Abstract
Background
The past three decades have witnessed a dramatic increase in interest in the whitefly Bemisia tabaci, owing to its nature as a taxonomically cryptic species, the damage it causes to a large number of herbaceous plants because of its specialized feeding in the phloem, and to its ability to serve as a vector of plant viruses. Among the most important plant viruses to be transmitted by B. tabaci are those in the genus Begomovirus (family, Geminiviridae). Surprisingly, little is known about the genome of this whitefly. The haploid genome size for male B. tabaci has been estimated to be approximately one billion bp by flow cytometry analysis, about five times the size of the fruitfly Drosophila melanogaster. The genes involved in whitefly development, in host range plasticity, and in begomovirus vector specificity and competency, are unknown.
Results
To address this general shortage of genomic sequence information, we have constructed three cDNA libraries from non-viruliferous whiteflies (eggs, immature instars, and adults) and two from adult insects that fed on tomato plants infected by two geminiviruses: Tomato yellow leaf curl virus (TYLCV) and Tomato mottle virus (ToMoV). In total, the sequence of 18,976 clones was determined. After quality control, and removal of 5,542 clones of mitochondrial origin 9,110 sequences remained which included 3,843 singletons and 1,017 contigs. Comparisons with public databases indicated that the libraries contained genes involved in cellular and developmental processes. In addition, approximately 1,000 bases aligned with the genome of the B. tabaci endosymbiotic bacterium Candidatus Portiera aleyrodidarum, originating primarily from the egg and instar libraries. Apart from the mitochondrial sequences, the longest and most abundant sequence encodes vitellogenin, which originated from whitefly adult libraries, indicating that much of the gene expression in this insect is directed toward the production of eggs.
Conclusion
This is the first functional genomics project involving a hemipteran (Homopteran) insect from the subtropics/tropics. The B. tabaci sequence database now provides an important tool to initiate identification of whitefly genes involved in development, behaviour, and B. tabaci-mediated begomovirus transmission.
Background
The past three decades have witnessed a dramatic increase in the economic importance of the whitefly Bemisia tabaci (Genn.) (Aleyrodidae; Hemiptera) in subtropical and mild temperate agriculture systems, owing to the damage it causes to plants when it feeds in the phloem, and its ability to transmit plant viruses. B. tabaci occupies tropical and subtropical habitats, producing 11–15 generations per year [1,2]. The B. tabaci complex consists of diverse biological 'types' with distinct genetic polymorphisms [3,4], and differences in host range, fecundity, dispersal behaviours, prokaryotic endosymbiont composition, and competency with respect to begomovirus transmission, a group of small circular ssDNA plant viruses (genus Begomovirus, family Geminiviridae) [5,6]. The highly fecund Old World B biotype can produce ~ 300 eggs/female, colonizes over 500 host species, while the New World A type colonizes about 200 species and has a lower fecundity (~ 100 eggs/female). In contrast, the Jatropha type colonizes only a few species within the genus Jatropha and exhibits low fecundity (~ 30–50 eggs/female) [3].
B. tabaci adults develop from eggs, after passing through four instars in approximately 2–3 wk and development is temperature dependent. Members of this complex are haplodiploid and thus unfertilized eggs give rise to haploid males; fertilized eggs develop into diploid females (arrhenotoky) [1,2].
The B type of B. tabaci transmits begomoviruses to a large number of crop, ornamental, and weed species [7]. Begomovirus have either one (monopartite) or two (bipartite) genomic components [8]. Those infecting tomato constitute a large group of begomoviruses. Among them the bipartite Tomato mottle virus (ToMoV) originated in the New World (Florida/Caribbean region), whereas, the monopartite Old World Tomato yellow leaf curl virus (TYLCV) is indigenous to the Old World (Middle East and Africa). TYLCV recently was introduced to the Caribbean Islands and has since spread into the South eastern states of the U.S.A. [9].
Begomoviruses are transmitted by B. tabaci in a circulative manner [10,11]. Virus particles ingested through the stylets enter the oesophagus and the filter chamber, are transported through the gut into the hemocoel, reach the salivary glands and are finally 'transmitted' during feeding, about 8–12 h after the beginning of an acquisition access period [10]. Velocity of translocation is reported to constitute an intrinsic property of the vector, not of the virus [12,13]. B. tabaci is able to transmit begomoviruses, and in particular TYLCV, for its lifetime, after the latent period has been achieved [14,15]. The ingestion of TYLCV by the whitefly vector is accompanied by a marked decrease in whitefly longevity and fertility [15]. In contrast whiteflies that have ingested ToMoV displayed higher fecundity when reared on virus-free tomato than whiteflies not exposed to the virus [16]. TYLCV transcripts have been found in B. tabaci harbouring this virus, whereas viral transcripts are not detected in whiteflies that have ingested ToMoV [17], suggesting a fundamental difference in interactions between these two begomoviruses and their whitefly vector.
At least one whitefly species that colonizes some of the same hosts as B. tabaci (e.g. the greenhouse whitefly, Trialeurodes vaporariorum) is known to be capable of ingesting, but does not transmit begomoviruses [12], and at least one barrier to transmission has been shown to occur at the gut/hemocoel interface [12,18]. The receptors that are hypothesized to mediate begomovirus translocation into the salivary glands of B. tabaci, which is a requisite to transmission, and their genes, are presently unidentified.
Surprisingly very little is known about the genetic make up of this insect. The nuclear DNA content of B. tabaci male and female was estimated as 1.04 and 2.06 pg respectively, using flow cytometry, indicating that the haploid genome of B. tabaci contains about one billion bp, which is approximately five times the size of the genome of the fruitfly Drosophila melanogaster [19]. However, it is still not clear if this size estimate will prove to be accurate and so a long-term goal is to determine the complete genome of this whitefly. Ultimately it is of interest to isolate and identify the genes expressed during the life cycle of the whitefly B. tabaci and to understand the genetic makeup of this pest. Of particular interest is the identification of specific genes and their functions, which are expressed during the development of B. tabaci, as well as those involved in circulative virus transmission, the detoxification of insecticides, and the determination of polyphagy or monophagy in different B. tabaci biotypes. Consequently, the construction of cDNA libraries and the analyses of the sequences for the widespread 'B' biotype of B. tabaci constitute a first step in this endeavour.
Results
Preparation of cDNA libraries
Five independent cDNA, or expressed sequence tag (EST), libraries were prepared from mRNA isolated from adult B. tabaci 'B' biotype whiteflies: (1) reared in Israel on cotton plants [a non-host of TYLCV/ToMoV (= HBT)], or reared in Florida, USA on (2) TYLCV-infected tomato plants (= TYLCV), and (3) ToMoV-infected tomato plants (= TOMOV). Libraries also were constructed from mRNA isolated from the eggs of non-viruliferous whiteflies (= EGG), and immature instars (crawler to pupae) of non-viruliferous insects (= INST). The libraries were not normalized or amplified. Among them the bipartite Tomato mottle virus (ToMoV) originated in the New World (Florida/Caribbean region), whereas, the monopartite Tomato yellow leaf curl virus (TYLCV) is indigenous to the Old World (Middle East and Africa).
