Abstract
Escherichia coli O157:H7 causes bloody diarrhea and potentially fatal systemic sequelae in humans. Cattle are most frequently identified as the primary source of infection, and E. coli O157:H7 generally colonizes the gastrointestinal tracts of cattle without causing disease. In this study, persistence and tropism were assessed for four different E. coli O157:H7 strains. Experimentally infected calves shed the organism for at least 14 days prior to necropsy. For the majority of these animals, as well as for a naturally colonized animal obtained from a commercial beef farm, the highest numbers of E. coli O157:H7 were found in the feces, with negative or significantly lower levels detected in lumen contents taken from the gastrointestinal tract. Detailed examination demonstrated that in these individuals the majority of tissue-associated bacteria were adherent to mucosal epithelium within a defined region extending up to 5 cm proximally from the recto-anal junction. The tissue targeted by E. coli O157:H7 was characterized by a high density of lymphoid follicles. Microcolonies of the bacterium were readily detected on the epithelium of this region by immunofluorescence microscopy. As a consequence of this specific distribution, E. coli O157:H7 was present predominately on the surface of the fecal stool. In contrast, other E. coli serotypes were present at consistent levels throughout the large intestine and were equally distributed in the stool. This is a novel tropism that may enhance dissemination both between animals and from animals to humans. The accessibility of this site may facilitate simple intervention strategies.
Enterohemorrhagic Escherichia coli (EHEC) has emerged as an important cause of human intestinal disease in developed countries over the past 20 years. In addition to bloody diarrhea, intestinal infection can lead to potentially fatal systemic sequelae resulting from the activity of Shiga toxins. The majority of these infections are caused by E. coli O157:H7 (21, 26). This serotype has been frequently isolated from cattle feces, and most human EHEC O157:H7 infections originate, either directly or indirectly, from this source (5, 8). A key step in protecting public health is to know how and where the bacterium persists in this major animal reservoir. Until now, no defined site of colonization by E. coli O157:H7 in the bovine gastrointestinal tract (GIT) has been described, beyond an affinity for the large intestine (17).
Enteropathogenic E. coli (EPEC) and most EHEC strains are known to carry a locus of enterocyte effacement (LEE) pathogenicity island (24). This locus encodes a type III secretion system that mediates attachment to mucosal epithelial cells. Injection of effector proteins results in intimate attachment and characteristic attaching and effacing (A/E) lesions dependent on intimin and Tir (translocated intimin receptor) (16, 22). E. coli O157:H7 intimately attaches to a variety of cell types and tissues in vitro, and a few studies have demonstrated that E. coli O157:H7 can form A/E lesions on explants of bovine intestinal tissue (3, 29). Studies by Dean-Nystrom et al. recovered E. coli O157:H7 from a range of intestinal sites with associated A/E lesions 3 days after challenge of neonatal calves (13) and 4 days after challenge of fasting weaned calves (14). However, it is not clear whether either of these experimental systems reflects natural carriage. The importance of A/E lesion formation in the colonization of ruminants has been shown by the deletion and complementation of intimin in a sheep persistence model (11). The same intimin-negative strain also had an impaired ability to colonize calves. Despite this evidence, no mucosal adherence of any form has been reported in naturally colonized cattle.
The experimental challenge of healthy weaned calves reproduces natural carriage and does not result in clinical disease; E. coli O157:H7 distribution in such animals has been investigated (9, 12, 17). Brown et al. (9) recovered E. coli O157:H7 from all sites sampled within the GIT, except the abomasum, with the highest recovery rate in the forestomachs. Cray and Moon (12) also demonstrated a ubiquitous distribution but found the highest numbers in large-intestinal sites. These studies suggest a variable distribution with no specific tropism for any site within the bovine GIT. However, Grauke et al. (17) used rumen and duodenal cannulae to sample from live calves, and beyond day 16 neither of these locations harbored E. coli O157:H7. This was despite the fact that E. coli O157:H7 was present in some of the corresponding fecal samples up to day 34, implying a large-intestinal location. None of these studies demonstrated any evidence of mucosal localization or A/E lesions in persistently shedding animals.
The objective of our study was to examine the patterns of fecal excretion and bacterial GIT localization following experimental challenge with different E. coli O157:H7 strains. Our early data revealed that E. coli O157:H7 frequently is absent or is isolated in insignificant numbers from the GIT contents at necropsy despite remaining easily demonstrable in the feces (103 to 105 CFU per g). Significantly, Grauke et al. made a similar observation for sheep colonized for more than 2 weeks (17). We therefore investigated the possibility that E. coli O157:H7 persists in either the very terminal portion of the rectum or the anal canal. Due to their intrapelvic location, these sites are not routinely sampled during necropsy and have been overlooked in early sampling procedures. Progressively more detailed analysis of shedding cattle, including a 12-month-old naturally colonized steer, has highlighted a unique tropism for the rectal mucosa adjacent to the recto-anal junction (RAJ). This is a significant and previously overlooked aspect of E. coli O157:H7 biology.
