Abstract
The genes that encode the enterotoxigenic Escherichia coli (ETEC) CS4 fimbriae, csaA, -B, -C, -E, and -D′, were isolated from strain E11881A. The csa operon encodes a 17-kDa major fimbrial subunit (CsaB), a 40-kDa tip-associated protein (CsaE), a 27-kDa chaperone-like protein (CsaA), a 97-kDa usher-like protein (CsaC), and a deleted regulatory protein (CsaD′). The predicted amino acid sequences of the CS4 proteins are highly homologous to structural and assembly proteins of other ETEC fimbriae, including CS1 and CS2, and to CFA/I in particular. The csaA, -B, -C, -E operon was cloned on a stabilized plasmid downstream from an osomotically regulated ompC promoter. pGA2-CS4 directs production of CS4 fimbriae in both E. coli DH5α and Shigella flexneri 2a vaccine strain CVD 1204, as detected by Western blot analysis and bacterial agglutination with anti-CS4 immune sera. Electron-microscopic examination of Shigella expressing CS4 confirmed the presence of fimbriae on the bacterial surface. Guinea pigs immunized with CVD 1204(pGA2-CS4) showed serum and mucosal antibody responses to both the Shigella vector and the ETEC fimbria CS4. Among the seven most prevalent fimbrial antigens of human ETEC, CS4 is the last to be cloned and sequenced. These findings pave the way for CS4 to be included in multivalent ETEC vaccines, including an attenuated Shigella live-vector-based ETEC vaccine.
Enterotoxigenic Escherichia coli (ETEC) is a major cause of diarrhea in infants and young children in developing countries, accounting for a high rate of infantile morbidity and mortality, and is also a major cause of traveler's diarrhea (4, 5, 12, 24, 31). ETEC infection is characterized by watery diarrhea, often accompanied by low-grade fever, abdominal cramps, malaise, and vomiting. Following ingestion of contaminated food or water by individuals, ETEC strains colonize the small-bowel mucosa by means of surface fimbriae called colonization factor antigens (CFA) and coli surface antigens (CS) (36). They subsequently elaborate heat-labile enterotoxins (LT) and or heat-stable enterotoxins (ST), which are responsible for the profuse watery diarrhea (36). ETEC fimbriae are proteinaceous filaments exhibiting different morphologies, such as rigid rod-like shapes, thin flexible wiry fibrillae, or bundle-forming and nonfimbrial structures (19, 32, 34). Among human ETEC strains, over 20 serologically distinct fimbriae have been described previously (9, 19, 32, 57), with most strains expressing either one or two antigenic types on their surfaces. The most common human ETEC isolates found in diverse geographic areas express CFA/I, CFA/II, and CFA/IV (3, 23, 31, 32, 43, 57). CFA/I is composed of a single antigenic type of fimbriae, while CFA/II and CFA/IV constitute families of antigens. For example, CFA/II strains always express CS3, either alone or in conjunction with CS1 or CS2 (19, 32, 34); similarly, CFA/IV strains always express CS6, either alone or with CS4 or CS5 (19, 32, 55, 57, 58). The current body of evidence indicates that antibody responses against these fimbriae provide protection against ETEC disease by inhibiting attachment of the bacteria to the human intestine (6, 10, 15, 30, 31, 33, 34). To achieve broad-spectrum protection, current fimbria-based ETEC vaccine strategies aim to incorporate the seven most prevalent human ETEC fimbrial types, including CFA/I and CS1 through CS6, in an ETEC vaccine (29, 32, 35, 51). The genetic loci encoding six of the seven critical fimbriae have been cloned and sequenced. Here we report the characterization and complete sequence of the seventh fimbria, CS4.
The genes required for the expression of functional fimbriae are characteristically linked in gene clusters that include the structural genes and assembly cassette genes (50). The assembly cassette genes include chaperone and usher genes responsible for folding and transport of the fimbrial subunits. The fimbriae are composed of multiple repeating fimbrial subunit proteins that are encoded by the structural subunit gene. Although the operon encoding CS4 had not heretofore been cloned, analysis of the purified fimbriae had demonstrated that they are composed of a 17-kDa structural subunit with an amino-terminal sequence that has homology to the CFA/I and CS2 structural subunits (58). Some fimbriae, such as CFA/I, CS1, and CS2, contain a minor fimbrial protein that is associated with the fimbrial tip and is believed to be involved in the attachment of the bacteria to the cell receptors (49). Fimbrial expression is controlled by regulatory genes, such as Rns and CfaD, that are similar to members of the AraC family of positive transcriptional activators (8, 11, 21, 52). Although not sequenced, a regulatory region from a CS4+ ETEC strain has been cloned which hybridized to cfaD and was able to promote fimbrial expression (23, 56).
The operons encoding most ETEC fimbriae, such as CFA/I (22, 26), CS1 (16), CS3 (25, 37), CS5 (13), and CS6 (59), have been located on large plasmids; in contrast, the CS2 operon is located on the chromosome (17). Early experiments to identify the location of the CS4-encoding genes produced disparate results (53, 54, 58). Here we report cloning of the operon encoding CS4 and elucidation of its entire nucleotide sequence and of its localization to an ETEC plasmid.
At the Center for Vaccine Development of the University of Maryland School of Medicine, a multivalent hybrid vaccine to prevent Shigella dysentery and ETEC diarrhea is being developed (1, 2, 27, 29). The approach employed consists of engineering attenuated Shigella live-vector strains to express ETEC fimbrial antigens and an antigen to elicit antibodies against LTh (the LT variant found in human ETEC strains). Heretofore, we have reported the immunogenicity of the ETEC antigens CFA/I, CS2, CS3, and LThK63 expressed in S. flexneri 2a strain CVD 1204. In this work we describe the expression of CS4 as intact fimbriae in Shigella vaccine strain CVD 1204 and the immunogenicity of the fimbria-expressing live vector in the guinea pig model with respect to the elicitation of mucosal immunoglobulin A (IgA) and serum IgG anti-CS4 antibody responses.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. ETEC strains were grown on CFA agar plates (14). DH5α transformants were grown in Luria broth in the presence of carbenicillin at 50 μg/ml or kanamycin at 50 μg/ml. Shigella strains were grown in Trypticase soy agar (TSA) plates containing 0.1% Congo red and 10 μg of guanine/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain | Relevant feature(s) (reference) | Reference or source |
|---|---|---|
| E. coli | ||
| E11881A | O25:H42, CS4+ CS6+ LT+ ST+ | CVD collectiona |
| E11881C | O25:H42, CS4− CS6+ LT+ ST+ | CVD collection |
| DS 9-1 | O25, CS4+ CS6+ ST+, nonmotile | M. Wolf |
| S. flexneri 2a CVD1204 | ΔguaB-A | 34 |
| Plasmids | ||
| pBluescript KS | Cloning vector, Apr | Stratagene |
| pEXO1 | Stabilized Kmr plasmid | 16 |
| pGA2 | Stabilized cloning vector, derived from pEXO1 containing the multiple cloning site | This work |
| pKS-CSA-I | A derivative of pKS containing csaB, csaC, csaE, and csaD::IS1E | This work |
| pKS-CSA-II | A derivative of pKS containing the promoter sites for csa, the csaA gene, and an IS21 element | This work |
| pGA2-CS4 | A derivative of pGA2 which contains the csaABCE genes | This work |
| pGA2-LThK63 | A derivative of pGA2 containing the genes encoding LThK63 as described previously (27) | This work |
| pGA2-CS4-LThK63 | A derivative of pGA2-CS4 containing the genes encoding LThK63 as described previously (27) | This work |
CVD, Center for Vaccine Development.