Assessment of library and sequence quality
From 18,976 sequencing attempts 9,110 sequences remained after quality, vector and adapter trimming, and removal of mitochondrial DNA sequences (see Materials and methods). The fraction of cleaned sequences from the total number of sequences from each library was between one half and one third (Table 1). The number of sequences from the various libraries that were assembled into contigs and singletons was as follows: EGG: 201, INST: 1816, HBT: 2093, TYLCV: 2704, and TOMOV: 2296 (Table 1).
Table 1.
Number of sequences from the various libraries and number of sequences assembled into contigs (sequences of mitochondrial origin were removed from the contig assembly process)
| Number of sequences | From EGG | From INST | From HBT | FromTYLCV | From TOMOV | |
| Total number of sequenced clones | 18,976 | 673 | 3,745 | 4,321 | 5,857 | 4,380 |
| Clones of mitochondrial origin | 5,542 | 59 | 866 | 1,576 | 1,465 | 1,576 |
| Sequences in contigs and singletons | 9,110 | 201 | 1,816 | 2,093 | 2,704 | 2,296 |
EST assembly into contigs
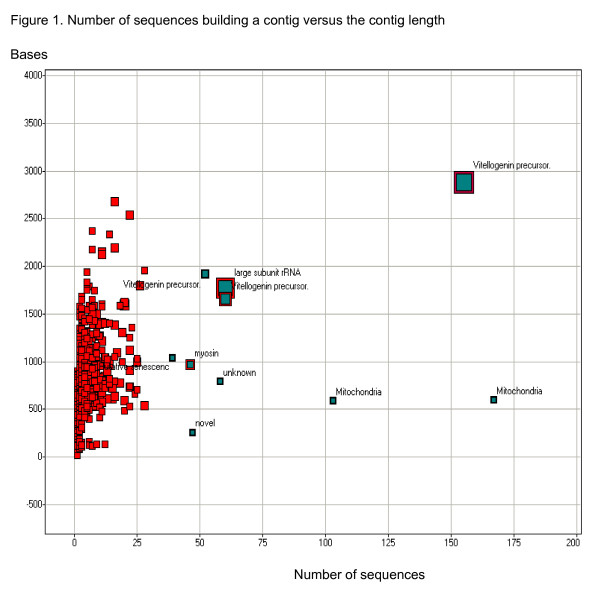
To identify clones belonging to the same gene, sequences were assembled into contigs [see Additional files 1 and 2] using the Staden Gap4 program [20]. The advantage of this program is that both the bases and their quality are used to assess overlap and contig consensus sequence. In the assembly process the genome of the whitefly primary endosymbiotic bacteria Candidatus Portiera aleyrodidarum [21] (AY268081.1) was included since a preliminary analysis revealed that some of the sequences were identical to bacterial DNA. In the assembly process 4,860 contigs and singletons were assembled from 9,109 sequences (Table 2). The number of singletons was 3,843. The GC content and the average length were higher in the contigs than in the singletons. The contigs with more than a single sequence resulted in sequences 1.5 fold longer than the average sequence length. Figure 1 shows that the contigs with up to 25 sequences had a linear relationship between sequence number and contig length, i.e. the assembly process produces longer sequences than the singletons. Table 3 shows that the largest contig (with respect to sequence number) was assembled with sequences from the genome of Candidatus Portiera aleyrodidarum (AY268081.1), and was derived primarily from egg (EGG) and instar (INST) libraries; endosymbiotic sequences were rare in adult whitefly libraries (Figure 2). Most clones that shared sequence homology with this bacterium aligned between nucleotide coordinates 23,000–24,000 of the partial sequence of the bacterial genome, which encodes a 16S ribosomal RNA gene. The region downstream was found to be rich in adenine, a feature which may have contributed to poly(T)-mediated capture intended to selectively bind poly(A)-containing eukaryotic mRNAs.
Table 2.
Contig and singleton statistics. Breakdown with respect to their number. GC content, average length and % annotated are shown. Annotation was determined by having a homolog in any of the databases searched with an E-value of 1.0e-06. Even though the Candidatus Portiera aleyrodidarum bacterial DNA was present in the assembly processes, the sequences were removed when statistical calculations were carried out (represented by *) to avoid distortion of the results
| Number of clones | Average GC | Average Length | Annotated | |
| All sequences | 4,860 | 0.34* | 515* | 45.5% |
| Number of contigs | 1,017 | 0.38* | 785.5* | 58% |
| Number of singletons | 3,843 | 0.34 | 443 | 42% |
*excluding sequences from the endosymbiont Candidatus Portiera aleyrodidarum
Figure 1.
Number of sequences building a contig versus the contig length Scatter plot of contigs sequence number (x axis; number of sequences that make up a certain contig) versus the contig length (y axis; bases). The colour scale represents the amount of sequences; the size of the square represents the number of HBT sequences. The annotations for the ten contigs, having the highest number of sequences is shown.
Table 3.
Information on the most abundant contigs. The number of sequences that compose the contigs and the source library, their length, GC content and annotation are indicated. The contig with the highest number of sequences was identified as part of the B. tabaci primary symbiont, Candidatus Portiera aleyrodidarum, based on a partial genomic sequence (gi|32423678|gb|AY268081.1|). This DNA sequence was included in the assembly process. Among the most the abundant contigs were B. tabaci mitochondrial genome sequences (most of them were removed during the preassembly stage)
| Contig name | Number of sequences | Length (bp) | GC content | EGG | INST | HBT | TYLCV | TOMOV | Annotation |
| Candidatus | 425 | 31,123* | 0.3 | 22 | 393 | 3 | 2 | 4 | Candidatus |
| Bt-HInst-045-1-B11-T3_B11 | 167 | 598 | 0.16 | 8 | 159 | 0 | 0 | 0 | Mitochondria |
| Bt_TYLCV004_B07 | 155 | 2,883 | 0.48 | 0 | 0 | 51 | 57 | 47 | Vitellogenin precursor |
| Bt-HInst-008-1-D2-T3_D02 | 103 | 589 | 0.15 | 0 | 103 | 0 | 0 | 0 | Mitochondria |
| Bt-ToMoV-020-1-D2-T3_D02 | 60 | 1,660 | 0.64 | 0 | 0 | 22 | 13 | 25 | Vitellogenin precursor |
| Bt-H-024-1-D9-T3_D09 | 60 | 1,775 | 0.47 | 0 | 0 | 43 | 15 | 2 | Large subunit rRNA |
| Bt-HInst-032-1-E6-T3_E06 | 58 | 792 | 0.46 | 2 | 56 | 0 | 0 | 0 | Unknown |
| Bt-TYLCV-043-1-D2-T3_D02 | 52 | 1,922 | 0.46 | 0 | 0 | 8 | 20 | 24 | Vitellogenin precursor |
| Bt-HInst-013-1-H10-T3_H10 | 47 | 260 | 0.38 | 1 | 46 | 0 | 0 | 0 | Novel |
| Bt-HInst-003-1-G5-T3_G05 | 46 | 970 | 0.4 | 0 | 19 | 13 | 9 | 5 | Myosin |
| Bt-ToMoV-034-1-B10-T3_B10 | 39 | 1,039 | 0.65 | 0 | 0 | 0 | 30 | 9 | Putative senescence |
| Bt-ToMoV-023-1-C12-T3_C12 | 28 | 542 | 0.46 | 0 | 0 | 7 | 12 | 9 | Novel (signal peptide & transmembranal domain) |
| Bt-TYLCV-030-1-C9-T3_C09 | 28 | 1,959 | 0.44 | 0 | 0 | 3 | 23 | 2 | Vitellogenin precursor |
* In the assembly process the sequence of the whitefly primary endosymbiotic bacteria Candidatus Portiera aleyrodidarum [21] (AY268081.1) was included (see Results, EST assembly into contigs)
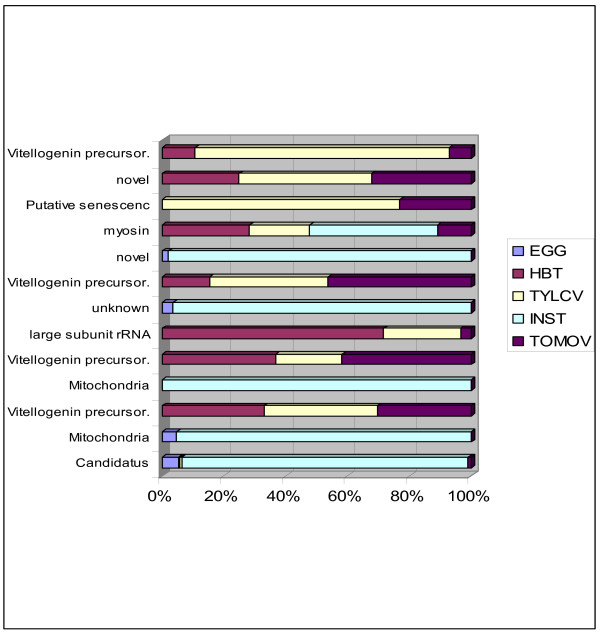
Figure 2.
