Abstract
Rolling-circle amplification (RCA) and ramification amplification (RAM, also known as hyperbranched RCA) are isothermal nucleic acid amplification technologies that have gained a great application in in situ signal amplification, DNA and protein microarray assays, single nucleotide polymorphism detection, as well as clinical diagnosis. Real-time detection of RCA or RAM products has been a challenge because of most real-time detection systems, including Taqman and Molecular Beacon, are designed for thermal cycling-based DNA amplification technology. In the present study, we describe a novel fluorescent probe construct, termed molecular zipper, which is specially designed for quantifying target DNA by real-time monitoring RAM reactions. Our results showed that the molecular zipper has very low background fluorescence due to the strong interaction between two strands. Once it is incorporated into the RAM products its double strand region is opened by displacement, therefore, its fluorophore releases a fluorescent signal. Applying the molecular zipper in RAM assay, we were able to detect as few as 10 molecules within 90 min reaction. A linear relationship was observed between initial input of targets and threshold time (R2 = 0.985). These results indicate that molecular zipper can be applied to real-time monitoring and qualification of RAM reaction, implying an amenable method for automatic RAM-based diagnostic assays.
INTRODUCTION
Rolling-circle amplification (RCA) (1–3) and ramification amplification (RAM) (4), also known as hyperbranched RCA (2) or exponential RCA (5), are isothermal nucleic acid amplification technologies that are based on rolling-circle DNA replication used by bacterial phages and plamids (6). Hybridization of a specially designed single-stranded DNA (ssDNA) probe to a target DNA brings the two ends of the probe close to each other. Ligation of these two ends by a DNA ligase (7,8) forms a circularized probe (C-probe) that subsequently can be amplified either by RCA with one primer (9–11) or by RAM with a pair of primers (one forward and one reverse primer) (2,4). These technologies are promising for target DNA and protein detections in various assay platforms, such as microplate, DNA and protein microarray and in situ signal amplification (12), and may provide a simple, cost-effective, fully automated high-throughput method for genotyping analysis (13).
It will be desirable to monitor and quantify the RCA and RAM products in a real-time fashion in a homogeneous reaction. Several fluorescence-based RCA and RAM assays have been reported. We have demonstrated that RAM products can be monitored in the presence of SYBR green (D. Y. Zhang, unpublished data). SYBR green is a double-stranded DNA (dsDNA) binding dye that emits fluorescent light when it binds to dsDNA and can be detected with a fluorometer. However, a major limitation of such intercalating dye is that its signal may also be interfered by non-specific polymerization products.
Several fluorescence resonance energy transfer (FRET)-based detection methods were used in RCA with a molecular beacon (14–16) and in RAM with a peptide nucleic acid (PNA) probe (17,18) or a quenched-PNA (Q-PNA) probe (5). The molecular beacon is a nucleic acid probe with a stem–loop structure (19) or without a loop (17,18), and this probe contains a quencher at its 3′ end and a fluorophore at the 5′ end (19). In the absence of the target, the quencher is in close contact with the fluorophore, and, therefore, no light is emitted. When the loop hybridizes to the target, and the quencher and the fluorophore are separated apart from each other, the light emission can be detected. However, since molecular beacon was initially designed for thermal cycling-based amplification, such as PCR, it was a challenge to apply molecular beacon in RCA (14) or RAM reaction [(17) and also D. Y. Zhang, unpublished data).
In this report, we developed a unique FRET-based probe, termed molecular zipper, for real-time RAM assay. In this probe, one of the two RAM primers is incorporated into a partially double stranded short DNA fragment, which contains a fluorophore at its 5′ end and a quencher at its 3′ end (Figure 1A). Initially, the fluorophore is in close contact with the quencher, therefore, it does not fluoresce. During RAM reaction, one of the strands is incorporated into the RAM product as a primer and the other strand is displaced from its complementary strand (Figure 2). As a consequence, the fluorophore and quencher are separated, the fluorecent signal can be detected which is in proportion to the RAM products.
