Abstract
A better understanding of mycobacterial gene regulation under certain stress conditions (e.g., low pH) may provide insight into mechanisms of adaptation during infection. To identify mycobacterial promoters induced at low pH, we adapted the recombinase-based in vivo expression technology (RIVET) promoter trap system for use with mycobacteria. Our results show that the TnpR recombinase of transposon γδ is active in Mycobacterium smegmatis and Mycobacterium tuberculosis. We developed a method to perform sequential double selection with mycobacteria by using RIVET, with a kanamycin preselection and a sucrose postselection. A library of M. tuberculosis DNA inserted upstream of tnpR was created, and using the double selection, we identified two promoters which are upregulated at low pH. The promoter regions drive the expression of a gene encoding a putative lipase, lipF (Rv3487c), as well as a PE-PGRS gene, Rv0834c, in a pH-dependent manner in both M. smegmatis and M. tuberculosis. The acid inducibility of lipF and Rv0834c was independent of the stress response sigma factor, SigF, as acid induction of the two genes in an M. tuberculosis sigF mutant strain was similar to that in the wild-type strain. No induction of lipF or Rv0834c was observed during infection of J774 murine macrophages, an observation which is in agreement with previous reports on the failure of phagosomes containing M. tuberculosis to acidify.
One-third of the world's population is infected with Mycobacterium tuberculosis, the causative agent of tuberculosis (15). M. tuberculosis survives for prolonged periods within phagosomes of infected macrophages (14). In addition, M. tuberculosis can persist within the host in an asymptomatic, latent state and can reactivate years later if the host's immune system wanes. The exact location of these latent bacilli within the host has been the subject of some speculation and controversy, but a possible site is within the calcified remnants of healed granulomas (21, 28). Considering the magnitude of the threat to public health posed by M. tuberculosis, research into the pathogenesis of the bacterium is of vital importance. Our present lack of understanding of how M. tuberculosis evades host defenses and causes disease remains a limitation in developing improved therapeutic and diagnostic tools against tuberculosis.
Recombinase-based in vivo expression technology (RIVET) is a type of promoter trap first described for use with Vibrio cholerae and later with Staphylococcus aureus (5, 19). RIVET uses the Escherichia coli transposon γδ recombinase, TnpR, which recombines DNA at specific res sites. DNA sequences between these res sites are lost upon recombination, and antibiotic resistance genes placed between res sites may be used as markers for the recombination event (4).
We adapted this system for use with mycobacteria by placing genes for kanamycin resistance and sucrose sensitivity between the DNA res sites. In the absence of tnpR expression, mycobacteria are kanamycin resistant and sucrose sensitive, but after tnpR expression from an active promoter, the strain becomes kanamycin sensitive and sucrose resistant. With this system, it is possible to identify differentially regulated promoters of mycobacteria within libraries of promoters cloned upstream of a tnpR gene. By preselecting with kanamycin for promoter inactivity, administering a stimulus, and then postselecting for sucrose resistance, promoters activated by the stimulus may be identified. We have used this double selection to search for genes of M. tuberculosis which are upregulated under conditions of low pH. Such genes may be important for the pathogenic mycobacteria to resist acid stress during infection, as caseating granulomas that form in an infected host may have significantly lowered the pH at their centers (11, 12).
MATERIALS AND METHODS
Bacterial strains and plasmids.
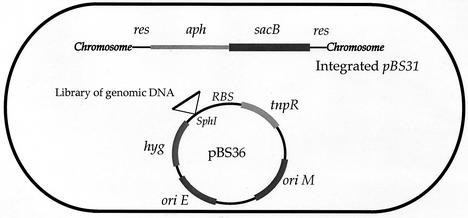
Mycobacterium smegmatis strain mc26 1-2C, M. tuberculosis strain CDC1551, and E. coli strain SH288 were the parent strains for all genetic manipulations and transformations. All incubations were carried out at 37°C. The tnpR promoter trap plasmid was constructed by inserting a promoterless tnpR gene amplified by PCR from pIVET6 (4) into the XbaI site of pMH94 (18) to create pBS14. tnpR was subsequently excised as an XbaI fragment, blunted, and ligated into the PvuII site of pNBV1 (17) to create pBS25. To create a terminator, oligonucleotides containing the sequence for the stem-loop terminator downstream of the M. tuberculosis whiB3 gene (Rv3416), an internal SphI site, and terminal SpeI and HindIII sites were annealed and ligated into the SpeI and HindIII sites of pBS25 to create pBS36 (see Fig. 1).
FIG. 1.
Schematic diagram of the mycobacterial RIVET system used in this study. The elements carried by integrating plasmid pBS31 and episomal tnpR promoter-probe plasmid pBS36 are as follows: res is the recombination site for TnpR resolvase; aph encodes aminoglycoside phosphotransferase, conferring kanamycin resistance; sacB encodes levan sucrase, which is toxic in the presence of sucrose; tnpR encodes the TnpR resolvase of transposon γδ; oriM is the origin of replication for propagation in mycobacteria; oriE is the origin of replication for propagation in E. coli; hyg confers resistance to hygromycin. RBS, ribosome-binding site for tnpR.
The indicator plasmid pBS31 was constructed by inserting the sequence for res-aph-res into pMH94. Oligonucleotides BS21 and BS26 containing a DNA res site and terminal HindIII and XmaI sites were annealed. Oligonucleotides BS23 and BS25 containing a DNA res site and terminal MfeI and HindIII sites were annealed. The aph gene of pMH94 was PCR amplified by using oligonucleotides containing MfeI and XmaI terminal restriction enzyme sites. The product was then ligated to the annealed BS21-BS26 and BS23-BS25 oligonucleotides, cut with HindIII, and inserted into the HindIII site of pMH94 to create pBS20. The resultant plasmid was cut with MfeI, and a sacB-containing fragment from pBS-SK+ was inserted to create pBS31.
