Abstract
The gram-negative bacterium Escherichia coli is the leading cause of urinary tract infection. The interaction between type 1 piliated E. coli and bladder epithelial cells leads to the rapid production of inflammatory mediators, such as interleukin-6 (IL-6) and IL-8. Conflicting reports have been published in the literature regarding the mechanism by which uroepithelial cells are activated by type 1 piliated E. coli. In particular, the role of lipopolysaccharide (LPS) in these responses has been an area of significant debate. Much of the data arguing against LPS-mediated activation of bladder epithelial cells have come from studies using a renal epithelial cell line as an in vitro model of the urinary epithelium. In this report, we analyzed three bladder epithelial cell lines and demonstrated that they all respond to LPS. Furthermore, the LPS responsivity of the cell lines directly correlated with their ability to generate IL-6 after E. coli stimulation. The LPS receptor complex utilized by the bladder epithelial cell lines included CD14 and Toll-like receptors, and signaling involved the activation of NF-κB and p38 mitogen-activated protein kinase. Also, reverse transcription-PCR analysis demonstrated that bladder epithelial cells express CD14 mRNA. Thus, the molecular machinery utilized by bladder epithelial cells for the recognition of E. coli is very similar to that described for traditional innate immune cells, such as macrophages. In contrast, the A498 renal epithelial cell line did not express CD14, was hyporesponsive to LPS stimulation, and demonstrated poor IL-6 responses to E. coli.
In order for a pathogen to cause disease at a mucosal surface, it must be able to adhere to host epithelial cells, establishing a focus of infection. This places epithelial cells in a unique position at the forefront of innate host defense. In addition to functioning as a barrier to pathogen entry, epithelial cells also play an active role in host defense through the production of cytokines, chemokines, and antimicrobial peptides (28). It is likely that the early interaction between bacteria and epithelial cells is a critical determinant in the outcome of an infection. In spite of this fact, the receptors utilized by epithelial cells to recognize pathogens and to initiate antipathogen responses are not completely understood.
The well-established in vitro and in vivo models for the interaction between Escherichia coli and uroepithelial cells make this system ideal for the investigation of pathogen recognition by epithelial cells. The ability of uropathogenic E. coli (UPEC) to establish infection in the urinary tract depends on its ability to express surface-adhesive organelles that facilitate colonization of the uroepithelium. P pili are produced by pyelonephritic strains of E. coli and mediate binding to globoseries glycolipids that predominate in the kidney (22). Consequently, P pili have been shown to be critical in the ability of E. coli to cause pyelonephritis (42). Type 1 pili bind to mannose-containing glycoproteins present on the bladder epithelial surface and are critical in the establishment of cystitis (31, 36). Type 1 pili are under an on-off phase variation control (6), and colonization of the bladder selects for type 1 piliated phase variants (11, 23).
Despite the distinct receptor specificities of P and type 1 pili, E. coli strains expressing either of these adhesive organelles have been demonstrated to augment bladder and kidney epithelial cell cytokine production compared to isogenic nonpiliated strains (14, 44). However, bacterial attachment mediated by these different adhesive pili results in the activation of distinct signaling pathways (16). The binding of P-piliated E. coli to globoside receptors present on kidney epithelial cells appears to activate interleukin-6 (IL-6) and IL-8 production via a predominantly lipopolysaccharide (LPS)-independent mechanism (7, 17). The role of Toll-like receptor 4 (TLR4) in this process is controversial. It has been reported that the LPS-independent activation of cytokine induction in kidney epithelial cells is TLR4 dependent (7, 17). In contrast, TLR4 has also been reported to be missing in kidney epithelial cells (3). The mechanism of activation of bladder epithelial cells by type 1 piliated E. coli is equally confusing. Several reports have argued that type 1 pili directly activate uroepithelial cytokine production and that LPS plays only a minor role (15, 47). These conclusions were based in large part on studies with A498 kidney cells. However, studies with bladder epithelial cells demonstrated that LPS is the primary bacterial factor activating cytokine production and that the role of type 1 pili is to augment the presentation of LPS to the LPS receptor complex on the bladder epithelial cells (3, 44). Thus, the discrepancies regarding the roles of type 1 pili and LPS in bladder cell activation may be explained by differences in the cell lines utilized for analysis.
LPS is the predominant component of the outer membrane of gram-negative bacteria, and its recognition by host cells requires an array of proteins. LPS-binding protein (LBP) and soluble CD14 (sCD14) are present in the serum and facilitate the transfer of LPS to membrane-bound CD14 (mCD14), a glycosylphosphatidylinositol-linked receptor on the surfaces of some host cells (24). It is thought that mCD14 subsequently interacts with TLR4, the signaling component of the LPS receptor (4). A secreted molecule known as MD-2 physically interacts with the extracellular domain of TLR4 and significantly enhances host cell responses to LPS (45, 51). TLR2 has also been reported to interact with certain lipid A structures, as well as with the lipoproteins that are intimately associated with LPS (18, 19). In host cells lacking expression of mCD14, sCD14 can partially compensate for the absence of this receptor under some circumstances (13, 32). Investigators using A498 cells as a model have reported that uroepithelial cells are CD14 negative (3, 15) and thus hyporesponsive to LPS stimulation.
The host signaling cascades that occur following TLR ligation involve a conserved cytoplasmic domain known as the Toll/IL-1 receptor (TIR) domain (38). The TIR domain of TLRs interacts with the adaptor proteins MyD88 and/or TIRAP, which subsequently recruits IL-1 receptor-associated kinases (IRAKs) to the receptor (21, 30, 50). Once an IRAK becomes activated, it dissociates from the receptor complex and leads to the activation of TRAF-6, an event that facilitates the downstream activation of I-κB kinase (IKK) and p38 mitogen-activated protein (MAP) kinase via a Tak-dependent mechanism (46, 49). The result of these signaling events is the nuclear translocation and transactivation of NF-κB, leading to the production of proinflammatory mediators and the up-regulation of costimulatory molecules (29). In addition, the activation of TLR2 has been shown to activate a phosphatidylinositol 3-kinase/Rac-1-dependent pathway, which leads to the transactivation of NF-κB by Akt (1).
In this study, we sought to definitively assess the role of LPS and type 1 pili in the activation of bladder epithelial cells and to clarify discrepancies in the literature by analyzing multiple cell lines of uroepithelial origin. In contrast to the A498 cell line, all of the bladder epithelial cell lines tested were responsive to LPS. Furthermore, CD14 is expressed by bladder epithelial cells and, together with TLRs, mediates cellular activation following challenge with type 1 piliated E. coli or LPS.
MATERIALS AND METHODS
Bacterial strains.
E. coli AAEC185 transformed with the entire type 1 pilus gene cluster (AAEC185/pSH2) or the type 1 pilus gene cluster minus the tip adhesin FimH (AAEC185/pUT2002) was grown static in Luria broth for 48 h at 37°C to induce the expression of type 1 pili. Before all experiments, expression of type 1 pili was confirmed by mannose-inhibitable yeast agglutination.
Cell culture.
5637 epithelial cells (derived from a bladder carcinoma; ATCC HTB-9), T24 epithelial cells (derived from a bladder carcinoma; ATCC HTB-4), J82 epithelial cells (derived from a bladder carcinoma; ATCC HTB-1), and A498 epithelial cells (derived from a kidney carcinoma; ATCC CRL-7908) were cultured in RPMI medium (Gibco BRL, Carlsbad, Calif.) plus 10% fetal bovine serum (FBS; Sigma, St. Louis, Mo.) at 37°C in a water-saturated atmosphere containing 95% air and 5% CO2. THP-1 monocyte/macrophage cells (derived from a monocytic leukemia; ATCC TIB-202) were cultured in RPMI medium containing 10% FBS, 1.5 g of sodium bicarbonate/liter, 4.5 g of glucose/liter, 1 mM sodium pyruvate, and 0.05 mM 2-mercaptoethanol.
