Abstract
The primary effector for Shigella invasion of epithelial cells is IpaC, which is secreted via a type III secretion system. We recently reported that the IpaC N terminus is required for type III secretion and possibly other functions. In this study, mutagenesis was used to identify an N-terminal secretion signal and to determine the functional importance of the rest of the IpaC N terminus. The 15 N-terminal amino acids target IpaC for secretion by Shigella flexneri, and placing additional amino acids at the N terminus does not interfere with IpaC secretion. Furthermore, amino acid sequences with no relationship to the native IpaC secretion signal can also direct its secretion. Deletions introduced beyond amino acid 20 have no effect on secretion and do not adversely affect IpaC function in vivo until they extend beyond residue 50, at which point invasion function is completely eliminated. Deletions introduced at amino acid 100 and extending toward the N terminus reduce IpaC's invasion function but do not eliminate it until they extend to the N-terminal side of residue 80, indicating that a region from amino acid 50 to 80 is critical for IpaC invasion function. To explore this further, the ability of an IpaC N-terminal peptide to associate in vitro with its translocon partner IpaB and its chaperone IpgC was studied. The N-terminal peptide binds tightly to IpaB, but the IpaC central hydrophobic region also appears to participate in this binding. The N-terminal peptide also associates with the chaperone IpgC and IpaB is competitive for this interaction. Based on additional biophysical data, we propose that a region between amino acids 50 and 80 is required for chaperone binding, and that the IpaB binding domain is located downstream from, and possibly overlapping, this region. From these data, we propose that the secretion signal, chaperone binding region, and IpaB binding domain are located at the IpaC N terminus and are essential for presentation of IpaC to host cells during bacterial entry; however, IpaC effector activity may be located elsewhere.
A common theme among many gram-negative bacterial pathogens is the presence of a type III secretion system (TTSS), which is composed of 25 to 30 genes that are collectively responsible for transporting effector proteins from the bacterial cytoplasm to a target host cell membrane and/or cytoplasm (18). The sequences of the component proteins of the TTSS apparatus are homologous to those that make up the gram-negative flagellar basal body (2, 18, 25). Likewise, the TTSS apparatus outwardly resembles a flagellar basal body with the filament replaced by a needle that serves as a channel for the movement of effectors out of the bacterial cytoplasm following contact with a target cell (5, 7). Prior to transport, most TTSS effector proteins are translated and held in the bacterial cytoplasm by their cognate chaperones (2). Following host cell contact, however, there is a release of the effectors from their chaperones and a temporal transport of these proteins to the host cell via the TTSS needle (7, 18, 23). Some of the TTSS effectors are inserted into the host cell membrane to form a conduit (translocon), which helps in the subsequent movement of other effectors into the host cytoplasm (5, 8, 33, 37). Most TTSS effectors that gain access to the host cytoplasm function in promoting cytoskeletal rearrangements and/or subverting host cell signaling processes (1, 13, 14, 18, 32, 43).
Although TTSS structural components from diverse gram-negative pathogens share significant sequence and structural similarities, the sequences and functions of the protein substrates targeted for TTSS export are quite diverse. In contrast, the organization of important functional regions within the primary structures of these effectors does appear to be conserved (18). Within the first 25 amino acids lies a sequence or structural component necessary for effector protein secretion (20). The amino acid sequence of this region varies greatly, not only between proteins from different organisms, but also between TTSS substrates within a single organism (18). Nevertheless, deletion of this region or mutations in the region expected to cause substantial changes in the biophysical characteristics of the region abolishes substrate secretion (20, 21). Downstream from the TTSS export signal lies a structure often referred to as a translocation region (18). Deletion of this region does not adversely affect secretion of the substrate, but it does abolish proper transport of the protein to the host cell. Though not located at a specific site, many TTSS substrates, especially those that are inserted into the host cell membrane to form the translocon, also have a hydrophobic segment having predicted transmembrane segments, along with predicted coiled-coil structures (8, 29). The most diverse component of TTSS effector substrates is the nature of the effector function itself, which is often located within the C-terminal half of the protein (18).
For Shigella flexneri, the TTSS and its substrates are required for invasion of human colonic epithelial cells, an early step in the development of bacillary dysentery (39). Enterocyte invasion is governed by a subset of proteins encoded by the mxi, spa, and ipa loci located within a 31-kb region of a large virulence plasmid (35). The mxi/spa operons encode the TTSS apparatus composed of a cytoplasmic bulb, a transmembrane neck domain, and an external needle (5, 7). The ipa operon encodes the effectors, IpaA, B, C, and D, along with IpgC, the cognate chaperone for IpaB and IpaC (39). IpaB and IpaC are stored within the bacterial cytoplasm and are prevented from forming a complex by interactions with IpgC (23). Following host cell contact by the needle complex of the Shigella TTSS, cytoplasmic IpaB and IpaC are transported through the needle complex and inserted into the host cell membrane to form a 2.5-nm pore through which other protein effectors are believed to pass (5). Interaction of the Ipa proteins at the host-pathogen interface leads to localized actin polymerization on the inner face of the host membrane. Activation of Cdc42 causes the formation of filopodia that mature into lamellopodia and eventually trap the bacterium in a membrane-bound vacuole (40). Following entry, the vacuolar membrane is lysed and the bacterium enters the host cell cytoplasm to replicate and directly invade neighboring cells (3, 34). While both IpaB and IpaC have been implicated as effectors for subverting host cell signaling events, IpaC possesses biochemical properties that are consistent with its having a major role in triggering S. flexneri entry into host cells. IpaC does not have a specific cellular receptor, but it does associate with phospholipid membranes (11, 38), trigger dramatic cytoskeletal changes in cultured cells (22, 40), form homopolymers (10, 27), and oligomerize with IpaB to form membrane pores (5, 27, 30).
Much of what is known about the structural components necessary for proper translocation of TTSS substrates has been elucidated in Yersinia species (6, 18). Meanwhile, the determinants needed for movement of TTSS targets into host cells by pathogens like S. flexneri remain unknown except by extrapolation and conjecture. With respect to IpaC, it has been shown recently that the N terminus is responsible for proper secretion and translocation, while the C terminus probably possesses effector functions (30). Using a yeast two-hybrid system, Page and colleagues suggested that IpgC interacts with a region near the N terminus of IpaC and of IpaB and that IpaC interacts with the C terminus of IpaB (28). The region of IpaC that interacts with IpaB remains unknown.
The use of Shigella to determine the molecular basis for the secretion of TTSS substrates is attractive because of the relative simplicity of this pathogen's invasion mechanism. Unlike Salmonella, Shigella possesses only one known TTSS (18), and IpaC appears to be the only Shigella effector molecule required for the up-regulation of Rho subfamily GTPases (39). Furthermore, IpaC and the related Salmonella invasion protein SipC are presently the only known translocon proteins that also have recognized effector functions (16, 39). It has been demonstrated previously that deletion of a large segment from the IpaC N terminus eliminates its ability to function as a TTSS substrate (30). In the work presented here, the IpaC N terminus is further dissected to identify the precise region needed for Shigella TTSS-dependent export, the region needed for IpaC binding to its cognate chaperone, and the region or regions involved in IpaB binding.
MATERIALS AND METHODS
Materials.
S. flexneri 2a strain 2457T was grown at 37°C on Trypticase soy agar containing 0.025% Congo red. The S. flexneri ipaC mutant strain SF621 was from P. J. Sansonetti (Unite de Pathogenie Microbienne Moleculaire, Institut Pasteur) (24). IpaC monoclonal antibodies were from E. V. Oaks (Department of Enteric Infections, Walter Reed Army Institute for Research) (41). Henle 407 cells (ATCC CCL6) were propagated as monolayers in Eagle's modified minimal essential medium (MEM) (Fisher Scientific, St. Louis, Mo.) supplemented with 10% newborn calf serum (Life Technologies, Gaithersburg, Md.) and grown in 5% CO2. PCR SuperMix High Fidelity and oligonucleotides were from Invitrogen (Carlsbad, Calif.), and NovaBlue and BL21(DE3) strains of Escherichia coli pET15b were from Novagen (Madison, Wis.). Fluorescein isothiocyanate (FITC), fluorescein maleimide (FM), 3-[4-(isothiocyanato)-7-diethylamino-4-methylcoumarin (CPI), and 5-((((2-iodoacetyl)amino)ethyl) amino) naphthalene-1-sulfonic acid (IAEDANS) were from Molecular Probes, Inc. (Eugene, Oreg.). All other chemicals were reagent grade.
