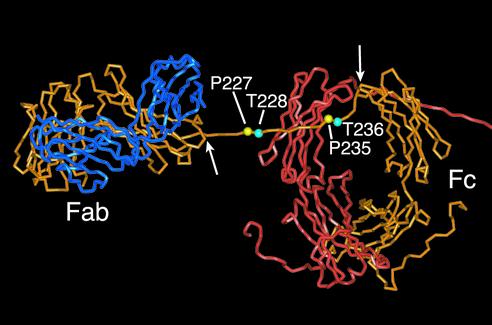
The locations of the residues under investigation are indicated on a molecular model of human IgA1 (6) shown in Fig. 8. A stretch of α heavy chain encompassing the hinge, running between residue Val222 (point of leaving the globular CH1 domain) and residue Cys241 (most N-terminal tethering point at the top of CH2) (Fig. 8), appears freely mobile and presumably allows the Fab arm to move into very many different positions relative to the Fc. The fact that the hinge is rich in proline is likely to maintain an extended hinge conformation. The model allows visualization of the ready access of IgA1 proteases to this region. Analysis of serum IgA1 has revealed that Thr228 has an O-linked sugar attached (24), whereas in a myeloma IgA1, O-linked sugars were found only on hinge serine residues (3). Although carbohydrate moieties are not depicted in the model, the O-linked sugars attached to each α chain hinge are likely to influence, to some as-yet-undefined degree, both conformation and access. It has been shown earlier that removal of sialic acid increased susceptibility to cleavage, whereas extensive deglycosylation decreased the susceptibility to streptococcal IgA1 proteases (38). Although it is clear that the glycosylation profile for the recombinant IgA1 antibodies used here probably closely emulates that in natural IgA1 (24), it is not known if the mutations introduced into the hinge have affected the extent or position of O-linked sugar attachment. Nevertheless, our observation that deglycosylation rendered the previously resistant P227T antibody susceptible to cleavage by the S. sanguis protease provides confirmation that hinge glycosylation can influence the efficiency of IgA1 protease cleavage.
FIG. 8.
Partial view of a molecular model of human IgA1 highlighting the hinge residues investigated in this study. Coordinates were taken from Brookhaven Protein Data Bank as PDB code 1iga (6). For clarity, one entire heavy chain (orange), one entire light chain (blue), and the Fc and partial hinge of the second heavy chain (red) are shown (the remainder of the heavy chain and the second light chain lie out of field to the right of the picture). Proline residues Pro227 and Pro235 are highlighted in yellow, and Thr228 and Thr236 are shown in cyan. This model, based on X-ray and neutron scattering data (6), predicts that the hinge of each heavy chain crosses over the CH2 domain of the other heavy chain and adopts an extended conformation. The arrows indicate residues Val222 and Cys241, the points of constraint at each end of the hinge region, between which the hinge is assumed to be relatively mobile.
The assembly of the different mutant antibodies into mature IgA1 molecules appeared, in the main, to be unaffected by the amino acid substitutions made. However, mutant antibody T228P had an apparently increased propensity to form molecules lacking the inter-heavy chain disulfide bridges, possibly as a result of the introduction of an extra proline in the hinge region affecting the efficiency of disulfide bridge formation between the two α chains. Nevertheless, all of the mutated antibodies retained reactivity with antigen and were recognized by specific antibody reagents. As discussed previously (32), these mutant proteins are unlikely to have undergone any gross structural aberrations, allowing conclusions to be drawn on the relative contributions of mutated residues to the interaction with IgA1 proteases. Indeed, the results of the rosette assays support the view that the amino acid substitutions in the hinge did not affect the overall conformation of the α chain, for all of the mutated IgA1 antibodies bound the Fcα receptor with comparable affinity to wild-type IgA1. This conclusion is further supported by the sensitivity of all of the mutant antibodies to cleavage by the type 1 and 2 IgA1 proteases of N. meningitidis and H. influenzae, which cleave at hinge sites distal to the mutations.