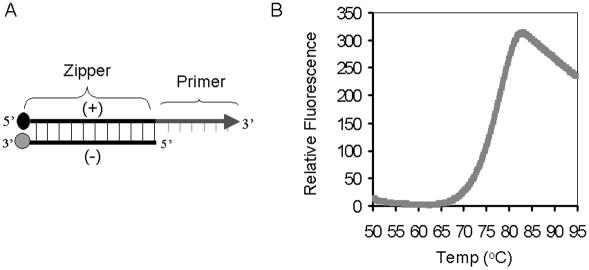
Figure 1.
Molecular zipper and its denaturation curve. (A) Molecular zipper is a short segment of dsDNA (25 bp), consisting of a positive strand with a quencher at its 5′ end and a negative strand with a fluorophore at its 3′ end. A short ssDNA (23 nt) is attached to the 3′ end of the positive strand, which serves as a reverse primer. The arrow indicates the direction of primer extension. (B) A preannealed molecular zipper with equal amount of negative and positive strands (0.4 μM) were incubated in 25 μl containing 100 mM Tris and 10 mM EDTA (pH 8.3) at 30°C for 10 min and the denaturation curve was determined by measuring the change in fluorescence intensity in a fluorometer (Smart Cycler; Cepheid).
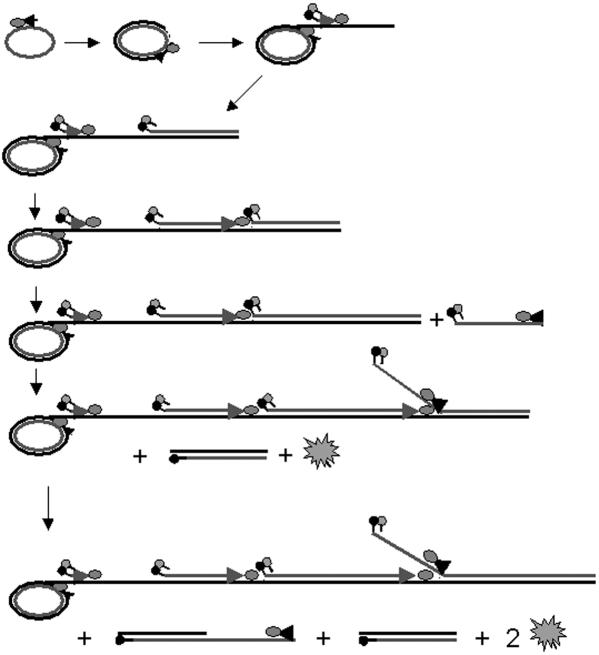
Figure 2.
Schematic representation of molecular zipper in RAM reaction: A forward primer binds to a DNA circle and is continuously extended along the circle by a DNA polymerase, resulting in a long ssDNA with multiple repeats of circle sequence. Multiple molecular zippers, which serve as reverse primers, bind to the growing ssDNA and are extended by DNA polymerases, generating multiple ssDNAs by displacing downstream molecular zippers and their associated ssDNAs. To these ssDNAs, many forward primers bind and are extended by DNA polymerases. The negative strands of the molecular zippers are displaced by the polymerases when encountered. As a consequence, fluorescence emitted from the fluorophores of the negative strands can be detected.