Generation of ipaC deletion mutants for expression in S. flexneri SF621.
The preparation of pWPsf and pWPsfc has been described in detail (30). pWPsfc was the original construct generated in this laboratory for expressing ipaC in S. flexneri SF621; however, IpaC synthesized from pWPsfc possesses eight additional amino acids from β-galactosidase (MTMITNSH) at its immediate N terminus. Even though these amino acids do not interfere with the ability of IpaC to restore invasiveness to SF621, pWPsf4c was designed to allow translation to begin with the first Met codon of the actual ipaC insert (see Table 1). Furthermore, the ipaC coding sequence used in pWPsfc is based on the DNA sequence reported by Venkatesan and coworkers (42), which actually encodes a protein that is 19 amino acids longer than the actual native form of IpaC. These residues (designated residues −19 to −1) are shown in italics in Table 1. pWPsfc' and pWPsf4c' (with and without the eight β-galactosidase residues, respectively) were designed to allow the synthesis of IpaC lacking the residues −19 to −1.
TABLE 1.
Plasmids used in this study and relative levels of secretion and invasion activity of the resulting IpaC proteins in S. flexneri SF621
| Plasmid | N-terminal IpaC sequencea | Secretion (%)b | Relative invasionc |
|---|---|---|---|
| pWPsf (and pWPsf4)d | No ipaC insert | 0 | 0 ± 0 |
| pWPsfc’ | MTMITNSHMEIQNTKPTQTLYTDISTKQ | 100 | 100 ± 3 |
| pWPsf4c’ | MEIQNTKPTQTLYTDISTKQ | 100 | 105 ± 4 |
| pWPsf4c’Δ1-20 | M | 0 | 0 ± 1 |
| pWPsf4c’Δ18-20 | MEIQNTKPTQTLYTDISLE | 80 | 76 ± 9 |
| pWPsf4c’Δ16-20 | MEIQNTKPTQTLYTLE | 100 | 88 ± 9 |
| pWPsf4c’Δ11-20 | MEIQNTKPTQTLE | 20 | 15 ± 6 |
| pWPsf4c’MAIAA | MAIAATKPTQTLYTDISTKQ | 20 | 25 ± 10 |
| pWPsfc | MTMITNSHMLQKQFCNKLLLDTNKENVMEIQNTKPTQTLYTDISTKQ | 99 | 77 ± 33 |
| pWPsf4c | MLQKQFCNKLLLDTNKENVMEIQNTKPTQTLYTDISTKQ | 97 | 90 ± 5 |
| pWPsfcΔ-6-20 | MTMITNSHMLQKQFCNKLLLDT | 100 | 85 ± 10 |
| pWPsf4cΔ-6-20 | MLQKQFCNKLLLDT | 0 | 0 ± 1 |
| pWPsf4cΔ-6-20HIT | MHQIQTCNKLLLDT | 100 | 100 ± 15 |
| pWPsf4c’Δ1-20ipaD | MNITTLTNSISTSSLE | 100 | n.d. |
| pWPsf4c’Δ1-20ipaB | MHNVSTTTTGFPLALE | 75 | n.d. |
| pWPsf4c’Δ1-20ipaA | MHNVNNTQADTFLYLE | 100 | n.d. |
| pWPsf4c’Δ1-20ipgD | MHITNLGLHQVSFQLE | 100 | n.d. |
The sequences shown are all the amino acids preceding what would be residue 21 of native IpaC. The remainder of the protein (amino acids 21 to 363) is not changed. Boldfaced amino acid sequences are native IpaC sequences; italicized amino acids are residues −19 to −1 of IpaC−19-363, which were originally described as representing the first 19 amino acids of native IpaC according to the DNA sequence data reported by Venkatesan and colleagues (42). Underlined amino acids are the eight β-galactosidase residues added to the N terminus when IpaC is generated from the plasmid pWPsf; normal text represents either amino acid substitutions in the native IpaC secretion signal or amino acids derived from other S. flexneri TTSS substrates used to replace amino acids 1 to 20 of IpaC.
Secretion of IpaC into culture supernatants was determined by ELISA by using monoclonal antibodies specific for sequences located in the C-terminal half of IpaC. The values shown are normalized for the secretion level of IpaC−19-363. The values given are averages of triplicate results, with an error of less than 10% in all cases.
Invasion of Henle 407 cells is shown relative to S. flexneri SF621 harboring pWPsfc’, as described in Materials and Methods. The invasion efficiency of IpaC possessing residues from other TTSS substrates at its N terminus was not determined (n.d.).
These plasmids are the backbones into which the ipaC coding sequence was inserted. Insertion of ipaC coding sequences into pWPsf results in the production of an IpaC protein possessing eight additional β-galactosidease residues (MTMITNSH), while insertion into pWPsf4 results in production of an IpaC protein with no β-galactosidase residues at the N terminus.
ipaC deletion mutants were made by inverse PCR by using pWPsfc or pWPsf4c (or their c' counterparts) as template and primers encoding GAGAGA, an XhoI or NdeI restriction site, and 18 nucleotides flanking the region to be deleted. The desired linear plasmid was amplified by PCR, digested with NdeI or XhoI, intramolecularly ligated, and transformed into E. coli NovaBlue. The resulting plasmid was introduced into S. flexneri SF621 by electroporation. Ampicillin selection ensured the presence of the recombinant plasmid, while kanamycin resistance and/or Congo red binding was used to ensure that the transformants still possessed the S. flexneri virulence plasmid. The changes introduced at the IpaC N terminus and the plasmids used to generate these IpaC mutants in S. flexneri SF621 are described in Table 1, which also describes the relative level of secretion and invasion by S. flexneri SF621 for each of the resulting IpaC derivatives.
Generation of ipaC/sipC chimeras for expression in S. flexneri SF621.
SipC/IpaC chimeric proteins were generated by amplifying specific sipC fragments by using a 5′ primer containing GAGAGA, an NdeI site, and 18 bases of sipC coding sequence. The 3′ primer contained GAGAGA, an XhoI site, and 18 bases upstream corresponding to the 3′ end of the sipC fragment being generated. These fragments were ligated with ipaC fragments designed to have XhoI sites at their 5′ ends. After insertion of the chimeric sipC/ipaC gene into pWPsf, S. flexneri SF621 was transformed for synthesis of the resulting chimeric proteins. The chimeras used in this experiment were IpaC−19-62/SipC82-409, IpaC−19-177/SipC197-409, SipC1-81/IpaC63-363, and SipC1-189/IpaC171-363.
Preparation of affinity-purified recombinant proteins.
Plasmids used to prepare recombinant IpaC (called IpaC−19-363 here), IpaCΔ−9-62 (IpaCΔI), IpaB, SipC, and SipB have been previously described (22, 27, 30, 31). pWP2916 (for generating IpaCΔ21-100) was prepared by subcloning the mutant ipaC gene from pWPsf2916C into pET15b. pWPC171-363 (lacking residues −19 to 170) was generated as described for pWPC15, except that the 5′ primer was designed to start ipaC translation at codon 171. pWPI' was designed to encode an IpaC peptide called region I′ (IpaC−19-100), which consists of amino acids −19 to 100. This plasmid was generated as described for pWPC15 except that the 3′ primer was designed to place a translation stop site at codon position 101. pWPipgC was produced in the same way as pWPC15 by copying the gene from the virulence plasmid of Shigella by using a 5′ primer containing an XhoI restriction site and a 3′ primer containing a BamHI site. All new plasmids were transformed into E. coli BL21(DE3) for high levels of protein production. Recombinant proteins were purified via the N-terminal His6 tag by nickel chelation chromatography as described in detail previously (31).
For some fluorescence analyses, an IpaC derivative was generated that consisted of amino acids 51 to 362 with Met and Cys residues at the immediate N terminus. The gene encoding this protein, called IpaCCys, was produced by inverse PCR by using a primer that contained GAGAGA, an XhoI restriction site, the TGC Cys anticodon, the NdeI restriction site, and 10 bases of the pET15b vector upstream from the NdeI site. The second primer contained GAGAGA, an XhoI restriction site, and 18 bases from IpaC starting at codon 51. The PCR fragment was digested with XhoI, intramolecularly ligated, and transformed into E. coli NovaBlue cells. The resulting plasmid was purified and used to transform E. coli BL21(DE3) to synthesize protein that could be affinity purified via an N-terminal His6 tag.