Library distribution for the most abundant contigs The figure represents for each of the 13 most abundant contigs as revealed by their sequence composition. The number of sequences building the contigs is from 28 to 425. The percentage in based on the total number of sequence forming the contig. The contigs are represented by their annotation.
The second largest contig was composed of sequences homologous to the published B. tabaci mitochondrial genome (AY521259). The number of mitochondrial DNA clones was extremely high. We performed an initial screening for mitochondrial sequences by running RepeatMasker against the published mitochondrial genome (see Methods: analysis of library quality) using a threshold of up to 10% substitutions in matching DNA region or a Smith – Waterman score of at least 2500. For initial selection we preferred not to use stringent criteria in order not to leave aside nuclear genes and to allow assembly of contigs, a process that would not succeed without this screening. This first screening allowed eliminating 4,631 mitochondrial clones. Analysis of the remaining clones revealed the presence of 911 additional mitochondrial sequences (E value smaller or equal to e-06), 521 singletons and 41 contigs. Out of a total of 5,542 sequences, 4869 matched with sequences located between nucleotides 9,450 and 11,700 of the mitochondrial genome. This region contains the large subunit ribosomal RNA and three tRNA genes. It is possible that these sequences allowed RNA:tRNA dimer formation and initiation of cDNA synthesis as described in some retroviruses [22]. The mitochondrial sequences could be used to study metabolism in microarray experiments. The number of sequences that are not mitochondrial or Candidatus Portiera aleyrodidarum from the various libraries is as follows: EGG: 357, HBT:1873, INST:1529, TOMOV:1985 and TYLCV:2367 (submitted to dbEST Genbank).
The third largest contig shared high homology to other insect vitellogenin genes; this contig is also the second longest (2,883 bp) and originated exclusively from the adult whitefly libraries (Figure 2). Since the libraries were not normalized, a high level of redundancy allowed appraising the level of expression of certain genes at certain developmental stages, or as a consequence of virus ingestion and/or transmission. Figure 3 shows that in the thirteen longest contigs (each containing 28 or more sequences) the INST library seems to have a different set of expressed genes than the adult libraries. Contig Bt-ToMoV-023-1-C12-T3_C12 was composed of 28 sequences which shared no significant homology with any of the sequence databases that were searched. However a search of the Interpro database [23] has revealed that it contains a signal peptide and a trans-membrane domain.
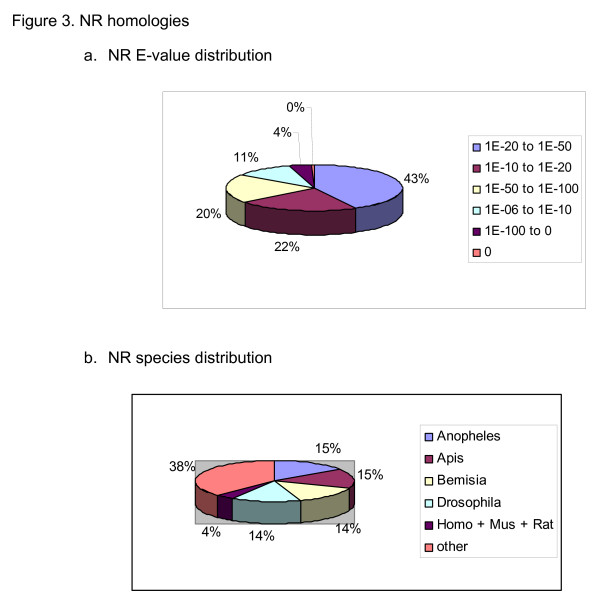
Figure 3.
NR-Homologies a. NR E-value distribution Nr E-value distribution is shown as a percent of the total top homologies. Represented are the 1,544 contigs and singletons that had a homology to a protein in the nr database with an E-value of at least 1.0e-06. b. NR species distribution Species distribution is shown as a percent of the total top homologies. Represented are the 1,544 contigs and singletons that had a homology to a protein in the nr database with an E-value of at least 1.0e-06 (abbreviations used: Homo for Homo sapiens, Mus for Mus musculus and Rat for Rattus norvegicus).
Identification of whitefly contigs and singletons by BLAST analysis
To identify homologies and identities of the contigs and singletons to known proteins, genes and/or genomes, contigs and singletons were subjected to blastn and blastx searches against the following databases: non-redundant protein database (nr, NCBI), non-redundant nucleotide database (nt, NCBI), Swiss-Prot [24], Flybase protein database [25] and EST other (non-human and non-mouse, NCBI) [see Additional file 3] Managing and parsing the BLAST outputs were carried out using the BioCloneDB application [26]. About 45% of the contigs and singletons had a match with an E value of at least 1.0e-06 to one of the above databases (Table 4). 1,544 contigs and singletons had a homology to a protein in the nr database. The E-value distributions of the top hits in the nr database (Figure 3a) showed that 43% of the homolog contigs and singletons ranged between 1.0e-20 to 1.0e-50, whereas 67% had a moderate to strong homology (smaller than 1.0e-20). The species distribution of the top hits (Figure 3b) showed that 58% of contigs and singletons had sequence homology to genomes of insects completely or partially sequenced, and were approximately evenly distributed between the mosquito Anopheles gambiae, the honeybee Apis mellifera, the whitefly B. tabaci and the fruitfly D. melanogaster.
Table 4.
Number of contigs (out of the 4,860) that had a significant hit (E-value equal or smaller than 1.0e-06) with the listed databases
| Database | Number of contigs and singletons |
| Drosophila | 1,053 |
| Nr | 1,544 |
| Nt | 1,207 |
| Swissprot | 1,224 |
| EST other | 1,224 |
| Contigs and singletons found in at least one of the above databases | 2,211 |
Sequences with no identifiable homology
No homologous sequences could be found for 2,649 (54.5%) of the contigs and singletons among the databases searched. The singletons showed a higher occurrence of lack of homology (58%). Because the library was poly(dT)-primed, some of these sequences may represent 3' untranslated regions (3' UTRs). It is also possible that the putative homologous regions are too short to produce a significant alignment.