Isolation of the csa operon.
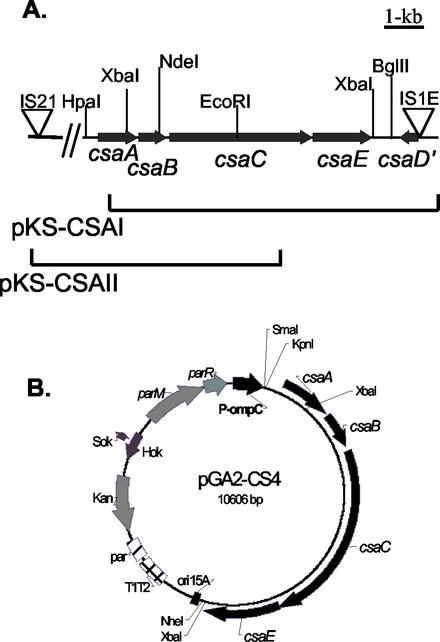
Genomic DNA from ETEC strain E11881A was isolated using GNOME DNA kit protocol (Bio 101, Vista, Calif.). The DNA was partially digested with Sau3AI. DNA fragments with sizes of 5 to >20 kb were isolated from agarose gels and ligated into pBluescript KS, digested with BamHI, and treated with shrimp alkaline phosphatase. The resulting DH5α transformants were harvested into 96-well microtiter plates and, using PCR with primers designed on the basis of the published sequence of csfA (the structural gene of the CS4 fimbriae [NCBI accession number X97493]), probed for the presence of csa genes. All primers used in these studies are listed in Table 2. The primers CS44 and CS45 were designed to amplify a 319-bp fragment. Circa 1,200 colonies were initially analyzed in pools of 96 colonies. Colonies from positive pools were subsequently assayed individually and yielded two positive clones. Both colonies were found to contain the same plasmid, pKS-CSA-I (Fig. 1A). Sequencing of the cloned DNA fragment indicated that pKS-CSA-I contained most of the csa operon, lacking only 430 bp from the 5′ end of csaA. Analysis of the sequence of the csa operon revealed a high degree of homology to the DNA sequences of the CFA/I operon. Based on this homology, two new primers were designed for screening the genomic library for the 5′ end of the operon by PCR. Primer CS433 is based on the csaA sequence at bp 718, and primer CS434 is based on the CFA/I sequences from bp 878. PCR assays using these primers amplified a 429-bp DNA fragment from the genomic DNA of strain E118811A. By screening the DNA library with these primers, a positive clone that contains the entire csaA gene as well as upstream sequences was isolated. The clone contains the 8,000-bp plasmid pKS-CSA-II (Fig. 1A).
TABLE 2.
Primers used in this study
| Primer | |
|---|---|
| CS44 | 5′-GTTGACCC TACAATTGATATTTTGCAAGC-3′ |
| CS45 | 5′-CGACC CCACTATAATTCCCG CC GTTGGTGC-3′ |
| CS433 | 5′-GTGATATGTTTTGTTCACTTGGTAA AGATC-3′ |
| CS434 | 5′-CTCATGGCTCCATTTGTTGCAAATGCAAACTTTATG-3′ |
| 4a | 5′-GGGATCGATCCC GGGGCGGCCGCGGGCCCGGTACCAGG CCTT CTAGAAAGCTTGACGTCG-3′ |
| 4b | 5′-CCCGCTAGCGGCGCGCCTCGCGAGGATCCGTCGACGACGTCAAGCTTTCT AGA AGGCCTGG-3′ |
| 4c | 5′-AAGCTTGACGTCGTCGACGG-3′ |
| 4d | 5′-CCCGCTAGC GGCGCGCCTCGCG-3′ |
| LTA (162) | 5′-CCGTGCTGACTCTACACCCCCAGATG-3′ |
| LTB (895) | 5′-GCACATAGAGAGGATAGTAACGCCG-3′ |
| GYRA (347) | 5′-CGGTCATTGTTGGCCGTGCGCTGCC-3′ |
| GYRA (1147) | 5′-CACGCAGCGCGCTGATGCCTTCCACGCG-3′ |
| CS4D3 | 5′-CATATTTGATATCTGAGATATCTGG-3′ |
| CS4D2 | 5′-TGTTGCATTCAGATTGAACGGAG-3′ |
FIG. 1.
Isolation of the csa operon. (A) The entire csa operon and flanking regions were cloned on two plasmids: pKS-CSA-I contains truncated csaA, csaB, csaC, csaE, and csaD′::IS1, and pKS-CSA-II contains upstream regions as well as full-length csaA. (B) The csaABCE genes were cloned into the expression vector pGA2, creating pGA2-CS4.
DNA sequence analysis.
DNA sequencing of CS4-encoding genes in plasmids pKS-CSA-I and pKS-CSA-II was performed on both strands. The primers were synthesized on a model 3948 Perkin-Elmer DNA Synthesizer at a synthesis scale of 40 nM. Using a Perkin-Elmer DNA Synthesizer 373 Stretch Sequencer and dye terminators from a BigDye kit (Perkin-Elmer, Emeryville, Calif.) in the University of Maryland at Baltimore Biopolymer Laboratory, the sequencing reactions were performed. The sequencing results identified the csa operon and its flanking genes, as is schematically presented in Fig. 1A.
Construction of pGA2.
The vector pGA2, used for expression of the CS4-encoding genes in Shigella, was derived from pEXO1 by replacing gfp with a 87-bp ClaI-NheI fragment that encodes multiple cloning sites constructed by PCR with primers 4a, 4b, 4c, and 4d (Fig. 1 and Table 2). pEXO1 is an expression plasmid derived from pGEN222 (20) which carries a two-component plasmid maintenance system comprised of the hok-sok postsegregational killing system plus the parA plasmid partitioning system; these two components have been shown to work in concert to minimize plasmid loss from a population of actively growing bacteria and to lyse any bacteria from which plasmids have segregated. pEXO1 was created from pGEN222 by replacing the bla gene (encoding β-lactamase) with an engineered aph allele encoding reduced levels of resistance to kanamycin (7). Transcription of this aph allele was modified by PCR such that the separation of the −35 and −10 regions within the promoter was increased from 18 to 19 bp; this reengineered allele was then introduced as an NheI fragment into pGEN222 cleaved with XbaI and SpeI to replace the bla gene. pGA2 is therefore expected to be present at approximately 15 copies per chromosomal equivalent and to drive expression of CS4-encoding genes from the osmotically responsive ompC promoter.
Construction of the CS4 expression plasmid.