IL-6 stimulation assay.
5637, T24, J82, and A498 cells were seeded into 24-well plates at 0.3 × 105 to 1 × 105 cells per well and grown to confluence over 2 days. THP-1 cells were seeded into 24-well plates at 5 × 105 cells per well in a medium containing 50 mM phorbol 12-myristate 13-acetate (PMA) and allowed to differentiate for 2 days. Forty-eight-hour bacterial cultures were pelleted by centrifugation, resuspended in phosphate-buffered saline (PBS), and diluted to a concentration of 108 CFU/ml (except for the dose curve experiments, where multiple bacterial stock dilutions were generated). Ten microliters of the bacterial suspension was added to appropriate wells containing 1 ml of fresh medium to achieve a final bacterial concentration of 106 CFU/ml. Titers of bacterial suspensions were determined at the outset of each experiment to verify the number of live bacteria added to the wells. The bacteria were spun onto the cells by low-speed centrifugation and incubated at 37°C for 6 h. After the incubation, the supernatants were collected, cell debris and bacteria were removed by centrifugation, and samples were frozen at −80°C until they were assayed by using a human IL-6 sandwich enzyme-linked immunosorbent assay (R & D Systems, Minneapolis, Minn.). A similar protocol was used for determining the effects of IL-1α, tumor necrosis factor alpha (TNF-α) (R & D Systems), LPS (Sigma), and the bacterial lipoprotein Pam3-Cys-Ser-Lys4-OH (Boehringer Mannheim, Indianapolis, Ind.) on cellular activation.
Signaling inhibition studies.
5637 cells were preincubated with 10 μM MG-132 or 5 μM SB203580 (both from Calbiochem, San Diego, Calif.) for 1 h, after which the cells were either left unstimulated or treated with the indicated stimulants. T24 cells were preincubated with 20 μM MG-132 or 50 μM SB203580 for 1 h, after which the cells were either left unstimulated or treated with the indicated stimulants. Supernatants were analyzed as described above.
Immunohistochemistry.
T24 cells were plated in 24-well plates on coverslips at 0.3 × 104 to 1 × 104 cells per well and were grown over 2 days. This lower cell density was utilized in order to better evaluate individual cells. After stimulation, cells were fixed with 3.7% paraformaldehyde-0.2% Triton X-100 in PBS for 10 min. After three 5-minute washes in PBS, coverslips were blocked with 1% bovine serum albumin and 0.2% milk in PBS (PBS-BB) for 15 min. Coverslips were then stained with 1 μg of anti-NF-κB p65 (BD Pharmingen, San Diego, Calif.)/ml in PBS-BB overnight at 4°C. As a negative control, coverslips were stained without a primary antibody. After three 5-min PBS washes, coverslips were incubated with 3 μg of Cy3-conjugated F(ab′)2 donkey anti-mouse immunoglobulin G (IgG) (Jackson Immunoresearch Laboratories, West Grove, Pa.)/ml for 1 h at room temperature.
CD14 inhibition.
To block CD14-mediated responses, cells were preincubated with My4 (Beckman-Coulter, Fullerton, Calif.), a monoclonal anti-CD14 antibody, at 5 μg/ml or with the same concentration of a mouse IgG2b isotype control (Sigma) for 30 min at 37°C. The cells were subsequently either left unstimulated or treated with the indicated stimulants, and supernatants were analyzed as described above.
Flow cytometry.
Cell suspensions were prepared from confluent cells growing as monolayers in T25 flasks and detached from the surface by using 0.02% EDTA in PBS. Between 2.5 × 105 and 5 × 105 cells were stained with My4 or the IgG2b isotype control at 5 μg/ml. Subsequently, the samples were incubated with biotinylated goat anti-mouse IgG (1:100) and finally phycoerythrin (PE)-conjugated streptavidin (1:600) (both from BD Pharmingen). Cells were analyzed by using a FACScalibur flow cytometer, and 12,000 live cells from each sample were collected. Data were analyzed by using Cell Quest software. The live cell population was defined by using propidium iodide staining.
CD14 mRNA reverse transcription-PCR (RT-PCR).
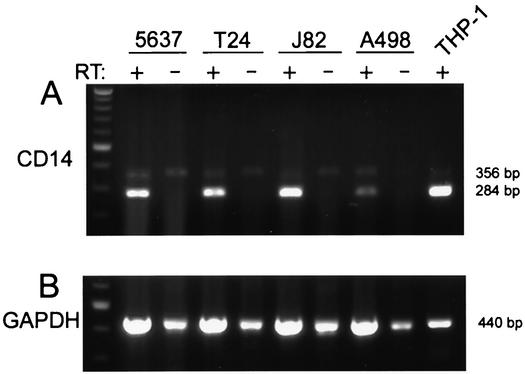
Following the protocol of Funda et al. (8), total RNA was isolated from cultured cells with TRIzol (Gibco BRL) according to the package protocol. No additional efforts were made to remove genomic DNA. First-strand cDNA was prepared from 4 μl of total RNA product by using 200 U of Superscript II reverse transcriptase (Gibco BRL) in a reaction mixture containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 1.5 μM oligo(dT)12-18 primers, and 0.125 mM each deoxynucleotide triphosphate (dNTP) at 42°C for 50 min, then 70°C for 15 min. Three microliters of the cDNA product was subjected to PCR amplification with 2.5 U of Taq polymerase (Invitrogen, Carlsbad, Calif.) in a solution containing 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 2 mM MgCl2, 0.2 mM each dNTP, and 0.8 mM each primer. Cycling parameters were as follows: 30 cycles of 94°C for 45 s, 57°C for 45 s, and 72°C for 2 min, followed by a 10-min final extension at 72°C. Primers for human CD14 (Invitrogen) were 5′-GCTGTGTAGGAAAGAAGCTA-3′ (sense) and 5′-TTTAGAAACGGCTCTAGGTTG-3′ (antisense). Since these primers span the single intron of the CD14 gene, expected product sizes are 356 bp from genomic DNA and 284 bp from mRNA (8). Primers for amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Invitrogen) were 5′-TCATCTCTGCCCCCTCTGCT-3′ (sense) and 5′-CGACGCCTGCTTCACCACCT-3′ (antisense), with an expected product size of 440 bp (from either mRNA or genomic DNA). PCR products were run on 1.5% agarose gels stained with ethidium bromide and viewed with a UV transilluminator.
LPS tolerance.
LPS tolerance was induced in 5637 and T24 bladder epithelial cells by incubating the cells with LPS from E. coli O55:B5 (Sigma) at 1 μg/ml for 16 to 20 h at 37°C. Following the incubation, cells were washed with PBS (plus Mg2+ and Ca2+), and fresh medium was added to the cells. Cells were subsequently either left unstimulated or treated with the indicated stimulants, and supernatants were analyzed as described above.
RESULTS
LPS responsiveness is common to bladder epithelial cell lines.