The Pace::tnpR plasmid was created by amplifying the acetamide-inducible gene block and promoter from pAGAN11 (23) by PCR and inserting the fragment into the SpeI and BspHI sites of pBS14 to create pBS16. The DraI-XbaI fragment of pBS16 containing the Pace::tnpR fragment was then cloned into EcoRV-XbaI-digested pNBV1, creating pBS18.
The library of CDC1551 genomic DNA cloned upstream of tnpR was created by using pBS36. Total genomic DNA was partially cut with the restriction enzyme NlaIII and size fractionated by using a 0.7% agarose gel. DNA with a size of 1.0 kb was extracted and ligated into pBS36 that had been cut with SphI and treated with calf intestinal alkaline phosphatase. Six separate ligations were used to transform E. coli SH288. Two thousand colonies were pooled from each transformation, and the plasmid DNA was isolated by using Wizard Minipreps (Promega). Plasmid DNA from the six individual preparations was then used to transform M. smegmatis mc26 1-2C containing integrated pBS31.
To analyze promoter regions, we used the gfp promoter-probe plasmid pFPV27 (2). The putative promoter region for lipF from −150 to −626 with respect to the putative start of translation was amplified by PCR. The putative promoter region for Rv0834c from −1 to −468 was similarly amplified. The promoter region from the constitutively expressed Rv2415c gene was used as a positive control. The fragments were cut with KpnI and BamHI (sites built into the PCR primers) and ligated into pFPV27 to create PlipF::gfp, PRv0834c::gfp, and PRv2415c::gfp, respectively.
Genetic selection.
To preselect the library of CDC1551 DNA, recombinant M. smegmatis containing the pBS36 library was grown in 7H9 medium plus kanamycin (25 μg/ml) at pH 7.0 for nine generations. The bacteria were diluted in the same medium, grown to mid-log phase, and pelleted. The bacteria were resuspended in 7H9 medium adjusted to pH 4.5 with HCl and grown for another 3 h. The bacteria were then washed and diluted in 7H9 medium at pH 7.0 and plated onto 7H10 agar-hygromycin (50 μg/ml)-10% sucrose to obtain sucrose-resistant colonies. This was repeated for two cycles, at which point 30 clones were analyzed for their acid induction.
Determination of TnpR activity.
To determine if the promoter trap plasmid pBS36 had background expression, the level of TnpR activity was determined. Both M. smegmatis and M. tuberculosis containing pBS36 and pBS31 or pNBV1 and pBS31 were grown for nine generations in 7H9 medium in the absence of kanamycin selection. The mycobacteria were washed and diluted in 7H9 medium and plated onto 7H10 agar to obtain single colonies. Colonies were then patched onto 7H10 agar-kanamycin plates to assay for the loss of the aph (kanamycin resistance) gene. All measurements were done in triplicate. To determine if tnpR is inducible in mycobacteria, M. smegmatis bacteria containing pBS18 and pBS31, pBS36 and pBS31, or pNBV1 and pBS31 were grown in 7H9 medium for one generation in the absence of kanamycin selection and in the presence or absence of acetamide. The bacteria were diluted and plated for single colonies onto 7H10 agar-hygromycin and onto 7H10 agar-hygromycin-10% sucrose. The colonies that grew on 7H10 agar-hygromycin were patched onto 7H10 agar-hygromycin-kanamycin to assay for the loss of the aph gene. All measurements were done in triplicate. To determine the inducibility of the PlipF::tnpR and PRv0834c::tnpR promoter fusions, M. smegmatis bacteria bearing these constructs as well as pBS36 were grown to mid-log phase in the presence of kanamycin. The bacteria were pelleted, washed, and resuspended in 7H9 medium at either pH 7.0 or pH 4.5 for 3 h. All bacterial samples were then washed, resuspended in 7H9 medium at pH 7.0, diluted to obtain single colonies, and plated onto 7H10 agar-hygromycin plates. Single colonies were patched onto 7H10 agar-hygromycin-kanamycin plates to assess the loss of kanamycin resistance and hence TnpR production. All measurements were done in triplicate.
Flow cytometry.
M. smegmatis and M. tuberculosis strains containing pBEN (Phsp60::gfp), pFPV27, PlipF::gfp, PRv0834c::gfp, and PRv2415c::gfp were grown to mid-log phase at 37°C in 7H9 medium at pH 7.0. The bacteria were then washed and resuspended in 7H9 medium at either pH 7.0 or pH 4.5 and grown at 37°C for 4, 8, and 24 h. Bacteria were then diluted in water and analyzed with a Becton Dickinson FACSCalibur flow cytometry system equipped with an argon laser at a wavelength of 488 nm. The bacteria were detected by side scatter and forward scatter. Fluorescence data were collected by using logarithmic amplifiers.
Macrophage infection and fluorescence microscopy.
Cell culture was performed by using Dulbecco modified Eagle medium (DMEM; high glucose formula) supplemented with 10% fetal bovine serum, 2 mM glutamine, 15 mM HEPES, and 0.02% NaHCO3 (all from Life Technologies). All incubations were performed at 37°C and 5% CO2 in humidified incubators. J774 cells were seeded at 105 cells per well in four-well Lab-Tek II chamber slides (Nalge Nunc International) and allowed to settle and adhere for 5 h prior to the addition of gamma interferon (IFN-γ) to some wells, followed by overnight incubation. Some cells were activated by treatment with 400 U of murine recombinant IFN-γ (Invitrogen)/ml for 24 h and 200 ng of lipopolysaccharide (LPS; Sigma)/ml for 2 h prior to infection. Macrophages were infected at a multiplicity of infection (MOI) of five bacilli per macrophage by using exponentially growing M. tuberculosis strains which had been passed through 5-μm-pore-size syringe filters to remove clumps. The concentrations of bacilli in the filtered samples were assessed by using a hemacytometer to count cells, and the suspensions were diluted with DMEM for infection. Bacteria diluted in DMEM were added to another set of chamber slides without macrophages. After 72 h, medium was removed from the wells, slides were fixed with 4% paraformaldehyde, and macrophage nuclei were stained with Hoechst 33342 (Molecular Probes). Slides were mounted with ProLong Anti-Fade mounting medium (Molecular Probes) and visualized by epifluorescence microscopy on a Nikon E-800 microscope.