Assay of bacterial entry into cultured epithelial cells.
S. flexneri invasion of Henle 407 cells was monitored by using a standard gentamicin protection assay as described previously (10). Preconfluent monolayers of Henle 407 cells were seeded into 24-well plates and grown overnight. SF621 harboring the desired plasmid was grown in Trypticase soy broth containing 100 μg of ampicillin/ml and 50 μg of kanamycin/ml to an A600 of 0.4 to 0.6. The bacteria were then diluted with serum-free MEM containing 0.45% glucose (MEM-glc), centrifuged onto the surface of semiconfluent Henle 407 monolayers, and incubated with the cells for 30 min at 37°C. Free bacteria were removed by aspiration, and the cells were washed with MEM containing 5% calf serum and 50 μg of gentamicin/ml. The cells were incubated in the final gentamicin wash for 2 h to kill adherent, noninternalized bacteria and rinsed with MEM-glc. The cells were lysed by overlaying them with 250 μl of 0.5% agarose in water. The agarose was then overlaid with 0.5% agar containing 2× Luria-Bertani medium. After overnight incubation at 37°C, internalized bacteria formed subsurface colonies that were quantified.
Measurement of IpaC secretion by ELISA.
Bacteria were grown to an A600 of 0.5 to 0.8, and 10 ml of the culture was centrifuged for 10 min at 4,500 × g. The bacterial pellet was resuspended in 400 μl of carbonate buffer, and the cells were lysed by sonication to release cytoplasmic IpaC antigen. The culture supernatant was the source of secreted IpaC antigen. Each antigen source was diluted 1:3 with carbonate buffer, and 100 μl was used to coat the wells of a polystyrene 96-well microtiter plate that was incubated overnight in preparation for a standard enzyme-linked immunosorbent assay (ELISA).
Fluorescence labeling of proteins.
Recombinant IpaC and its derivatives were labeled at primary amines by using FITC. Briefly, the purified proteins were dialyzed against 0.1 M carbonate (pH 8.0) containing 2 M urea and labeled with FITC by combining 1/10 volume of 1 mg of FITC/ml in DMF with 1 ml of protein. The concentration of protein used for fluorescence labeling varied somewhat from one preparation to the next, but it was always maintained between 1 and 2 mg/ml in the reaction mixture. After incubating at 4°C, the labeled protein was separated from unreacted dye by gel filtration on Sephadex G25 equilibrated with Tris-buffered saline containing 1 M urea. The fluorescent protein was rapidly diluted in Tris-buffered saline lacking urea to promote folding into its final tertiary structure. The FITC labeling efficiency for each IpaC derivative was between 2 and 3 probes per protein based on an FITC extinction coefficient of 70,000 at 490 nm. IpaC region I' was also labeled with CPI by using the same protocol except that the dye concentration was reduced by half and the reaction mixture was made to 50% DMF. IpgC was labeled with FITC by using the same reaction conditions used to make FITC-IpaC except without urea, as IpgC is more soluble in aqueous solutions. FITC-IpgC labeled at a lower stoichiometry than IpaC (0.26 FITC/protein). Alternatively, some IpaC derivatives were site-specifically labeled at a single Cys residue with FM or IAEDANS in Tris (pH 7.5)-150 mM NaCl-2 M urea.
Monitoring protein-protein interactions by using fluorescence polarization.
After excitation with polarized light, the photons emitted by fluorescent probes are also polarized; however, rotational diffusion of a fluorophore over its excited-state lifetime causes depolarization of the emitted photons (19). Fluorescence polarization measures the average angular displacement of a fluorophore over its excited-state lifetime, which is affected by the molecule's rate of rotational diffusion, as described by the relationship P = (I| − I⊥)/(I| + I⊥), where P is the polarization value (ranging from 0 to 0.5), I| is the fluorescence intensity with the excitation beam polarized vertically and the emission beam polarized in a parallel fashion, and I⊥ is the fluorescence intensity when the excitation beam is polarized vertically and the emission beam is polarized in a perpendicular fashion (19). Because the rate of rotational diffusion of a molecule is inversely related to its size, polarization provides a sensitive measure of changes in molecular volume that occur when the fluorescent protein is bound by a nonfluorescent protein (19). Fluorescence polarization was used to monitor the association of FITC-labeled IpaC (or its recombinant derivatives) with nonfluorescent IpaB. Polarization was measured by using a Beacon 2000 variable-temperature fluorescence polarization system (PanVera Corp., Madison, Wis.) with a 480-nm bandpass excitation filter and a 530-nm bandpass emission filter. Samples (0.5 ml each) were measured at 25°C in a 10- by 75-mm borosilicate glass tube, with three average cycles read per sample. Polarization values are presented in millipolarization (mP) units.
Monitoring the effect of IpaB and IpgC binding on the solvent accessibility of site-specific probes on IpaC by using fluorescence quenching analysis.
Steady-state fluorescence quenching was used to determine solute accessibility to an FM probe linked to the single Cys residue (position −13) of IpaC−19-363 or the Cys at position 2 of IpaCCys in the presence and absence of IpaB or IpgC. The association of either nonfluorescent protein near the label would be expected to shield the site-specific fluorescent probe from interaction with solute molecules such as acrylamide, which quenches FM fluorescence according to the relationship first described by Stern and Volmer (36) and modified by Eftink (12). To quantify quenching, the emission spectrum of the sample was taken in the absence and then the presence of increasing concentrations of acrylamide. A graph of Fo/F versus acrylamide concentration was then plotted where Fo is the starting fluorescence intensity and F is the observed intensity at a given concentration of acrylamide. The Stern-Volmer steady-state quenching constant (Ksv) is given by the slope of this plot according to the relationship Fo/F = 1 + Ksv[Q], where [Q] is the concentration of acrylamide in the sample. The larger the Ksv value, the greater accessibility acrylamide has to the fluorescent probe.
Monitoring the relationship between IpgC and IpaB binding to region I' (IpaC−19-100) by using energy transfer.
Another method for sensitively detecting protein-protein interactions is fluorescence resonance energy transfer (FRET), as described by Lakowicz (19). For FRET, region I' (IpaC−19-100) was labeled with a coumarin donor probe, CPI, while a second protein was labeled with a fluorescein acceptor probe, FITC. The donor probe is one whose emission spectrum overlaps the absorption spectrum of the acceptor probe. This property allows nonradiative transfer of donor excitation energy to the acceptor in a distance-sensitive manner. FRET efficiency depends on the degree of overlap of donor emission and acceptor absorption and their relative dipole orientations. These properties provide a characteristic Ro distance (giving 50% FRET efficiency) for different donor-acceptor pairs (19). More importantly, for distances less or greater than Ro, energy transfer efficiency changes by an inverse sixth power relationship, making FRET extremely useful for monitoring the approach of one macromolecule to another (19). The coumarin protein used here serves as a donor for pairing with fluorescein-labeled Ipa protein acceptors (Ro, near 4.5 nm).
To determine FRET from CPI-I' to FITC-IpgC, a set concentration of CPI-I' was mixed with either a known concentration of FITC-IpgC (FDa) or the same concentration of unlabeled IpgC (Fd). A fluorescence emission spectrum was then collected on a FluoroMax spectrofluorometer (JY/Spex, Edison, N.J.) by using an excitation wavelength of 385 nm with emission scanned from 430 to 500 nm. FRET efficiency was then determined from the relationship E = 1 − (FDa/Fd). The advantage of using FRET analysis over fluorescence polarization in this case is that additional nonfluorescent proteins can be added to determine whether they are competitive for the observed binding event. In this case, nonfluorescent IpaB was used as a potential competitor for the association between region I' and IpgC. In other experiments, FRET was used to monitor the distance from an IAEDANS bound to Cys-13 of IpaC−19-363 or the N-terminal Cys of IpaCCys to FITC-IpgC.
Measurement of contact-mediated hemolysis.
Contact-mediated hemolysis was measured as described by Sansonetti (34). Briefly, S. flexneri were grown overnight on Trypticase soy agar-Congo red plates, and a single red colony was used to inoculate Trypticase soy broth. The bacteria were grown to mid-log phase, and the cells were collected by centrifugation and resuspended in phosphate-buffered saline (PBS) at 1/40 of the original volume. Sheep red blood cells were washed and resuspended in PBS at a concentration of 1 × 1010 cells/ml. Red blood cells and bacteria (50 μl of each) were added to wells of microtiter plates, and the plates were centrifuged at 2,200 × g for 15 min at 20°C. The plates were then incubated at 37°C for 2 h. The cells were resuspended by adding 90 μl of cold PBS, and the plates were centrifuged again at 2,200 × g at 15°C for 15 min. The supernatant fraction (100 μl) was transferred to a second microtiter plate, and the absorbance of the released hemoglobin was measured by absorbance by using a microtiter plate reader.