It was remarkable that despite the cleavage site in the IgA1 hinge being identical for all of the different streptococcal IgA1 proteases, their site requirements for cleavage appeared to differ. In general, those for the IgA1 proteases of S. sanguis, S. mitis, and S. oralis appeared to be more stringent than those for the S. pneumoniae protease (Table 1). The protease of S. pneumoniae had no absolute requirement for proline at residue 227, with threonine being an acceptable alternative. In P227T, the substitution has the effect of generating a Pro-Thr bond moved one residue towards the N terminus relative to its position in wild-type IgA1 (Fig. 1). It is possible that the S. pneumoniae protease may cleave at this site in the absence of the usual Pro227-Thr228 bond. Indeed, in a study of the cleavage sites for the S. pneumoniae protease in ape IgAs with differing hinge sequences, the enzyme was found always to cleave a Pro-Thr bond even though the position of the bond in the hinge varied by up to two amino acids (37). However, our study shows that the enzyme also displayed no absolute requirement for threonine as the post-Pro residue 228, with either valine or even proline being an acceptable alternative, permitting IgA1 cleavage. Mutants T228P and T228V lack a Pro-Thr bond in the first half of the hinge, suggesting that in these cases the enzyme either cleaves the Pro-Pro or Pro-Val bonds, respectively, lying at the normal cleavage site or is instead forced to cleave at the Pro-Ser bond lying two residues further towards the C terminus. Localization of the precise cleavage sites awaits peptide sequencing data, but whatever the outcome, it is clear that the S. pneumoniae IgA1 protease active site can accommodate residues other than Pro and Thr adjacent to the scissile bond and efficiently catalyze the cleavage of such peptide bonds.
The proteases of S. sanguis and S. mitis, and to a lesser degree that of S. oralis, were more stringent in their IgA1 cleavage site requirements despite having a common site of action in the IgA1 hinge (Table 1). This is probably a reflection of diversity within the streptococcal proteases at the molecular level, for it is known, for example, that although the iga genes of S. sanguis strains are very homogeneous, they have low homology to other streptococcal iga genes sequenced (35) and do not hybridize with DNA from protease-producing S. pneumoniae (11). Thus, it was not surprising to find that the proteases of the S. sanguis strains acted differently from the proteases of the other streptococcal species. Most notably, when the wild-type cleavage peptide bond at Pro227-Thr228 was mutated to the less acceptable cleavage bond of Pro227-Val 228, as in antibody T228V, the protease of S. sanguis strain SK1 was unable to cleave it, whereas that of S. sanguis strain SK49 cleaved it and that of strain SK4 was able also to cleave at a second site presumed to be at or near the Pro-Thr peptide bond at the equivalent position (235 and 236) in the other half of the duplicated hinge of IgA1 (Fig. 1). Such cleavage indicates that regard for the context of the particular half hinge within the whole hinge may be overruled in certain circumstances. Our earlier work supports such a possibility (40). We showed that when only one-half of the IgA1 hinge was inserted into protease-resistant IgA2, the hybrid antibody produced was sensitive to cleavage by many different IgA1 proteases, including the proteases of S. sanguis and representatives of other bacteria which cleave in the different duplicated halves of the IgA1 hinge (40). However, in this present study, the activity of the IgA1 proteases of the other streptococcal species on the mutant antibodies P227T and T228P cannot be explained on the basis of cleavage at the alternative Pro235-Thr236 site in the hinge because the mass of the Fab fragment formed was different from that produced by the type 2 IgA1 protease of N. meningitidis, which cleaves preferentially at this Pro235-Thr236 peptide bond.
The amino acid sequences of the IgA1 proteases of different strains and species of streptococci are very similar, except for the N-terminal third of the protein in which there are several repeat sequences differing in length, sequence, and number (35). It is thought that these repeat sequences are not essential for enzyme activity but that they contribute to antigenic diversity among the proteases (14). This antigenic multiplicity is greatest among the IgA1 proteases of S. mitis and S. pneumoniae and least among those of S. sanguis and S. oralis (23, 38), and this is consistent with the structural diversity of the enzymes from nucleotide sequence data (35). Thus, the known antigenic differences between the IgA1 proteases of S. sanguis strains of biovar 4 and those of biovars 1 to 3 (38) may be the reason for the different actions of S. sanguis strain SK49 (biovar 4) and S. sanguis strain SK1 (biovar 1) on antibodies T228V and T228/236V. However, it is unlikely that the unusual activity of the protease of S. sanguis strain SK4 (biovar 2) on these mutant antibodies can be explained on this basis.