RESULTS
RIVET with mycobacteria.
RIVET was adapted for use with mycobacteria. The reporter gene tnpR encodes a recombinase from the E. coli transposon γδ that catalyzes recombination between two DNA res sites, excising the intervening DNA (5). We placed res sites flanking both the aminoglycoside phosphotransferase (aph) gene, providing kanamycin resistance, and the sacB gene, conferring sucrose sensitivity, on a mycobacterial integrating plasmid called pBS31 (25). pBS31 is a derivative of pMH94 and is able to integrate into the mycobacterial attB locus as a single chromosomal copy (18, 26). tnpR was placed on a mycobacterial shuttle plasmid derived from pNBV1 (17) and electroporated into an M. smegmatis strain containing an integrated pBS31 (Fig. 1).
To test whether tnpR expression can be induced in this mycobacterial system, we placed the promoter and regulatory gene block driving expression of M. smegmatis acetamidase (Pace) upstream of tnpR (23). This promoter together with its upstream regulatory genes, cloned as a 4.2-kb cassette, confers chemical inducibility by the substrate acetamide. M. smegmatis bacteria containing integrated pBS31 and the episomal acetamide-inducible Pace::tnpR fusion were grown in hygromycin-kanamycin-7H9 medium to mid-log phase. The cells were then spun down, washed, and resuspended in either 7H9 medium-hygromycin-0.2% acetamide or 7H9 medium-hygromycin. The cells were allowed to grow for 2 h, at which point they were washed with 7H9 medium and plated onto 7H10 agar-hygromycin plates to obtain single colonies. Single colonies were then patched onto 7H10 agar plates containing kanamycin to determine if, in the absence of antibiotic selection and in the presence of acetamide, kanamycin resistance had been lost. Indeed, as seen in Table 1, acetamide induction of tnpR resulted in a 79% loss of kanamycin resistance as opposed to a 2% loss in the absence of acetamide. We confirmed that this loss was due to the excision of the res-aph-sacB-res sequence by monitoring the sequence via PCR with oligonucleotides that amplify the aph gene. Indeed, more than 95% of those bacteria that were unable to grow on kanamycin had lost the aph gene from their chromosomes (data not shown).
TABLE 1.
Acetamide-inducible transposase activity in recombinant M. smegmatis
| Plasmid | Resolution (%) resulting in:
|
|||
|---|---|---|---|---|
| Loss of kanamycin resistance
|
Gain of sucrose resistance
|
|||
| Without acetamide | With acetamide | Without acetamide | With acetamide | |
| Empty vector | 0 | 0 | 0.3 | NDa |
| pBS36 (promoterless tnpR) | 1 | 0 | 1 | ND |
| pBS18 (Pace::tnpR) | 2 | 79 | 4 | 92 |
ND, not done.
To determine if selection on sucrose was feasible, we repeated the above procedure by inducing the M. smegmatis Pace::tnpR fusion strain with acetamide for 2 h. The cells were washed and diluted to allow the isolation of single colonies and were plated onto 7H10 agar-hygromycin plates with or without 10% sucrose. The 7H10 agar plates should support the growth of all bacteria, while the 7H10 agar-sucrose plates should support the growth of only those bacteria which have lost the sacB gene due to the resolution of the res sites. Indeed, from the uninduced culture, the frequency of sucrose-resistant colonies was 4%. After acetamide induction, the frequency of sucrose-resistant colonies increased to 92%, indicating that the majority of cells had lost the sacB gene in response to acetamide induction (Table 1).
A promoterless expression plasmid, pBS25, containing tnpR was tested for residual expression of tnpR in M. smegmatis. In the absence of any promoter, this plasmid expressed some tnpR such that after nine generations of growth in the absence of any antibiotic selection approximately 20% of the bacteria had lost kanamycin resistance (data not shown). In order to decrease the background expression of tnpR, we cloned a 54-bp stem-loop transcriptional terminator upstream of the SphI cloning site in pBS25 to create pBS36. Background expression of this plasmid after insertion of the terminator was reduced and resulted in only 1% of M. smegmatis bacteria having lost kanamycin resistance after nine generations in the absence of antibiotic selection. The background expression was tested in M. tuberculosis bacteria as well and was found to be 1% after nine generations of growth in the absence of antibiotics (data not shown).
Library construction and selection.
The usefulness of tnpR as a reporter gene to isolate differentially regulated promoters from a library was assessed by using low pH as a stress condition. We created a library of M. tuberculosis DNA cloned upstream of tnpR within the mycobacterial shuttle vector pBS36. This library was then used to transform M. smegmatis containing an integrated copy of pBS31 and to isolate M. tuberculosis promoters upregulated upon exposure to pH 4.5. Restriction analysis of 25 representative E. coli clones from the DNA library revealed that 90% of the clones contained M. tuberculosis chromosomal DNA inserts. The average size of the insert was approximately 1 kb.