MATERIALS AND METHODS
E. coli O157:H7 strains.
Strains used for oral inoculation of calves were ZAP 196, ZAP 198, and ZAP 3. ZAP 196 and ZAP 198 were both isolated from the same human patient in Washington state (27). Cattle were established as the source of this outbreak. ZAP 196 possesses Shiga toxin 2 (Stx2), while ZAP 198 has been naturally cured of the Stx2 bacteriophage. They are otherwise indistinguishable as determined by phage typing (both type 32), LEE protein secretion level (25), and pulsed-field gel electrophoresis (both nontypeable). ZAP 3 (Scottish E. coli O157:H7 reference laboratory number 659) is a bovine isolate from Red House Dairy (Blackburn, West Lothian, Scotland) that caused a milk-borne human outbreak (2). The strain used for challenge via cohabitation with a shedding calf at Washington State University was WSU 2043. This was originally isolated from an asymptomatic calf and possesses the genes for Stx1, Stx2, and Stx2c. All these strains were selected for spontaneous resistance to nalidixic acid in order to facilitate recovery from GIT contents and tissues. The strain isolated from a naturally colonized 12-month-old steer on a farm in Inverness-shire (Scotland) has been designated ZAP 278. It possesses genes for Stx2 and Stx2c (4). All strains used have been shown by PCR using published primers to possess genes for enterohemolysin (15), intimin-γ (1), EspA (25), and EspB (25).
Calf colonization.
Separate experimental calf challenges used either ZAP 3, ZAP 196, or ZAP 198 and were performed at the Moredun Research Institute in either containment level 2 or containment level 3 large-animal housing facilities under Home Office license number 60/02105. Calves were reared conventionally on a farm until at least 2 weeks postweaning and were transported to the Moredun Research Institute, where they were acclimatized for at least 3 days prior to challenge. Prior to challenge fecal samples were taken at least twice from each calf and confirmed negative for E. coli O157:H7 by immunomagnetic separation (IMS). At the time of challenge the calves' ages ranged from 8 to 14 weeks. The challenge E. coli O157:H7 strain was grown overnight in Luria-Bertani (LB) broth (37°C, with aeration) and diluted in sterile phosphate-buffered saline (PBS) to achieve an inoculum of 109 CFU per animal in a total volume of 10 ml. The inoculum was administered to the calves via stomach tube and washed down with 500 ml of sterile PBS. Feces were caught or collected per rectum for culture and bacterial enumeration.
Experimental calf challenges with WSU 2043 were performed at Washington State University by a direct-contact infection model system (6). Briefly, groups of calves were first confirmed to be free of detectable E. coli O157:H7 shedding as described above. Then a single calf was removed from the group and experimentally challenged with an inoculum prepared as described for the above calves but administered orally by syringe. The reintroduction of the inoculated calf into the group pen constituted the challenge, and the subject calves were monitored for acquisition of E. coli O157:H7 infection.
The 12-month-old steer that was naturally shedding >104 CFU of E. coli O157:H7 per g of feces was identified on a farm in Inverness-shire by field epidemiology work.
Necropsy sampling.
The precise procedure varied; more-detailed analysis of the terminal rectum was performed as the tropism of E. coli O157:H7 became better defined with each necropsy. The most complete necropsy procedure is described. As close as possible to euthanasia, a sample of naturally passed feces was caught with a fresh glove and was split into surface and core components by removing the surface layer with a sterile scalpel and exposing the core. For certain animals it was not possible to split feces into surface and core components. In these cases whole feces containing an undetermined mixture of surface and core were used for enumeration.
Following euthanasia with intravenous pentobarbital, the abdomen was opened. The terminal 20 to 30 cm of rectum and anus were removed as a single piece after the rectum was double ligated and transected, the anus circumsected, and the pubic bone reflected. This length of gut was opened longitudinally in a proximal-to-distal direction. When present, 10-g samples of lumen contents were taken from between 30 and 20 cm proximal to the RAJ. This junction is grossly visible and is the interface between the colorectal epithelium and anal stratified squamous epithelium. Samples of tissue visibly free of feces were taken from the following portions of rectum relative to the RAJ (in centimeters): 20 to 30, 10 to 20, 5 to 10, 3 to 5, 1 to 3, and −1 to +1. Lumen contents and tissue were also taken from the following locations: rumen (cranioventral sac), ileum (10 cm proximal to the ileo-ceco-colic junction, including both normal absorptive mucosa and Peyer's patches), cecum (apex), and colon (point of inflection of spiral colon). All lumen contents and tissue were processed for microbiological analysis (described below). A sample of each tissue was fixed in 4% paraformaldehyde for cutting of sections to be used for immunofluorescent detection of E. coli O157.
Microbiology.
Ten-gram quantities of feces or GIT contents were suspended in 90 ml of sterile PBS and serially diluted in 10-fold steps in PBS. Tissue samples were placed in sterile PBS (1 cm2 in 5 ml) and vortexed vigorously for 60 s, and the supernatant was diluted in a 10-fold dilution series. These serial dilutions were cultured as 100-μl aliquots spread in duplicate or triplicate onto sorbitol MacConkey agar (SMAC; Oxoid) plates containing 15 μg of nalidixic acid (Sigma-Aldrich) ml−1. All inoculated media were incubated overnight at 37°C. Non-sorbitol-fermenting colonies on SMAC plates were counted, and one to three colonies from each sample were tested for O157 lipopolysaccharide antigen by using a latex agglutination test kit (Oxoid). The most probable number of CFU was determined as described under “Statistical methods” below. For enrichment cultures 1 ml of the undiluted suspension or tissue washing supernatant was added to 9 ml of LB broth containing 15 μg of nalidixic acid ml−1. The enrichment cultures for samples negative on direct plating were spread onto fresh SMAC plates and incubated overnight at 37°C. These procedures are more sensitive than IMS for detecting marked strains (6). Some samples were also spread on tryptone bile X-glucuronide medium (TBX; Oxoid) to enumerate glucuronidase-positive, non-O157:H7 E. coli strains (18). Samples for enumeration from the naturally colonized animal were cultured on sorbitol MacConkey agar by using cefixime-tellurite (0.05 and 25 μg ml−1, respectively; Oxoid) instead of nalidixic acid selection. Similarly, enrichment was performed with LB broth containing cefixime-tellurite in place of nalidixic acid.