There is evidence that the IgA1 protease gene in S. mitis strains is a complex mosaic of sequences from fragments of the iga genes of S. oralis and S. pneumoniae because of the horizontal transfer of DNA between species (35). This most probably is the reason for S. mitis strains having IgA1 proteases of much greater antigenic diversity, and it may account for the different sequence requirements of the IgA1 protease of S. mitis strain SK597 from those of the other S. mitis strains for cleavage of antibody T228P.
This study describes the first example of an IgA1 molecule (P227T) in which resistance to IgA1 proteases resulted from the mutation of a single amino acid. Given the increasing interest in the use of IgA as a therapeutic agent, this structure-function information could be of considerable significance. Despite a growing appreciation of active-site structural components in zinc metalloproteases through use of known crystal structures as standards of reference (42), there remains a lack of three-dimensional structural information on the active sites of the IgA1 metalloproteases. In the meantime, the approach described here is a first stage in understanding these enzyme-substrate interactions, which in turn may permit the development of reagents engineered to inhibit the action of such enzymes.
Acknowledgments
This work was supported by the Wellcome Trust.
We thank G. Carlone and K. Poulsen for the generous gifts of antibody HP6116 and the S. pneumoniae IgA1 protease-negative strain, respectively.
Editor: J. T. Barbieri
REFERENCES
- 1.Atkin, J. D., R. J. Pleass, R. J. Owens, and J. M. Woof. 1996. Mutagenesis of the human IgA1 heavy chain tailpiece that prevents dimer assembly. J. Immunol. 157:156-159. [PubMed] [Google Scholar]
- 2.Bachovchin, W. W., A. G. Plaut, E. R. Flentke, M. Lynch, and C. A. Kettner. 1990. Inhibition of IgA1 proteinases from Neisseria gonorrhoeae and Haemophilus influenzae by peptide prolyl boronic acids. J. Biol. Chem. 265:3738-3743. [PubMed] [Google Scholar]
- 3.Baenziger, J., and S. Kornfeld. 1974. Structure of the carbohydrate units of IgA1 immunoglobulin. II. Structure of the O-glycosidically linked oligosaccharide units. J. Biol. Chem. 249:7270-7281. [PubMed] [Google Scholar]
- 4.Beck, S. C., and T. F. Meyer. 2000. IgA1 protease from Neisseria gonorrhoeae inhibits TNFα-mediated apoptosis of human monocytic cells. FEBS Lett. 472:287-292 [DOI] [PubMed] [Google Scholar]
- 5.Blake, M., K. K. Holmes, and J. Swanson. 1979. Studies on gonococcus infection. XVII. IgA1-cleaving protease in vaginal washings from women with gonorrhea. J. Infect. Dis. 139:89-92. [DOI] [PubMed] [Google Scholar]
- 6.Boehm, M. K., J. M. Woof, M. A. Kerr, and S. J. Perkins. 1999. The Fab and Fc fragments of IgA1 exhibit a different arrangement from that in IgG: a study by X-ray and neutron solution scattering and homology modelling. J. Mol. Biol. 286:1421-1447. [DOI] [PubMed] [Google Scholar]
- 7.Brooks, G. F., C. J. Lammel, M. S. Blake, B. Kusecek, and M. Achtman. 1992. Antibodies against IgA protease are stimulated both by clinical disease and asymptomatic carriage of serogroup A Neisseria meningitidis. J. Infect. Dis. 166:1316-1321. [DOI] [PubMed] [Google Scholar]
- 8.Carayannopoulos, L., J. M. Hexham, and J. D. Capra. 1996. Localization of the binding site for the monocyte immunoglobulin (Ig) A-Fc receptor (CD89) to the domain boundary between Cα2 and Cα3 in human IgA1. J. Exp. Med. 183:1579-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, E. Y., and P. H. Seeburg. 1985. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA 4:165-170. [DOI] [PubMed] [Google Scholar]