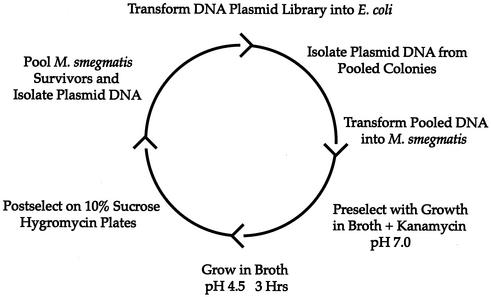
Six pools of 2,000 E. coli clones were used to prepare six amplified DNA plasmid libraries. The DNA from the six pools was then used to transform M. smegmatis, generating six pools of M. smegmatis recombinant reporter strains with each pool consisting of approximately 10,000 clones. In order to screen for acid-inducible promoters, we subjected these M. smegmatis pools to low-pH stress. We first preselected the library to eliminate promoters which were expressed during vegetative growth at neutral pH. To accomplish this, we isolated clones that grew on hygromycin-kanamycin-7H10 agar plates and grew these clones in pools to mid-log phase in the presence of kanamycin and hygromycin in 7H9 medium at neutral pH. Only those clones devoid of promoters or with promoters that are inactive at neutral pH can survive in the presence of kanamycin. The M. smegmatis pools were then spun down and resuspended in 7H9 medium at pH 4.5 without kanamycin. After growth for 3 h in acidic medium, the bacteria were washed with 7H9 medium, pH 7.0, and plated onto 7H10 agar containing 10% sucrose and hygromycin. This allowed the growth of any bacteria that had lost the aph and sacB genes due to an upregulated promoter in response to acid induction. Bacteria that grew on the sucrose plates were pooled, and plasmid DNA was prepared and used to transform E. coli to amplify the plasmid DNA. The amplified plasmid DNA was isolated and used to transform M. smegmatis to undergo another round of selection (Fig. 2). After the second round, the bacteria growing on the sucrose plates were pooled and the DNA was recovered and used to transform E. coli as before. We focused on 30 clones, each of which was retested for acid induction by the same RIVET assay system by using the single pure plasmid to transform M. smegmatis harboring an integrated pBS31. Two clones were found to be reproducibly upregulated in response to medium at pH 4.5.
FIG. 2.
Strategy for the identification of acid-inducible promoters of M. tuberculosis by using RIVET-based sequential double selection. The M. tuberculosis chromosomal DNA library cloned upstream of tnpR in pBS36 was first used to transform E. coli, and plasmid DNA was isolated from clones in six pools of 2,000. The plasmid DNA was used to transform M. smegmatis to create six pools. The mycobacteria were preselected by growth in 7H9 medium, pH 7.0, in the presence of kanamycin. The bacteria were then washed and resuspended in 7H9 medium, pH 4.5, containing no kanamycin for 3 h. The mycobacteria were then postselected by growth on 7H10 agar plates containing 10% sucrose. The plasmid DNA was then isolated from M. smegmatis bacteria growing on the 10% sucrose plates. Two rounds of sequential double selection were performed.
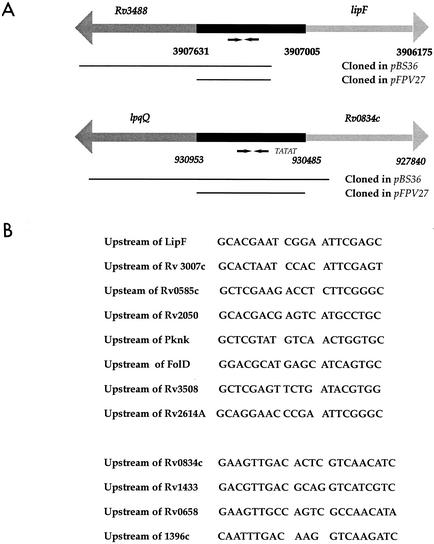
These two clones were sequenced and found to contain M. tuberculosis DNA corresponding to regions upstream of the genes lipF (Rv3487c) and Rv0834c (Fig. 3A). The levels of resolution induced by exposure to acidic medium were quantitated for the two clones designated lipF-tnpR and Rv0834c-tnpR. Each clone was tested in triplicate. In the presence of acid, lipF-tnpR had a resolution rate of 85%, whereas in the presence of neutral pH it had a resolution frequency of 2%. Rv0834c-tnpR had a resolution rate of 55% in the presence of acid and a 22% resolution rate in the presence of neutral pH. As expected, M. smegmatis bearing the promoterless pBS36 had no increase in the level of resolution during acid induction.
FIG. 3.
(A) Map of the M. tuberculosis lipF and Rv0834c genetic regions. The numbers denote the locations of the genes within the M. tuberculosis H37Rv genome sequence. Putative promoter regions are shown in black and coding sequences are in gray. Inverted repeat sequences are identified with arrows below the putative promoter regions. Bars below the genetic regions indicate DNA that was cloned upstream of tnpR in pBS36 or upstream of gfp in pFPV27. A TATAT putative −10 promoter element was identified upstream of Rv0834c. (B) The sequences of the inverted repeats preceding the lipF and Rv0834c genes are shown. Seven inverted repeat sequences similar to the repeat upstream of lipF were identified elsewhere in the M. tuberculosis genome, and three sequences similar to the Rv0834c repeat were identified.
Confirmation of acid inducibility by gfp reporter analysis.