Immunofluorescence microscopy.
E. coli O157 was detected by fluorescence microscopy of paraformaldehyde-fixed sections after incubation with a rabbit anti-O157 polyclonal antibody (Mast-Assure) (1:100 for 30 min at room temperature), followed by a fluorescein isothiocyanate-conjugated goat anti-rabbit antibody (Sigma-Aldrich) (1:1,000 for 30 min at room temperature). Both antibodies were diluted in PBS containing 0.1% bovine serum albumin. Stained sections were viewed on a Leica DMLB epifluorescent microscope with a 40× objective. As a negative control, certain samples were also stained with normal rabbit serum or a polyclonal anti-O26 antibody (Mast-Assure). For confocal microscopy, tissue was counterstained with tetramethyl rhodamine isothiocyanate (TRITC)-phalloidin and viewed with a Zeiss 510 confocal microscope with a 63× objective lens.
Statistical methods.
The most probable number of CFU was determined by fitting generalized linear models with a Poisson error distribution and logarithmic link function while incorporating the logarithm of dilution as an offset variable (23). Bacterial counts at different sites were compared by fitting a generalized linear mixed model (10), with the same error and link functions, while fitting “animal” and “animal site” as random effects. The unbalanced nature of the data did not always allow this model to converge to stable values, in which case “animal” was fitted as the sole random effect. These models typically had a moderate to large overdispersion estimate, and these values were always used to estimate the standard errors of the model estimates.
Of the 15 animals studied, the first 4 were not sampled at the full range of sites, and therefore complete data sets were not available for analysis (a random dropout from the sample). A further two animals were identified as exhibiting a colonization pattern qualitatively different from that seen in the others, in that the contents and fecal results suggested that the terminal rectum was not the principal site of colonization (animals 607 and 364). Therefore, only samples from the nine remaining animals were used in the statistical analysis of tissue-associated E. coli O157:H7.
RESULTS
Shedding of E. coli O157:H7 from orally challenged cattle.
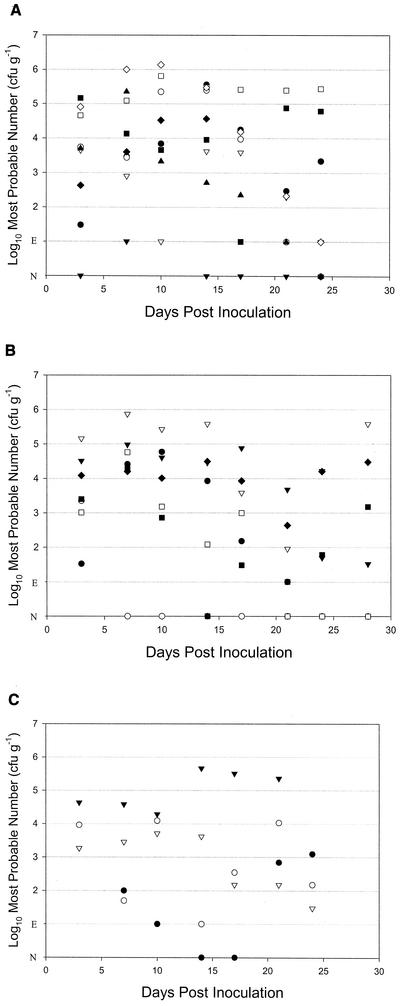
In order to investigate the localization of E. coli O157:H7 within the bovine host, it was essential to establish a model that resulted in persistent shedding from healthy weaned animals. By using a number of different strains, persistent shedding for more than 3 weeks was consistently obtained with an oral dose of 109 CFU. The shedding patterns for the separate challenges are shown in Fig. 1. Animals for necropsy were chosen from those shedding at 14 to 28 days postinfection.
FIG. 1.
Scattergraphs illustrating the shedding profiles of three strains, ZAP 196 (A), ZAP 198 (B), and ZAP 3 (C). Each strain was inoculated separately into different calf groups by oral challenge of 109 CFU of E. coli O157 (Nalr). Each plot represents the most probable number of E. coli O157 per gram of feces. Some of the animalsrepresented in these graphs were not shedding at the time of necropsy and therefore were not included in the tables of necropsy results. Samples yielding no colonies on direct culture have been labeled E if enrichment positive and N if enrichment negative to facilitate graphical representation.
Distribution of E. coli O157:H7 in GIT contents and feces at necropsy.