RESULTS
Identifying the minimum sequence needed at the IpaC N terminus for secretion.
Consistent with TTSS substrates from other systems (18), deletion of the first 20 amino acids of IpaC abolishes its ability to be secreted into S. flexneri culture supernatants (see Table 1). The absence of secretion is reflected in the loss of IpaC's ability to restore invasiveness to the ipaC mutant S. flexneri SF621 (Table 1). In contrast, deletion of amino acids 21 to 50 did not adversely affect IpaC secretion (data not shown), invasiveness, or contact hemolysis activity (Table 2). To determine the minimum number of amino acids needed for efficient IpaC secretion, different deletions were introduced that ended at amino acid 20 (Table 1). Deleting amino acids 18 to 20 (replacing TKQ with LE of the XhoI restriction site; see Materials and Methods) had only a minor effect on IpaC secretion, and deletion of residues 15 to 20 had even less effect on IpaC secretion. In contrast, deletion of amino acids 12 to 20 resulted in an 80% reduction in IpaC secretion from SF621 (Table 1), indicating that the information needed for IpaC secretion lies within the first 15 amino acids.
TABLE 2.
Effects of deletions near the IpaC N terminus on in vivo function
| IpaC derivative in pWPsf | Relative invasion (%)a | Contact hemolysis (%)b |
|---|---|---|
| None | 0 ± 0 | 8 ± 2 |
| ipaC | 100 ± 10 | 100 ± 5 |
| ipaCΔ21-50 | 100 ± 5 | 100 ± 10 |
| ipaCΔ21-55 | 5 ± 1 | 10 ± 2 |
| ipaCΔ63-100 | 5 ± 1 | 12 ± 6 |
| ipaCΔ81-100 | 25 ± 5 | 85 ± 5 |
| ipaCΔ92-100 | 75 ± 8 | 70 ± 10 |
Invasion of Henle 407 cell monolayers was tested by using a standard gentamicin protection assay, and the values shown are relative to that of SF621 expressing the native form of ipaC. Bacteria were added to monolayers at a multiplicity of infection of approximately 50.
The values shown for the contact hemolysis assay are the percent lysis of sheep red blood cells relative to that with the addition of water, which causes complete hemolysis.
Identification of amino acids important to IpaC secretion.
Recently, it was shown that replacing the secretion signal of YopE from Yersinia enterocolitica with an amphipathic peptide consisting of alternating Ser and Ile restored the protein's secretion function (20, 21). Those data suggest that there is a structural feature at the N terminus of YopE, rather than a specific amino acid sequence, that is needed to trigger type III secretion. However, when the first 20 amino acids of IpaC were replaced with a 15-amino-acid poly(I-S) sequence, secretion and invasion functions were not restored to IpaC (data not shown). Because the N terminus of IpaC, unlike that of YopE, contains several charged amino acids, the first three polar residues of IpaC were replaced with Ala (1MEIQN5 → 1MAIAA5). In this case, IpaC secretion was reduced 80%, which was accompanied by a nearly 80% decrease in invasion function (Table 1), showing that amino acids that disrupt the physical and/or chemical properties of the IpaC N terminus eliminate a feature necessary for IpaC secretion. Individual Ala replacements for these three polar residues had no negative effect on IpaC secretion (data not shown).
This laboratory was the first to purify recombinant IpaC for biochemical analysis (22, 31); however, it was originally cloned based on the sequence reported by Venkatesan (42), which encodes a protein that is 19 amino acids longer than native IpaC. Thus, the recombinant protein consisted of amino acids −19 to 363 (IpaC−19-363) based on the numbering system of Turbyfill (41). For expression in the nonpolar ipaC mutant S. flexneri SF621, ipaC−9-363 was inserted into the modified pUC18 plasmid pWPsf. The resulting plasmid construct (pWPsfc) also led to the addition of eight lacZ codons at the 5′ end of ipaC−9-363 (Table 1). The resulting IpaC derivative restored invasiveness and contact hemolysis activity (data not shown) to SF621, demonstrating that additions at the N terminus do not interfere with IpaC function. Introduction of ipaC−9-363 into the plasmid pWPsf4 eliminates the extra eight lacZ codons (Table 1). As expected, pWPsf4c encodes an IpaC−19-363 derivative that is also secreted and restores invasiveness to SF621. In contrast, deletion of amino acids −6 to 20 (pWPsf4cΔ-6-20) eliminates the putative secretion signal and abolishes IpaC secretion. However, when this ipaC sequence is subcloned into pWPsf (pWPsfcΔ-6-20), thereby introducing the eight LacZ residues at the IpaC N terminus, IpaC is once again secreted by SF621 and invasiveness is restored (Table 1). More intriguing is the fact that if the first six amino acids of pWPsf4cΔ-6-20 are changed from MLQKQF to MHQIQT, secretion is restored to 100% of that seen for native IpaC. As in Yersinia spp., none of these constructs allows identification of a consensus secretion sequence, suggesting that there is no consensus sequence for the secretion but that instead, a structural feature is responsible for signaling export. This concept is supported by four constructs in which the first 20 amino acids encoded by pWPsf4c' are replaced by the first 14 amino acids of IpaA, IpaB, IpaD, and IpgD. All four of these constructs encode an IpaC chimera that is efficiently secreted, with only the IpaB chimera being secreted to a lesser extent (Table 1).
Identification of an important functional region near the N terminus of IpaC.
Small deletions beyond amino acid 20 do not adversely affect IpaC's ability to restore invasiveness to S. flexneri SF621 (Table 2). However, when these deletions extend beyond amino acid 50, they immediately eliminate IpaC invasion and contact hemolysis functions in vivo (Table 2) without affecting secretion (data not shown). From these data, a region essential to IpaC activity is believed to have a sharp N-terminal boundary between amino acids 50 and 55. To determine the C-terminal boundary of this essential internal region, deletions were introduced at amino acid 100 that extended upstream. When amino acids 92 to 100 were deleted from IpaC, there was a relatively minor negative effect on function (Table 2). Deletion of amino acids 81 to 100 still did not completely eliminate IpaC function, but IpaCΔ63-100 was almost completely inactive in vivo for both invasion and contact hemolysis functions, yet it was still efficiently secreted (data not shown). These findings indicate that the region between amino acids 50 and 80 is essential for IpaC function in S. flexneri. The location of this region is consistent with that of the chaperone-binding domain described by Page et al. (28). It may also be required for association with IpaB within the host cell cytoplasmic membrane.
IpgC binds near the N terminus of IpaC.
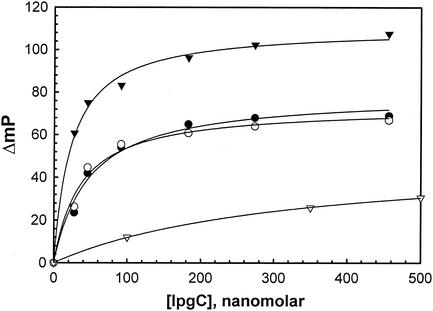
IpgC is a Shigella cytoplasmic chaperone that binds to both IpaB and IpaC to prevent their premature association in the bacterial cytoplasm (23) and to queue them for secretion. By using fluorescence polarization, the interaction between IpgC and IpaC−19-363 that is site-specifically labeled with thiol-reactive FM at Cys−13 can be demonstrated in vitro, with half-maximal binding occurring near 45 nM (Fig. 1). This binding can be localized to the N terminus of IpaC, since this interaction still occurs for region I' (IpaC−19-100) labeled with FITC, with a dissociation constant of about 36 nM (Fig. 1). Furthermore, it seems likely that the region of IpaC involved in IpgC binding requires the region between amino acids 50 and 80, based on deletion mutagenesis data (Table 2) and the fact that a truncated form of IpaC which consists of amino acids 50 to 363 possesses a MetCys pair at its immediate N terminus (IpaCCys) and is labeled with FM binds very tightly to IpgC (Fig. 1). Moreover, the magnitude of the polarization changes presented in Fig. 1 are consistent with IpgC binding relatively close to the site-specific FM probe on IpaCCys. In contrast, IpaCΔ−9-62 does not appear to bind IpgC nearly as well (half-maximal binding at 320 nM IpgC) as those IpaC derivatives that possess an intact region between amino acids 50 and 80 (Fig. 1).