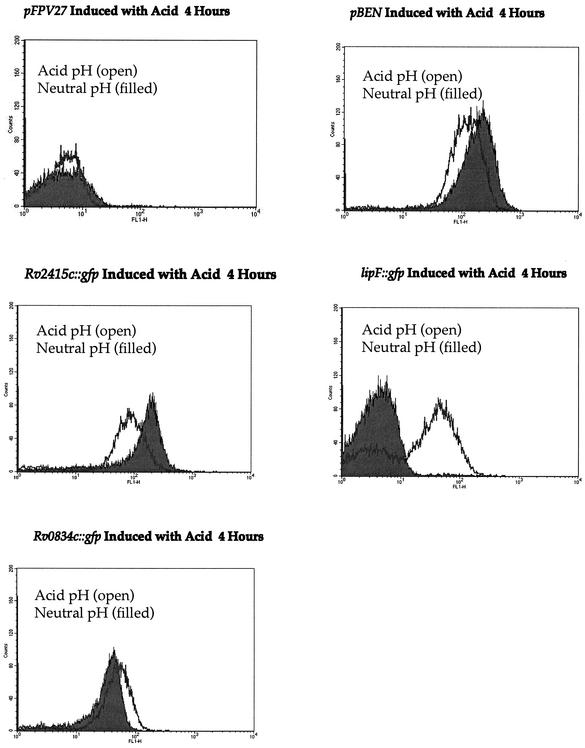
In order to confirm the acid inducibility of lipF and Rv0834c, we used gfp, encoding the green fluorescent protein, as a reporter gene (9, 24). We PCR amplified ∼500-bp fragments from the 5′ untranslated regions of lipF and Rv0834c and cloned the products upstream of gfp in the promoter-probe plasmid pFPV27 (2) (Fig. 3A). In addition, the putative promoter region of the constitutively active gene Rv2415c was placed upstream of gfp and a positive control clone called pBEN containing the Mycobacterium bovis BCG hsp60 promoter (Phsp60) cloned into pFPV27 was also used (27). These plasmids were each used to transform M. smegmatis. The bacteria were grown to mid-log phase and acid induced as previously described. The level of acid-induced gfp expression in these strains was assessed by the amount of fluorescence produced by using a Becton Dickinson FACSCalibur flow cytometer. As expected, both lipF and Rv0834c were induced with 4 h of acid exposure, with lipF being more dramatically induced. The induction of lipF increased more than 10-fold in response to acid, while the induction of Rv0834c increased approximately 2-fold (Fig. 4), in general agreement with the tnpR-derived results. The constitutive controls, Phsp60 and the promoter for Rv2415c, were both downregulated in response to acid shock, thus indicating a possible general response to acid shock to reduce transcription of unneeded genes (Fig. 4). The promoterless control reporter plasmid (pFPV27) showed no significant change in expression in response to acid induction. Incubation in medium at pH 4.5 for more than 4 h did not cause an increase in induction of Rv0834c; however, the level of gfp expression continued to increase slightly up until 22 h for lipF (data not shown). M. smegmatis containing the PlipF::gfp fusion was visualized under phase-contrast and fluorescence microscopy after exposure to acidic and neutral pH. Fluorescence was detected after exposure to low pH but not after exposure to neutral pH, indicating significant acid induction of the lipF promoter (Fig. 5).
FIG. 4.
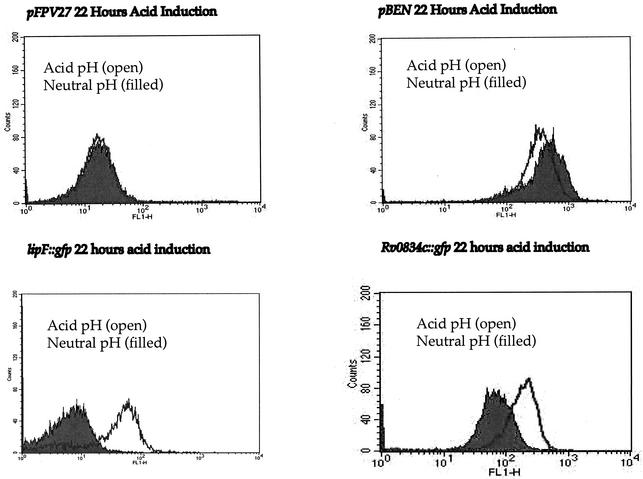
Flow cytometry traces for M. smegmatis transformed with gfp reporter constructs pFPV27 (promoterless gfp), pBEN (Phsp60::gfp), PRv2415c::gfp, PRv0834c::gfp, and PlipF::gfp. The bacteria were induced by growth in 7H9 medium at pH 7.0 or pH 4.5 for 4 h and analyzed with a Becton Dickinson FACSCalibur flow cytometry system. The x axis represents increasing fluorescence (FL), while the y axis represents the number of events. Filled curves are results for cells at pH 7.0 and open curves are results for cells at pH 4.5.
FIG. 5.
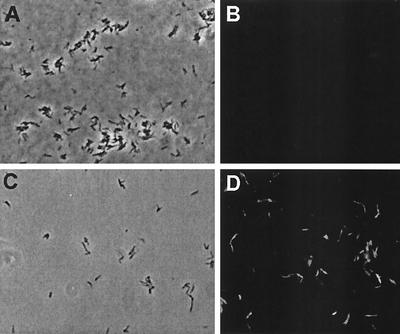
Microscopic analysis of acid-inducible gfp expression in recombinant M. smegmatis. M. smegmatis harboring the PlipF::gfp fusion was grown to mid-log phase, pelleted, and grown in 7H9 medium at either pH 4.5 or pH 7.0 for 4 h. The bacteria were then visualized by phase-contrast and fluorescence microscopy. (A and C) Phase-contrast images of M. smegmatis PlipF::gfp bacteria in 7H9 medium at neutral pH (A) and at pH 4.5 (C). (B and D) Fluorescence images of M. smegmatis PlipF::gfp bacteria in 7H9 medium at neutral pH (B) and at pH 4.5 (D). Images are not identical fields.
Analysis of promoter regions.