METHODS
Synthetic DNAs
A molecular zipper is a dsDNA composed of a positive strand [5′-Fluorescein-GCTGAGGACCCGGATGCGAATGCGGATGCGGATGCCGAACCAAGAGCAACTACACGAATTC-3′(61 nt)] and a negative strand [5′-TCGGCA TCCGCATCCGCATTCGCATCCGGGTCCTCAGC-DABCYL-3′ (38 nt)], where underlined nucleotides indicate the primer sequence. Both strands were obtained from Integrated DNA Technology, Inc., Coralville, IA. The sequence of the molecular zipper was designed with minimal secondary structure and a melting temperature (Tm) for the dsDNA portion was ∼77.5°C, above the RAM reaction temperature (63°C). The double-stranded molecular zipper was formed by mixing equal amount (20 μM) of positive and negative strands in a buffer containing 100 mM Tris and 10 mM EDTA (pH 8.3), heating at 95°C for 5 min, and cooling slowly to room temperature. The C-probe [5′-GGTTTTGTCTTCGTAACTCGCTCCGGATGTCTGTGTATCTGCT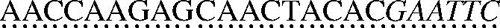 TCG
TCG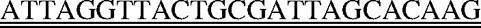 CTCTACAAGAGTACATCGGTCAACGAAGA-3′(124 nt)] was obtained from Genelink, Hawthorne, NY, where the single underline indicates the target complementary regions at the 5′ and 3′ ends, the double underline indicates the forward primer binding site, the dotted underline indicates the reverse primer binding site and italic letters indicate EcoRI restriction site. The synthetic target DNA [5′-TCCGGAGCGAGTTACGAAGACAAAACCTCTTCGTTGACCGATGTACTCTTGTAGAAAG TTATAATAATCCTCTTTTCTGTCTGACGGTTCTTAAGC-3′ (96 nt)] also was obtained from Genelink, where the underlined nucleotides indicate the C-probe binding region. The forward primer [5′-CTTGTGCTAATCGCAGTAACCTAAT-3′ (25 nt)] and the reverse primer [5′-ACCAAGAGCAACTACACGAATTC-3′ (23 nt)] were synthesized in the DNA Core Facility at Mount Sinai School of Medicine, New York, NY.
CTCTACAAGAGTACATCGGTCAACGAAGA-3′(124 nt)] was obtained from Genelink, Hawthorne, NY, where the single underline indicates the target complementary regions at the 5′ and 3′ ends, the double underline indicates the forward primer binding site, the dotted underline indicates the reverse primer binding site and italic letters indicate EcoRI restriction site. The synthetic target DNA [5′-TCCGGAGCGAGTTACGAAGACAAAACCTCTTCGTTGACCGATGTACTCTTGTAGAAAG TTATAATAATCCTCTTTTCTGTCTGACGGTTCTTAAGC-3′ (96 nt)] also was obtained from Genelink, where the underlined nucleotides indicate the C-probe binding region. The forward primer [5′-CTTGTGCTAATCGCAGTAACCTAAT-3′ (25 nt)] and the reverse primer [5′-ACCAAGAGCAACTACACGAATTC-3′ (23 nt)] were synthesized in the DNA Core Facility at Mount Sinai School of Medicine, New York, NY.
Ligation reaction
Ligation of two ends of the C-probe was carried out by incubating the C-probe with target DNA in 20 μl containing 20 mM Tris–HCl (pH 7.6), 25 mM potassium acetate, 10 mM magnesium acetate, 10 mM DTT, 1 mM NAD, 0.1% Triton X-100 and 12 U Taq DNA ligase (New England BioLabs, Beverly, MA) at 65°C for 15 min. The reaction was further incubated at 95°C for 30 min to inactivate the ligase.
RAM assay
RAM reaction was performed in 50 μl containing 20 mM Tris–HCl (pH 8.8), 300 μM each of dATP, dCTP, dGTP and dTTP, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100, 1.2 μM forward primer, 0.8 μM reverse primer, 0.4 μM molecular zipper, 6% Dimethylsulfoxide and 6.4 U of exo− Bst DNA polymerase (large fragment) (New England Biolabs). The reaction was incubated at 63°C for 2–3 h in a fluorometric thermal cycler (iCycler; Bio-Rad Laboratories, Hercules, CA, or Smart Cycler; Cepheid, Sunnyvale, CA).