Comparison to existing B. tabaci sequences in NCBI
There were 448 contigs and singletons presenting high similarity (E-value equal or smaller than 1.e-40) with B. tabaci DNA sequences. The majority of these hits (399) were to the whitefly mitochondria genome (AY521257.1), and represented mitochondrial sequences that were not removed in the preassembly process. The BLAST search against the EST database did not reveal any ESTs originating from B. tabaci (currently there are no B. tabaci ESTs in Genbank). Thus, homology searches indicated that the majority of the contigs and singletons described herein are novel B. tabaci genes, which are not known in the NCBI sequence databases.
Assignment of the whitefly contigs and singletons to common Gene Ontology terms
Based on homologies with the Swiss-Prot database, the contigs and singletons were assigned a biological process, molecular function and cellular component from the Gene Ontology (GO) terminology [27]. The GO was extracted electronically using the FatiGO tool [28]. The top hit of 1,224 contigs and singletons with an E-value equal or smaller than 1.0e-06 was to 922 different Swiss-Prot entries (Figure 4). According to the FatiGo analysis on this list of Swiss-Prot entries only 35 did not have a GO annotation. The most dominant Biological process GO annotation at level 2 was physiological process (97% of the proteins) and 95% were annotated as cellular process. The most dominant molecular function GO category was catalytic activity (51%) and binding (50%) in level 2. The most dominant cellular component was cell (97%) and the second largest was organelle (78%).
Figure 4.
Ontology using Swiss-Prot homologies The Swiss-Prot homologs were used as a query for the FatiGO tool. The output of FatiGO is summarized here in three main categories in level 2: biological process, molecular function and cellular component.
Comparing whitefly Gene Ontology to Drosophila
To evaluate how similar B. tabaci is to Drosophila, a blastx search was carried out against all Drosophila proteins. Here, 1,053 contigs and singletons had a top hit with an E-value equal, or smaller, than 1.0e-06. This set of genes was used to compare B. tabaci to Drosophila in respect to their GO profile. The overrepresented terms were associated with ribosome and protein biosynthesis as well as mitochondria and generation of precursor metabolites and energy (Table 5). The underrepresented terms were 'unknowns', or were identified as receptors, or as having a role in signal transduction (Table 6).
Table 5.
Drosophila homologs used to discover over-represented ontologies in the B. tabaci contigs and singletons
| Best GOs | Term | Count | Total | P-Value |
| (Max: 50) | 732 | 10,309 | ||
| GO:0005840 | Ribosome | 106 | 202 | 0 |
| GO:0005737 | cytoplasm | 357 | 1,800 | 0 |
| GO:0030529 | ribonucleoprotein complex | 124 | 370 | 0 |
| GO:0005830 | cytosolic ribosome (sensu Eukaryota) | 74 | 98 | 0 |
| GO:0003735 | structural constituent of ribosome | 106 | 194 | 0 |
| GO:0043234 | protein complex | 306 | 1623 | 0 |
| GO:0015934 | large ribosomal subunit | 58 | 103 | 6.50E-81 |
| GO:0015935 | small ribosomal subunit | 44 | 74 | 3.73E-65 |
| GO:0005829 | Cytosol | 122 | 440 | 2.52E-63 |
| GO:0005739 | mitochondrion | 131 | 509 | 2.87E-60 |
| GO:0005622 | intracellular | 467 | 3,678 | 2.12E-58 |
| GO:0044249 | cellular biosynthesis | 205 | 1,077 | 1.06E-55 |
| GO:0043229 | intracellular organelle | 394 | 2,919 | 2.21E-54 |
| GO:0043226 | Organelle | 394 | 2,919 | 2.21E-54 |
| GO:0043228 | non-membrane-bound organelle | 160 | 744 | 3.87E-54 |
| GO:0043232 | intracellular non-membrane-bound organelle | 160 | 744 | 3.87E-54 |
| GO:0044237 | cellular metabolism | 562 | 5,047 | 1.27E-52 |
| GO:0009058 | biosynthesis | 208 | 1,145 | 2.01E-51 |
| GO:0008152 | metabolism | 584 | 5,438 | 7.70E-50 |
| GO:0006412 | protein biosynthesis | 151 | 731 | 4.65E-47 |
| GO:0009059 | macromolecule biosynthesis | 154 | 760 | 2.74E-46 |
| GO:0005743 | mitochondrial inner membrane | 64 | 187 | 3.25E-45 |
| GO:0019866 | inner membrane | 64 | 187 | 3.25E-45 |
| GO:0006119 | oxidative phosphorylation | 56 | 152 | 6.10E-44 |
| GO:0005746 | mitochondrial electron transport chain | 38 | 82 | 1.29E-40 |
| GO:0005740 | mitochondrial membrane | 68 | 222 | 1.37E-40 |
| GO:0042773 | ATP synthesis coupled electron transport | 35 | 72 | 8.45E-40 |
| GO:0050875 | cellular physiological process | 645 | 6,729 | 3.39E-39 |
| GO:0005759 | mitochondrial matrix | 52 | 151 | 7.69E-37 |
| GO:0005842 | cytosolic large ribosomal subunit (sensu Eukaryota) | 42 | 57 | 1.12E-34 |
| GO:0043170 | macromolecule metabolism | 354 | 2,902 | 2.19E-34 |
| GO:0005198 | Structural molecule activity | 140 | 760 | 2.73E-34 |
| GO:0044238 | primary metabolism | 512 | 4,908 | 4.18E-34 |
| GO:0016282 | eukaryotic 43S preinitiation complex | 43 | 64 | 8.21E-33 |
| GO:0044260 | cellular macromolecule metabolism | 330 | 2,676 | 1.98E-32 |
| GO:0015077 | monovalent inorganic cation transporter activity | 49 | 151 | 1.09E-31 |
| GO:0009987 | cellular process | 661 | 7,297 | 2.03E-31 |
| GO:0015078 | Hydrogen ion transporter activity | 48 | 149 | 1.10E-30 |
| GO:0007582 | physiological process | 674 | 7,566 | 1.67E-30 |
| GO:0005623 | Cell | 512 | 5,045 | 3.62E-30 |
| GO:0006091 | generation of precursor metabolites and energy | 106 | 532 | 4.41E-30 |
| GO:0031090 | Organelle membrane | 82 | 382 | 1.28E-26 |
| GO:0005843 | cytosolic small ribosomal subunit (sensu Eukaryota) | 32 | 43 | 1.28E-26 |
| GO:0016283 | eukaryotic 48S initiation complex | 32 | 43 | 1.28E-26 |
| GO:0044267 | cellular protein metabolism | 293 | 2,433 | 1.48E-25 |
| GO:0015399 | primary active transporter activity | 52 | 192 | 3.21E-25 |
| GO:0019538 | protein metabolism | 293 | 2,452 | 7.25E-25 |
| GO:0043231 | intracellular membrane-bound organelle | 291 | 2,524 | 1.50E-21 |
| GO:0043227 | membrane-bound organelle | 291 | 2,524 | 1.50E-21 |
| GO:0005761 | mitochondrial ribosome | 28 | 76 | 1.69E-21 |
| Ribosome mitochondrial andmetabolism enriched |
Table 6.