FIG. 1.
IpgC associates with a region near the N terminus of IpaC in vitro. Fluorescence polarization was used to monitor the interaction between fluorescein-labeled IpaC derviatives and nonfluorescent IpgC. The increase in polarization caused by the addition of increasing concentrations of IpgC is shown for FM-labeled IpaC−19-363 (•), FITC-labeled region I' (IpaC−19-100) (○), FM-labeled IpaCCys (▴), and FITC-labeled IpaCΔ1-62 (▵). IpaCCys is composed of IpaC50-363 with a MetCys pair at the immediate N terminus. The fluorescent proteins were used at approximately 20 nM.
In a related experiment, IpaC−19-363 was labeled with IAEDANS, and the effect of IpgC binding was compared to that of IAEDANS-labeled IpaCCys. IAEDANS fluorescence is very sensitive to changes in the probe's microenvironment (26). Therefore, the approach of a protein to a site labeled with IAEDANS should cause a change in the fluorescence properties of this probe. When the emission maximum and intensity of IAEDANS-labeled IpaC−19-363 and IpaCCys are compared, the latter undergoes greater change, indicating that IpgC influences the IAEDANS microenvironment much more for IpaCCys (Table 3). This is consistent with the observation that IAEDANS linked to IpaCCys is more efficient at transferring its excitation energy to FITC-IpgC than is IAEDANS linked to position −13 of IpaC−19-363 (Table 3).
TABLE 3.
Effect of IpgC binding on the fluorescence of IAEDANS-labeled IpaC
| IAEDANS-labeled proteina | IpgC additionb | Intensityc | Emmax (nm)d | FRET to FITC-IpgC (%)e |
|---|---|---|---|---|
| IpaC−19-363 | − | 1.00 | 485 | n.a. |
| + | 1.23 | 482 | 20 | |
| IpaCCys | − | 1.00 | 480 | n.a. |
| + | 1.42 | 475 | 26 |
IpaCCys is composed of IpaC50-363 with a Met-Cys pair at the immediate N terminus.
+, nonfluorescent IpgC (100 nM) was added to approximately 50 nM IAEDANS-labeled protein in the absence of any other proteins; −, IpgC was not added.
Fluorescence intensity is shown relative to that of IAEDANS-labeled protein in the absence of added IpgC. The intensity of labeled IpaCCys relative to that of IpaC−19-363 was 0.87, indicating that the starting microenvironments of the two probes are different.
The wavelength at which the peak of fluorescence occurred was determined in the presence and absence of nonfluorescent IpaC.
FRET efficiency was determined by using IAEDANS-labeled protein with nonfluorescent IpgC to determine Fd and by using IAEDANS-labeled protein with FITC-IpgC (in the absence of additional nonfluorescent IpgC) to determine Fda. n.a., not applicable.
An N-terminal fragment from IpaC associates with IpaB.
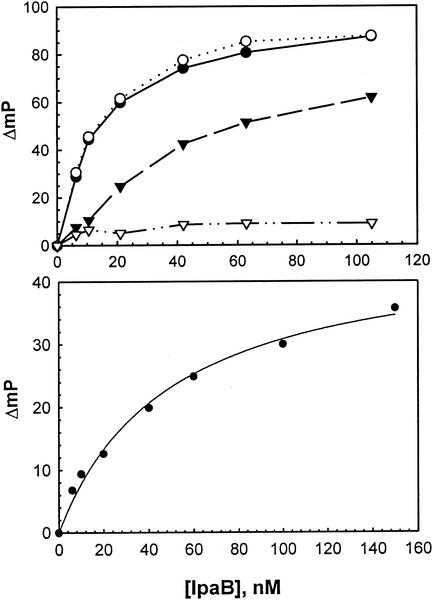
It was previously shown that IpaB and IpaC interact in vitro (10), and the ability of IpaC region I' to interact with IpaB by fluorescence polarization was analyzed. Region I' (IpaC−19-100) was labeled with FITC (FITC-I') so that its interaction with IpaB could be monitored in vitro and compared to the ability of FITC-labeled IpaC−19-363, IpaCΔ21-100, and IpaC171-363 to interact with nonfluorescent IpaB. As shown in Fig. 2A, FITC-I' and FITC-IpaC−19-363 are bound by IpaB, with half-maximal polarization increases at approximately 15 nM IpaB in each case. IpaCΔ21-100 also associates with IpaB, with the half-maximal polarization increase for this binding occurring at approximately 50 nM IpaB (Fig. 2A). However, IpaC171-363, which lacks both the IpgC binding domain and the central hydrophobic region, does not appear to associate with IpaB (Fig. 2A). These data show that while the entire N-terminal half of IpaC contributes to the formation of IpaB/IpaC complexes, the N-terminal hydrophilic fragment contains a sequence that allows the tightest association between IpaC and IpaB. As expected, the IpgC-IpaB interaction could also be demonstrated in vitro by using FITC-labeled IpgC (Fig. 2B).
FIG. 2.
IpgC binds to both IpaC and IpaB in vitro. (A) IpaC and proteins derived from IpaC were purified and labeled with FITC for fluorescence polarization measurements. The increase in polarization caused by the addition of increasing concentrations of IpaB is shown for FITC-labeled IpaC−19-363 (○), region I' (IpaC−19-100) (•), IpaCΔ21-100 (▴), and IpaC171-363 (▵). The values shown are an average of three measurements of a single sample and are representative of multiple (three or more) independent experiments. (B) The increase in the polarization of FITC-labeled IpgC (20 nM) was monitored following the addition of increasing concentrations of nonfluorescent IpaB.
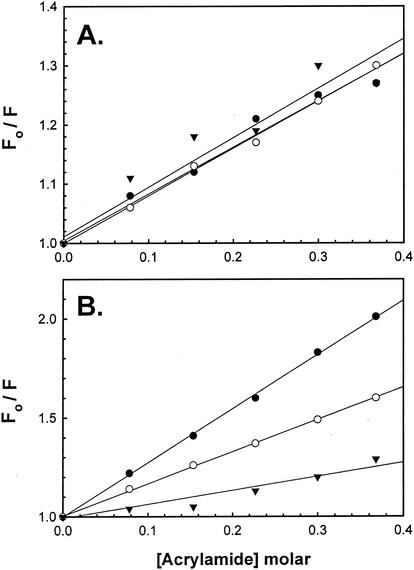
Unlike the random labeling that occurs with FITC, IpaC−19 −363 and IpaCCys can be site specifically labeled with FM. IpaC−19-363 possesses a single Cys residue at position −13, and IpaCCys lacks amino acids up to residue 50 but contains a MetCys pair at the immediate N terminus. Each protein can be labeled with thiol-reactive FM, and solute accessibility then can be estimated by using fluorescence quenching analysis with acrylamide as the quenching agent. When IpaB or IpgC is incubated with FM-IpaC−19-363, there is relatively little change in the steady-state Stern-Volmer quenching constant or Ksv, with all Ksv values being near 0.8 M−1 (Fig. 3A). These data indicate that the interaction of IpaB and IpgC with FM-IpaC−19-363 has little effect on acrylamide accessibility to the fluorescein probe. In contrast, IpaB binding to FM-IpaCCys resulted in a 40% decrease in acrylamide accessibility (Fig. 4B), with the Ksv decreasing from 2.73 M−1 to 1.63 M−1. Similarly, IpgC binding to FM-IpaCCys resulted in a 75% decrease in Ksv from 2.73 M−1 to 0.71 M−1 (Fig. 4B). These data indicate that, unlike with the fluorescein probe at position −13, the probe near amino acid 50 is more intimately influenced by IpaB and IpgC binding, again indicating that these proteins bind to a region of IpaC beginning at amino acid 50, with IpgC binding closer to amino acid 50 than IpaB.
FIG. 3.