- 10.Devenyi, A. G., A. G. Plaut, F. J. Grundy, and A. Wright. 1993. Post-infectious human serum antibodies inhibit IgA1 proteinases by interaction with the cleavage site specificity determinant. Mol. Immunol. 30:1243-1248. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert, J. V., A. G. Plaut, Y. Fishman, and A. Wright. 1988. Cloning of the gene encoding streptococcal immunoglobulin A protease and its expression in Escherichia coli. Infect. Immun. 56:1961-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert, J. V., A. G. Plaut, B. Longmaid, and M. E. Lamm. 1983. Inhibition of microbial IgA proteases by human secretory IgA and serum. Mol. Immunol. 20:1039-1049. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert, J. V., A. G. Plaut, and A. Wright. 1991. Analysis of the immunoglobulin A protease gene of Streptococcus sanguis. Infect. Immun. 59:7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert, J. V., J. P. Ramakrishna, A. Wright, and A. G. Plaut. 1993. Streptococcal IgA protease tandem repeat influences antigenicity but not activity. J. Dent. Res. 72:327. [Google Scholar]
- 15.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 16.Insel, R. A., P. Z. Allen, and I. D. Berkowitz. 1982. Types and frequency of Haemophilus influenzae IgA1 proteases. Semin. Infect. Dis. 4:225-231. [Google Scholar]
- 17.Jerry, L. M., and H. G. Kunkel. 1972. The unique reassociation of human IgA2 immunoglobulins from dimer subunits. J. Immunol. 109:982-991. [PubMed] [Google Scholar]
- 18.Kilian, M., and K. Holmgren. 1981. Ecology and nature of immunoglobulin A1 protease-producing streptococci in the human oral cavity and pharynx. Infect. Immun. 31:868-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kilian, M., S. Husby, A. Høst, and S. Halken. 1995. Increased proportions of bacteria capable of cleaving IgA1 in the pharynx of infants with atopic disease. Pediatr. Res. 38:182-186. [DOI] [PubMed] [Google Scholar]
- 20.Kilian, M., J. Mestecky, R. Kulhavy, M. Tomana, and W. T. Butler. 1980. IgA1 proteases from Haemophilus influenzae, Streptococcus pneumoniae, Neisseria meningitidis, and Streptococcus sanguis: comparative immunochemical studies. J. Immunol. 124:2596-2600. [PubMed] [Google Scholar]
- 21.Kilian, M., J. Reinholdt, H. Lomholt, K. Poulsen, and E. V. G. Frandsen. 1996. Biological significance of IgA1 proteases in bacterial colonization and pathogenesis: critical evaluation of experimental evidence. APMIS 104:321-338. [DOI] [PubMed] [Google Scholar]
- 22.Krugmann, S., R. J. Pleass, J. D. Atkin, and J. M. Woof. 1997. Structural requirements for assembly of dimeric IgA probed by site-directed mutagenesis of J chain and a cysteine residue of the α chain CH2 domain. J. Immunol. 159:244-249. [PubMed] [Google Scholar]
- 23.Lomholt, H. 1995. Evidence of recombination and an antigenically diverse immunoglobulin A1 protease among strains of Streptococcus pneumoniae. Infect. Immun. 62:3178-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattu, T. S., R. J. Pleass, A. C. Willis, M. Kilian, M. R. Wormald, A. C. Lellouch, P. M. Rudd, J. M. Woof, and R. A. Dwek. 1998. The glycosylation and structure of human serum IgA1, Fab, and Fc regions and the role of N-glycosylation on Fcα receptor interactions. J. Biol. Chem. 273:2260-2272. [DOI] [PubMed] [Google Scholar]
- 25.Mestecky, J., R. G. Hamilton, C. G. Magnusson, R. Jefferis, J. P. Vaerman, M. Goodall, G. G. de Lange, I. Moro, P. Aucouturier, J. Radl, C. Cambiaso, C. Silvain, J. L. Preud'homme, K. Kusama, G. M. Carlone, J. Biewenga, K. Kobayashi, F. Skvaril, and C. B. Reimer. 1996. Evaluation of monoclonal antibodies with specificity for human IgA, IgA subclasses and allotypes and secretory component. Results of an IUIS/W.H.O. collaborative study. J. Immunol. Meth. 193:103-148. [DOI] [PubMed] [Google Scholar]
- 26.Morton, H. C., J. D. Atkin, R. J. Owens, and J. M. Woof. 1993. Purification and characterization of chimeric human IgA1 and IgA2 expressed in COS and Chinese hamster ovary cells. J. Immunol. 151:4743-4752. [PubMed] [Google Scholar]