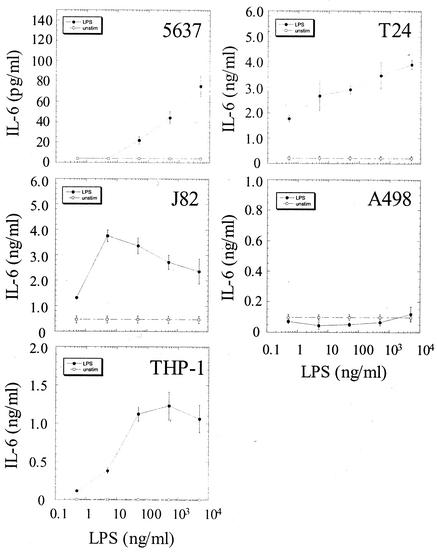
Several studies using the A498 epithelial cell line as a model system have argued that the uroepithelium is not responsive to LPS(9, 15). To test this hypothesis, multiple uroepithelial cell lines, including 5637 cells (bladder), T24 cells (bladder), J82 cells (bladder), A498 cells (kidney), and THP-1 cells (macrophage/monocyte, differentiated with PMA), were stimulated with LPS in log increments from 0.5 ng/ml to 5 μg/ml. All of the bladder epithelial cell lines were capable of responding to LPS, but the 5637 cells were significantly less responsive than the other two bladder epithelial cell lines (Fig. 1). Both T24 and J82 bladder cells responded to LPS at concentrations as low as 0.5 ng of LPS/ml, with maximal responses occurring at 5 μg/ml and 5 ng/ml, respectively. The threshold of activation for 5637 cells occurred at an LPS concentration between 5 and 50 ng/ml. A498 cells, which have been shown to respond poorly to soluble LPS, failed to produce IL-6 even when stimulated with the highest concentration of LPS (3, 16). PMA-differentiated THP-1 cells responded to LPS concentrations as low as 0.5 ng/ml and reached their maximal response at 50 ng/ml. Thus, the bladder epithelial cell lines appear to be uniformly LPS responsive, in contrast to the A498 cell line.
FIG. 1.
Bladder epithelial cells uniformly respond to LPS. 5637 (bladder), T24 (bladder), J82 (bladder), A498 (kidney), and THP-1 (monocyte/macrophage) cells were stimulated with log increments of LPS between 0.5 ng/ml and 5 μg/ml. Supernatant IL-6 concentrations were determined 6 h after the addition of LPS. Experiments were performed at least three times in triplicate.
LPS responsiveness is correlated with cellular responses to E. coli.
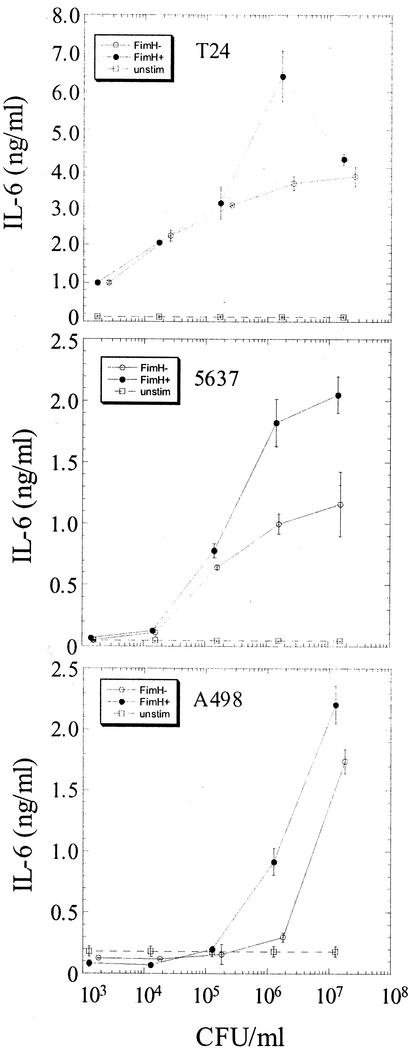
The type 1 mannose-binding adhesin, FimH, is located in specialized tip fibrillae found at the distal ends of pili (27). The nonpiliated K-12 E. coli strain AAEC185 was transformed with a plasmid encoding the type 1 pilus gene cluster (pSH2) or with a fimH deletion variant of the type 1 plasmid (pUT2002). To determine the relationship between responses to purified LPS and to E. coli, T24, 5637, and A498 cells were stimulated with FimH+ or FimH− E. coli at doses of bacteria ranging from 103 to 107 CFU/ml in log increments. T24 cells were activated at doses of E. coli as low as 103 CFU/ml, 5637 cells at a dose of 105 CFU/ml, and A498 cells at a dose of 106 CFU/ml (Fig. 2). At the lower doses of bacteria, FimH+ and FimH− E. coli strains elicited similar levels of IL-6 production from either T24 or 5637 cells; however, at approximately 106 CFU/ml, FimH+ E. coli stimulated significantly more cytokine than did its isogenic FimH− counterpart (Fig. 2). In A498 cells, FimH+ E. coli stimulated substantially more IL-6 at 106 CFU/ml than the FimH− strain or the nonpiliated strain; however, these differences were less prominent at bacterial doses of 107 CFU/ml (Fig. 2 and data not shown). IL-6 production was decreased in all cell lines if more than 107 CFU of E. coli/ml was used to stimulate the cells (data not shown). Thus, the threshold of activation following E. coli challenge directly correlates with the LPS responsiveness of the cell type (Fig. 1 and 2). In contrast, cellular stimulation with FimH+ E. coli characteristically leads to an increase in IL-6 production at 106 CFU/ml irrespective of the baseline LPS responsiveness of the cell type. The observation that the expression of adhesive (FimH+) type 1 pili did not shift the threshold of activation for these uroepithelial cell lines is consistent with previous reports demonstrating that type 1 pilus-mediated invasion augments bladder epithelial responses at this bacterial dose (44). FimH+ E. coli binds and invades T24, 5637, and A498 cells to equivalent levels (data not shown).
FIG. 2.
Uroepithelial responses to E. coli directly correlate with LPS responsiveness. T24 (high LPS responder), 5637 (intermediate LPS responder) and A498 (low LPS responder) cells were infected with increasing concentrations of FimH+ (filled circles) or FimH− (open circles) E. coli. Open squares, constitutive IL-6 production. IL-6 levels in the supernatant were determined 6 h after infection. Experiments were performed at least three times in triplicate.
NF-κB and p38 MAP kinase activation is required for bladder epithelial responses to LPS and E. coli.
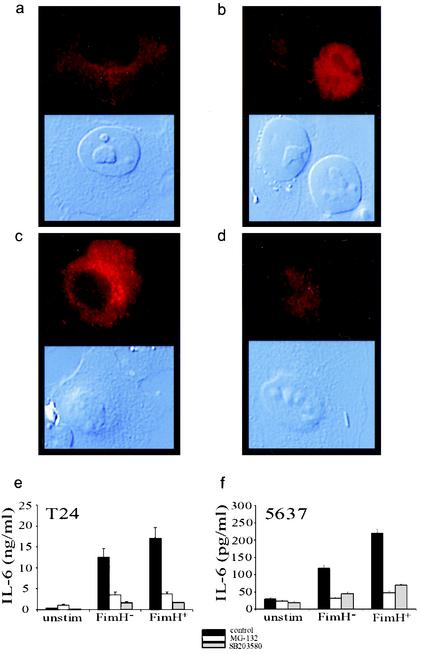
The production of IL-6 following LPS stimulation generally involves the activation of NF-κB and p38 MAP kinase; however, NF-κB-independent pathways have been described (5). Stimulation of T24 bladder cells with type 1 piliated E. coli led to a redistribution of NF-κB p65 from the cytoplasm to the nucleus, consistent with the activation of this transcription factor (Fig. 3a and b). Proteasome inhibitors have been shown to block NF-κB translocation to the nucleus by preventing the degradation of IκB (39), whereas p38 MAP kinase inhibitors appear to block the transactivation of NF-κB in the nucleus (46). Preincubation of T24 bladder cells with the proteasome inhibitor MG-132 blocked NF-κB p65 translocation and IL-6 production following challenge with FimH+ E. coli (Fig. 3c and e). Preincubation of bladder cells with the p38 MAP kinase inhibitor SB203580 blocked IL-6 production despite translocation of NF-κB p65 to the nucleus (Fig. 3d and e). Similar results were observed when 5637 cells infected with FimH+ E. coli were pretreated with these inhibitors (Fig. 3f). As expected, both inhibitors blocked bladder cell IL-6 responses to IL-1α and LPS (data not shown). These results argue that the nuclear translocation of NF-κB and the activation of p38 MAP kinase-dependent signal transduction cascades are required for the induction of bladder epithelial IL-6 following stimulation with inflammatory molecules and E. coli.