Necropsies were performed on experimentally and naturally colonized animals that were persistently shedding E. coli O157. All animals had positive feces, with levels ranging from 103 to 105 CFU g−1 (Table 1). For 13 of the 15 animals, concentrations of E. coli O157:H7 were at least 10-fold higher in the feces (antemortem) than in the midrectal contents collected 20 to 30 cm proximal to the RAJ (Table 1). For 11 of the 15 animals, levels of E. coli O157:H7 in the feces were at least 100-fold higher than those in either the colon or the midrectal contents. E. coli O157:H7 was not recovered from the GIT contents of 6 ruminal, 13 ileal, 7 colonic, and 7 midrectal samples. Statistical analysis showed no evidence of significant differences between samples collected from the colon and the midrectum (P = 0.99). However, counts were significantly higher in feces than in both colonic and midrectal contents (P < 0.001).
TABLE 1.
E. coli O157:H7 concentrations in gastrointestinal contents and feces from shedding cattle
| Calfa | Strain | Dayb | Infectionc |
E. coli O157:H7 concnd (CFU g−1) in:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GIT contents
|
Feces
|
||||||||||
| Rumen | Ileum | Cecum | Colon | Rectum, 20-30 cme | Whole | Surface | Core | ||||
| 351 | ZAP 198 | 21 | OD | 33 | 0 | 0 | 0 | 0 | NA | 2,400 | 0 |
| 364 | ZAP 198 | 21 | OD | 0 | 0 | 670 | 1,100 | 130 | NA | 1,400 | 860 |
| 360 | ZAP 198 | 20 | OD | E+ | 0 | 0 | E+ | 0 | NA | 5,100 | E+ |
| 325 | ZAP 278 | NDf | NC | E+ | 0 | 0 | 0 | 0 | 33,000 | NA | NA |
| 607 | WSU 2043 | 22 | CH | E+ | 550 | ND | 750 | 4,200 | 3,200 | NA | NA |
| 609 | WSU 2043 | 22 | CH | 0 | 0 | ND | 0 | 0 | 5,300 | NA | NA |
| 611 | WSU 2043 | 22 | CH | E+ | 0 | ND | 4,700 | 5,400 | 16,000 | NA | NA |
| 612 | WSU 2043 | 22 | CH | 300 | 0 | ND | 50 | 150 | 30,000 | NA | NA |
| 295 | ZAP 196 | 25 | OD | 0 | 0 | ND | 0 | 0 | NA | 70,000 | 0 |
| 299 | ZAP 196 | 24 | OD | E+ | 0 | ND | E+ | 150 | NA | 280,000 | 390 |
| 307 | ZAP 196 | 14 | OD | 0 | 0 | ND | 520 | E+ | 36,000 | NA | NA |
| 310 | ZAP 198 | 28 | OD | 45 | 0 | ND | 0 | 0 | 380,000 | NA | NA |
| 323 | ZAP 198 | 28 | OD | E+ | 0 | ND | 0 | 0 | 330,000 | NA | NA |
| 118 | ZAP 3 | 28 | OD | 0 | 0 | ND | 0 | E+ | 2,600 | NA | NA |
| 121 | ZAP 3 | 23 | OD | 0 | E+ | ND | 50 | E+ | 230,000 | NA | NA |
Calves are listed in the reverse chronological order in which necropsies were performed.
Number of days postinoculation or postexposure at which necropsy was performed. For calves in the cohabitation model, necropsy was performed 22 days after initial exposure to the shedding animal.
Source of infection; OD, oral dose of 109 CFU g−1; NC, naturally colonized; CH, cohabitation with shedding individual.
Values represent most probable numbers rounded to 2 significant figures. NA, not applicable; either whole feces was used for enumeration or it was split into surface and core components. E+, positive by enrichment culture but not by direct plating.
Contents of the rectum within a region 20 to 30 cm proximal to the RAJ.
ND, not determined. The naturally colonized animal acquired E. coli O157:H7 at an undetermined time point.
Distribution of E. coli O157:H7 on GIT mucosae at necropsy.
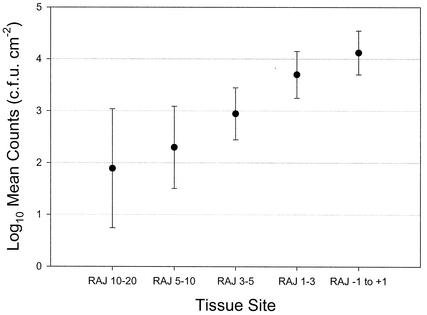
Tissue-associated E. coli O157:H7 bacteria were recovered and enumerated from defined mucosal sites for 11 animals (Table 2). Levels of mucosa-associated E. coli O157:H7 on the terminal 5 cm of the rectum were at least 10 times higher than those on any other mucosal surface examined for 9 of the 11 animals. The rumen, ileum, Peyer's patches, colon tissue, and samples 20 to 30 cm from the RAJ, when sampled, typically were negative. Calculation of the mean count for E. coli O157:H7 CFU per square centimeter allowed differences between defined tissue sites to be compared statistically. There is clear evidence of an upward trend in counts as samples are taken closer to the RAJ, and the increases are particularly strong between the zones 3 to 5 cm and 1 to 3 cm from the RAJ (P < 0.001) and between the zones 1 to 3 cm and −1 to +1 cm from the RAJ (P = 0.001) (Fig. 2).
TABLE 2.