Plasmid pGA2-CS4 (Fig. 1B) was constructed in three steps. The 5′ end of the csaA gene was cloned as a 710-bp HpaI/XbaI fragment from plasmid pKS-CSA-II into pBluescript KS digested with SmaI and XbaI. The HpaI site is located 241 bp upstream from the ATG codon of the csaA gene. This DNA fragment was subsequently cloned as a KpnI/XbaI fragment into pGA2 to construct pGA2-ΔcsaA. The remaining CS4-encoding genes, including the 3′ ends of csaA, csaB, csaC, and csaE, were cloned from pKS-CSA-I as a 4,526 bp XbaI fragment into pGA2-ΔcsaA to create pGA2-CS4. The XbaI sites used for cloning are located in csaA and in the stop codon of csaE.
To express CS4 and LThK63 from a single plasmid, the operon encoding LThK63 was cloned downstream of the CS4 operon in plasmid pGA2-CS4, creating pGA2-CS4-LThK63. The genes encoding LThK63 (as described previously [27]) were cloned as a BamHI-AvrII fragment into pGA2-CS4 digested with the same enzymes.
Additional DNA primers for PCR assays.
For location of LT, csa genes, and (as a control) gyrA genes, the following primers were constructed. For LT, primers LTA and LTB amplify a DNA fragment from bp 162 to 895 (National Center for Biotechnology Information [NCBI] accession number AB01167). For gyrA primers, GYRA and GYRB amplify a fragment from bp 347 to 1147 (NCBI accession number X57174). For the csaD′ gene, primers CS4D3 and CS4D2 amplify a fragment from bp 5540 to 6030 of the csa operon.
Transformation of Shigella strains.
Competent cells of S. flexneri 2a CVD 1204 were prepared by growing the bacteria in Luria broth supplemented with guanine to an optical density at 600 nm (OD600) of 0.6. The cells were centrifuged, washed twice with cold H2O and once with cold 10% glycerol, and resuspended in the same buffer at 1/100 of the original volume. A mixture of 150 μl of cells and plasmid DNA was electroporated in 0.2-cm-path-length cuvettes in a Gene Pulser (Bio-Rad Laboratories, Hercules, Calif.) at 2.5 kV, 200 Ω, and 25 μF or at 1.75 kV, 600 Ω, and 25 μF. Transformants were selected on TSA plates containing kanamycin, guanine, and Congo red.
Detection of fimbrial synthesis.
ETEC strains were grown on CFA agar plates at 37°C overnight, followed by resuspension in phosphate-buffered saline (PBS). Shigella strains that contained the plasmid pGA2-CS4 were grown to logarithmic phase in TS broth (1.5% tryptone, 0.5% Soytone) containing 150 mM NaCl. The bacteria were assayed by bacterial agglutination assays, and by immunoblotting of cell extracts, for fimbriae production. For immunoblotting, the bacterial cultures were adjusted to an OD600 of 10 and boiled for 10 min in Laemmli sample buffer. The cell extract proteins were separated by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis, transferred to nitrocellulose filters, and probed with anti-CS4 antibody. Following immunization of rabbits with ETEC strain E11881A (a CS4+ CS6+-producing strain) and absorption of the sera with strain EI1881C, a CS4− CS6+ strain, the specific anti-CS4 antiserum was produced.
Hemagglutination.
For hemagglutination tests, ETEC and Shigella strains were grown overnight on CFA plates and TSA-Congo red-guanine-kanamycin-containing plates, respectively, and were resuspended in PBS to an OD600 of 10. The slide hemagglutination tests were performed by mixing 20 μl of bacterial suspension with 20 μl of PBS containing 0.1 M d-mannose and 20 μl of washed group A human erythrocytes, as described by Sakellaris et al. (49). For hemagglutination inhibition assays, the bacterial suspension was incubated with fourfold-diluted antibodies for 1 h at 37°C prior to the hemagglutination tests.
Electron microscopy.
Bacterial cells, harvested from overnight broth cultures of CVD 1204 or CVD 1204(pGA2-CS4), were washed in PBS and placed onto 300-mesh copper grids coated with carbon Formvar for 2 min. The grids were then stained for 45 s with 1% phosphotungstic acid (pH 7.2) and examined in a JOEL JEM 1200 EXII transmission electron microscope at 80 kV.
RESULTS
Cloning and sequencing of the CS4 fimbriae-encoding genes.
The csa operon, consisting of the genes necessary for the synthesis of the CS4 fimbriae, was isolated from a genomic DNA library of ETEC strain E11881A in two overlapping clones, pKS-CSA-I and pKS-CSA-II (Fig. 1A). The genomic library was screened using primers designed from the sequence of csfA, the structural gene for CS4 (NCBI no. X97493), and from the sequence of cfaB of CFA/I. Fragments from each plasmid were ligated together in the expression plasmid pGA2 to create pGA2-CS4, containing the contiguous CS4 operon (Fig. 1B).
The entire region was sequenced on both strands (NCBI accession no. AF296132), revealing five open reading frames with a G+C content of 34.8% and with homology to genes encoding other ETEC fimbriae. The csa operon is located on a 7,239-bp DNA fragment that is flanked by insertion elements, in similarity to a pathogenicity island (Fig. 1A). Four open reading frames, csaA, csaB, csaC, and csaE, are transcribed in the same direction downstream from a predicted promoter site, and a fifth gene, csaD′, is transcribed from the opposite strand. The promoter site for the csa operon was predicted using the promoter prediction by neural network method (45) and is proposed to be located between bp 145 and 194, 89 bp upstream of the ATG start codon for csaA. This region had 90% homology to the CFA/I fimbrial operon promoter area. The location of each gene is described in Table 3. Upstream from csaA is an IS21 element. At the opposite end, the start of csaD′ is disrupted by integration of an IS1E element.
TABLE 3.
Open reading frames of the CS4 operon
| Open reading frame | Location (bp) | Protein | Properties of the predicted CsaA-E protein
|
||
|---|---|---|---|---|---|
| No. of amino acids | Molecular weight | Signal peptide (no. of amino acids) | |||
| csaA | 283-999 | CsaA | 238 | 27,305.6 | 19 |
| csaB | 1028-1529 | CsaB | 167 | 17,343.9 | 23 |
| csaC | 1589-4190 | CsaC | 867 | 97,686.93 | 22 |
| csaE | 4196-5279 | CsaE | 361 | 40,102.4 | 23 |
| csaD′a | 5448-6068 | CsaD′ | 100 | ||
The CsaD′ protein is predicted to be truncated.
A homology search using the BLAST program with the csa operon DNA sequence (bases 1 to 6096) revealed homology to ETEC CFA/I, CS1, CS2, CS14, and CS19 fimbrial genes, with the highest degree of homology being to the CFA/I operon. The genes encoding these fimbriae are contained in operons encoding chaperone, structural subunit, usher, and tip adhesin proteins. The csa operon contained homologues to each of these genes, in the same order and orientation, as shown in Table 3. Sequence analysis of the 3′ end of the CS4 operon revealed an open reading frame, csaD′, oriented oppositely from the other CS4 genes and homologous to cfaD′ of the CFA/I operon, which is also oriented oppositely in CFA/I ETEC strains. In CFA/I strains, the cfaD′ gene contains deletions at approximately the middle of the gene which result in a frameshift causing translation termination and a presumably nonfunctional product (18). The open reading frame identified in the csa operon, named csaD′, also contained a deletion at the middle of the gene which resulted in the same frameshift and premature termination. In addition, csaD′ is interrupted at its 5′ end with the insertion of the IS1E element, resulting in deletion of the first 144 bp of the gene.