Association of IpaB and IpgC to FM-IpaCCys results in protection of the probe from solute quenching. (A) The addition of increasing concentrations of acrylamide results in quenching of FM-IpaC−19-100 fluorescence (•). The degree of acrylamide quenching is not greatly affected by the addition of IpaB (○) or IpgC (▴). The fluorescent protein was used at 30 nM in each case, and the results are from a single representative experiment. Acrylamide quenching of FM-IpaCCys alone is shown by the closed circles, and quenching with IpgC is shown by the closed triangles. (B) The same experiment was carried out with FM-IpaCCys at approximately 30 nM. The data shown are from a single representative experiment. The average Ksv value in each case (n ≥ 3) is given in the text.
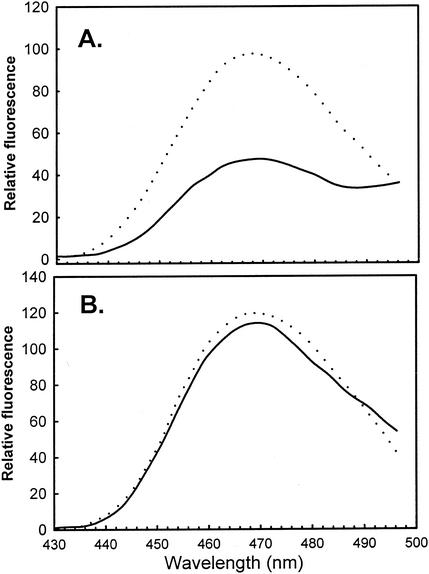
FIG. 4.
Region I' (IpaC−19-100) associates with IpgC and IpaB in a competitive manner. (A) The emission spectrum of 230 nM CPI-labeled region I' is shown in the presence of an equimolar amount of IpgC (dashed line) or FITC-labeled IpgC (solid line). The decrease in emission with FITC-IpgC is due to FRET of CPI excitation energy from the coumarin to the fluorescein moiety. (B) The same experiment was carried out with CPI-labeled region I' and IpgC (dashed line) and FITC-labeled IpgC (solid line), except that 250 nM IpaB is also present in the sample. CPI fluorescence was measured by using an excitation wavelength of 385 nM. The data are from a single representative experiment, which was performed three times.
IpgC and IpaB bind competitively to the N terminus of IpaC.
Region I' (IpaC−19-100) was next labeled with a coumarin isothiocyanate derivative, CPI, to give CPI-I', which was incubated with purified recombinant IpgC labeled with FITC (FITC-IpgC). Their association was then detected by using FRET (see Materials and Methods), as shown in Fig. 4. The CPI donor probe used here paired with FITC is useful for monitoring distances of less than 50 Å, meaning that FRET is detected only if CPI-I' interacts intimately with FITC-IpgC. As shown in Fig. 4 and Table 4, mixing CPI-I' with a one- or tenfold concentration of FITC-IpgC results in more than 50% energy transfer. The fact that nearly maximum FRET is achieved at equimolar concentrations of CPI-I' and FITC-IpgC suggests a strong association. The observation of 50% energy transfer is particularly significant, since IpgC labels with FITC with a relatively low (∼30%) efficiency, which means that the acceptor probes on FITC-IpgC must be in close proximity to the donor probes to give rise to such high levels of FRET. When this FRET experiment is modified to include nonfluorescent IpaB (at a concentration equal to that of CPI-I'), the increase in the level of energy transfer to FITC-IpgC is almost completely eliminated (Fig. 4). The fact that the IpaC-IpaB interaction is stronger than the IpgC interaction with IpaB or IpaC indicates that the decrease in FRET is due to competition between IpaB and FITC-IpgC for association with CPI-I' and not to IpaB simply stripping the IpgC away from this IpaC fragment. This competition could be partially relieved with the addition of a tenfold greater concentration of FITC-IpgC (Table 4), showing that IpaB and IpgC are competitive for interactions with region I'.
TABLE 4.
Effect of IpaB on the interaction between CPI-I’ and FITC-IpgC
| FITC-IpgC concna | IpaB concn (nM) | FRET efficiency (%)b |
|---|---|---|
| 230 nM | 0 | 52 |
| 2.3 μM | 0 | 53 |
| 230 nM | 250 | 5 |
| 2.3 μM | 250 | 27 |
The concentration of CPI-I’ was 230 nM.
FRET efficiency is calculated by the equation FRET = 1 − (Fda/Fd) where Fda is CPI-I’ fluorescence with FITC-IpgC and Fd is CPI-I’ fluorescence with unlabeled IpgC.
The N terminus of IpaC can be replaced by the N terminus of Salmonella SipC.
SipC, the Salmonella homologue of IpaC, is efficiently secreted by S. flexneri but is incapable of replacing IpaC in restoring invasiveness to SF621 (27), and the portion of SipC corresponding to the IpaC region displays significant sequence divergence. Interestingly, polarization experiments show that FITC-labeled SipC binds IpaB with a half-maximal polarization change that is similar to that of IpaCΔ21-100 (Table 5), and this IpaC mutant is also incapable of restoring invasiveness to SF621. Furthermore, FITC-labeled IpaC binds to IpaB and Salmonella SipB with what appear to be similar affinities, and ipaC does restore invasiveness to both S. flexneri SF621 and the Salmonella sipC mutant SB220. Meanwhile, FITC-labeled SipC binds to SipB with an affinity that is similar to that seen for IpaC binding to either IpaB or SipB (Table 5). These data indicate that differences in IpaC and SipC function may stem from differences in the binding of the latter with IpaB. SipC also binds to FITC-IpgC, although the affinity appears to be less than that seen between FITC-IpaC and unlabeled IpgC (data not shown).
TABLE 5.
Strength of IpaC/SipC binding relative to IpaB/SipB binding
| Compound A: fluorescent protein | Compound B: nonfluorescent protein | Concn of compound B needed for half-maximal ΔmPa |
|---|---|---|
| FITC-IpaC | IpaB | 19.0 ± 5.1 |
| SipB | 23.1 ± 5.3 | |
| FITC-SipC | IpaB | 77.2 ± 15.6 |
| SipB | 15.5 ± 2.5 |
The values given are in millipolarization (mP) units. ΔmP is the change in polarization as a function of IpaB/SipB concentration. The fluorescent protein was used at 25 nM, indicating that the values given are not true dissociation constants.
We recently reported that the difference between IpaC and SipC was most likely related to effector functions located at the C terminus of IpaC, since both proteins associate with IpaB, SipB, and IpgC (27). However, sequence differences at the N terminus of these proteins and the observation that SipC binds with reduced affinity to IpaB (Table 5) and IpgC relative to IpaC suggest that functional differences associated with the N terminus of these proteins (chaperone binding and/or translocon formation) could account for the inability of sipC to restore invasiveness to S. flexneri SF621. To test this, IpaC-SipC chimeras were generated and their ability to restore invasiveness to SF621 was determined. In all cases, when N-terminal portions of IpaC are fused with C-terminal portions of SipC, no invasiveness is restored to SF621 (data not shown). In contrast, when the amino acids of SipC corresponding to amino acids −19 to 62 of IpaC are fused with amino acids 63 to 363 of IpaC, SF621 invasiveness is restored to 90% (±15%; n = 3). Interestingly, when larger regions of the SipC N terminus are used to replace the corresponding portion of IpaC (e.g., SipC residues 1 to 189 replacing residues −19 to 170 of IpaC), invasiveness is not restored to SF621 (data not shown). These data suggest that in Shigella, IpaC and SipC function may differ as a result of both effector function and translocation; however, at least part of the putative chaperone binding site, corresponding to IpaC amino acids 50 to 80, is interchangeable. This is consistent with previous findings that coexpression of sipC and its cognate Salmonella chaperone sicE in SF621 is still incapable of restoring invasiveness.
DISCUSSION
Diarrheal diseases are among the most important global public health problems, and S. flexneri is responsible for significant morbidity and mortality for young children in developing regions of the world (15). IpaC mediates S. flexneri entry into the cytoplasm of enterocytes, an essential step in the pathogenesis of shigellosis (39). Understanding the mechanism of S. flexneri invasion (both its uniqueness and its similarity to other TTSS substrates) requires answering detailed questions about the basic protein biochemistry of IpaC. Elucidation of the requirements for secretion located at the N terminus of IpaC is the first step toward finding a common denominator in the secretion mechanism of members of the Ipa protein family along with providing new information relevant to the export of TTSS substrates from diverse gram-negative bacteria that communicate with a wide variety of eukaryotic cells (18). A unique feature of IpaC and the related Salmonella protein SipC, however, is the fact that they are the only known TTSS substrates that simultaneously (i) are part of a conduit or translocon through the host cytoplasmic membrane used to transport other proteins into the host cell and (ii) possess effector activities that subvert the host cell actin cytoskeleton (5, 17, 18). In other systems, including that of Salmonella, additional specialized effector proteins (not part of the pore complex) must be injected directly into the host cytoplasm to subvert host signaling mechanisms (18).