Tissue-associated E. coli O157:H7 levels on mucosae taken from regions of the GIT at necropsy
| Calfa | Strain | Dayb | Infectionc |
E. coli O157:H7 concn (CFU cm−2) in the following mucosal sample:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rumen | Ileum | Peyer's patches | Colon | RAJe
|
|||||||||
| 20 to 30 | 10 to 20 | 5 to 10 | 3 to 5 | 1 to 3 | −1 to +1 | ||||||||
| 351 | ZAP 198 | 21 | OD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | E+ | 1,100 | 1,600 |
| 364 | ZAP 198 | 21 | OD | 0 | 0 | 0 | E+ | 0 | 0 | 0 | E+ | 0 | E+ |
| 360 | ZAP 198 | 20 | OD | 0 | 0 | 0 | 0 | 0 | 0 | 0 | E+ | 2,100 | 9,800 |
| 325 | ZAP 278 | NDf | NC | 0 | 0 | 0 | 0 | 0 | 0 | 61 | 83,000 | 21,000 | 5,800 |
| 607 | WSU 2043 | 22 | CH | 0 | 200 | ND | 230 | ND | 17 | 250 | 33 | 33 | 33 |
| 609 | WSU 2043 | 22 | CH | 0 | 0 | ND | ND | ND | 0 | 0 | 33 | 50 | 1,900 |
| 611 | WSU 2043 | 22 | CH | 0 | 0 | ND | ND | ND | 950 | 720 | 57,000 | 300,000 | 29,000 |
| 612 | WSU 2043 | 22 | CH | 0 | 0 | ND | ND | ND | 17 | 1,600 | 3,300 | 4,000 | 16,000 |
| 295 | ZAP 196 | 25 | OD | ND | ND | ND | ND | ND | 30 | 210 | E+ | 680 | 100,000 |
| 299 | ZAP 196 | 24 | OD | ND | ND | ND | ND | ND | 30 | 45 | 15 | 620 | 84,000 |
| 307 | ZAP 196 | 14 | OD | ND | ND | ND | ND | ND | ND | 2,500 | ND | ND | 190,000 |
Calves are listed in the reverse chronological order in which necropsies were performed.
Number of days postinoculation or postexposure at which necropsy was performed.
Source of infection: OD, oral dose of 109 CFU g−1; NC, naturally colonized; CH, cohabitation with shedding individual.
Values represent most probable numbers rounded to 2 significant figures. E+, positive by enrichment culture but not by direct plating.
Numbers indicate proximal distance (in centimeters) from this anatomical boundary.
ND, not determined. The naturally colonized animal acquired E. coli O157:H7 at an undetermined time point.
FIG. 2.
Mean counts of tissue-associated E. coli O157:H7 on five regions of the rectum defined by distance from the RAJ (in centimeters). Error bars indicate upper and lower 95% confidence intervals.
Distribution of E. coli O157:H7 within feces.
Freshly produced feces from five animals were obtained as the animals defecated and were dissected into surface and core components. E. coli O157:H7 bacteria were unevenly distributed in the feces of four animals. For these, the mean surface count was 2.2 × 104 CFU g−1, approximately 1,000-fold higher than the mean core count (2.5 × 101 CFU g−1) (Table 1). This difference is highly statistically significant (P < 0.001). In contrast, there was no significant difference between mean counts in the core feces and midrectal contents (P = 0.82). In calf 364, E. coli O157:H7 was widely distributed throughout the large intestine and was not localized specifically at the terminal rectal mucosa. There was little difference (<2-fold) between the surface and core fecal E. coli O157:H7 counts in this individual (Table 1). Fecal samples from the remaining animals were too fluid to be split into surface and core components.
Distribution of non-O157:H7 E. coli at necropsy.
Numbers of glucuronidase-positive E. coli bacteria were counted in a random selection of the above samples to ensure that the majority of E. coli bacteria within the bovine GIT do not exhibit the same tropism. E. coli O157:H7 is excluded from these data because most strains of this serotype, including all those in this study, are glucuronidase negative (30). The lowest mean counts of tissue-associated non-O157 E. coli occurred in the rumen, and a statistically significant increase was seen in the ileum (P = 0.01). Similar population sizes were maintained throughout the length of the large intestine, although there is some evidence that counts are lower nearer the RAJ (P < 0.001). A similar pattern was seen in contents, with the counts increasing between the rumen and the ileum and between the ileum and the cecum. There was no statistical evidence of any difference in mean non-O157 E. coli numbers between the cecum, colon, rectum, and feces (all P > 0.05) and no difference between surface and core fecal samples (P = 0.47).
Detection of adherent E. coli O157 by microscopy.
Adherent E. coli O157 bacteria were readily detected on the epithelial surface of the terminal rectal mucosa by fluorescence microscopy using anti-O157 immunostaining. Microcolonies of E. coli O157 were detected on sections with corresponding tissue-associated bacterial counts of 2 × 103 CFU cm−2 or greater. This included demonstration of E. coli O157 microcolonies on the rectal mucosa from the naturally colonized animal (calf 325) (Fig. 3). No fluorescent bacteria were observed when either normal rabbit serum or an anti-O26 antibody was used in place of the anti-O157 antibody.
FIG. 3.
Confocal micrographs of a microcolony of E. coli O157 on the epithelium of the terminal rectum from calf 325. (A) Pseudocrypt lined with adherent E. coli O157:H7 (green), with tissue counterstained with TRITC-phalloidin (red) (63× objective). (B) A different Z-plane from a region of the sample featured in panel A that has been digitally magnified by a factor of 6 by using the Zeiss 510 confocal image software.