The predicted amino acid sequences of the CsaABCED′ proteins exhibit a high degree of identity with those of the proteins of the CFA/I, CS1, and CS2 fimbriae (19, 57). The csa operon is predicted to encode four proteins, CsaA, CsaB, CsaC, and CsaE. As determined according to the sequence similarity of the CsaA-E proteins to other ETEC fimbrial proteins, the predicted functions of the CsaA-E proteins are as follows: CsaA, a 27.3-kDa protein, functions as a periplasmic chaperone-like protein; CsaB is the 17.3-kDa major pilin subunit; CsaC is a 97.6-kDa membrane usher-like protein; and CsaE is the fimbrial tip adhesin. Each protein has a predicted signal sequence at its 5′ terminus. A greater degree of identity exists between the CS4 usher and chaperone proteins and those of CFA/I than is evident for the structural and tip adhesin proteins of the CS4 and CFA/I. Identity between CS4 and CFA/I usher proteins CsaA and CfaA amino acid sequences was 93% and between chaperone proteins CsaC and CfaC was 94%. In contrast, there was only 67% identity between the structural proteins CsaB and CfaB and 81% identity between the tip adhesin proteins CsaE and CfaE. The lower degree of identity between structural and tip adhesin proteins might reflect sequence divergence which resulted in immunologic distinctiveness.
Localization of the csa operon in strain E11881A.
To identify the location of the csa operon, total genomic DNA was isolated from strain E11881A and subjected to agarose gel electrophoresis. The electrophoresis results indicated the presence of three plasmids, including a large plasmid visible above the chromosomal band and two smaller plasmids located under the chromosomal DNA band. Each form of DNA, including the plasmids as well as the chromosomal bands, was isolated by gel elution and tested by PCR analysis for the presence of the csa operon by using primers CS44 and CS45 for amplification of the csaB gene. The results indicate that the csa operon is located on the large plasmid. The location of the LTh gene was also detected on the large plasmid by PCR assays using primers LTA162 and LTB895 for amplification of a 708-bp DNA fragment (data not shown). Using primers GYRA347 and GYRA1147 for amplification of gyrA as a control confirmed the location of this known chromosomal gene.
Expression of CS4 fimbriae in DH5α and CVD 1204.
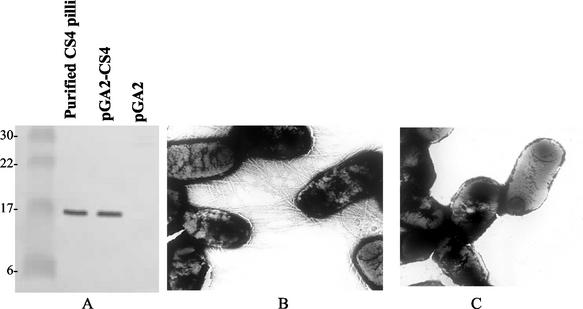
The expression of CS4 fimbriae was assayed in E. coli and S. flexneri 2a strain CVD 1204, following electroporation with pGA2-CS4 containing csaABCE. CS4 fimbriae production by strain CVD 1204(pGA2-CS4) was detected by Western immunoblotting of cell extracts (Fig. 2). Western blot results indicated that the cloned csaABCE gene cluster in pGA2-CS4 in CVD 1204 encodes a 17-kDa protein that corresponds to the CS4 fimbrial structural subunit, as detected with anti-CS4 antisera.
FIG. 2.
Expression of CS4 in S. flexneri strain CVD 1204. (A) Western blot analysis of CS4 fimbriae or whole-cell lysates probed with anti-CS4 antisera. Far left lane, molecular weight markers; center-left lane, purified CS4 fimbriae; center-right lane, whole-cell lysates of CVD 1204(pGA2-CS4); far right lane, whole-cell lysates of CVD 1204(pGA2). (B) Electron microscopy of whole cells of CVD 1204(pGA2-CS4). (C) Electron microscopy of CVD 1204(pGA2).
Bacterial agglutination assays with anti-CS4 polyclonal sera resulted in agglutination of ETEC strains E11881A, DS 9-1, DH5α(pGA2-CS4), and CVD 1204(pGA2-CS4), indicating surface expression of CS4 fimbriae (Table 4). Negative-control strains, including ETEC strain E11881C (a CS4-negative derivative), DH5α, and CVD 1204(pGA2), failed to agglutinate.
TABLE 4.
Bacterial agglutination and hemagglutination assay results
| Strain | Phenotype | Hemagglutinationa | Bacterial agglutinationb |
|---|---|---|---|
| E11881A | CS4+ CS6+ | ++ | + |
| E11881C | CS4− CS6+ | − | − |
| DS 9-1 | CS4+ CS6+ | ++++ | ++++ |
| DH5α(pGA2-CS4) | CS4+ | + | ++++ |
| DH5α | CS4− | − | − |
| CVD 1204(pGA2-CS4) | CS4+ | + | ++++ |
| CVD1204(pGA2) | CS4− | − | − |
Hemagglutination was scored as the time for agglutination of red blood cells to occur following the addition of the bacterial strain. A score of ++++ represents immediate (<1 min) hemagglutination, a score of +++ represents 1 to <5 min to hemagglutination, a score of ++ represents 5 to <10 min to hemagglutination, a score of + indicates 10 to 15 min to agglutination, and a − score indicates no hemagglutination after 30 min.
Bacterial agglutination was scored, as the time to agglutination of the bacteria following addition of the sera, as described for hemagglutination.
ETEC strains that produce CS4 fimbriae cause mannose-resistant hemagglutination of human type A red blood cells. ETEC-CS4 strains and DH5α and CVD 1204 strains expressing CS4 were tested for their hemagglutination properties. The results presented in Table 4 indicate that all strains bearing CS4 fimbriae, including E11881A, DS 9-1, DH5α(pGA2-CS4), and CVD 1204(pGA2-CS4), caused hemagglutination; in contrast, strains lacking CS4, including E11881C, DH5α, and CVD 1204(pGA2), did not result in hemagglutination. ETEC CS4+ strains demonstrated more intense hemagglutination than strains expressing the cloned csa operon.
Agglutination with CS4-specific antisera and hemagglutination of red blood cells indicated surface expression of functional fimbriae. Surface expression and typical fimbrial morphology were confirmed by electron microscopy (Fig. 2). CVD 1204(pGA2-CS4) cells displayed a profuse amount of typical fimbrial structures on their surfaces, while bacterial cells of the control strain CVD 1204(pGA2) did not.
Immunogenicity of CS4 expressed in the live vector CVD 1204.