Drosophila homologs used to discover under-represented ontologies in the B. tabaci contigs and singletons
| Best GOs | Count | Total | P-Value | |
| 732 | 10309 | |||
| GO:0005554 | molecular_function unknown | 8 | 939 | 3.31E-13 |
| GO:0000004 | biological_process unknown | 8 | 810 | 9.08E-11 |
| GO:0008372 | cellular_component unknown | 17 | 983 | 2.48E-10 |
| GO:0004871 | signal transducer activity | 39 | 1,092 | 3.28E-05 |
| GO:0004872 | receptor activity | 14 | 584 | 0.000109 |
| GO:0004888 | transmembrane receptor activity | 9 | 452 | 0.000286 |
| GO:0007165 | signal transduction | 58 | 1,303 | 0.000994 |
Mapping whitefly contigs and singletons to pathways
From the top nr homologies the additional information extracted are the KEGG EC numbers [29]. In total, out of the 1,544 nr hits, 48 had an EC number, 37 of which were unique. The EC numbers were mapped to their respective pathway using the KEGG tools (gpath) (Table 7).
Table 7.
Mapping the pathways for the 37 unique EC numbers extracted by the BiocloneDB application for the nr homologs. The output was produced by the 'KEGG gpath tool'.
| Pathway | EC accession |
| map00010 Glycolysis / Gluconeogenesis | EC 1.2.1.3 |
| EC 3.1.3. | |
| EC 5.3.1. | |
| map00030 Pentose phosphate pathway | EC 1.1.1.44 |
| EC 1.1.99.10 | |
| EC 3.1.3.11 | |
| map00031 Inositol metabolism | EC 5.3.1.1 |
| map00040 Pentose and glucuronate interconversions | EC 2.4.1.17 |
| map00051 Fructose and mannose metabolism | EC 3.1.3.11 |
| EC 5.3.1.1 | |
| map00052 Galactose metabolism | EC 2.7.7.12 |
| map00053 Ascorbate and aldarate metabolism | EC 1.2.1.3 |
| map00071 Fatty acid metabolism | EC 1.2.1.3 |
| map00120 Bile acid biosynthesis | EC 1.2.1.3 |
| EC 1.2.1.3 | |
| map00130 Ubiquinone biosynthesis | EC 1.6.5.3 |
| map00150 Androgen and estrogen metabolism | EC 2.4.1.17 |
| map00190 Oxidative phosphorylation | EC 1.6.5.3 |
| EC 1.9.3.1 | |
| EC 1.10.2.2 | |
| EC 3.6.3.14 | |
| map00193 ATP synthesis | EC 3.6.3.14 |
| map00195 Photosynthesis | EC 3.6.3.14 |
| map00220 Urea cycle and metabolism of amino groups | EC 2.6.1.11 |
| EC 6.3.4.5 | |
| map00230 Purine metabolism | EC 2.7.4.6 |
| EC 2.7.7.6 | |
| map00240 Pyrimidine metabolism | EC 2.7.4.6 |
| EC 2.7.4.9 | |
| EC 2.7.7.6 | |
| map00251 Glutamate metabolism | EC 6.3.1.2 |
| map00252 Alanine and aspartate metabolism | EC 6.3.4.5 |
| map00260 Glycine, serine and threonine metabolism | EC 1.1.1.95 |
| EC 2.3.1.37 | |
| EC 2.7.1.32 | |
| map00280 Valine, leucine and isoleucine degradation | EC 1.2.1.3 |
| map00310 Lysine degradation | EC 1.2.1.3 |
| map00330 Arginine and proline metabolism | EC 1.2.1.3 |
| EC 6.3.4.5 | |
| map00340 Histidine metabolism | EC 1.2.1.3 |
| map00380 Tryptophan metabolism | EC 1.2.1.3 |
| map00400 Phenylalanine, tyrosine and tryptophan biosynthesis | EC 4.2.3.4 |
| EC 6.1.1.20 | |
| map00410 beta-Alanine metabolism | EC 1.2.1.3 |
| map00480 Glutathione metabolism | EC 2.5.1.18 |
| map00500 Starch and sucrose metabolism | EC 2.4.1.1 |
| EC 2.4.1.17 | |
| map00520 Nucleotide sugars metabolism | EC 2.7.7.12 |
| map00530 Aminosugars metabolism | EC 3.2.1.14 |
| map00550 Peptidoglycan biosynthesis | EC 6.3.1.2 |
| map00561 Glycerolipid metabolism | EC 1.2.1.3 |
| map00564 Glycerophospholipid metabolism | EC 2.7.1.32 |
| map00620 Pyruvate metabolism | EC 1.2.1.3 |
| EC 2.7.9.2 | |
| map00630 Glyoxylate and dicarboxylate metabolism | EC 4.1.1.39 |
| map00631 1,2-Dichloroethane degradation | EC 1.2.1.3 |
| map00640 Propanoate metabolism | EC 1.2.1.3 |
| map00650 Butanoate metabolism | EC 1.2.1.3 |
| map00710 Carbon fixation | EC 3.1.3.11 |
| map00720 Reductive carboxylate cycle (CO2 fixation) | EC 2.7.9.2 |
| map00860 Porphyrin and chlorophyll metabolism | EC 2.4.1.17 |
| EC 4.99.1.1 | |
| map00903 Limonene and pinene degradation | EC 1.2.1.3 |
| map00910 Nitrogen metabolism | EC 6.3.1.2 |
| map00970 Aminoacyl-tRNA biosynthesis | EC 6.1.1.20 |
| map02040 Flagellar assembly | EC 3.6.3.14 |
| map03020 RNA polymerase | EC 2.7.7.6 |
| map03050 Proteasome | EC 3.4.25.1 |
| map03070 Type III secretion system | EC 3.6.3.14 |
Multiple alignments of vitellogenin-like contigs
Nine contigs shared the same vitellogenin homolog from the butterfly Athalia rosae; (BAA22791.1) as revealed in a blastx search against nr-database. Table 8 shows homologies to vitellogenin ranging from an E-value of 6.0e-13 to 2.0e-83; the location of homology was not the same for the various contigs. To evaluate whether the contigs encoded a protein family or the same protein, a multiple alignment of the contigs having homology to the carboxyl terminus of vitellogenin (amino acids 1371 to 1770, BAA22791.1) was performed (Figure 5). The multiple alignments clearly demonstrated that at least two different types of proteins were represented (2, 6 and 8 versus 4 and 7). The contigs shared a region rich in serine codons flanked by an AAC repeat in the DNA sequence, two features not found in the C-terminal moiety of the A. rosae vitellogenin homolog (Figure 6). In A. rosae vitellogenin, two serine repeats can be found in the N-terminal moiety of the protein, between amino acids 344 and 367 and between amino acids 402 and 434.
Table 8.