FIG. 3.
Activation of NF-κB and p38 MAP kinase is required for bladder epithelial cytokine responses to E. coli. (a through d) Immunohistochemical staining (in red) for the p65 subunit of NF-κB is shown in the upper panels, and differential interference microscopy is shown in the lower panels. (a) NF-κB in unstimulated T24 cells is localized to the cytoplasm. (b) After stimulation with type 1 piliated E. coli, NF-κB is translocated to the nuclei of T24 cells. AAEC185/pUT2002 (FimH−) also induces nuclear localization (data not shown). (c) Preincubation of T24 cells with MG-132 inhibits nuclear localization of NF-κB in response to E. coli. (d) Preincubation with the p38 MAP kinase inhibitor SB203580 does not inhibit NF-κB translocation. (e and f) T24 and 5637 cell responses to FimH+ or FimH− E. coli (solid bars) are blocked by preincubation with MG-132 (open bars) or SB203580 (shaded bars).
Bladder epithelial cells express CD14 on their surfaces.
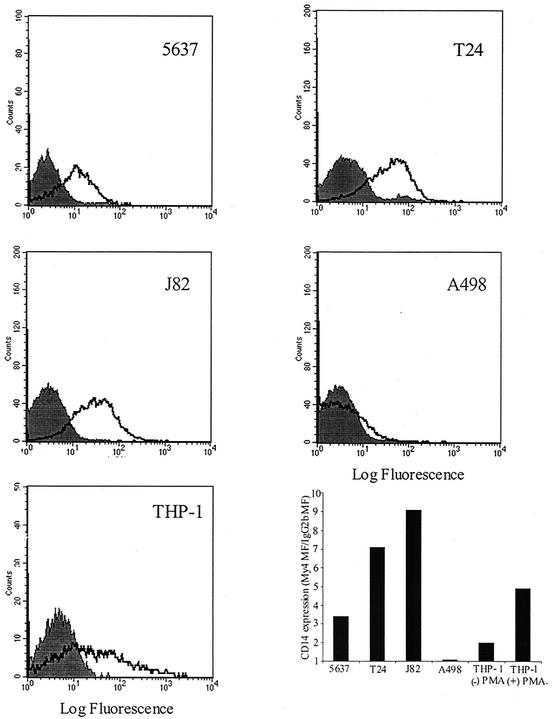
CD14 is a major component of the LPS receptor complex on macrophages; however, several reports have stated that this molecule is not involved in uroepithelial responses to E. coli (15, 47). Given the data implicating LPS recognition in bladder epithelial cytokine responses, uroepithelial expression of cell surface CD14 was evaluated. To assess CD14 expression, 5637, T24, J82, A498, and PMA-differentiated THP-1 cells were analyzed by flow cytometry using the well-characterized anti-CD14 monoclonal antibody My4. The results showed a clear My4-mediated fluorescence shift over the fluorescence level observed with the isotype control antibody in 5637, T24, and J82 cells (Fig. 4). In contrast, no significant fluorescence shift was detected with A498 cells. The positive control THP-1 cells also showed a significant induction of CD14 cell surface expression following PMA differentiation, as has been previously reported (10). The relative level of CD14 on the cell surface was estimated by determining the fold induction of My4-mediated fluorescence over the antibody isotype control fluorescence level (the geometric means of the fluorescence intensities were used for this purpose). Notably, cell lines of higher LPS responsivity had the most mCD14 by fluorescence-activated cell sorting (FACS) (J82, ratio 9.1; T24, 7.1; THP-1 [plus PMA], 4.9), the intermediate LPS responder cell line had lower levels of CD14 (5637, 3.4), and the LPS-hyporesponsive cell line had undetectable levels of CD14 (A498, 1.08) (Fig. 4). Thus, three independent bladder epithelial cell lines express mCD14, in contrast to previous suggestions that uroepithelial cells are CD14 negative (17). Bladder cells are capable of endogenous CD14 production; RT-PCR analysis of total RNA from 5637, T24, and J82 bladder cells demonstrated the presence of CD14 mRNA, by using differentiated THP-1 cells as a positive control (Fig. 5). CD14 transcript was also detectable in A498 cells.
FIG. 4.
Bladder epithelial cell lines express mCD14. Expression of CD14 was assessed by indirect immunofluorescence using the anti-CD14 monoclonal antibody My4 (open histograms) or the IgG2b isotype control antibody (shaded histograms). The fluorescence signal was amplified by using a biotinylated goat anti-mouse secondary antibody followed by PE-conjugated streptavidin. Relative CD14 expression is shown (lower right graph) as fold induction in mean fluorescence for My4-stained cells, by using the isotype control-stained cells as the baseline fluorescence value.
FIG. 5.
Uroepithelial cells contain CD14 transcript. (A) RT-PCR amplification of total RNA from 5637, T24, and J82 cells demonstrates CD14 products, from both mRNA (284 bp) and from genomic DNA (356 bp). Omission of reverse transcriptase (RT) in the reaction mixture results in the absence of the mRNA-related band. A498 cells also contain CD14 transcript. PMA-differentiated THP-1 cells were used as a positive control. (B) Amplification of the GAPDH sequence was utilized to generally relate the amounts of mRNA used in the CD14 RT-PCR experiments. The 440-bp band arises from both mRNA and genomic DNA. All amplifications, including reverse transcriptions, were performed at least three times.
CD14 is required for bladder epithelial cell responses to LPS and E. coli.
The presence of CD14 on the surfaces of bladder epithelial cells argues that this receptor likely plays a role in the recognition of E. coli and LPS by this cell type. To assess the contribution of CD14 to bladder cytokine responses, 5637, T24, or J82 cells were preincubated with My4 or the isotype control antibody at 5 μg/ml for 30 min. Subsequently, the cells were stimulated with increasing concentrations of LPS ranging from 0.5 ng/ml to 5 μg/ml. The response to LPS in each of these bladder cell lines was inhibited by My4, indicating an important role for CD14 in bladder epithelial cell recognition of LPS. To determine the role of CD14 in the cellular response to E. coli, 5637 cells were either left unstimulated or stimulated with either IL-1α, LPS (500 ng/ml), or FimH+ or FimH− E. coli. My4 had no effect on unstimulated or IL-1α-stimulated cells but significantly reduced IL-6 production induced by LPS (65% inhibition), AAEC185/pUT2002 (FimH−) (89% inhibition), or AAEC185/pSH2 (FimH+) (77% inhibition) (Fig. 6). My4 had no effect on binding or invasion by AAEC185/pSH2 (data not shown).