- 27.Mulligan, R. C., and P. Berg. 1981. Selection for animal cells that express the E. coli gene coding for xanthine-guanine phosphoribosyltransferase. Proc. Natl. Acad. Sci. USA 78:2072-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:369-380. [DOI] [PubMed] [Google Scholar]
- 29.Plaut, A. G. 1978. Microbial IgA proteases. N. Engl. J. Med. 298:1459-1463. [DOI] [PubMed] [Google Scholar]
- 30.Plaut, A. G. 1983. The IgA1 proteases of pathogenic bacteria. Annu. Rev. Microbiol. 37:603-622. [DOI] [PubMed] [Google Scholar]
- 31.Plaut, A. G., R. J. Genco, and T. B. Tomasi, Jr. 1974. Isolation of an enzyme from Streptococcus sanguis which specifically cleaves IgA. J. Immunol. 113:289-291. [PubMed] [Google Scholar]
- 32.Pleass, R. J., J. I. Dunlop, C. M. Anderson, and J. M. Woof. 1999. Identification of residues in the CH2/CH3 domain interface of IgA essential for interaction with the human Fcα receptor (FcαR). J. Biol. Chem. 274:23508-23514. [DOI] [PubMed] [Google Scholar]
- 33.Pleass, R. J., P. D. Andrews, M. A. Kerr, and J. M. Woof. 1996. Alternative splicing of the human IgA Fc receptor CD89 in neutrophils and eosinophils. Biochem. J. 318:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pohlner, J., T. Klauser, E. Kuttler, and R. Halter. 1992. Sequence-specific cleavage of protein fusions using a recombinant Neisseria type 2 IgA protease. Bio/Technology 10:799-804. [DOI] [PubMed] [Google Scholar]
- 35.Poulsen, K., J. Reinholdt, C. Jespersgaard, K. Boye, T. A. Brown, M. Hauge, and M. Kilian. 1998. A comprehensive genetic study of streptococcal immunoglobulin A1 proteases: evidence for recombination within and between species. Infect. Immun. 66:181-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poulsen, K., J. Reinholdt, and M. Kilian. 1996. Characterization of the Streptococcus pneumoniae immunoglobulin A1 protease gene (iga) and its translation product. Infect. Immun. 64:3957-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu, J., G. P. Brackee, and A. G. Plaut. 1996. Analysis of the specificity of bacterial immunoglobulin A (IgA) proteases by a comparative study of ape serum IgAs as substrates. Infect. Immun. 64:933-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reinholdt, J., M. Tomana, S. B. Mortensen, and M. Kilian. 1990. Molecular aspects of immunoglobulin A1 degradation by oral streptococci. Infect. Immun. 58:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell, M. W., M. Kilian, and M. E. Lamm. 1999. Biological activities of IgA. p. 225-240. In P. L. Ogra, J. Mestecky, M. E. Lamm, W. Strober, J. Bienenstock, and J. R. McGhee (ed.), Mucosal immunology, 2nd ed. Academic Press, San Diego, Calif.
- 40.Senior, B. W., J. I. Dunlop, M. R. Batten, M. Kilian, and J. M. Woof. 2000. Cleavage of a recombinant human immunoglobulin A2 (IgA2)-IgA1 hybrid antibody by certain bacterial IgA1 proteases. Infect. Immun. 68:463-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senior, B. W., W. W. Stewart, C. Galloway, and M. A. Kerr. 2001. Cleavage of the hormone human chorionic gonadotropin, by the type 1 IgA1 protease of Neisseria gonorrhoeae, and its implications. J. Infect. Dis. 184:922-925. [DOI] [PubMed] [Google Scholar]
- 42.Valle, B. L., and D. S. Aude. 1990. Active-site zinc ligands and activated H2O of zinc enzymes. Proc. Natl. Acad. Sci. USA 87:220-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker, M. R., B. M. Kumpel, K. Thompson, J. M. Woof, D. R. Burton, and R. Jefferis. 1988. Immunogenic and antigenic epitopes of immunoglobulins: binding of human monoclonal anti-D antibodies to FcRI on the monocyte-like U937 cell line. Vox Sang. 55:222-228. [DOI] [PubMed] [Google Scholar]
- 44.Wani, J. H., J. V. Gilbert, A. G. Plaut, and J. N. Weiser. 1996. Identification, cloning, and sequencing of the immunoglobulin A1 protease gene of Streptococcus pneumoniae. Infect. Immun. 64:3967-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]