RESULTS AND DISCUSSION
A molecular zipper (Figure 1A) contains a generic, dsDNA sequence (38 bp) with a quencher moiety at the 5′ end of one strand (positive) and a fluorophore moiety at the 3′ end of the other strand (negative). The positive strand of the molecular zipper also contains an additional sequence (24 nt) at its 3′ end, which is identical to a portion of the C-probe and serves as a reverse primer for the RAM reaction. In its double-stranded form, the fluorophore of the negative strand is held close to the quencher of the positive strand, therefore, no fluorescence is emitted. However, when the two strands are separated from each other, for example, by increasing temperature, the quencher is separated from fluorophore and light is emitted. Figure 1B shows a typical denaturation curve (or unzipping) of a molecular zipper. At temperatures below 70°C, low background fluorescence was noticed, indicating that both strands were bound together, thereby efficiently quenching the fluorophore. However, there was a sharp increase of fluorescence above 72°C (the Tm of the molecular zipper is ∼77°C) that reached plateau at 81°C, indicating a rapid unzipping of the molecular zipper. There was a decrease of fluorescence above 82°C. This is probably due to random interactions of the fluorophores with the quenchers in solution. Another explanation is that increasing of temperature induces the decreasing in pH of Tris buffer, which causes the decreasing in fluorescence of pH-sensitive fluorophore (i.e. fluorescein in this study).
In a RAM reaction, a forward primer bound to the C-probe is extended along the DNA circle by the large fragment of Bst DNA polymerase that has high processivity and displacement activity (12,20). The DNA polymerase displaces the initiating forward primer from the C-probe and continues copying around the circle, generating a long ssDNA with multiple repeats of the C-probe sequence (Figure 2). Multiple molecular zippers bind to the growing ssDNA under isothermal conditions (2,4) and are extended simultaneously by the DNA polymerases. The polymerases displace upstream molecular zippers and their associated sequences from the growing ssDNA when they encounter the molecular zippers, generating ssDNAs identical to the sequence of the original C-probe. To these ‘second-round products,’ multiple forward primers bind and are likewise extended by the polymerases. When the polymerases encounter the molecular zippers, they displace the negative strands of the molecular zippers (containing the fluorophores) from the positive strands, thereby separating fluorophore and quencher pairs and resulting in an increase of fluorescence (Figure 2).
To determine the actual performance of the molecular zipper in RAM reaction, we carried out two reactions, both containing target DNA, C-probe and molecular zipper. One reaction included Taq DNA ligase and the other contained no ligase (Methods). In the presence of the ligase, two ends of C-Probe were linked to form a DNA circle, while the C-probe remained linear in the absence of DNA ligase. As expected, the DNA circles formed in the presence of DNA ligase were utilized as templates in RAM reactions, and fluorescence was increasing as the RAM reaction proceeds (Figure 3A). The change in fluorescence in the RAM reaction was similar to that observed in conventional real-time exponential amplification (i.e. PCR), confirming the exponential nature of the RAM reaction (12). In the other reaction, no circle was formed in the absence of the DNA ligase, thus, no fluorescence was detected even after a long period of incubation (Figure 3A), indicating a circle-dependent amplification in RAM reaction. Our results also indicate that the presence of dsDNA sequence in the molecular zipper does not interfere with the RAM reaction.
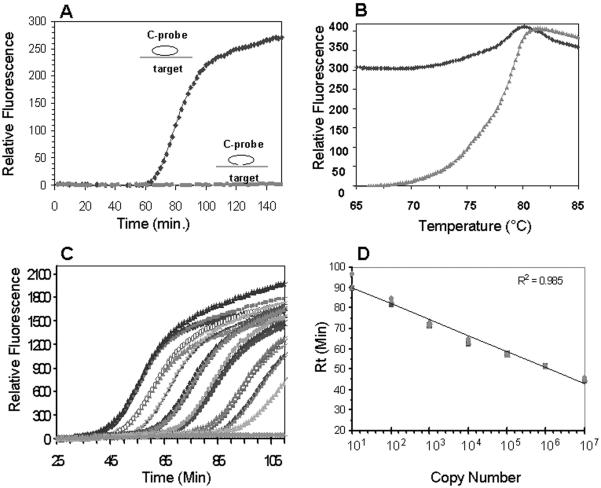
Figure 3.