Results of nine contigs BLAST searched against gi|2522237|dbj|BAA22791.1| vitellogenin from the butterfly Athalia rosae
| Contig Name | % identity | Alignment length | Contig start | Contig end | Vitellogenin start | Vitellogenin end | E-value |
| BT-TYLCV-021-1-E5-T3_E05_1 | 50.72 | 207 | 618 | 1 | 1 | 207 | 9.20E-60 |
| BT-TOMOV-035-1-E6-T3_E06_1 | 58.33 | 84 | 595 | 344 | 241 | 324 | 4.10E-25 |
| BT-TYLCV-039-1-F7-T3_F07_1 | 46.97 | 132 | 398 | 3 | 478 | 607 | 3.90E-31 |
| BT_TYLCV004_B07_1 | 30.97 | 649 | 5 | 1,885 | 1,141 | 1,755 | 9.80E-89 |
| BT-TYLCV-024-1-F5-T3_F05_1 | 39.02 | 123 | 372 | 4 | 1,256 | 1,378 | 3.60E-20 |
| BT-TYLCV-030-1-C9-T3_C09_1 | 30.3 | 396 | 3 | 1,187 | 1,391 | 1,755 | 6.80E-54 |
| BT-TYLCV-043-1-D2-T3_D02_1 | 34.14 | 331 | 8 | 997 | 1,463 | 1,759 | 2.90E-49 |
| BT-TOMOV-020-1-D2-T3_D02_1 | 35.25 | 244 | 46 | 777 | 1,550 | 1,759 | 1.50E-35 |
| TMVBT002_D07_T3_061_1 | 48.68 | 76 | 20 | 247 | 1,684 | 1,759 | 2.80E-18 |
Figure 5.
Multiple alignment of vitellogenin-like contig Protein multiple alignments of five contigs with BAA22791.1 (amino acid 1371 to 1770) used as a profile. The numbers in the figure designate the following sequences :(1) BAA22791.1 (2) BT-TOMOV-020-1-D2-T3_D02 (4) BT-TYLCV-030-1-C9-T3_C09 (6) BT-TYLCV-043-1-D2-T3_D02 (7) BT_TYLCV004_B07 (8) TMVBT002_D07. Mview parameters: Identities computed with respect to: (1) gi_2522237_dbj_BAA22791.1 and coloured by: consensus/50% and property
Figure 6.
Repeats in the vitellogenin-like contig Translation of BT-TOMOV-020-1-D2-T3_D02 from 1 to 1611. The region of the AAC repeats is highlighted in yellow. The region rich in serine codons is highlighted in blue.
The whitefly contig and EST database
A relational database with a web-based front end (WhiteFlyDB) was created to store, navigate, annotate and retrieve sequence and contig information [30]. This database is based on the BioCloneDB application [26]. The database contains all the relevant contig information such as the names of the sequence that compose it, the top hits against the described databases and the information extracted for these top hits (GO, EC, cellular location) as well as information on the homology itself. The sequences in fasta format and the tab delimited BLAST reports can be easily extracted and imported to Excel files.
Discussion
The whitefly B. tabaci is a major pest to agricultural crops because it causes damage due to feeding and because it transmits many important viruses to plant species cultivated for food and fiber nearly worldwide. Previous to the present research and despite the importance of B. tabaci, the sequence of only a handful of mRNAs (mostly partial) encoding a handful of nuclear protein-coding genes has been published in Genbank. They include sequences encoding actins, a para-sodium channel, putative knottins, a NADP-dependent ketose reductase, two heat shock proteins, a nicotinic acetylcholine receptor alpha subunit, an acetyl cholinesterase-like protein, and a diffusible secreted glycoprotein. The results described in this communication represent the first attempt to develop a functional genomics program involving a homopteran species.
Since the amount of total RNA that could be extracted from eggs and instars was extremely low, we have not isolated polyA+-RNA, which has inevitably reduced the mRNA representation in the sample. Instead we have used total RNA as template for synthesizing cDNA. Libraries have been prepared from another insect pest, the brown citrus aphid Toxoptera citricida starting from RNA samples enriched in polyA+-RNA [31]. However, it has to be noted that an adult aphid weighs approximately 300 micrograms, while an adult whitefly weighs approximately 30 micrograms. Moreover, the weight of a whitefly egg is approximately 1/1000 that of an adult. We have not normalized the libraries, a fact that allowed us to roughly estimate and compare the levels of expression of major genes in the different libraries.
The fraction of the expressed whitefly genes present in our database can be roughly estimated. Although the genome size of B. tabaci was estimated to be approximately five times that of Drosophila [19], it is logical to speculate that the two insect species may have approximately the same number of protein-encoding nuclear genes. The whitefly database contains the sequences of 975 contigs and 3,322 singletons (non-mitochondrial and non-bacterial). If we take into account that each contig represents a transcript of a single protein-coding nuclear gene our sequences represent 4,297 genes. The number of gene families (protein families) in Drosophila has been estimated as 674 and the number of genes not member of a gene family has been estimated as 10,786; altogether 11,460 protein-encoding genes [32]. Hence the B. tabaci database may represent approximately one third of the insect nuclear protein-encoding genes. Additional sequencing from the 3' end of the clones may provide a more accurate estimation.
Within this whitefly database approximately half of the sequences had a match with an E value of at least 1.0e-06 to one of the databases; 1,544 sequences had a homology to a protein in the nr database. Approximately 60% of the whitefly contigs presented homologies with sequenced genomes of other insect species. No homologous sequence could be found for 2,649 contigs and singletons (54.5%) with any of the databases searched.
It was notable that the most abundant contig was vitellogenin. This ancient protein is the major yolk protein of eggs, where it is used as a food source during embryogenesis [33]. There are three vitellogenin genes in Drosophila [25]. The whitefly vitellogenin sequences were found exclusively in libraries from adult whiteflies, indicating that a relatively large amount of resources transcriptional activity is mobilized towards the production of eggs.
The database developed in this study provides a large source of information for studies of whitefly development, circulative transmission of begomoviruses, and choice of host plant. Comparing the sequences present in the various libraries may provide preliminary information on genes expressed during acquisition and transmission of begomoviruses, and ultimately those involved in B. tabaci development.
Conclusion
The set of sequences developed in this study makes available the first DNA sequence database for an important hemipteran (homopteran) pest of agricultural crops for the scientific community. Its availability will allow the investigation of important questions regarding whitefly biology, development, gene expression, and comparative biology. It will also facilitate studies to elucidate the genetics underlying gene expression in pest- and non-pest biotypes, and the basis for virus-vector specificity, resistance to insecticides, and plant host preferences for this cryptic species. This sequence set has been arrayed in a microchip format and enables biologically-based questions to be addressed by examining gene functionalities and expression patterns of the whitefly genome.
Methods
Libraries from adult viruliferous and non-viruliferous whiteflies
Directional cDNA libraries were constructed in the Lambda Uni-ZAP® XR vector using 5 μg of total RNA according to the manufacturers' instructions (Stratagene, La Jolla, CA). Whiteflies (B. tabaci, B biotype) were reared on TYLCV-infected tomato plants, on ToMoV-infected tomato plants and on non-infected tomato plants. The plants were kept in insect-proof cages. Insects were collected randomly from each of the colonies and placed at -80°C. Total RNA was isolated from pools of approximately 200 insects using the RNeasy Midi kit (QIAGEN). Contaminating DNA was discarded using RNAase-free DNAse I. Double stranded cDNA was synthesized using the cDNA Synthesis Kit (Stratagene) with primers containing each EcoRI and XhoI sites. Blunt ends were created, ligated to EcoRI adaptors, phosphorylated and digested with EcoRI. The cDNAs were size-fractionated using Drip Columns to obtain molecules longer than 300 bp. The cDNA was ligated to the Zap Express vector and used to transform E. coli cells in the presence of kanamycin. The phagemid pBK-CMV was packaged using the Gigapack III Gold packaging (Stratagene). Phages were used to inoculate XL1-Blue MRF cells. A library was generated with a titer of 5.2 × 1010 plaque-forming units per ml. The library was mass excised using Ex-Assist® helper phage (Stratagene) and bacterial clones containing excised pBluescript SK+ phagemids were recovered by random colony selection.