To assess the immunogenicity of ETEC CS4 expressed in the S. flexneri vaccine strain CVD 1204, a guinea pig model was employed. In addition to testing the immunogenicity of CS4 alone, another plasmid was constructed expressing both CS4 and LThK63, a modified version of LT with a single-amino-acid substitution, rendering the molecule almost devoid of enzymatic activity, as previously described in detail (27, 42, 44). Expression vector pGA2-CS4-LThK63, encoding a fimbrial and mutant LTh antigen, was assessed for its ability to elicit immune responses to the two ETEC antigens expressed from a single plasmid. An additional control strain expressing LThK63 alone was included to detect whether immune responses were diminished between single- and double-antigen expression plasmids. Groups of five animals each were immunized intranasally with a 100-μl suspension containing approximately 109 bacteria. Immunization groups received CVD 1204(pGA2) (the vector alone), CVD 1204(pGA2-CS4) expressing CS4, CVD 1204(pGA2-CS4-LThK63) expressing CS4 and LThK63, or CVD 1204(pGA2-LThK63) expressing LThK63.
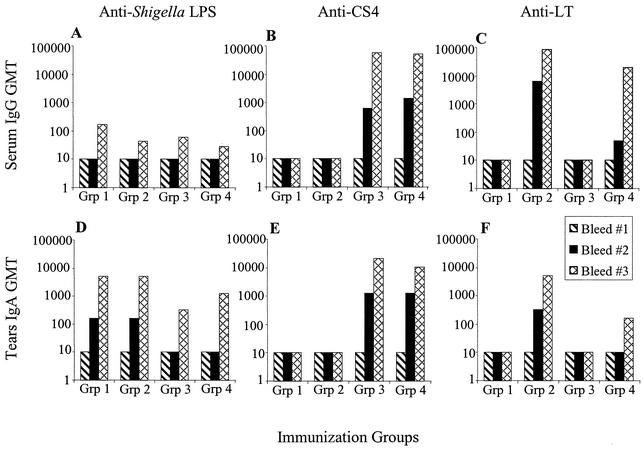
Animals immunized with CVD 1204(pGA2-CS4) or CVD 1204(pGA2-CS4-LThK63) showed high-titer anti-CS4 responses in both serum and tears following a single intranasal dose (Fig. 3B and E). Titers were boosted to reach much higher levels following a second immunization. Those animals immunized with CVD 1204(pGA2-LThK63) or CVD 1204(pGA2-CS4-LThK63) responded with both serum and mucosal anti-LT antibodies. The animals immunized with CVD 1204(pGA2-CS4-LThK63) were able to mount strong immune responses to both CS4 and LThK63 ETEC antigens.
FIG. 3.
Immunogenicity of S. flexneri 2a derivatives expressing CS4 in the guinea pig model. Groups of guinea pigs were immunized with two doses of live bacteria as follows: group 1 (Grp 1), CVD 1204(pGA2); group 2 (Grp 2), CVD 1204(pGA2-LThK63); group 3 (Grp 3), CVD 1204(pGA2-CS4); and group 4 (Grp 4), CVD 1204(pGA2-CS4-LThK63). Serum IgG and tears IgA titers are represented as the geometric mean titers (GMT) from the five animals per group. Titers are shown for samples taken prior to immunization (Bleed #1), 2 weeks following the first dose (Bleed #2), and 1 week following the second dose (Bleed #3).
In addition to eliciting high-titer anti-CS4 antibodies, CVD 1204(pGA2-CS4) immunization elicited antibodies capable of inhibiting hemagglutination of red blood cells by wild-type CS4-expressing ETEC strains. Serum from guinea pigs immunized with two doses of CVD 1204(pGA2-CS4) was able to inhibit hemagglutination of CS4-expressing ETEC strains E11881A (at a dilution of 1:256) and DS 9-1 (at a dilution of 1:32); in contrast, sera from control animals did not inhibit hemagglutination by these ETEC strains.
As shown in Fig. 3A and D, all animals immunized with CVD 1204 responded with strong serum and mucosal antibody responses to the Shigella vector itself. Every group achieved high titers of anti-lipopolysaccharide antibodies, demonstrating that the expression of a heterologous surface antigen did not diminish the immunogenicity of the Shigella vector strain. Furthermore, all animals immunized with CVD 1204 (either expressing an ETEC antigen or not) were protected against challenge with wild-type S. flexneri 2a strain 2457T in the Sereny test. Whereas negative-control animals immunized with E. coli or PBS exhibited full-blown keratoconjunctivitis within 3 days following conjunctival challenge with wild-type S. flexneri 2a, animals immunized with any CVD 1204 derivative were protected.
DISCUSSION
Two critical steps in the pathogenesis of human ETEC diarrheal infection, fimbria-mediated attachment to enterocytes of the proximal small intestine and enterotoxin production, offer targets for immunoprophylaxis. The most prevalent antigenic types of colonization factor fimbriae found on human ETEC strains include CFA/I and CS1 through CS6. All seven of these fimbrial types are considered essential for inclusion in an ETEC vaccine to elicit broad-spectrum protection (29, 32, 35, 51). Heretofore, however, full sequence particulars and information on organization of the operon remained unavailable for one fimbria, CS4, thereby impeding the development of a broad-spectrum, engineered, fimbria-based live-vector ETEC vaccine. Herein we fill a knowledge gap by reporting the complete sequence of the operon encoding the CS4 ETEC fimbria. This removes another obstacle on the road to development of a live ETEC vaccine that can include all seven of the most prevalent fimbrial antigens found on human ETEC isolates.
The DNA sequence of the four open reading frames comprising the CS4 operon and their predicted amino acid sequences display a high degree of homology to those of members of a family of ETEC fimbriae that includes CFA/I, CS1, and CS2. This finding is consistent with studies demonstrating some degree of cross-reactivity among antibodies against CS4, CFA/I, CS1, CS2, and CS17 fimbriae (38) and N-terminal sequence homology of structural subunits for CFA/I, CS1, and CS4 (58). The genetic determinants of this family consist of genes encoding usher, structural subunit, chaperone, and tip adhesin proteins (50). The predicted amino acid sequences of the CS4 proteins show the highest degree of identity with those of CFA/I, with identities of 67, 81, 93, and 95% for CsaA, CsaB, CsaC, and CsaE, respectively, with their Cfa homologues. The structural subunit CsaB and tip adhesin CsaE sequences display a greater degree of diversity from their CFA/I homologues than those of the usher and chaperone proteins. This might reflect the evolution of the fimbrial determinants into antigenically distinct structures. Since subunit and tip adhesin proteins make up the fimbrial structure against which immune responses are mounted, more divergence in sequence is expected for these proteins. Moreover, the maintenance of identity between usher and chaperone sequences of CS4 and CFA/I might reflect conservation of function in fimbrial assembly and biogenesis. While CS4 and CFA/I remain antigenically distinct, common immunorecessive epitopes have been postulated, following the demonstration that immunization with purified CFA/I or CS4 fimbriae can prime and boost immune response against the homologous as well as heterologous fimbriae in animal models (46-48). The amino acid sequence identity confirmed by our studies provides evidence for the presence of such epitopes.