Characterization of molecular zipper in RAM reaction. (A) Molecules (1 × 107) of the C-probe were hybridized to 1 × 1010 molecules of the DNA target in a 20 μl reaction at 65°C for 15 min in the presence or absence of DNA ligase. The ligation products (2 μl) were used to initiate 25 μl RAM reactions in the presence of 0.4 μM molecular zipper and the change in fluorescence was analyzed with a Smart Cycler. (B) After completion of the RAM reaction, the denaturation curves were determined by measuring the fluorescence change as a function of temperature change (up to 95°C). (C) Known concentrations of ligated C-probes (108–10 molecules) were used to initiate the RAM reaction in the presence of 0.4 μM molecular zipper in 50 μl RAM reactions and the kinetics of fluorescence change were determined in a fluorometric thermal cycler (iCycler). Each concentration was carried out in triplicates. For negative control (gray line), no ligated C-probe was added. (D) Rt values obtained from Figure 3C are plotted against initial copy number of the C-probe (three replicates per concentration). The linear curve was added from a linear fitting.
After completion of the RAM reaction, we further measured the fluorescence change by denaturing the RAM products (Figure 3B). Our results showed that the denaturation curve of the RAM products initiated with unligated C-probe was the same as that of the molecular zipper alone (compare Figure 3B with Figure 1B), indicating that the molecular zipper was intact during the incubation. In contrast, the fluorescence intensity of the RAM products derived from the ligated C-probe increased only mildly (25%) as compared with that derived from unligated C-probe, indicating that the majority of the molecular zippers were in an unzipped state. These results indicate that the positive strand of the molecular zipper was incorporated into the RAM products and the negative strand was displaced from the positive strand (i.e. unzipped).
To test the ability of the molecular zipper to quantify the copy number of C-probes present in a RAM reaction, we initiated the RAM reaction with known concentrations of ligated C-probes. Figure 3C presented real-time observation of triplicates for each concentration of C-probe (from 108 to 10 molecules), demonstrating a reasonably high reproducibility of RAM reactions monitored by molecular zipper. Most importantly, the figure showed that the time at which fluorescence raised above the baseline (response time, or Rt) correlated with the number of C-probes initially present. The greater the number of the C-probes, the earlier the response time occurred. A linear relationship (R2 = 0.985) existed between the threshold times (Rt values) and initial number of the C-probe (Figure 3D), demonstrating the ability of the molecular zipper for the real-time quantitative analysis in RAM reactions. Our results also demonstrated that RAM, in combination with the molecular zipper, detected a large dynamic range of C-probe (101–108 molecules) with an excellent analytical sensitivity.
Because of its sensitivity (i.e. exponential amplification), specificity (i.e. ligation-dependent discrimination of single nucleotide difference) and isothermal nature, the RAM technology becomes an important tool in research and clinical diagnosis (12). However, its applications can be further broadened if the RAM products can be monitored in a real-time fashion as in real-time PCR. Several FRET-based DNA probes, such as molecular beacons (19,21), Taqman probe (22) and Scorpion probe (23), have been described. However, the use of these FRET probes in RAM reactions is problematic and challenging. Owing to the lack of exonuclease activity of the DNA polymerase that is required for the displacement in RAM reaction (20), Taqman cannot be used in RAM reaction. The use of molecular beacons in RCA also encountered some difficulties because of the interaction of neighboring beacons, which resulted in insufficient fluorescent signal (14). Our attempts to directly use molecular beacons in RAM reactions also were discouraged because of the high background and low signal-to-noise ratios (data not shown), most likely due to the poor accessibility of molecular beacons to dsDNA product under isothermal conditions. On the other hand, a FRET probe linked to a primer, such as a molecular beacon (21) and a Q-PNA (5), has been used successfully in real-time monitoring of isothermal DNA amplification. The molecular beacon (21), in this case, was designed to have a specific sequence to restrict enzyme recognition and digestion. This digestion, similar to the case of a stem–loop Scorpion (23), separates the fluorophore from the quencher for maximum fluorescence release.