FIG. 6.
CD14 is required for optimal bladder epithelial cytokine responses to LPS and type 1 piliated E. coli. T24 (a) and J82 (b) bladder epithelial cells were preincubated for 30 min with an isotype control antibody (filled bars) or the anti-CD14 antibody My4 (open bars) at 5 μg/ml. IL-6 production was determined 6 h after stimulation with LPS at 0.5, 5, or 50 ng/ml. (c) 5637 cells require higher doses of LPS to generate IL-6 responses, but these responses are similarly inhibited by an anti-CD14 antibody. (d) Antibody-treated 5637 cells were challenged with 10 ng of IL-1α/ml, 500 ng of LPS/ml, or 106 CFU of AAEC185/pUT2002 (FimH−) or AAEC185/pSH2 (FimH+)/ml. The IL-1α response is independent of CD14 activity, but responses to bacteria were blocked by My4. Experiments were performed at least three times in triplicate.
Activation of bladder epithelial cells requires TLR signaling.
TLRs are pathogen pattern recognition receptors that recognize a variety of pathogen-associated molecular products (PAMPs), including LPS, and initiate the activation of NF-κB and p38 MAP kinase. Therefore, TLRs would be predicted to be important signaling receptors in bladder epithelial cells, which have been shown to express tlr4 and tlr2 mRNA (3, 44). Bäckhed et al. transiently transfected a dominant-negative TLR4 construct into T24 cells and observed a modest decrease in LPS responsiveness (2). Prestimulation of macrophages with LPS renders them refractory to subsequent LPS stimulation, a phenomenon termed LPS tolerance. Though the precise molecular mechanism of LPS tolerance is unclear, it has been shown that LPS-tolerant macrophages have decreased TLR4 surface expression and also fail to activate IRAKs following LPS restimulation (34, 37). Cross-tolerance between IL-1 and LPS has been demonstrated, adding further support to the idea that LPS tolerance is the consequence of disrupted TIR domain-mediated signaling (33, 34). To further address the role of TLR signaling in bladder epithelial responses, we investigated the ability of 5637 bladder cells to become tolerant to LPS stimulation.
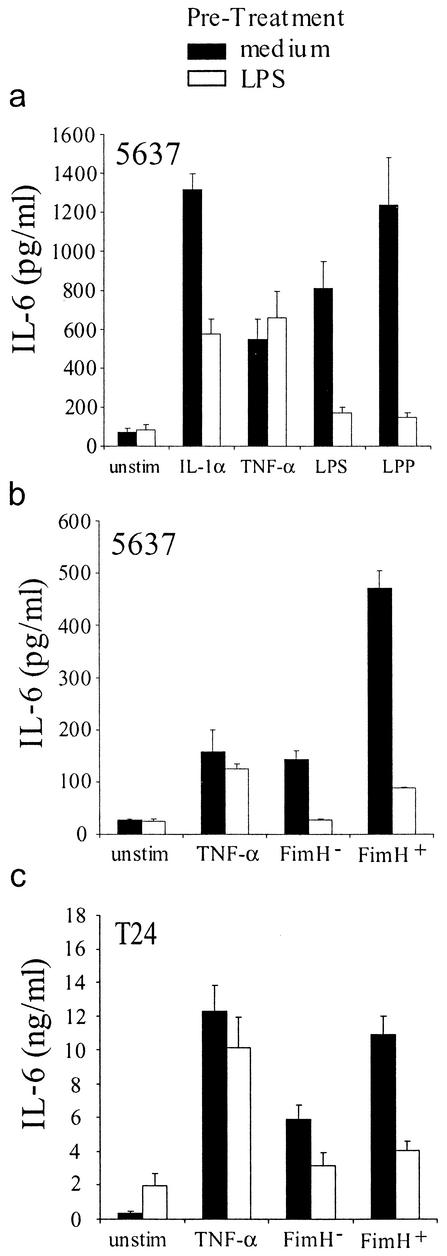
To induce tolerance, bladder epithelial cells were incubated with LPS for 16 to 20 h, after which time the cells were washed and either left unstimulated or stimulated with IL-1α, TNF-α, LPS, or bacterial lipoprotein (LPP). 5637 cells had defective responses to IL-1α (60% inhibition), LPS (87% inhibition), and LPP (94% inhibition) following tolerance induction (Fig. 7a). However, the responses to TNF-α were unaffected in tolerant 5637 cells. These results demonstrate that LPS tolerance can be induced in bladder epithelial cells and argue that LPS-tolerant 5637 cells have a defect in the MyD88/IRAK pathway, as this signaling pathway is shared by IL-1R and TLRs but not by TNF receptors.
FIG. 7.
Toll-like receptors mediate bladder epithelial recognition of LPS and type 1 piliated E. coli. (a) 5637 cells were prestimulated with medium alone (filled bars) or 1 μg of LPS/ml (open bars) and were subsequently challenged with 10 ng of IL-1α/ml, 10 ng of TNF-α/ml, 500 ng of LPS/ml, or 500 ng of LPP/ml. (b) LPS-tolerant or nontolerant 5637 cells were challenged with 10 ng of TNF-α/ml or 106 CFU of AAEC185/pUT2002 (FimH−) or AAEC185/pSH2 (FimH+)/ml. (c) LPS-tolerant or nontolerant T24 cells were challenged with 10 ng of TNF-α/ml or 103 CFU of AAEC185/pUT2002 or AAEC185/pSH2/ml. Experiments were performed at least three times in triplicate.
The effect of LPS tolerance and the accompanying TLR signaling defect on the responses of 5637 and T24 bladder cells to type 1 piliated E. coli was determined. Following induction of LPS tolerance, 5637 and T24 cells produced significantly lower levels of IL-6 in response to FimH+ or FimH− E. coli, arguing that TLR signaling is required for epithelial activation in response to infection (Fig. 7b and c). Binding and invasion by the FimH+ strain of E. coli was unaffected by the induction of LPS tolerance (data not shown).
DISCUSSION
The role of LPS in the activation of uroepithelial cells has been a controversial area of research. Several reports have claimed that uroepithelial cells are refractory to LPS stimulation and have argued that, instead, bacterial pili, such as the type 1 pilus, drive the induction of cytokines in these cells (47). In contrast, other studies have suggested an important role for LPS in the stimulation of bladder epithelial cells by type 1 piliated E. coli (3, 44). The data arguing against a role for LPS in uroepithelial cell activation have come largely from investigations utilizing the A498 kidney epithelial cell line as a model of the uroepithelium (15). To clarify the controversy over the role of LPS in bladder epithelial cell activation and to assess the molecular basis of bladder cell responses, we compared three bladder epithelial cell lines to the A498 cell line with respect to their responses to LPS and type 1 piliated E. coli.