Libraries from eggs and instars
Directional cDNA libraries were constructed using the Creator SMART cDNA Library Construction Kit (Clontech). Eggs and instars were collected from leaves of cotton plants caged with whiteflies (B. tabaci, B biotype) and held in insect-proof cages. RNA was isolated using TRIZOL and Phase Lock Gel-Heavy tubes. The first strand cDNA was synthesized using the total RNA and the CDS III/3' PCR Primer which contains a Sfi IB site. The cDNA was amplified by PCR: the first-strand cDNA was used together with the 5' PCR primer which contains a Sfi IA site and the CDS III/3' PCR primer. Following phenol treatment, the DNA was digested with SfiI and size-fractionated using CHROMA SPIN-400 Column. The high molecular weight cDNA fractions (with sticky ends) were pooled together and ligated with dephosphorylated pDNR-LIB vector treated Sfi IA and Sfi IB. The recombinant plasmids were electroporated into DH5α and 10G competent cells, and plated on LB agar plate containing chloramphenicol.
Sequencing
Plasmid clones were isolated from 1.7 ml overnight Luria-Bertani broth cultures using a Qiagen 9600 robot and Qiaprep 96 turbo plasmid isolation kits (Qiagen, Valencia, CA). Plasmid DNA (80 ng) was used as a template for ABI Prism® BigDye™ terminator cycle sequencing (PE Applied Biosystems, Foster City, CA.). Sequencing of the adult whitefly libraries was from the 5' end of the cloned cDNA using a T3 promoter universal primer: 5'ATTAACCCTCACTAAAGGGA3'. Sequencing of the egg and instar whitefly libraries was from the 5' end of the cloned cDNA using a M13 primer: 5'GAAGTTATCAGTCGACGG-3'. Reactions were concentrated and washed by ethanol precipitation. Pellets were resuspended in 15 μl of formamide prior to separation on an ABI Prism 3700 Sequencer (USHRL/ARS/USDA Genomics Laboratory, Fort Pierce FL).
Analysis of library quality
Mitochondrial sequences were not considered for clustering. Significant homology to the B. tabaci mitochondrion, complete genome (NC_006279.1) was found using cross_match [34]. Of the 18,900 sequences analyzed 5,542 were found to contain significant mitochondrial sequences. However, as further analysis revealed the sequences that passed this filtering still contained some mitochondrial contamination.
Analysis and assembly of sequence data
Analysis of the chromatograms was done using Staden pregap4 [20] and the following integrated programs: Phred, cross_match and RepeatMasker [34]. Passed sequences were required to be longer than 75 bases after the trimming procedure. Cross_match and RepeatMasker were used to detect additional vector and adapter contamination after the sequence vector clip. A script was written for trimming the vector from the sequence ends, according to the information obtained by cross_match. Assembly of the contigs was carried out using the Staden gap4 normal assembly feature. Assembled contigs were used to perform BLAST searches (BLASTX and BLASTN) locally using the NCBI-BLAST [35] against the non-redundant protein sequence database, Drosophila proteins (dmel-translation), Swiss-Prot and EST_other (released March-May 2005), which were downloaded from the NCBI [36] or the FlyBase [25] databases.
The database application BiocloneDB [26] was used to manage the BLAST run, and to parse homologue alignment information, whereby an E-value of 1e-06 was used as a maximum cut-off. This information was stored in the SQL database. The DB application also supports sequence and contig queries and down-loads through the web interface [30].
Electronic annotation of contigs and singletons
The BiocloneDB application was used to extract from the EBI SRS information regarding the closest annotated homolog of contigs and singletons, including the GO annotation [27], EC number [29] and cellular location. The ontology distribution, according to the Swiss-Prot homologous proteins, was determined using the FatiGO tool [28]. KEGG tool [37] was used to find the pathway distribution with the nr homologies, which contain an EC number. Over- and under-represented ontologies, with respect to the Drosophila genome, were Drosophila melanogaster homologies using the GOstat tool [38].
Multiple alignment
Multiple sequence alignment of the 5 translated contigs which their best homology was to Vitellogenin precursor from A. rosae (BAA22791.1) and their homology was to the same region in the A. rosae protein ranging from amino acid 1400-1920. The alignment was done by VectorNTI AlignX tool using the A. rosae protein as a profile. The MSF file produced is displayed using the Mview tool [39].
Other bioinformatic tools
Translation view was accomplished using the Prettyseq tool and percent GC calculations were carried out using Geecee. Both tools are part of the EMBOSS suite [40].
Authors' contributions
HC, CM, RLS, JKB, overall project supervision; SG, MG, JKB, preparation of libraries; CM, RLS, sequencing of clones; DL, ER, sequence processing, assembly, annotation and bioinformatic analyses; ER, DL, development of whitefly database search engine; DL, HC, CM, RLS, JKB, preparation of manuscript; ER, JKB preparation of web site.
Supplementary Material
This file contains sequence information regarding the sequences and the contigs they assembled.
This file contains contigs and singletons information such as a list of sequence names assembling the contig and their library count, contig length, GC content and more.
This file contains the top BLAST hit for each contig and singleton against the databases searched.
Acknowledgments
Acknowledgements
This research was supported by research grant number 593/02-1 from The Israel Science Foundation (ISF) to HC and by research grant IS-3479-03 from The United States – Israel Binational Agricultural Research and Development Fund (BARD) to HC, JKB, RS, and CM. DL was supported by grant #1424 of the Israeli Ministry of Science to COBI (Center of Knowledge Bioinformatics Infrastructure), as part of the Bioinformatics unit of the Hebrew University of Jerusalem. We thank Dvorah Weisman for computer resource support.
Contributor Information
Dena Leshkowitz, Email: dena@agri.huji.ac.il.
Shirley Gazit, Email: gazits@agri.huji.ac.il.
Eli Reuveni, Email: reuveni@embl.it.
Murad Ghanim, Email: ghanim@agri.gov.il.
Henryk Czosnek, Email: czosnek@agri.huji.ac.il.
Cindy McKenzie, Email: cmckenzie@ushrl.ars.usda.gov.
Robert L Shatters, Jr, Email: rshatters@saa.ars.usda.gov.
Judith K Brown, Email: jbrown@ag.arizona.edu.