IpaC secretion by S. flexneri requires amino acid sequences located at its immediate N terminus; however, no specific consensus sequence can be proposed. Instead, the accumulated data seem to indicate that there is a physical or structural determinant that directs effector protein export. This is in agreement with data on TTSS substrates in Yersinia, where a poly(I-S) sequence is sufficient for directing Yop secretion (20) and changing the overall hydrophobicity of the region can have an effect on secretion (21). Unlike in Yersinia, however, replacement of the IpaC secretion signal with poly(I-S) is not sufficient to direct its export from S. flexneri SF621. On the other hand, when the first five amino acids of IpaC are changed from highly polar residues to Ala, secretion is drastically reduced even though individual Ala substitutions have no effect. Furthermore, when the N-terminal MLQKQFC sequence of the IpaC derivative generated from pWPsf4cΔ6-20 was changed to MHQIQTC to place a charged residue at the second position, a hydrophobic residue at the fourth position, and a Thr in place of Phe, secretion is fully restored. As in Yersinia, no specific secretion signal can be identified for IpaC in Shigella, but physical and/or structural features appear to have a major role in targeting this protein for type III secretion. Further study of the IpaC secretion signal will require a more thorough analysis of the structural and biophysical nature of the peptide(s) present at the immediate N terminus.
Downstream from the IpaC secretion signal is a region required for IpaC's ability to direct invasion and contact hemolysis but which is not required for IpaC secretion via the Shigella TTSS apparatus. Amino acids 50 to 80 appear to contribute substantially to the tight association of IpaC to both the cytoplasmic chaperone IpgC and the other Shigella translocon component IpaB. By fluorescence polarization analysis, the probable dissociation constants for IpaB-IpaC, IpgC-IpaC, and IpgC-IpaB interactions are in the low nanomolar range, which is consistent with that observed for the SycE-YopE interaction in Yersinia (9); however, the location of the SycE binding region on YopE appears to be nearer to the N terminus, beginning at amino acid 15 (4), than is the IpgC binding region on IpaC. Page and colleagues assigned IpgC binding to amino acids 58 to 72 of IpaB and to amino acids 73 to 122 of IpaC by using yeast two-hybrid analysis (28). The latter IpgC binding site differs slightly from what is reported here; however, the different biophysical methods used here clearly indicate that IpgC occurs very near amino acid 50 of IpaC, with IpaB probably binding at sequences downstream from, but possibly overlapping, this position. Also, deletions that include amino acids 51 to 55 of IpaC abruptly eliminate IpaC invasion and hemolysis functions, suggesting that a region critical for IpaC activity butts up against amino acid 50.
Page also suggested that deletion of a segment of IpaC that includes the chaperone binding region (amino acids 56 to 143) may reduce its stability in S. flexneri but not in E. coli (28). We did not observe reduced levels of IpaC in the cytoplasm of SF621 for mutations within the region encompassed by amino acids 50 to 100; however, deletion of amino acids 41 to 100 and 21 to 100 did result in slightly less IpaC being detected by ELISA in S. flexneri cell extracts. This could be due to instability, but the data presented here don't appear to be consistent with chaperone binding being required for IpaC stability in the Shigella cytoplasm. In other work, we have found that deletion of an internal portion of IpaC that includes the central hydrophobic domain (amino acids 63 to 170) results in relatively low levels of the protein (IpaCΔ63-170) in the S. flexneri cytoplasm (unpublished observation) despite the fact that the protein is very efficiently secreted (30). In contrast, IpaCΔ63-170 is quite stable when made in E. coli, which is consistent with the data reported for the 56-143 IpaC deletion mutant by Page (28). In examining the protein generated in E. coli, we have found that removal of the IpaC hydrophobic region results in a significant loss of tertiary structure (unpublished data), which could provide a signal for the destruction of IpaC in the S. flexneri cytoplasm.
Interestingly, the interaction of an IpaC N-terminal fragment with IpaB and IpgC is competitive, which is consistent with the role of IpgC in preventing the premature association of IpaC and IpaB in the Shigella cytoplasm. Based on fluorescence emission and quenching data, it appears that IpgC binds near amino acid 50, which shields the fluorescent probe on FM-IpaCCys from quenching agents in the surrounding solvent, while IpaB may bind further downstream and is therefore less efficient at protecting FM-IpaCCys fluorescence from acrylamide quenching (Fig. 3). Since the binding regions on IpaC recognized by IpgC and IpaB appear to overlap but are not identical, simultaneous binding of the proteins is precluded without requiring identical binding sequences. Because IpgC appears to bind more closely to amino acid 50 than does IpaB and deletions that extend downstream of amino acid 50 prevent IpaC function in S. flexneri, chaperone binding appears to be required for proper IpaC function. The observation that SipC sequences up to the equivalent of IpaC residue 62 can replace the corresponding sequence in IpaC suggests that chaperone binding between Shigella and Salmonella is interchangeable; however, important sequences downstream of the chaperone binding site (required for proper translocon formation with IpaB) are not readily swapped without loss of function (data not shown).
This study represents the most detailed description of the functional importance of the N terminus of IpaC to date. An important aspect of the data presented here is the apparent close relationship between disruption of the chaperone and IpaB binding regions of IpaC and the loss of both invasion function and contact-mediated hemolysis activity. The inability to separate these activities by introducing mutations within the N-terminal quarter of IpaC suggests that these mutations cause defects in the presentation of the protein to target cells (whether enterocytes or erythrocytes) in the context of IpaB. The ability to lyse red blood cells is probably a function of IpaC and IpaB association with and disruption of phospholipid membranes. Such a function would not require specific interaction with host cell signaling molecules such as Cdc42. Improper translocation could result in incorrect placement of the protein in the host cell membrane in association with IpaB. This would prevent IpaC from carrying out its effector activity and prevent the IpaB/IpaC complex from disrupting the membrane as needed for contact-mediated hemolysis. Therefore, the N terminus of IpaC serves primarily in the proper secretion, transport, and presentation of the protein to target cells, while the IpaC effector activity resides within the C-terminal half of the protein (unpublished data).
Acknowledgments
We acknowledge important technical assistance from Kelly Flentie and Susan Burkit and useful discussions with Russ Middaugh, Lisa Kueltzo (University of Kansas), and Ariel Blocker (University of Oxford, United Kingdom).
This work was supported by an NIH grant (AI 034428) to W.D.P. and by PHS grant 1-P20-RR017708.
Editor: J. T. Barbieri
REFERENCES
- 1.Barbieri, J. T., M. J. Riese, and K. Aktories. 2002. Bacterial toxins that modify the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 18:315-344. [DOI] [PubMed] [Google Scholar]
- 2.Bennett, J. C., and C. Hughes. 2000. From flagellum assembly to virulence: the extended family of type III export chaperones. Trends Microbiol. 8:202-204. [DOI] [PubMed] [Google Scholar]
- 3.Bernardini, M. L., J. Mounier, H. d'Hauteville, M. Coquis-Rondon, and P. J. Sansonetti. 1989. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. USA 86:3867-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birtalan, S. C., R. M. Phillips, and P. Ghosh. 2002. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol. Cell 9:971-980. [DOI] [PubMed] [Google Scholar]
- 5.Blocker, A., P. Gounon, E. Larquet, K. Niebuhr, V. Cabiaux, C. Parsot, and P. Sansonetti. 1999. The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J. Cell Biol. 147:683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blocker, A., D. Holden, and G. Cornelis. 2000. Type III secretion systems: what is the translocator and what is translocated? Cell Microbiol. 2:387-390. [DOI] [PubMed] [Google Scholar]
- 7.Blocker, A., N. Jouihri, E. Larquet, P. Gounon, F. Ebel, C. Parsot, P. Sansonetti, and A. Allaoui. 2001. Structure and composition of the Shigella flexneri “needle complex,” a part of its type III secreton. Mol. Microbiol. 39:652-663. [DOI] [PubMed] [Google Scholar]
- 8.Buttner, D., and U. Bonas. 2002. Port of entry—the type III secretion translocon. Trends Microbiol. 10:186-192. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, L. W., and O. Schneewind. 1999. Yersinia enterocolitica type III secretion. On the role of SycE in targeting YopE into HeLa cells. J. Biol. Chem. 274:22102-22108. [DOI] [PubMed] [Google Scholar]
- 10.Davis, R., M. E. Marquart, D. Lucius, and W. D. Picking. 1998. Protein-protein interactions in the assembly of Shigella flexneri invasion plasmid antigens IpaB and IpaC into protein complexes. Biochim. Biophys. Acta 1429:45-56. [DOI] [PubMed] [Google Scholar]
- 11.De Geyter, C., B. Vogt, Z. Benjelloun-Touimi, P. J. Sansonetti, J. M. Ruysschaert, C. Parsot, and V. Cabiaux. 1997. Purification of IpaC, a protein involved in entry of Shigella flexneri into epithelial cells and characterization of its interaction with lipid membranes. FEBS Lett. 400:149-154. [DOI] [PubMed] [Google Scholar]
- 12.Eftink, M. R. 1991. Fluorescence quenching reactions: probing biological macromolecular structures. In T. G. Dewey (ed.), Biophysical and biochemical aspects of fluorescence spectroscopy. Plenum Press, New York, N.Y.