In this study, 5637 (bladder), T24 (bladder) and J82 (bladder) cells were all shown to respond to LPS, whereas A498 cells (kidney) failed to respond to even the maximal dose of LPS tested. The ability to recognize LPS directly correlated with the production of IL-6 following challenge with both FimH+ and FimH− E. coli. Specifically, the threshold of activation for the LPS-hyporesponsive A498 cells by E. coli was 1,000 times and 10 times higher than it was for T24 and 5637 bladder cells, respectively. This is consistent with previous reports arguing that T24 and 5637 cells recognize and respond to LPS on the surfaces of type 1 piliated E. coli cells (3, 44). At a stimulatory dose of 106 CFU/ml, FimH-expressing E. coli enhanced IL-6 production from the uroepithelial cell lines over production with the isogenic FimH− mutant strain. This augmentation has been shown previously to be a consequence of type 1 pilus-mediated bacterial invasion (44). However, FimH− bacteria still activate a significant IL-6 response in 5637, T24, and A498 cells, indicating that functional (adhesive) type 1 pili are not required to induce such a response. It is not known which bacterial product(s) is recognized by A498 cells; however, the present data would suggest that LPS may not mediate the activation of these cells (7). It is possible that the lack of a functional LPS receptor complex in A498 kidney cells unmasks the stimulatory capacity of other bacterial molecules, as has been demonstrated using a lipid A mutant strain of Neisseria meningitidis (25, 41).
The present data support the model that bladder epithelial cell lines are LPS responsive and that the ability to recognize LPS is a critical event in the induction of innate responses against FimH+ and FimH− E. coli. In macrophages, the primary host cell receptor complex for LPS consists of mCD14 and TLR4/MD-2, which lead to the activation of p38 MAP kinase and the nuclear translocation of the transcription factor NF-κB (35, 38). It has been proposed that epithelial cells recognize pathogens through pathways that are fundamentally different from those in macrophages (9, 40). Moreover, if one were to assume (based on examination of the A498 cells alone) that uroepithelial cells do not express mCD14 or utilize sCD14 for anti-E. coli responses, then an alternate pathway for LPS recognition would have to exist (9, 17, 47).
To define the LPS receptor complex utilized by bladder epithelial cells, the signal transduction cascades leading to IL-6 production were investigated. Stimulation of bladder epithelial cells with E. coli led to the nuclear translocation of NF-κB p65. In addition, preincubation of bladder epithelial cells with the proteasome inhibitor MG-132 (which blocks NF-κB translocation) or with the p38 MAP kinase inhibitor SB203580 completely blocked the induction of IL-6 following bacterial challenge. These findings demonstrate a role for these signal transduction cascades in the activation of bladder epithelial cells by LPS on type 1 piliated E. coli.
To further characterize the receptors involved in the generation of LPS-mediated responses, the expression and role of CD14 were evaluated. The responses of 5637, T24, and J82 bladder epithelial cell lines to LPS were inhibitable with the monoclonal anti-CD14 blocking antibody My4, demonstrating that CD14 is a component of the LPS receptor complex on all of these bladder cell lines. It was also shown that bladder epithelial responses to FimH+ and FimH− E. coli require CD14. Cell surface expression of CD14 was shown by using flow cytometry with an anti-CD14 antibody for 5637, T24, and J82 bladder cells; however, consistent with previous results, A498 cells did not express CD14 (17). RT-PCR analysis confirmed the presence of CD14 mRNA in all three bladder epithelial cell lines. In contrast, Bäckhed et al. were unable to demonstrate amplification of CD14 mRNA from T24 cells (2). The reason for this discrepancy is unclear, but it may result from growth conditions or cell passage number, as CD14 expression on human nonmyeloid cells in vitro has been shown to decrease with time in culture (26). Since our functional experiments were performed in the presence of serum, a role for sCD14 in contributing to the activation of these mCD14-expressing cells cannot be excluded, although no CD14 was detected on the surfaces of A498 kidney cells. In addition, our bladder cell cultures for flow cytometry and RT-PCR contained no LPS, and sCD14 has been shown to require the presence of LPS in order to bind to the surface of CD14-negative nonmyeloid cells (48). Thus, our data argue that at least a portion of the surface CD14 on bladder cells is likely to be endogenously produced.
TLRs are pathogen pattern recognition receptors that play a critical role in innate immunity. TLR4 and TLR2 have been implicated in LPS recognition by macrophages, but other cell types also express these receptors, including 5637 and T24 bladder epithelial cells (3, 44). LPS tolerance is a phenomenon first described for macrophages where following a prestimulation with low to intermediate concentrations of LPS, cells become refractory to a second LPS stimulation. The current understanding of LPS tolerance would argue that the primary mechanism underlying the nonresponsiveness of tolerant cells is the disruption of signal transduction cascades initiated by TIR domain-containing receptors, potentially superimposed on a loss of TLR4 expression on the cell surface (37). Bladder epithelial cells that were prestimulated with LPS had significantly reduced responses to IL-1α, LPS, and LPP but had normal responses to TNF-α. These results demonstrate that LPS tolerance can be induced in uroepithelial cells. Furthermore, the observation that the response to TNF-α is unaffected in LPS-tolerant cells indicates that the cells are not globally suppressed but instead have a specific signaling defect that maps to TIR domain-containing receptors (such as TLRs) or early downstream signaling molecules. When LPS-tolerant bladder cells were stimulated with FimH+ or FimH− E. coli, IL-6 production by these cells was significantly inhibited. These data argue that bladder epithelial activation following bacterial stimulation is mediated by TLR activation, consistent with the findings of Bäckhed et al. (2).
Since signaling via multiple TLRs is likely rendered nonfunctional in the LPS-tolerant state, it is not possible to conclusively state which member(s) of this receptor family mediates E. coli recognition on bladder epithelial cells. However, the importance of LPS for bladder epithelial cytokine responses, the inability of C3H/HeJ mice (TLR4 mutant) to effectively clear E. coli-induced bladder infections, and the effect of the dominant-negative TLR4 mutant demonstrated by Bäckhed et al. (2) suggest that TLR4 is critical in E. coli recognition (20, 44). TLR2 has also been implicated in the recognition of LPS and its associated lipoproteins, suggesting a potential role for this receptor as well (19). To more closely mimic the LPS that would be encountered by host cells infected with whole bacteria, the LPS used in this study was not repurified to eliminate the trace lipoproteins that are intimately associated with commercially purified LPS preparations (18). Thus, it is possible that TLR2 activation is contributing to the LPS-driven stimulation we observed in bladder epithelial cells, particularly at higher doses of LPS. Ongoing studies aim to determine the respective roles of TLR4 and TLR2 in bladder epithelial responses to LPS and E. coli.
Epithelial cells are uniquely positioned to coordinate early host defenses against invading pathogens. However, in order to study the contribution of epithelial activation to the innate immune response, the molecules utilized by these cells to recognize pathogens must be defined. Significant progress has been made in understanding these details by using in vitro uroepithelial cell systems. However, this study further illustrates the fact that cell cultures are an imperfect representation of the in vivo epithelium and that it is often difficult to determine which, if any, of the cell lines provides the best representation of the in vivo epithelial surface. In this analysis, we utilized three independent bladder epithelial cell lines to gain a more comprehensive understanding of bladder epithelial responses to LPS and E. coli. The data from this investigation demonstrated that all of these bladder cell lines are LPS responsive and that this response is facilitated by the activation of NF-κB and p38 MAP kinase through a CD14-TLR complex. The present data are consistent with those of Bäckhed et al. showing a role for TLR4 in LPS responsiveness in bladder epithelial cells. Also, the significantly impaired ability of C3H/HeJ mice to clear an experimental bladder infection (12, 20, 44) and a recent report that the murine bladder mucosa expresses numerous inflammatory genes following LPS challenge in vivo (43) are consistent with our findings. Thus, we argue that the bladder mucosal surface is primed to recognize and respond to LPS. In contrast, the A498 cell line is hyporesponsive to LPS and substantially less efficient at producing anti-E. coli responses than the bladder cell lines. Based on these results, we suggest that the bladder epithelium should be considered a CD14-expressing, LPS-responsive entity that is not accurately depicted by the A498 cell line. Alternatively, published results with the A498 cell line may in fact argue that there are fundamental differences in innate responses of bladder and kidney epithelia, as has been proposed by Bäckhed et al. (3). Future studies using in vivo analysis and primary cell lines will help to further characterize CD14-TLR interactions and bladder epithelial responses to UPEC during an acute urinary tract infection. The methods employed here may be applied to other epithelial cell types to determine whether the CD14-TLR receptor complex is employed by these epithelia to recognize and respond to LPS.