Characterization of the mucosa of the terminal rectum.
In gross appearance, the smooth raised mucosa adjacent to the RAJ contrasted subtly with the folded surface of the normal absorptive rectal mucosa. This characteristic was reminiscent of the Peyer's patches of the bovine ileum. Histologically the RAJ (Fig. 4A) is the clear junction between the stratified squamous epithelium of the anal canal and the columnar epithelial mucosa of the rectum. Large aggregates of submucosal lymphoid follicles were visible in hematoxylin-and-eosin-stained sections of the rectal mucosa adjacent to the RAJ (Fig. 4). These follicles extended for 2 to 5 cm proximal to the RAJ and, at their apexes, penetrated the muscularis mucosa to contact areas of epithelium.
FIG. 4.
Hematoxylin-and-eosin-stained sections. (A) The RAJ, defined as the junction between stratified squamous epithelium (SSE) and the columnar epithelium folded into pseudocrypts (PC) which lines the large intestine. Two lymphoid follicles (LF) are also visible (2.5× objective). (B and C) Terminal rectal mucosa from within 1 cm of the RAJ. Panel B demonstrates the typical density of lymphoid follicles found in this region (2.5× objective). Panel C highlights a lymphoid follicle in contact with an area of epithelium notable by the absence of goblet cells (10× objective).
DISCUSSION
This research demonstrates that E. coli O157:H7 exhibits a novel tropism for the terminal rectum in the bovine host. In almost all persistently colonized animals, the majority of tissue-associated bacteria were identified in a region within 5 cm, or in many cases 3 cm, proximal to the RAJ (Fig. 4A). The tropism for the terminal rectum was demonstrated in orally inoculated calves, calves colonized by cohabitation with an animal shedding E. coli O157:H7, and a naturally colonized shedding animal identified on a commercial beef farm. This tropism was demonstrated for four E. coli O157:H7 strains, ZAP 3, ZAP 196/198, ZAP 278, and WSU 2043, therefore indicating that this is a characteristic of the O157:H7 serotype and not of any particular strain.
Though similar, long-term experimental studies have been carried out before, this site of persistence has not been previously described. In this study, animals colonized experimentally by oral inoculation were allowed to shed bacteria for at least 2 weeks before necropsy. Our shedding data as well as those of others (12-14, 17) show that the levels of E. coli O157:H7 excreted following an oral challenge of at least 109 bacteria are initially very high (Fig. 1). As a consequence, there is an increased probability of observing a widespread distribution if examination of bacterial localization is carried out early after challenge. Waiting for at least 2 weeks allowed the bacteria to establish a long-term persistence that is similar in duration to the natural carriage described by Besser et al. (7). Interestingly, Grauke et al. (17) stated that for 10 sheep from whose feces the bacteria were easily cultured, after more than 2 weeks following inoculation, the organism could not be found in any GIT tissue or other digesta. In another experiment within the same study, the only tissue sample from a sheep persistently colonized beyond day 15 from which E. coli O157:H7 was cultured was taken from the rectum, although it is not stated whether it was from the terminal rectum. It is important that in no previous studies has there been sampling specifically from the terminal rectum, and presumably the possibility of carriage at this site has been overlooked.
For 13 of the 15 animals persistently shedding E. coli O157:H7, there was a significant increase in bacterial numbers present in the feces over those in samples of contents taken from various sites in the GIT (Table 1). There were also high levels of tissue-associated bacteria immediately proximal to the RAJ (Table 2). As a consequence there was an uneven distribution of E. coli O157:H7 in the fecal stool, since the bacteria coat only the surface as it is released from the rectum. E. coli O157:H7 microcolonies could be readily detected on sections taken from the terminal rectum, with bacteria adherent to the apical surfaces of epithelial cells (Fig. 3). Of the 15 animals examined in the study, only 3 (animals 364, 607, and 611) (Table 1) demonstrated evidence that E. coli O157:H7 bacteria from sites other than the terminal rectum were contributing to more than 10% of the fecal load of the organism. These animals may have been colonized by E. coli O157:H7 at a mucosal site(s) other than the terminal rectum, or the organism could simply be present within the GIT digesta. Reingestion from a contaminated environment may contribute to persistence in either case. Colonization at the terminal rectum has the greatest impact on the numbers of bacteria shed, but as the dynamics of persistence at the terminal rectum have not been explored, there is a possibility that this persistence is dependent on recolonization. This may account for the fluctuations in numbers of shed E. coli O157:H7 bacteria observed in our shedding data and demonstrated in other studies (12).
E. coli O157:H7 was present at the terminal rectum in the naturally colonized animal (animal 325) (Tables 1 and 2) in which mucosally adherent E. coli O157 was demonstrated (Fig. 3). This animal was taken from a straw court containing 35 cattle, 16 of which had feces positive for E. coli O157:H7 by IMS, a sensitive detection method, but only 3 of these were shedding high numbers (>104 CFU g−1). One hypothesis is that among an E. coli O157:H7-positive group of animals there are a small number of “supershedders” that greatly enhance transmission and persistence within a herd. Our data show that high shedding results principally from colonization at the terminal rectum, and therefore rectal colonization is likely to be a prerequisite for supershedders on the farm. Intervention aimed primarily at this animal subset may prove successful in reducing E. coli O157:H7 levels in positive herds.