References
- Gerling D. Whiteflies: Their bionomics, pest status and management. Andover: Intercept; 1990. [Google Scholar]
- Gerling D, Mayers R. Bemisia 1995: Taxonomy, Biology, Damage, Control and Management. Andover: Intercept; 1995. [Google Scholar]
- Brown JK, Frohlich DR, Rosell RC. The sweetpotato or silverleaf whiteflies: Biotypes of Bemisia tabaci or a species complex? Ann Rev Entomol. 1995;40:511–534. [Google Scholar]
- Frohlich D, Torres-Jerez I, Bedford ID, Markham PG, Brown JK. A phylogeographic analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers. Mol Ecol. 1999;8:1593–1602. doi: 10.1046/j.1365-294x.1999.00754.x. [DOI] [PubMed] [Google Scholar]
- Goodman RM. Single-stranded DNA genome in a whitefly-transmitted plant virus. Virology. 1977;83:171–179. doi: 10.1016/0042-6822(77)90220-3. [DOI] [PubMed] [Google Scholar]
- Harrison BD, Barker H, Bock KR, Guthrie EJ, Meredith G, Atkinson M. Plant viruses with circular single-stranded DNA. Nature. 1977;270:760–762. [Google Scholar]
- Bedford ID, Briddon RW, Brown JK, Rosell RC, Markham PG. Geminivirus transmission and biological characterisation of Bemisia tabaci (Gennadius) biotypes from different geographic regions. Ann Appl Biol. 1994;125:311–325. [Google Scholar]
- van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB. Virus Taxonomy: The Classification and Nomenclature of Viruses. San Diego: Academic Press; 2000. [Google Scholar]
- Brown JK. The Molecular Epidemiology of Begomoviruses. In: Khan JA and Dykstra J, editor. Trends in Plant Virology. New York: The Haworth Press; 2001. pp. 279–316. [Google Scholar]
- Brown JK, Czosnek H. Advances in Botanical Research. Vol. 36. New York: Academic Press; 2002. Whitefly Transmitted Viruses; pp. 65–100. [Google Scholar]
- Ghanim M, Rosell RC, Campbell LR, Czosnek H, Brown JK, Ullman DE. Digestive, salivary and reproductive organs of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) Biotype B. J Morphology. 2001;248:22–40. doi: 10.1002/jmor.1018. [DOI] [PubMed] [Google Scholar]
- Rosell RC, Torres-Jerez I, Brown JK. Tracing the geminivirus-whitefly transmission pathway by polymerase chain reaction in whitefly extracts, saliva, hemolymph, and honeydew. Phytopathology. 1999;89:239–246. doi: 10.1094/PHYTO.1999.89.3.239. [DOI] [PubMed] [Google Scholar]
- Ghanim M, Morin S, Czosnek H. Rate of Tomato Yellow Leaf Curl Virus (TYLCV) translocation in the circulative transmission pathway of its vector, the whitefly Bemisia tabaci. Phytopathology. 2001;91:188–196. doi: 10.1094/PHYTO.2001.91.2.188. [DOI] [PubMed] [Google Scholar]
- Caciagli P, Bosco D, Al-Bitar L. Relationships of the Sardinian isolate of tomato yellow leaf curl geminivirus with its whitefly vector Bemisia tabaci Gen. European J Plant Pathol. 1995;101:163–170. [Google Scholar]
- Rubinstein G, Czosnek H. Long-term association of tomato yellow leaf curl virus (TYLCV) with its whitefly vector Bemisia tabaci : effect on the insect transmission capacity, longevity and fecundity. J Gen Virol. 1997;78:2683–2689. doi: 10.1099/0022-1317-78-10-2683. [DOI] [PubMed] [Google Scholar]
- McKenzie CL. Effect of Tomato mottle virus (ToMoV) on Bemisia tabaci biotype B (Hemoptera: Aleyrodidae) oviposition and adult survivorship on healthy tomato. Florida Entomol. 2002;85:367–368. [Google Scholar]
- Sinisterra XH, McKenzie CL, Hunter WB, Powell CA, Shatters RG., Jr Differential transcriptional activity of plant-pathogenic begomoviruses in their whitefly vector (Bemisia tabaci, Gennadius: Hemiptera Aleyrodidae) J Gen Virol. 2005;86:1525–1532. doi: 10.1099/vir.0.80665-0. [DOI] [PubMed] [Google Scholar]
- Czosnek H, Ghanim M, Ghanim M. Circulative pathway of begomoviruses in the whitefly vector Bemisia tabaci – insights from studies with Tomato yellow leaf curl virus. Ann Appl Biol. 2002;140:215–231. [Google Scholar]
- Brown JK, Lambert GM, Ghanim M, Czosnek H, Galbraith DW. Nuclear DNA content of the whitefly Bemisia tabaci (Genn.) (Aleyrodidae: Homoptera/Hemiptera) estimated by flow cytometry. Bull Entomol Res. 2005;95:309–312. doi: 10.1079/ber2005361. [DOI] [PubMed] [Google Scholar]
- Staden R, Judge DP, Bonfield JK. Sequence assembly and finishing methods. Meth Biochem Anal. 2001;43:303–22. doi: 10.1002/0471223921.ch13. http://staden.sourceforge.net/ [DOI] [PubMed] [Google Scholar]
- Thao ML, Baumann P. Evolutionary relationships of primary prokaryotic endosymbionts of whiteflies and their hosts. Appl Environ Microbiol. 2004;70:3401–3406. doi: 10.1128/AEM.70.6.3401-3406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N, Berkhout B. The tRNA primer activation signal in the Human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription. J Virology. 2002;76:2329–2339. doi: 10.1128/jvi.76.5.2329-2339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, Durbin R, Falquet L, Fleischmann W, Gouzy J, Hermjakob H, Hulo N, Jonassen I, Kahn D, Kanapin A, Karavidopoulou Y, Lopez R, Marx B, Mulder NJ, Oinn TM, Pagni M, Servant F, Sigrist CJ, Zdobnov EM. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter M-C, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, Pilbout S, Schneider M. The Swiss-Prot protein knowledgebase and its supplement TrEMBL. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale RA, Crosby MA, The FlyBase Consortium FlyBase:genes and gene models. Nucleic Acids Research. 2005;33:D390–D395. doi: 10.1093/nar/gki046. http://flybase.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuveni E, Leshkowitz D, Yarden O. BioCloneDB: a database application to manage DNA sequence and gene expression data. Appl Bioinformatics. 2005;4:277–280. doi: 10.2165/00822942-200504040-00007. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology: The gene ontology consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F, Díaz-Uriarte R, Dopazo J. FatiGO – A web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitefly functional genomics database. WhiteFlyDB. http://whitefly.agri.huji.ac.il/ password: guests.
- Hunter WB, Dang PM, Bausher MG, Chaparro JX, Mckendree WL, Jr, Shatters RG, Jr, Mckenzie CL, Sinisterra X. Aphid biology: expressed genes from alate Toxoptera citricida, the brown citrus aphid. J Insect Sci. 2003;3:23–29. doi: 10.1093/jis/3.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Cavalcanti A, Chen F-C, Bouman P, Wen-Hsiung Li W-H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol. 2002;19:256–262. doi: 10.1093/oxfordjournals.molbev.a004079. [DOI] [PubMed] [Google Scholar]
- Hagedorn HH, Kunkel JG. Vitellogenin and vitellin in insects. Ann Rev Entomol. 1979;24:475–505. [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information 1997. http://www.ncbi.nlm.nih.gov/
- KEGG: Kyoto Encyclopedia of Genes and Genomes. http://www.genome.jp/kegg/
- Beissbarth T, Speed TP. GOstat: Find statistically overrepresented gene ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Brown NP, Leroy C, Sander C. MView: A Web compatible database search or multiple alignment viewer. Bioinformatics. 1998;14:380–381. doi: 10.1093/bioinformatics/14.4.380. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden L, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains sequence information regarding the sequences and the contigs they assembled.
This file contains contigs and singletons information such as a list of sequence names assembling the contig and their library count, contig length, GC content and more.
This file contains the top BLAST hit for each contig and singleton against the databases searched.