Acknowledgments
This work was supported by NIH grants R01-DK51406 and R01-A129549 (S.J.H.) and R21-DK57936 (R.G.L.). D.A.H. is an NICHD Fellow of the Pediatric Scientist Development Program (K12-HD00850).
We thank M. Chapman for helpful discussions throughout the generation of this paper.
Editor: A. D. O'Brien
REFERENCES
- 1.Arbibe, L., J. P. Mira, N. Teusch, L. Kline, M. Guha, N. Mackman, P. J. Godowski, R. J. Ulevitch, and U. G. Knaus. 2000. Toll-like receptor 2-mediated NF-κB activation requires a Rac1-dependent pathway. Nat. Immunol. 1:533-540. [DOI] [PubMed] [Google Scholar]
- 2.Bäckhed, F., L. Meijer, S. Normark, and A. Richter-Dahlfors. 2002. TLR4-dependent recognition of lipopolysaccharide by epithelial cells requires sCD14. Cell. Microbiol. 4:493-501. [DOI] [PubMed] [Google Scholar]
- 3.Bäckhed, F., M. Soderhall, P. Ekman, S. Normark, and A. Richter-Dahlfors. 2001. Induction of innate immune responses by Escherichia coli and purified lipopolysaccharide correlate with organ- and cell-specific expression of Toll-like receptors within the human urinary tract. Cell. Microbiol. 3:153-158. [DOI] [PubMed] [Google Scholar]
- 4.Beutler, B. 2000. Tlr4: central component of the sole mammalian LPS sensor. Curr. Opin. Immunol. 12:20-26. [DOI] [PubMed] [Google Scholar]
- 5.Dlaska, M., and G. Weiss. 1999. Central role of transcription factor NF-IL6 for cytokine and iron-mediated regulation of murine inducible nitric oxide synthase expression. J. Immunol. 162:6171-6177. [PubMed] [Google Scholar]
- 6.Eisenstein, B. I. 1981. Phase variation of type 1 fimbriae in Escherichia coli is under transcriptional control. Science 214:337-339. [DOI] [PubMed] [Google Scholar]
- 7.Frendeus, B., C. Wachtler, M. Hedlund, H. Fischer, P. Samuelsson, M. Svensson, and C. Svanborg. 2001. Escherichia coli P fimbriae utilize the Toll-like receptor 4 pathway for cell activation. Mol. Microbiol. 40:37-51. [DOI] [PubMed] [Google Scholar]
- 8.Funda, D. P., L. Tuckova, M. A. Farre, T. Iwase, I. Moro, and H. Tlaskalova-Hogenova. 2001. CD14 is expressed and released as soluble CD14 by human intestinal epithelial cells in vitro: lipopolysaccharide activation of epithelial cells revisited. Infect. Immun. 69:3772-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godaly, G., G. Bergsten, L. Hang, H. Fischer, B. Frendeus, A. C. Lundstedt, M. Samuelsson, P. Samuelsson, and C. Svanborg. 2001. Neutrophil recruitment, chemokine receptors, and resistance to mucosal infection. J. Leukoc. Biol. 69:899-906. [PubMed] [Google Scholar]
- 10.Golenbock, D. T., R. Y. Hampton, N. Qureshi, K. Takayama, and C. R. Raetz. 1991. Lipid A-like molecules that antagonize the effects of endotoxins on human monocytes. J. Biol. Chem. 266:19490-19498. [PubMed] [Google Scholar]
- 11.Gunther, N. I., V. Lockatell, D. E. Johnson, and H. L. Mobley. 2001. In vivo dynamics of type 1 fimbria regulation in uropathogenic Escherichia coli during experimental urinary tract infection. Infect. Immun. 69:2838-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagberg, L., R. Hull, S. Hull, J. R. McGhee, S. M. Michalek, and C. Svanborg Eden. 1984. Difference in susceptibility to gram-negative urinary tract infection between C3H/HeJ and C3H/HeN mice. Infect. Immun. 46:839-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haziot, A., E. Ferrero, F. Kontgen, N. Hijiya, S. Yamamoto, J. Silver, C. L. Stewart, and S. M. Goyert. 1996. Resistance to endotoxin shock and reduced dissemination of gram-negative bacteria in CD14-deficient mice. Immunity 4:407-414. [DOI] [PubMed] [Google Scholar]
- 14.Hedges, S., M. Svensson, and C. Svanborg. 1992. Interleukin-6 response of epithelial cell lines to bacterial stimulation in vitro. Infect. Immun. 60:1295-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedlund, M., B. Frendeus, C. Wachtler, L. Hang, H. Fischer, and C. Svanborg. 2001. Type 1 fimbriae deliver an LPS- and TLR4-dependent activation signal to CD14-negative cells. Mol. Microbiol. 39:542-552. [DOI] [PubMed] [Google Scholar]
- 16.Hedlund, M., M. Svensson, A. Nilsson, R. D. Duan, and C. Svanborg. 1996. Role of the ceramide-signaling pathway in cytokine responses to P-fimbriated Escherichia coli. J. Exp. Med. 183:1037-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedlund, M., C. Wachtler, E. Johansson, L. Hang, J. E. Somerville, R. P. Darveau, and C. Svanborg. 1999. P fimbriae-dependent, lipopolysaccharide-independent activation of epithelial cytokine responses. Mol. Microbiol. 33:693-703. [DOI] [PubMed] [Google Scholar]
- 18.Hirschfeld, M., Y. Ma, J. H. Weis, S. N. Vogel, and J. J. Weis. 2000. Repurification of lipopolysaccharide eliminates signaling through both human and murine Toll-like receptor 2. J. Immunol. 165:618-622. [DOI] [PubMed] [Google Scholar]
- 19.Hirschfeld, M., J. J. Weis, V. Toshchakov, C. A. Salkowski, M. J. Cody, D. C. Ward, N. Qureshi, S. M. Michalek, and S. N. Vogel. 2001. Signaling by Toll-like receptor 2 and 4 agonists results in differential gene expression in murine macrophages. Infect. Immun. 69:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopkins, W. J., A. Gendron-Fitzpatrick, E. Balish, and D. T. Uehling. 1998. Time course and host responses to Escherichia coli urinary tract infection in genetically distinct mouse strains. Infect. Immun. 66:2798-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horng, T., G. M. Barton, R. A. Flavell, and R. Medzhitov. 2002. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature 420:329-333. [DOI] [PubMed] [Google Scholar]
- 22.Hultgren, S. J., C. H. Jones, and S. N. Normark. 1996. Bacterial adhesins and their assembly, p. 2730-2756. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 23.Hultgren, S. J., W. R. Schwan, A. J. Schaeffer, and J. L. Duncan. 1986. Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect. Immun. 54:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingalls, R. R., H. Heine, E. Lien, A. Yoshimura, and D. Golenbock. 1999. Lipopolysaccharide recognition, CD14, and lipopolysaccharide receptors. Infect. Dis. Clin. N. Am. 13:341-353. [DOI] [PubMed] [Google Scholar]