A molecular zipper, either a dsDNA as described in this study or a PNA/DNA hybrid as in previous study (5) can be used and the release of fluorescence was achieved by displacement of the quencher in RAM. No specific sequence of molecular zipper is required for additional enzymatic digestion for real-time monitoring. The workability of molecular zipper was demonstrated in real-time RAM for genotyping detections with a Q-PNA (5). Our DNA molecular zipper further demonstrated its utility for quantitative determination of targeted DNA in a real-time fashion. Furthermore, molecular zipper offers several advantages over Taqman and molecular beacons for real-time detection of amplification products. First, the long double-stranded region of the molecular zipper is more stable. Therefore, the background signal is much lower as compared with molecular beacons in which the short-stem region is less stable (19). Second, the generic sequence of the molecular zipper allows it to be used universally for different targets. Third, the fluorophore and quencher are attached to two separate strands and the positions of fluorophore and quencher can be exchanged between the two strands. For example, in multiplex RAM reactions the quencher can be attached to the negative strand that can be used as a common strand (generic quencher) for different positive strands, each with a different fluorophore. This makes the synthesis of the FRET probe less costly and permits the convenient design of different probes with different fluorophores, thus increasing the capability for multiplexing. Finally, the molecular zipper can be used in other DNA amplification technologies that involve strand separation, such as PCR (24,25), transcription-mediated amplification (NASBA) (26), loop-mediated DNA amplification (27) and strand displacement amplification (28).
Acknowledgments
The authors thank Josephine Wu, Fei Ye and David Lane for critical comment. This work was supported by a grant from Hamilton-Thorne Bioscience, Inc. Funding to pay the Open Access publication charges for this article was provided by Hamilton-Thorne Bioscience, Inc.
Conflict of interest statement. None declared.
REFERENCES
- 1.Liu D., Daubendiek S.L., Zillman M.A., Ryan K., Kool E.T. Rolling circle DNA Synthesis: small circular oligonucleotides as efficient templates for DNA polymerases. J. Am. Chem. Soc. 1996;118:1587–1694. doi: 10.1021/ja952786k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lizardi P.M., Huang X., Zhu Z., Bray-Ward P., Thomas D.C., Ward D.C. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 3.Fire A., Xu S. Rolling Replication of Short DNA Circles. Proc. Natl Acad. Sci. USA. 1995;92:4641–4645. doi: 10.1073/pnas.92.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang D.Y., Brandwein M., Hsuih T.C.H., Li H. Amplification of target-specific, ligation-dependent circular probe. Gene. 1998;211:277–285. doi: 10.1016/s0378-1119(98)00113-9. [DOI] [PubMed] [Google Scholar]
- 5.Alsmadi O.A., Bornarth C.J., Song W., Wisniewski M., Du J., Brockman J.P., Faruqi A.F., Hosono S., Sun Z., Du Y., et al. High accuracy genotyping directly from genomic DNA using a rolling circle amplification based assay. BMC Genomics. 2003;4:21. doi: 10.1186/1471-2164-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novick R.P. Contrasting lifestyles of rolling-circle phages and plasmids. Trends Biochem. Sci. 1998;23:434–438. doi: 10.1016/s0968-0004(98)01302-4. [DOI] [PubMed] [Google Scholar]