The four genes (csaABCE) cloned in an expression vector in both E. coli DH5α and S. flexneri 2a vaccine strain CVD 1204 were sufficient to direct expression of CS4 fimbriae on the surface of the bacteria, as demonstrated by agglutination with CS4-specific antisera, hemagglutination of human type A erythrocytes, and electron microscopy. In the plasmid expression vector pGA2, control of gene expression is directed by the osmotically sensitive ompC promoter, with wild-type regulation circumvented by the deletion of upstream regulatory regions, allowing high-level fimbrial expression to be achieved.
Immunization of guinea pigs with CVD 1204(pGA2-CS4) elicited strong anti-CS4 serum and mucosal immune responses. Since different CFA fimbriae show preferences in the ability to agglutinate erythrocytes of different origins and blood groups, hemagglutination has been used as a simple, reproducible proxy for attachment of fimbriae to enterocyte receptors that share properties with the receptors on erythrocytes (49). Thus, the specificity and biological activity of the anti-CS4 antibody response was verified by demonstrating that serum from the immunized animals was able to inhibit hemagglutination of human erythrocytes by CS4-expressing ETEC strains.
To further extend the utility of these vectors by including an antigen that can stimulate LT antitoxin, LThK63 was cloned downstream from CS4 in pGA2-CS4-LThK63. The immunogenicity of this mutant LTh in CVD 1204 alone and in tandem with CFA/I was previously demonstrated (2, 27). Herein we report the immunogenicity of a double-construction CVD 1204(pGA2-CS4-LThK63) strain that stimulated anti-LT responses.
The fact that Shigella live-vector constructs expressing ETEC fimbrial antigens can also elicit anti-Shigella lipopolysaccharide O antibodies and mediate protection against Shigella in animal models provides encouragement that it might be possible, by using multiple carefully selected Shigella serotypes, to construct a complex live-vector vaccine that provides broad-spectrum protection against shigellosis as well as against ETEC. The strategy we are pursuing is to use attenuated strains of Shigella to express and deliver ETEC antigens, thus creating a combined vaccine against Shigella and ETEC (1, 27, 29, 40, 41). The final vaccine is to utilize five attenuated Shigella strains as live vectors of serotypes that are of epidemiologic and immunologic importance. These include S. dysenteriae 1 (cause of epidemic and pandemic Shiga dysentery), S. sonnei (an important cause of traveler's shigellosis and of disease in day care and other institutional settings in industrialized countries), and S. flexneri 2a, 3a, and 6 (which share type- or group-specific epitopes with the remaining 12 flexneri serotypes) (28, 39). Each of these five attenuated Shigella strains is being engineered to express two fimbrial antigens and an antigen that stimulates anti-LT. The cloning and expression of CFA/I, CS2, CS3, and mutant LT in attenuated S. flexneri 2a strain CVD 1204 has been previously reported (1, 2, 27). In the guinea pig model, mucosal immunization with these strains elicited serum and mucosal immune responses against the Shigella live-vector O antigen (a correlate of protection) as well as against the ETEC antigens. Here, for the first time, we have cloned and sequenced CS4, allowing its inclusion in our vaccine strategy. This work extends the feasibility of constructing an efficient multivalent Shigella-based oral ETEC vaccine.
Acknowledgments
This work was supported by grants from the Rockefeller Foundation to M.M.L.
We gratefully acknowledge the assistance of Nick Ambulos from the UMB Biopolymer Lab for DNA sequencing, Marcia Wolf for providing ETEC-CS4 strains, John Czeczulin for the EM studies, and David R. Maneval for antiserum production.
Editor: D. L. Burns
REFERENCES
- 1.Altboum, Z., E. M. Barry, G. Losonsky, J. E. Galen, and M. M. Levine. 2001. Attenuated Shigella flexneri 2a guaBA strain CVD 1204 expressing enterotoxigenic Escherichia coli (ETEC) CS2 and CS3 fimbriae as a live mucosal vaccine against Shigella and ETEC infection. Infect. Immun. 69:3150-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry, E. M., Z. Altboum, G. A. Losonsky, and M. M. Levine. 2003. Immune responses elicited against multiple fimbriae and mutant LT from enterotoxigenic E. coli expressed in attenuated Shigella vaccine strains. Vaccine 21:333-340. [DOI] [PubMed] [Google Scholar]
- 3.Binsztein, N., M. J. Jouve, G. I. Viboud, L. L. Moral, M. Rivas, I. Ørskov, C. Åhrén, and A.-M. Svennerholm. 1991. Colonization factors of enterotoxigenic Escherichia coli isolated from children with diarrhea in Argentina. J. Clin. Microbiol. 29:1893-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, R. E. 1986. Pathogens that cause travelers' diarrhea in Latin America and Africa. Rev. Infect. Dis. 8S:S131-S135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, R. E., M. H. Merson, I. Huq, A. R. M. A. Alim, and M. Yunus. 1981. Incidence and severity of rotavirus and Escherichia coli diarrhoea in rural Bangladesh. Lancet i:141-143. [DOI] [PubMed] [Google Scholar]
- 6.Black, R. E., M. H. Merson, B. Rowe, P. R. Taylor, A. R. Abdul Alim, R. J. Gross, and D. A. Sack. 1981. Enterotoxigenic Escherichia coli diarrhoea: acquired immunity and transmission in an endemic area. Bull. W. H. O. 59:263-268. [PMC free article] [PubMed] [Google Scholar]
- 7.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisenstein. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447-1457. [DOI] [PubMed] [Google Scholar]
- 8.Caron, J., L. M. Coffield, and J. R. Scott. 1989. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. USA 86:963-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassels, F. J., and M. K. Wolf. 1995. Colonization factors of diarrheagenic E. coli and their intestinal receptors. J. Ind. Microbiol. 15:214-226. [DOI] [PubMed] [Google Scholar]
- 10.Cravioto, A., R. E. Reyes, R. Ortega, G. Fernandez, R. Hernandez, and D. Lopez. 1988. Prospective study of diarrhoeal disease in a cohort of rural Mexican children: incidence and isolated pathogens during the first two years of life. Epidemiol. Infect. 101:123-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Haan, L. A., G. A. Willshaw, B. A. van der Zeijst, and W. Gaastra. 1991. The nucleotide sequence of a regulatory gene present on a plasmid in an enterotoxigenic Escherichia coli strain of serotype O167:H5. FEMS Microbiol. Lett. 67:341-346. [DOI] [PubMed] [Google Scholar]