Non-O157:H7 E. coli bacteria were enumerated to eliminate the possibility that the association of E. coli O157:H7 with the terminal rectum was a result of a rapid multiplication of enteric bacteria in response to environmental factors. The non-O157:H7 E. coli organisms were present throughout the gut and reached their highest levels in the large intestine. Importantly, no increases were observed at the RAJ and fecal surface. Therefore, the majority of E. coli bacteria in the bovine gut do not share the E. coli O157:H7 tropism for the terminal rectum.
Bacterial tropism for this site has not been previously described, and the factors that drive it are unknown. It is likely to be based on the interaction between a bacterial adhesin and an epithelial cell receptor, access or expression of which occurs predominantly in the terminal rectum. Our analysis of this region has shown it to contain a high concentration of lymphoid follicles. For both sheep (31) and humans (28) the RAJ has also been described as an area rich in lymphoid follicles. The apparent tropism for this site exhibited by E. coli O157:H7 may be related to this feature, since other members of the Enterobacteriaceae, e.g., Salmonella enterica serovar Typhimurium, Listeria monocytogenes, Shigella spp., and Yersinia spp. (20, 32), have well-described tropisms for Peyer's patches, another region of gut-associated lymphoid tissue. There is some evidence that intimin-γ, which is expressed by EHEC O157:H7, mediates a specificity for follicle-associated epithelium of human Peyer's patches (29) and shows more-restricted tissue affinity than do other intimin types (19). A recent study (11) has shown the importance of intimin to the persistence of E. coli O157:H7 in ruminant colonization, and our ongoing studies do support a role for type III secretion and intimate attachment at the terminal rectum of the bovine host. With the knowledge of the site of persistence in the bovine host, it will be feasible to examine the host-pathogen interactions directly. An understanding of the cellular interactions that allow E. coli O157:H7 to persist on the rectal mucosae should lead to the development of rational control strategies.
Worldwide, among shiga-toxigenic E. coli (STEC) serotypes, E. coli O157:H7 is the predominant human pathogen. Why this serotype is so relatively successful at causing disease has been an enigma, since the virulence factors identified to date are shared by many other STEC serotypes. Our preliminary studies with E. coli serotypes O26, O111, and O5, also associated with human disease, indicate that they do not share a tropism for the bovine terminal rectum. However, abattoir surveys are required to confirm the contribution of this tropism to the maintenance of E. coli O157:H7 and other STEC serotypes in cattle populations. These results will have substantial implications for research aimed at understanding the biology of this important food-borne organism. It is possible to envisage treatments aimed at eradicating the bacteria from the site, and such control strategies, whether applied at a farm or an abattoir level, could significantly benefit human health.
Acknowledgments
We acknowledge funding of S.W.N. from a DEFRA Veterinary Fellowship to D.L.G. J.C.L. and I.J.M. acknowledge the financial support of SEERAD. G.J.G. and M.C.P. acknowledge the support of the Wellcome Foundation via the IPRAVE project.
We also thank Alison Baird for processing of the sections, Linda Sharp for assistance with confocal microscopy, Carol Currie for assistance with necropsy sample processing, and the staff of the Clinical Department at the Moredun Research Institute for providing expert animal care and assistance.
Editor: A. D. O'Brien
REFERENCES
- 1.AduBobie, J., G. Frankel, C. Bain, A. G. Goncalves, L. R. Trabulsi, G. Douce, S. Knutton, and G. Dougan. 1998. Detection of intimins alpha, beta, gamma, and delta, four intimin derivatives expressed by attaching and effacing microbial pathogens. J. Clin. Microbiol. 36:662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, L., A. Stirrat, and F. M. Thompson-Carter. 1998. Genetic heterogeneity of Escherichia coli O157:H7 in Scotland and its utility in strain subtyping. Eur. J. Clin. Microbiol. Infect. Dis. 17:844-848. [DOI] [PubMed] [Google Scholar]
- 3.Baehler, A. A., and R. A. Moxley. 2000. Escherichia coli O157:H7 induces attaching-effacing lesions in large intestinal mucosal explants from adult cattle. FEMS Microbiol. Lett. 185:239-242. [DOI] [PubMed] [Google Scholar]
- 4.Bastian, S. N., I. Carle, and F. Grimont. 1998. Comparison of 14 PCR systems for the detection and subtyping of stx genes in Shiga-toxin-producing Escherichia coli. Res. Microbiol. 149:457-472. [DOI] [PubMed] [Google Scholar]
- 5.Besser, R. E., P. M. Griffin, and L. Slutsker. 1999. Escherichia coli O157:H7 gastroenteritis and the hemolytic uremic syndrome: an emerging infectious disease. Annu. Rev. Med. 50:355-367. [DOI] [PubMed] [Google Scholar]
- 6.Besser, T. E., B. L. Richards, D. H. Rice, and D. D. Hancock. 2001. Escherichia coli O157:H7 infection of calves: infectious dose and direct contact transmission. Epidemiol. Infect. 127:555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Besser, T. E., D. D. Hancock, L. C. Pritchett, E. M. McRae, D. H. Rice, and P. I. Tarr. 1997. Duration of detection of fecal excretion of Escherichia coli O157:H7 in cattle. J. Infect. Dis. 175:726-729. [DOI] [PubMed] [Google Scholar]
- 8.Borczyk, A. A., M. A. Karmali, H. Lior, and L. M. C. Duncan. 1987. Bovine reservoir for verotoxin-producing Escherichia coli O157:H7. Lancet i:98. [DOI] [PubMed] [Google Scholar]
- 9.Brown, C. A., B. G. Harmon, T. Zhao, and M. P. Doyle. 1997. Experimental Escherichia coli O157:H7 carriage in calves. Appl. Environ. Microbiol. 63:27-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, H., and R. Prescott. 1999. Applied mixed models in medicine. Wiley and Sons, Chichester, England.