- 13.Ehrbar, K., S. Mirold, A. Friebel, S. Stender, and W. D. Hardt. 2002. Characterization of effector proteins translocated via the SPI1 type III secretion system of Salmonella typhimurium. Int. J. Med. Microbiol. 291:479-485. [DOI] [PubMed] [Google Scholar]
- 14.Fraylick, J. E., M. J. Riese, T. S. Vincent, J. T. Barbieri, and J. C. Olson. 2002. ADP-ribosylation and functional effects of Pseudomonas exoenzyme S on cellular RalA. Biochemistry 41:9680-9687. [DOI] [PubMed] [Google Scholar]
- 15.Hale, T. L. 1991. Genetic basis of virulence in Shigella species. Microbiol. Rev. 55:206-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayward, R. D., and V. Koronakis. 1999. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 18:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayward, R. D., and V. Koronakiss. 2002. Direct modulation of the host cell cytoskeleton by Salmonella actin-binding proteins. Trends Cell Biol. 12:15-20. [DOI] [PubMed] [Google Scholar]
- 18.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lakowicz, J. R. 1983. Principles of fluorescence spectroscopy. Plenum Press, New York, N.Y.
- 20.Lloyd, S. A., M. Norman, R. Rosqvist, and H. Wolf-Watz. 2001. Yersinia YopE is targeted for type III secretion by N-terminal, not mRNA, signals. Mol. Microbiol. 39:520-531. [DOI] [PubMed] [Google Scholar]
- 21.Lloyd, S. A., M. Sjostrom, S. Andersson, and H. Wolf-Watz. 2002. Molecular characterization of type III secretion signals via analysis of synthetic N-terminal amino acid sequences. Mol. Microbiol. 43:51-59. [DOI] [PubMed] [Google Scholar]
- 22.Marquart, M. E., W. L. Picking, and W. D. Picking. 1996. Soluble invasion plasmid antigen C (IpaC) from Shigella flexneri elicits epithelial cell responses related to pathogen invasion. Infect. Immun. 64:4182-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menard, R., P. Sansonetti, C. Parsot, and T. Vasselon. 1994. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell 79:515-525. [DOI] [PubMed] [Google Scholar]
- 24.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Namba, K., I. Yamashita, and F. Vonderviszt. 1989. Structure of the core and central channel of bacterial flagella. Nature 342:648-654. [DOI] [PubMed] [Google Scholar]
- 26.Odom, O. W., W. D. Picking, and B. Hardesty. 1990. Movement of tRNA but not the nascent peptide during peptide bond formation on ribosomes. Biochemistry 29:10734-10744. [DOI] [PubMed] [Google Scholar]
- 27.Osiecki, J. C., J. Barker, W. L. Picking, A. B. Serfis, E. Berring, S. Shah, A. Harrington, and W. D. Picking. 2001. IpaC from Shigella and SipC from Salmonella possess similar biochemical properties but are functionally distinct. Mol. Microbiol 42:469-481. [DOI] [PubMed] [Google Scholar]
- 28.Page, A. L., M. Fromont-Racine, P. Sansonetti, P. Legrain, and C. Parsot. 2001. Characterization of the interaction partners of secreted proteins and chaperones of Shigella flexneri. Mol. Microbiol. 42:1133-1145. [DOI] [PubMed] [Google Scholar]
- 29.Pallen, M. J., G. Dougan, and G. Frankel. 1997. Coiled-coil domains in proteins secreted by type III secretion systems. Mol. Microbiol. 25:423-425. [DOI] [PubMed] [Google Scholar]
- 30.Picking, W. L., L. Coye, J. C. Osiecki, A. Barnoski Serfis, E. Schaper, and W. D. Picking. 2001. Identification of functional regions within invasion plasmid antigen C (IpaC) of Shigella flexneri. Mol. Microbiol 39:100-111. [DOI] [PubMed] [Google Scholar]
- 31.Picking, W. L., J. A. Mertz, M. E. Marquart, and W. D. Picking. 1996. Cloning, expression, and affinity purification of recombinant Shigella flexneri invasion plasmid antigens IpaB and IpaC. Protein Expr. Purif. 8:401-408. [DOI] [PubMed] [Google Scholar]
- 32.Riese, M. J., A. Wittinghofer, and J. T. Barbieri. 2001. ADP ribosylation of Arg41 of Rap by ExoS inhibits the ability of Rap to interact with its guanine nucleotide exchange factor, C3G. Biochemistry 40:3289-3294. [DOI] [PubMed] [Google Scholar]
- 33.Rosqvist, R., C. Persson, S. Hakansson, R. Nordfeldt, and H. Wolf-Watz. 1995. Translocation of the Yersinia YopE and YopH virulence proteins into target cells is mediated by YopB and YopD. Contrib. Microbiol. Immunol. 13:230-234. [PubMed] [Google Scholar]
- 34.Sansonetti, P. J., A. Ryter, P. Clerc, A. T. Maurelli, and J. Mounier. 1986. Multiplication of Shigella flexneri within HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect. Immun 51:461-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sasakawa, C., K. Kamata, T. Sakai, S. Makino, M. Yamada, N. Okada, and M. Yoshikawa. 1988. Virulence-associated genetic regions comprising 31 kilobases of the 230-kilobase plasmid in Shigella flexneri 2a. J. Bacteriol. 170:2480-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stern, D., and M. Volmer. 1919. On the quenching-time of fluorescence. Phys. Z. 20:183-188. [Google Scholar]
- 37.Tardy, F., F. Homble, C. Neyt, R. Wattiez, G. R. Cornelis, J. M. Ruysschaert, and V. Cabiaux. 1999. Yersinia enterocolitica type III secretion-translocation system: channel formation by secreted Yops. EMBO J. 18:6793-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran, N., A. B. Serfis, J. C. Osiecki, W. L. Picking, L. Coye, R. Davis, and W. D. Picking. 2000. Interaction of Shigella flexneri IpaC with model membranes correlates with effects on cultured cells. Infect. Immun. 68:3710-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran Van Nhieu, G., R. Bourdet-Sicard, G. Dumenil, A. Blocker, and P. J. Sansonetti. 2000. Bacterial signals and cell responses during Shigella entry into epithelial cells. Cell. Microbiol. 2:187-193. [DOI] [PubMed] [Google Scholar]
- 40.Tran Van Nhieu, G., E. Caron, A. Hall, and P. J. Sansonetti. 1999. IpaC induces actin polymerization and filopodia formation during Shigella entry into epithelial cells. EMBO J. 18:3249-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turbyfill, K. R., S. W. Joseph, and E. V. Oaks. 1995. Recognition of three epitopic regions on invasion plasmid antigen C by immune sera of rhesus monkeys infected with Shigella flexneri 2a. Infect. Immun. 63:3927-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venkatesan, M. M., J. M. Buysse, and D. J. Kopecko. 1988. Characterization of invasion plasmid antigen genes (ipaBCD) from Shigella flexneri. Proc. Natl. Acad. Sci. USA 85:9317-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Von Pawel-Rammingen, U., M. V. Telepnev, G. Schmidt, K. Aktories, H. Wolf-Watz, and R. Rosqvist. 2000. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol. Microbiol. 36:737-748. [DOI] [PubMed] [Google Scholar]