- 12.DuPont, H. L., J. Olarte, D. G. Evans, L. K. Pickering, E. Galindo, and D. J. Evans. 1976. Comparative susceptibility of Latin American and United States students to enteric pathogens. N. Engl. J. Med. 285:1520-1521. [DOI] [PubMed] [Google Scholar]
- 13.Duthy, T. G., L. H. Staendner, P. A. Manning, and M. W. Heuzenroeder. 1999. CS5 pilus biosynthesis genes from enterotoxigenic Escherichia coli O115:H40. J. Bacteriol. 181:5847-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans, D. G., D. J. Evans, Jr., W. S. Tjoa, and H. L. DuPont. 1978. Detection and characterization of colonization factor of enterotoxigenic Escherichia coli isolated from adults with diarrhea. Infect. Immun. 19:727-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, D. G., T. K. Satterwhite, D. J. Evans, Jr., and H. L. DuPont. 1978. Differences in serological responses and excretion patterns of volunteers challenged with enterotoxigenic Escherichia coli with and without the colonization factor antigen. Infect. Immun. 19:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Froehlich, B. J., A. Karakashian, L. R. Melsen, J. C. Wakefield, and J. R. Scott. 1994. CooC and CooD are required for assembly of CS1 pili. Mol. Microbiol. 12:387-401. [DOI] [PubMed] [Google Scholar]
- 17.Froehlich, B. J., A. Karakashian, H. Sakellaris, and J. R. Scott. 1995. Genes for CS2 pili of enterotoxigenic Escherichia coli and their interchangeability with those for CS1 pili. Infect. Immun. 63:4849-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaastra, W., B. J. Jordi, E. M. Mul, A. M. Hamers, M. M. McConnell, G. A. Willshaw, H. R. Smith, and B. A. van der Zeijst. 1990. A silent regulatory gene cfaD′ on region 1 of the CFA/I plasmid NTP 113 of enterotoxigenic Escherichia coli. Microb. Pathog. 9:285-291. [DOI] [PubMed] [Google Scholar]
- 19.Gaastra, W., and A. M. Svennerholm. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4:444-452. [DOI] [PubMed] [Google Scholar]
- 20.Galen, J. E., J. Nair, J. Y. Wang, S. S. Wasserman, M. K. Tanner, M. B. Sztein, and M. M. Levine. 1999. Optimization of plasmid maintenance in the attenuated live-vector vaccine strain Salmonella typhi CVD 908-htrA. Infect. Immun. 67:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grewal, H. M., W. Gaastra, A. M. Svennerholm, J. Roli, and H. Sommerfelt. 1993. Induction of colonization factor antigen I (CFA/I) and coli surface antigen 4 (CS4) of enterotoxigenic Escherichia coli: relevance for vaccine production. Vaccine 11:221-226. [DOI] [PubMed] [Google Scholar]
- 22.Hamers, A., H. Pel, G. Willshaw, J. Kusters, B. Van der Zeijst, and W. Gaastra. 1989. Nucleotide sequence of the first two genes of the fimbrial operon of human enterotoxigenic Escherichia coli. Microb. Pathog. 6:297-309. [DOI] [PubMed] [Google Scholar]
- 23.Hibberd, M. L., M. M. McConnell, G. A. Willshaw, H. R. Smith, and B. Rowe. 1991. Positive regulation of colonization factor antigen I (CFA/I) production by enterotoxigenic Escherichia coli producing the colonization factors CS5, CS6, CS7, CS17, PCFO9, PCFO159-H4, and PCFO166. J. Gen. Microbiol. 137:1963-1970. [DOI] [PubMed] [Google Scholar]
- 24.Hyams, K. C., A. L. Bourgeois, B. R. Merrell, R. Rozmahel, J. Escamilla, S. A. Thornton, G. M. Wasserman, A. Burke, P. Echeverria, K. Y. Green, A. Z. Kapikian, and J. N. Woody. 1991. Diarrheal disease during Operation Desert Shield. N. Engl. J. Med. 325:1423-1428. [DOI] [PubMed] [Google Scholar]
- 25.Jalajakumari, M. B., C. J. Thomas, R. Halter, and P. A. Manning. 1989. Genes for biosynthesis and assembly of CS3 pili of CFA/II enterotoxigenic Escherichia coli: novel regulation of pilus production by bypassing an amber codon. Mol. Microbiol. 3:1685-1695. [DOI] [PubMed] [Google Scholar]
- 26.Jordi, B. J., B. Dagberg, L. A. de Haan, A. M. Hamers, B. A. van der Zeijst, W. Gaastra, and B. E. Uhlin. 1992. The positive regulator CfaD overcomes the repression mediated by histone-like protein H-NS (H1) in the CFA/I fimbrial operon of Escherichia coli. EMBO J. 11:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koprowski, H., II, M. M. Levine, R. J. Anderson, G. Losonsky, M. Pizza, and E. M. Barry. 2000. Attenuated Shigella flexneri 2a vaccine strain CVD 1204 expressing colonization factor antigen I and mutant heat-labile enterotoxin of enterotoxigenic Escherichia coli. Infect. Immun. 68:4884-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotloff, K. L., J. P. Winickoff, B. Ivanoff, J. D. Clemens, D. L. Swerdlow, P. J. Sansonetti, G. K. Adak, and M. M. Levine. 1999. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. W. H. O. 77:651-666. [PMC free article] [PubMed] [Google Scholar]
- 29.Levine, M. M. 2000. Immunization against bacterial diseases of the intestine. J. Pediatr. Gastroenterol. Nutr. 31:336-355. [DOI] [PubMed] [Google Scholar]
- 30.Levine, M. M., R. E. Black, M. L. Clements, C. R. Young, C. P. Cheney, P. Schad, H. Collins, and E. C. Boedeker. 1984. Prevention of enterotoxigenic Escherichia coli diarrheal infection by vaccines that stimulate antiadhesion (antipili) immunity, p. 223-244. In E. C. Boedeker (ed.), Attachment of organisms to the gut mucosa. CRC Press, Boca Raton, Fla.
- 31.Levine, M. M., C. Ferreccio, V. Prado, M. Cayazzo, P. Abrego, J. Martinez, L. Maggi, M. Baldini, W. Martin, D. Maneval, B. Kay, L. Guers, H. Lior, S. S. Wasserman, and J. P. Nataro. 1993. Epidemiologic studies of Escherichia coli infections in a low socioeconomic level periurban community in Santiago, Chile. Am. J. Epidemiol. 138:849-869. [DOI] [PubMed] [Google Scholar]
- 32.Levine, M. M., J. A. Giron, and F. Noriega. 1994. Fimbrial vaccines. In P. Klemm (ed.), Fimbriae: adhesion, biogenics, genetics and vaccines. CRC Press, Boca Raton, Fla.
- 33.Levine, M. M., D. R. Nalin, D. L. Hoover, E. J. Bergquist, R. B. Hornick, and C. R. Young. 1979. Immunity to enterotoxigenic Escherichia coli. Infect. Immun. 23:729-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine, M. M., P. Ristaino, G. Marley, C. Smyth, S. Knutton, E. Boedeker, R. Black, C. Young, M. L. Clements, C. Cheney, and R. Patnaik. 1984. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect. Immun. 44:409-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levine, M. M., and A.-M. Svennerholm. 2000. Enteric vaccines: present and future. In H. L. DuPont and R. Steffen (ed.), The textbook of travel medicine and health. B.C. Decker, New York, N.Y.