In analyzing the putative promoter regions of lipF and Rv0834c, we identified two different inverted repeat sequences in the 5′ untranslated regions of the acid-induced genes. An inverted repeat was found within the putative promoter region for lipF and consisted of 8-bp half sites (5′-GCACGAAT-3′) separated by a 4-bp spacer (Fig. 3B). A different 9-bp inverted repeat containing one mismatch and separated by a 4-bp spacer was identified in the 5′ untranslated region preceding Rv0834c (5′-GAaGTTGAC-3′ [mismatch denoted by lowercase]). In searching the M. tuberculosis genome, we found several other instances of near-perfect inverted repeat sequences similar to those preceding the lipF and Rv0834c genes in the 5′ untranslated regions of other genes, as may be seen in Fig. 3B.
Survival of M. smegmatis under low pH conditions.
To assess the extent of killing during acid induction, M. smegmatis was exposed to pH 4.5 for 3, 8, and 22 h. At 3 h there was no appreciable killing of the bacteria (based on comparison of CFU per milliliter in acid-treated and control suspensions of M. smegmatis). However, at 8 h approximately 30% of the bacteria were killed due to the acidic conditions. At 22 h after acid induction, there was a significant level of killing, as 70% of the bacteria were nonviable. Therefore, while acidified medium does not have an immediate effect on the viability of the mycobacteria, over time it becomes bactericidal. Hence, under the selection conditions used in our RIVET assay, there was no appreciable bacterial killing by low pH.
Low-pH inducibility of the lipF and Rv0834c promoters in M. tuberculosis.
We assessed the level of acid-induced transcription within M. tuberculosis by using the PlipF::gfp and PRv0834c::gfp promoter-probe constructs to transform M. tuberculosis. The M. tuberculosis transformants were grown to mid-log phase and acid induced as previously described, and their fluorescence was monitored by using flow cytometry. Again, both promoter regions were induced at pH 4.5. Induction was found to be slower than that in M. smegmatis. While there was induction at 4 h postacidification, the greatest increase in acid induction occurred at 22 h (Fig. 6). Again, PlipF was upregulated approximately 10-fold, while PRv0834c, being semiconstitutive, was upregulated about 5-fold, an increase similar to that seen in M. smegmatis.
FIG. 6.
Flow cytometry traces for M. tuberculosis transformed with gfp reporter constructs pFPV27 (promoterless gfp), pBEN (Phsp60::gfp), PRv0834c::gfp, and PlipF::gfp. The bacteria were induced by growth in 7H9 medium at pH 7.0 or pH 4.5 for 22 h and analyzed with a Becton Dickinson FACSCalibur flow cytometry system. The x axis represents fluorescence (FL) intensity, while the y axis represents the number of events. Filled curves are results for bacteria at pH 7.0 and open curves are results for bacteria at pH 4.5.
Acid-induced expression of lipF and Rv0834c is not dependent on the M. tuberculosis sigma factor, SigF.
To test whether the M. tuberculosis genes lipF and Rv0834c are acid regulated by the stress response sigma factor, SigF, we used the promoter-gfp fusions to transform both an M. tuberculosis strain with a deletion of sigF and its isogenic wild-type parent strain, CDC1551 (6, 13, 20). When the M. tuberculosis transformants were acid induced, there were equivalent levels of induction in the wild-type strains and the sigF mutant strains bearing the PlipF::gfp and PRv0834c::gfp fusions. These data indicate that SigF does not mediate the acid-inducible expression of these promoters. As expected, there was no difference between the pre- and post-acid induction expression of gfp in either the wild-type strain or the sigF mutant strains bearing the vector control pFPV27 or the Phsp60::gfp plasmid pBEN (data not shown).
lipF and Rv0834c are not induced within the phagosome of J774 murine macrophages.
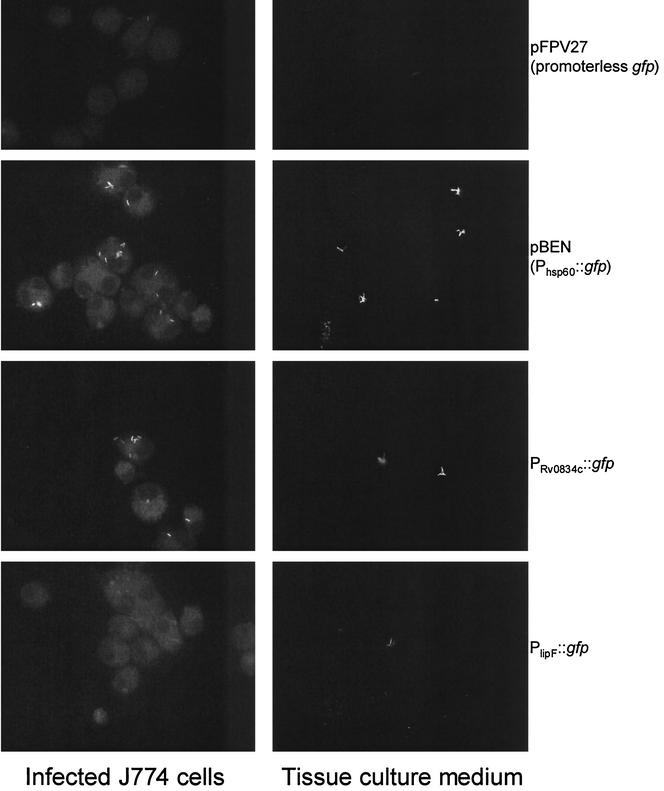
To investigate whether lipF or Rv0834c is induced within the intracellular environment of the macrophage phagosome, M. tuberculosis CDC1551 bacteria harboring pFPV27 (promoterless gfp), pBEN (Phsp60::gfp), PlipF::gfp, or PRv0834c::gfp were used to infect J774 cells which were either untreated or pretreated with IFN-γ and LPS. For comparison, the bacteria were also exposed to tissue culture medium alone. Infections were carried out at an MOI of five bacilli per J774 cell, and all samples were incubated at 37°C and 5% CO2 in a humidified incubator. After 72 h, cells were fixed with 4% paraformaldehyde and visualized by epifluorescence microscopy. By comparing the bacilli exposed to tissue culture medium alone to those within either resting or activated macrophages, we were unable to detect any change in the level of green fluorescent protein fluorescence in either the lipF or the Rv0834c fusion strain (Fig. 7).