- 11.Cornick, N. A., S. L. Booher, and H. W. Moon. 2002. Intimin facilitates colonization by Escherichia coli O157:H7 in adult ruminants. Infect. Immun. 70:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cray W. C., Jr., and H. W. Moon. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:1586-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean-Nystrom, E. A., B. T. Bosworth, W. C. Cray, and H. W. Moon. 1997. Pathogenicity of Escherichia coli O157:H7 in the intestines of neonatal calves. Infect. Immun. 65:1842-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean-Nystrom, E. A., B. T. Bosworth, and H. W. Moon. 1999. Pathogenesis of Escherichia coli O157:H7 in weaned calves. Adv. Exp. Med. Biol. 473:173-177. [DOI] [PubMed] [Google Scholar]
- 15.Fagan, P. K., M. A. Hornitzky, K. A. Bettelheim, and S. P. Djordjevic. 1999. Detection of Shiga toxin (stx1 and stx2), intimin (eaeA), and enterohemorrhagic Escherichia coli (EHEC) hemolysin (EHEC hlyA) genes in animal feces by multiplex PCR. Appl. Environ. Microbiol. 65:868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 17.Grauke, L. J., I. T. Kudva, J. W. Yoon, C. W. Hunt, C. J. Williams, and C. J. Hovde. 2002. Gastrointestinal tract location of Escherichia coli O157:H7 in ruminants. Appl. Environ. Microbiol. 68:2269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen, W., and E. Yourassowsky. 1984. Detection of beta-glucuronidase in lactose-fermenting members of the family Enterobacteriaceae and its presence in bacterial urine cultures. J. Clin. Microbiol. 20:1177-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartland, E. L., V. Huter, L. M. Higgins, N. S. Goncalves, G. Dougan, A. D. Phillips, T. T. MacDonald, and G. Frankel. 2000. Expression of intimin γ from enterohemorrhagic Escherichia coli in Citrobacter rodentium. Infect. Immun. 68:4637-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen, V. B., J. T. Harty, and B. D. Jones. 1998. Interactions of the invasive pathogens Salmonella typhimurium, Listeria monocytogenes, and Shigella flexneri with M cells and murine Peyer's patches. Infect. Immun. 66:3758-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karmali, M., M. Petric, C. Lim, P. Fleming, and B. Steele. 1983. Escherichia coli cytotoxin, hemolytic uremic syndrome and hemorrhagic colitis. Lancet ii:1299-1300. [DOI] [PubMed] [Google Scholar]
- 22.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 23.McCullagh, P., and J. A. Nelder. 1989. Generalised linear models, 2nd ed. Chapman and Hall, London., England.
- 24.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 25.McNally, A., A. J. Roe, S. Simpson, F. M. Thomson-Carter, D. E. E. Hoey, C. Currie, T. Chakraborty, D. G. E. Smith, and D. L. Gally. 2001. Differences in levels of secreted locus of enterocyte effacement proteins between human disease-associated and bovine Escherichia coli O157:H7. Infect. Immun. 69:5107-5114. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostroff, S. M., P. M. Griffin, R. V. Tauxe, L. D. Shipman, K. D. Greene, J. G. Wells, J. H. Lewis, P. A. Blake, and J. M. Kobayashi. 1990. A statewide outbreak of Escherichia coli O157:H7 infections in Washington State. Am. J. Epidemiol. 132:239-247. [DOI] [PubMed] [Google Scholar]
- 28.Parks, A. G., and B. C. Morson. 1962. Fistula-in-ano. Proc. R. Soc. Med. 55:751-754. [Google Scholar]
- 29.Phillips, A. D., S. Navabpour, S. Hicks, G. Dougan, T. Wallis, and G. Frankel. 2000. Enterohaemorrhagic Escherichia coli O157:H7 target Peyer's patches in humans and cause attaching/effacing lesions in both human and bovine intestine. Gut 47:377-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratnam, S., S. B. March, R. Almed, G. S. Bezanson, and S. Kasatiya. 1988. Characterisation of Escherichia coli serotype O157:H7. J. Clin. Microbiol. 26:2006-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sedgmen, B. J., S. A. Lofthouse, J. P. Y. Scheerlinck, and E. N. T. Meeusen. 2002. Cellular and molecular characterisation of the ovine rectal mucosal environment. Vet. Immunol. Immunopathol. 86:215-220. [DOI] [PubMed] [Google Scholar]
- 32.Vazquez-Torres, A., and F. C. Fang. 2000. Cellular routes of invasion by enteropathogens. Curr. Opin. Microbiol. 3:54-59. [DOI] [PubMed] [Google Scholar]