- 36.Levine, M. M., J. Kaper, R. E. Black, and M. Clements. 1983. New knowledge on pathogenesis of bacterial enteric infections as applied to vaccine development. Microbiol. Rev. 47:510-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manning, P. A., K. N. Timmis, and G. Stevenson. 1985. Colonization factor antigen II (CFA/II) of enterotoxigenic Escherichia coli: molecular cloning of the CS3 determinant. Mol. Gen. Genet. 200:322-327. [DOI] [PubMed] [Google Scholar]
- 38.McConnell, M. M., H. Chart, and B. Rowe. 1989. Antigenic homology within human enterotoxigenic Escherichia coli fimbrial colonization factor antigens: CFA/I, coli-surface-associated antigens (CS)1, CS2, CS4 and CS17. FEMS Microbiol. Lett. 52:105-108. [DOI] [PubMed] [Google Scholar]
- 39.Noriega, F. R., F. M. Liao, D. R. Maneval, S. Ren, S. B. Formal, and M. M. Levine. 1999. Strategy for cross-protection among Shigella flexneri serotypes. Infect. Immun. 67:782-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noriega, F. R., G. Losonsky, C. Lauderbaugh, F. M. Liao, J. Y. Wang, and M. M. Levine. 1996. Engineered ΔguaB-A ΔvirG Shigella flexneri 2a strain CVD 1205: construction, safety, immunogenicity, and potential efficacy as a mucosal vaccine. Infect. Immun. 64:3055-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noriega, F. R., G. Losonsky, J. Y. Wang, S. B. Formal, and M. M. Levine. 1996. Further characterization of ΔaroA ΔvirG Shigella flexneri 2a strain CVD 1203 as a mucosal Shigella vaccine and as a live-vector vaccine for delivering antigens of enterotoxigenic Escherichia coli. Infect. Immun. 64:23-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pizza, M., M. M. Giuliani, M. R. Fontana, G. Douce, G. Dougan, and R. Rappuoli. 2000. LTK63 and LTR72, two mucosal adjuvants ready for clinical trials. Int. J. Med. Microbiol. 290:455-461. [DOI] [PubMed] [Google Scholar]
- 43.Qadri, F., S. K. Das, A. S. G. Faruque, G. J. Fuchs, M. J. Albert, R. B. Sack, and A.-M. Svennerholm. 2000. Prevalence of toxin types and colonization factors in enterotoxigenic Escherichia coli isolated during a 2-year period from diarrheal patients in Bangladesh. J. Clin. Microbiol. 38:27-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rappuoli, R., M. Pizza, G. Douce, and G. Dougan. 1999. Structure and mucosal adjuvanticity of cholera and Escherichia coli heat-labile enterotoxins. Immunol. Today 20:493-500. [DOI] [PubMed] [Google Scholar]
- 45.Reese, M. G., N. L. Harris, and F. H. Eeckman. 1996. Large scale sequencing specific neural networks for promoter and splice site recognition, p. 737-738. In L. Hunter and T. E. Klein (ed.), Biocomputing: proceedings of the 1996 Pacific Symposium. World Scientific Publishing Company, Singapore.
- 46.Rudin, A., M. M. McConnell, and A.-M. Svennerholm. 1994. Monoclonal antibodies against enterotoxigenic Escherichia coli colonization factor antigen I (CFA/I) that cross-react immunologically with heterologous CFAs. Infect. Immun. 62:4339-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudin, A., and A. M. Svennerholm. 1994. Colonization factor antigens (CFAs) of enterotoxigenic Escherichia coli can prime and boost immune responses against heterologous CFAs. Microb. Pathog. 16:131-139. [DOI] [PubMed] [Google Scholar]
- 48.Rudin, A., G. Wiklund, C. Wenneras, and F. Qadri. 1997. Infection with colonization factor antigen I-expressing enterotoxigenic Escherichia coli boosts antibody responses against heterologous colonization factors in primed subjects. Epidemiol. Infect. 119:391-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakellaris, H., G. P. Munson, and J. R. Scott. 1999. A conserved residue in the tip proteins of CS1 and CFA/I pili of enterotoxigenic Escherichia coli that is essential for adherence. Proc. Natl. Acad. Sci. USA 96:12828-12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakellaris, H., and J. R. Scott. 1998. New tools in an old trade: CS1 pilus morphogenesis. Mol. Microbiol. 30:681-687. [DOI] [PubMed] [Google Scholar]
- 51.Savarino, S. J., E. R. Hall, S. Bassily, T. F. Wierzba, F. G. Youssef, L. F. Peruski, Jr., R. Abu-Elyazeed, M. Rao, W. M. Francis, H. El Mohamady, M. Safwat, A. B. Naficy, A. M. Svennerholm, M. Jertborn, Y. J. Lee, and J. D. Clemens. 2002. Introductory evaluation of an oral, killed whole cell enterotoxigenic Escherichia coli plus cholera toxin B subunit vaccine in Egyptian infants. Pediatr. Infect. Dis. J. 21:322-330. [DOI] [PubMed] [Google Scholar]
- 52.Savelkoul, P. H., G. A. Willshaw, M. M. McConnell, H. R. Smith, A. M. Hamers, B. A. van der Zeijst, and W. Gaastra. 1990. Expression of CFA/I fimbriae is positively regulated. Microb. Pathog. 8:91-99. [DOI] [PubMed] [Google Scholar]
- 53.Scotland, S. M., G. A. Willshaw, H. R. Smith, and B. Rowe. 1990. Properties of strains of Escherichia coli O26:H11 in relation to their enteropathogenic or enterohemorrhagic classification. J. Infect. Dis. 162: 1069-1074. [DOI] [PubMed] [Google Scholar]
- 54.Sommerfelt, H., H. M. S. Grewal, W. Gaastra, A.-M. Svennerholm, and M. K. Bhan. 1992. Use of nonradioactive DNA hybridization for identification of enterotoxigenic Escherichia coli harboring genes for colonization factor antigen I, coli surface antigen 4, or putative colonization factor O166. J. Clin. Microbiol. 30:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas, L. V., M. M. McConnell, B. Rowe, and A. M. Field. 1986. The possession of three novel coli surface antigens by enterotoxigenic Escherichia coli strains positive for the putative colonization factor PCF8775. J. Gen. Microbiol. 131:2319-2326. [DOI] [PubMed] [Google Scholar]
- 56.Willshaw, G. A., H. R. Smith, M. M. McConnell, and B. Rowe. 1991. Cloning of regulator genes controlling fimbrial production by enterotoxigenic Escherichia coli. FEMS Microbiol. Lett. 66:125-129. [DOI] [PubMed] [Google Scholar]
- 57.Wolf, M. K. 1997. Occurrence, distribution, and associations of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin. Microbiol. Rev. 10:569-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolf, M. K., G. P. Andrews, B. D. Tall, M. M. McConnell, M. M. Levine, and E. C. Boedeker. 1989. Characterization of CS4 and CS6 antigenic components of PCF8775, a putative colonization factor complex from enterotoxigenic Escherichia coli E8775. Infect. Immun. 57:164-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolf, M. K., L. A. de Haan, F. J. Cassels, G. A. Willshaw, R. Warren, E. C. Boedeker, and W. Gaastra. 1997. The CS6 colonization factor of human enterotoxigenic Escherichia coli contains two heterologous major subunits. FEMS Microbiol Lett. 148:35-42. [DOI] [PubMed] [Google Scholar]