FIG. 7.
Expression analysis of lipF and Rv0834c in J774 murine macrophages. Macrophages, activated with IFN-γ and LPS, were infected at an MOI of five bacilli per cell as described in Materials and Methods. After 72 h, cells were fixed, mounted, visualized by epifluorescence microscopy, and compared to bacilli exposed to tissue culture medium alone. Shown are corresponding images of M. tuberculosis Oshkosh strains in activated macrophages (left panels) and in tissue culture medium alone (right panels). Similar levels of fluorescence were observed in the unstimulated macrophages (data not shown).
DISCUSSION
In order to facilitate the isolation of differentially regulated genes in mycobacteria, we have adapted RIVET for use with M. smegmatis and other mycobacteria. The RIVET system has previously been used with V. cholerae and S. aureus to isolate in vivo-induced genes (5, 19) and may also be used to identify genes conditionally upregulated by in vitro stress. For V. cholerae and S. aureus, conditionally induced genes were selected first for inactivity in rich medium, after which a second screening process identified genes upregulated by stress or in vivo passage. In our present adaptation of the RIVET system we have added a positive selection (resistance to sucrose) to aid in the identification of conditionally regulated genes of mycobacteria.
In this study, we were able to isolate two putative promoter regions which are specifically upregulated in acidic medium. After two rounds of selection for promoters transcriptionally silent at pH 7.0 but transcriptionally upregulated at pH 4.5, 30 clones were identified and were subsequently screened individually. On repeat testing by RIVET, two clones proved to be reproducibly upregulated during exposure to acidic medium. The gene fragments identified by the procedure corresponded to putative promoter sequences for the M. tuberculosis lipF and Rv0834c genes. The promoter regions for lipF and Rv0834c were cloned upstream of a promoterless gfp gene and confirmed to be upregulated in the presence of acid. The lipF gene is annotated in the M. tuberculosis genome as being a lipase or esterase. As lipF is so dramatically upregulated at low pH, a possible function of lipF could be to hydrolyze toxic fatty acids present in caseous necrotic debris during tuberculosis; alternatively, it may modify the external cell wall as an adaptive response to acid damage. Rv0834c is annotated in the M. tuberculosis genome as a PE-PGRS gene that may be involved in modulating the immune system (8, 27). In a recent study using microarray analysis, several M. tuberculosis genes involved in fatty acid metabolism were shown to be induced in response to acid shock (16). Neither lipF nor Rv0834c had an increase in induction of more than 1.5-fold, the cutoff used in that study. It is possible that differences in the techniques between that study and the present study, e.g., induction at pH 5.5 versus induction at pH 4.5, account for the discrepancy. Alternatively, the inherent amplification of translation (i.e., one mRNA translated into multiple copies of protein) may have allowed us to detect acid-induced genes with greater sensitivity. It is also unclear why only 2 out of the 30 clones we investigated demonstrated reproducible acid induction. The most likely reason for this would be “leaky” expression of tnpR. We measured the background of our RIVET system to be around 1%. Even this low level of background could generate a large number of false positives. Further modification of the system may reduce the level of background tnpR expression.
We were unable to observe the induction of lipF or Rv0834c during macrophage infection. It is possible that the phagosomal compartment fails to acidify to the degree required for induction. Indeed, one of the tenets of mycobacterial pathobiology has been the prevention of phagosome maturation and subsequent acidification (7, 10, 14, 30, 31). However, it has been suggested that the centers of caseating granulomas can be quite acidic in vivo (11, 12). Therefore, acid-induced genes of M. tuberculosis could play a role in survival in such an environment.
In analyzing the putative promoter regions of lipF and Rv0834c, we identified two unique inverted repeat sequences—one upstream of lipF and the other upstream of Rv0834c. Inverted repeats similar to that preceding lipF were found in seven other 5′ untranslated regions in the M. tuberculosis genome, and repeats resembling that upstream of Rv0834c were found preceding three other M. tuberculosis genes. Bacterial transcriptional regulators are known to recognize and bind to DNA inverted repeats (1, 22, 29), and this raises the possibility that the repeats in the lipF and Rv0834c promoter regions may play a role in the transcriptional response to acid stress.
DNA-binding transcriptional regulators may either repress or activate transcription. For example, Rv0834c is semiconstitutive and a TATAT sequence can be found 59 bp upstream of the putative start site of translation and 15 bp downstream of the inverted repeat (Fig. 3A). This TATAT sequence is identical to the −10 region of a constitutive promoter identified in a study by Bashyam et al. and could explain the basal level of transcription of Rv0834c (3). Binding of a transcriptional regulator in response to acidic stress may then serve to increase the level of transcription beyond the constitutive level. The lipF gene is not expressed to any appreciable level in the absence of acidic stress; a transcriptional regulator may serve either to repress transcription in the absence of acidic stress or to activate transcription in the presence of acidic stress. Acid-induced genes such as lipF and Rv0834c may play a role in the pathogenesis of human tuberculosis because of the association between low pH and the core of granulomatous lesions. Further investigation is warranted to determine whether the inverted repeats play a role in the regulation of lipF and Rv0834c and to further define the role of these low-pH-responsive genes in infection.