- 25.Ingalls, R. R., E. Lien, and D. T. Golenbock. 2001. Membrane-associated proteins of a lipopolysaccharide-deficient mutant of Neisseria meningitidis activate the inflammatory response through Toll-like receptor 2. Infect. Immun. 69:2230-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jersmann, H. P., C. S. Hii, G. L. Hodge, and A. Ferrante. 2001. Synthesis and surface expression of CD14 by human endothelial cells. Infect. Immun. 69:479-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, C. H., J. S. Pinkner, R. Roth, J. Heuser, A. V. Nicholes, S. N. Abraham, and S. J. Hultgren. 1995. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 92:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kagnoff, M. F., and L. Eckmann. 1997. Epithelial cells as sensors for microbial infection. J. Clin. Investig. 100:6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaisho, T., and S. Akira. 2000. Critical roles of Toll-like receptors in host defense. Crit. Rev. Immunol. 20:393-405. [PubMed] [Google Scholar]
- 30.Kawai, T., O. Adachi, T. Ogawa, K. Takeda, and S. Akira. 1999. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity 11:115-122. [DOI] [PubMed] [Google Scholar]
- 31.Langermann, S., S. Palaszynski, M. Barnhart, G. Auguste, J. S. Pinkner, J. Burlein, P. Barren, S. Koenig, S. Leath, C. H. Jones, and S. J. Hultgren. 1997. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 276:607-611. [DOI] [PubMed] [Google Scholar]
- 32.Loppnow, H., F. Stelter, U. Schonbeck, C. Schluter, M. Ernst, C. Schutt, and H. D. Flad. 1995. Endotoxin activates human vascular smooth muscle cells despite lack of expression of CD14 mRNA or endogenous membrane CD14. Infect. Immun. 63:1020-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medvedev, A. E., P. Henneke, A. Schromm, E. Lien, R. Ingalls, M. J. Fenton, D. T. Golenbock, and S. N. Vogel. 2001. Induction of tolerance to lipopolysaccharide and mycobacterial components in Chinese hamster ovary/CD14 cells is not affected by overexpression of Toll-like receptors 2 or 4. J. Immunol. 167:2257-2267. [DOI] [PubMed] [Google Scholar]
- 34.Medvedev, A. E., K. M. Kopydlowski, and S. N. Vogel. 2000. Inhibition of lipopolysaccharide-induced signal transduction in endotoxin-tolerized mouse macrophages: dysregulation of cytokine, chemokine, and Toll-like receptor 2 and 4 gene expression. J. Immunol. 164:5564-5574. [DOI] [PubMed] [Google Scholar]
- 35.Medzhitov, R., P. Preston-Hurlburt, and C. A. Janeway, Jr. 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388:394-397. [DOI] [PubMed] [Google Scholar]
- 36.Mulvey, M. A., Y. S. Lopez-Boado, C. L. Wilson, R. Roth, W. C. Parks, J. Heuser, and S. J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494-1497. [DOI] [PubMed] [Google Scholar]
- 37.Nomura, F., S. Akashi, Y. Sakao, S. Sato, T. Kawai, M. Matsumoto, K. Nakanishi, M. Kimoto, K. Miyake, K. Takeda, and S. Akira. 2000. Endotoxin tolerance in mouse peritoneal macrophages correlates with down-regulation of surface Toll-like receptor 4 expression. J. Immunol. 164:3476-3479. [DOI] [PubMed] [Google Scholar]
- 38.O'Neill, L. 2000. The Toll/interleukin-1 receptor domain: a molecular switch for inflammation and host defence. Biochem. Soc. Trans. 28:557-563. [DOI] [PubMed] [Google Scholar]
- 39.Palombella, V. J., O. J. Rando, A. L. Goldberg, and T. Maniatis. 1994. The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell 78:773-785. [DOI] [PubMed] [Google Scholar]
- 40.Philpott, D. J., S. E. Girardin, and P. J. Sansonetti. 2001. Innate immune responses of epithelial cells following infection with bacterial pathogens. Curr. Opin. Immunol. 13:410-416. [DOI] [PubMed] [Google Scholar]
- 41.Pridmore, A. C., D. H. Wyllie, F. Abdillahi, L. Steeghs, P. van der Ley, S. K. Dower, and R. C. Read. 2001. A lipopolysaccharide-deficient mutant of Neisseria meningitidis elicits attenuated cytokine release by human macrophages and signals via Toll-like receptor (TLR) 2 but not via TLR4/MD2. J. Infect. Dis. 183:89-96. [DOI] [PubMed] [Google Scholar]
- 42.Roberts, J. A., B. I. Marklund, D. Ilver, D. Haslam, M. B. Kaack, G. Baskin, M. Louis, R. Mollby, J. Winberg, and S. Normark. 1994. The Gal(α-1,4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc. Natl. Acad. Sci. USA 91:11889-11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saban, M. R., H. Hellmich, N. B. Nguyen, J. Winston, T. G. Hammond, and R. Saban. 2001. Time course of LPS-induced gene expression in a mouse model of genitourinary inflammation. Physiol. Genomics 5:147-160. [DOI] [PubMed] [Google Scholar]
- 44.Schilling, J. D., M. A. Mulvey, C. D. Vincent, R. G. Lorenz, and S. J. Hultgren. 2001. Bacterial invasion augments epithelial cytokine responses to Escherichia coli through a lipopolysaccharide-dependent mechanism. J. Immunol. 166:1148-1155. [DOI] [PubMed] [Google Scholar]
- 45.Shimazu, R., S. Akashi, H. Ogata, Y. Nagai, K. Fukudome, K. Miyake, and M. Kimoto. 1999. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 189:1777-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shuto, T., H. Xu, B. Wang, J. Han, H. Kai, X. X. Gu, T. F. Murphy, D. J. Lim, and J. D. Li. 2001. Activation of NF-κB by nontypeable Haemophilus influenzae is mediated by toll-like receptor 2-TAK1-dependent NIK-IKKα/β-IκBα and MKK3/6-p38 MAP kinase signaling pathways in epithelial cells. Proc. Natl. Acad. Sci. USA 98:8774-8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svanborg, C., B. Frendeus, G. Godaly, L. Hang, M. Hedlund, and C. Wachtler. 2001. Toll-like receptor signaling and chemokine receptor expression influence the severity of urinary tract infection. J. Infect. Dis. 183(Suppl. 1):S61-S65. [DOI] [PubMed] [Google Scholar]
- 48.Tapping, R. I., and P. S. Tobias. 1997. Cellular binding of soluble CD14 requires lipopolysaccharide (LPS) and LPS-binding protein. J. Biol. Chem. 272:23157-23164. [DOI] [PubMed] [Google Scholar]
- 49.Wang, C., L. Deng, M. Hong, G. R. Akkaraju, J. Inoue, and Z. J. Chen. 2001. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412:346-351. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto, M., S. Sato, H. Hemmi, H. Sanjo, S. Uematsu, T. Kaisho, K. Hoshino, O. Takeuchi, M. Kobayashi, T. Fujita, K. Takeda, and S. Akira. 2002. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature 420:324-329. [DOI] [PubMed] [Google Scholar]
- 51.Yang, H., D. W. Young, F. Gusovsky, and J. C. Chow. 2000. Cellular events mediated by lipopolysaccharide-stimulated Toll-like receptor 4. MD-2 is required for activation of mitogen-activated protein kinases and Elk-1. J. Biol. Chem. 275:20861-20866. [DOI] [PubMed] [Google Scholar]