Acknowledgments
We thank A. Camilli for supplying pIVET6 and T. Parish for pAGAN11.
This work was supported by a postdoctoral fellowship to B.S. from the Heiser Foundation, an NRSA fellowship to S.C.W. (AI50360), and NIH grants AI36973, AI37856, and AI43846.
Editor: J. N. Weiser
REFERENCES
- 1.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker, L. P., D. M. Brooks, and P. L. Small. 1998. The identification of Mycobacterium marinum genes differentially expressed in macrophage phagosomes using promoter fusions to green fluorescent protein. Mol. Microbiol. 29:1167-1177. [DOI] [PubMed] [Google Scholar]
- 3.Bashyam, M. D., D. Kaushal, S. K. Dasgupta, and A. K. Tyagi. 1996. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J. Bacteriol. 178:4847-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camilli, A., D. T. Beattie, and J. J. Mekalanos. 1994. Use of genetic recombination as a reporter of gene expression. Proc. Natl. Acad. Sci. USA 91:2634-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camilli, A., and J. J. Mekalanos. 1995. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol. Microbiol. 18:671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, P., R. E. Ruiz, Q. Li, R. F. Silver, and W. R. Bishai. 2000. Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene, sigF. Infect. Immun. 68:5575-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemens, D. L., and M. A. Horwitz. 1995. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J. Exp. Med. 181:257-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 9.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 10.Crowle, A. J., R. Dahl, E. Ross, and M. H. May. 1991. Evidence that vesicles containing living, virulent Mycobacterium tuberculosis or Mycobacterium avium in cultured human macrophages are not acidic. Infect. Immun. 59:1823-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dannenberg, A. M., Jr. 1993. Immunopathogenesis of pulmonary tuberculosis. Hosp. Pract. 28:51-58. [DOI] [PubMed] [Google Scholar]
- 12.Dannenberg, A. M., and J. F. Tomashefski. 1997. Pulmonary diseases and disorders. In A. P. Fishman (ed.), Pathogenesis of pulmonary tuberculosis. McGraw-Hill, New York, N.Y.
- 13.DeMaio, J., Y. Zhang, C. Ko, D. B. Young, and W. R. Bishai. 1996. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 93:2790-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deretic, V., and R. A. Fratti. 1999. Mycobacterium tuberculosis phagosome. Mol. Microbiol. 31:1603-1609. [DOI] [PubMed] [Google Scholar]
- 15.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 16.Fisher, M. A., B. B. Plikaytis, and T. M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howard, N. S., J. E. Gomez, C. Ko, and W. R. Bishai. 1995. Color selection with a hygromycin-resistance-based Escherichia coli-mycobacterial shuttle vector. Gene 166:181-182. [DOI] [PubMed] [Google Scholar]
- 18.Lee, M. H., L. Pascopella, W. R. Jacobs, Jr., and G. F. Hatfull. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc. Natl. Acad. Sci. USA 88:3111-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe, A. M., D. T. Beattie, and R. L. Deresiewicz. 1998. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol. Microbiol. 27:967-976. [DOI] [PubMed] [Google Scholar]
- 20.Michele, T. M., C. Ko, and W. R. Bishai. 1999. Exposure to antibiotics induces expression of the Mycobacterium tuberculosis sigF gene: implications for chemotherapy against mycobacterial persistors. Antimicrob. Agents Chemother. 43:218-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Opie, E. L., and J. D. Aronson. 1927. Tubercle bacilli in latent tuberculous lesions and in lung tissue without tuberculous lesions. Arch. Pathol. Lab. Med. 4:1. [Google Scholar]
- 22.Pabo, C. O., and R. T. Sauer. 1984. Protein-DNA recognition. Annu. Rev. Biochem. 53:293-321. [DOI] [PubMed] [Google Scholar]
- 23.Parish, T., E. Mahenthiralingam, P. Draper, E. O. Davis, and M. J. Colston. 1997. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology 143(Pt. 7):2267-2276. [DOI] [PubMed] [Google Scholar]
- 24.Parker, A. E., and L. E. Bermudez. 1997. Expression of the green fluorescent protein (GFP) in Mycobacterium avium as a tool to study the interaction between mycobacteria and host cells. Microb. Pathog. 22:193-198. [DOI] [PubMed] [Google Scholar]
- 25.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Expression of the Bacillus subtilis sacB gene confers sucrose sensitivity on mycobacteria. J. Bacteriol. 178:1197-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pena, C. E., J. E. Stoner, and G. F. Hatfull. 1996. Positions of strand exchange in mycobacteriophage L5 integration and characterization of the attB site. J. Bacteriol. 178:5533-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288:1436-1439. [DOI] [PubMed] [Google Scholar]
- 28.Robertson, H. E. 1933. Persistence of tuberculous infection. Am. J. Pathol. 9:711. [PMC free article] [PubMed] [Google Scholar]
- 29.Segal, G., and E. Z. Ron. 1998. Regulation of heat-shock response in bacteria. Ann. N. Y. Acad. Sci. 851:147-151. [DOI] [PubMed] [Google Scholar]
- 30.Sturgill-Koszycki, S., P. H. Schlesinger, P. Chakraborty, P. L. Haddix, H. L. Collins, A. K. Fok, R. D. Allen, S. L. Gluck, J. Heuser, and D. G. Russell. 1994. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 263:678-681. [DOI] [PubMed] [Google Scholar]
- 31.Xu, S., A. Cooper, S. Sturgill-Koszycki, T. van Heyningen, D. Chatterjee, I. Orme, P. Allen, and D. G. Russell. 1994. Intracellular trafficking in Mycobacterium tuberculosis and Mycobacterium avium-infected macrophages. J. Immunol. 153:2568-2578. [PubMed] [Google Scholar]