- 7.Nilsson M., Malmgren H., Samiotaki M., Kwiatkowski M., Chowdhary B.P., Landegren U. Padlock Probes: circularizing oligonucleotides for localized DNA detection. Science. 1994;265:2085–2088. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- 8.Baner J., Nilsson M., Isaksson A., Mendel-Hartvig M., Antson D.O., Landegren U. More keys to Padlock Probes: mechanisms for high-throughput nucleic acid analysis. Curr. Opin. Biotechnol. 2001;12:11–15. doi: 10.1016/s0958-1669(00)00174-9. [DOI] [PubMed] [Google Scholar]
- 9.Baner J., Nilsson M., Mendel-Hartvig M., Landegren U. Signal amplification of Padlock Probes by rolling circle replication. Nucleic Acids Res. 1998;26:5073–5078. doi: 10.1093/nar/26.22.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schweitzer B., Roberts S., Grimwade B., Shao W., Wang M., Fu Q., Shu Q., Laroche I., Zhou Z., Tchernev V.T., et al. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat. Biotechnol. 2002;20:359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nallur G., Luo C., Fang L., Cooley S., Dave V., Lambert J., Kukanskis K., Kingsmore S., Lasken R., Schweitzer B. Signal amplification by rolling circle amplification on DNA microarrays. Nucleic Acids Res. 2001;29:e118. doi: 10.1093/nar/29.23.e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang D.Y., Liu B. Detection of target nucleic acids and proteins by amplification of circularizable probes. Expert Rev. Mol. Diagn. 2003;3:237–248. doi: 10.1586/14737159.3.2.237. [DOI] [PubMed] [Google Scholar]
- 13.Demidov V.V. Rolling-circle amplification in DNA diagnostics—power of simplicity. Expert Rev. Mol. Diagn. 2002;2:542–548. doi: 10.1586/14737159.2.6.542. [DOI] [PubMed] [Google Scholar]
- 14.Nilsson M., Gullberg M., Dahl F., Szuhai K., Raap A.K. Real-time monitoring of rolling-circle amplification using a Modified Molecular Beacon Design. Nucleic Acids Res. 2002;30:e66. doi: 10.1093/nar/gnf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faruqi A.F., Hosono S., Driscoll M.D., Dean F.B., Alsmadi O., Bandaru R., Kumar G., Grimwade B., Zong Q., Su Z., et al. High-throughput genotyping of single nucleotide polymorphisms with rolling circle amplification. BMC Genomics. 2001;2:4. doi: 10.1186/1471-2164-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pickering J., Bamford A., Godbole V., Briggs J., Scozzafava G., Roe P., Wheeler C., Ghouze F., Cuss S. Integration of DNA ligation and rolling circle amplification for the homogeneous, end-point detection of single nucleotide polymorphisms. Nucleic Acids Res. 2002;30:e60. doi: 10.1093/nar/gnf060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smolina I.V., Demidov V.V., Cantor C.R., Broude N.E. Real-time monitoring of branched rolling-circle DNA amplification with peptide nucleic acid beacon. Anal. Biochem. 2004;335:326–329. doi: 10.1016/j.ab.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 18.Kuhn H., Demidov V.V., Gildea B.D., Fiandaca M.J., Coull J.C., Frank-Kamenetskii M.D. PNA beacons for duplex DNA. Antisense Nucleic Acid Drug. Dev. 2001;11:265–270. doi: 10.1089/108729001317022269. [DOI] [PubMed] [Google Scholar]
- 19.Tyagi S., Kramer F.R. Molecular Beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 20.Zhang D.Y., Brandwein M., Hsuih T., Li H.B. Ramification amplification: a novel isothermal DNA amplification method. Mol. Diagn. 2001;6:141–150. doi: 10.1054/modi.2001.25323. [DOI] [PubMed] [Google Scholar]
- 21.Nadeau J.G., Pitner J.B., Linn P., Schram J.L., Dean C.H., Nycz C.M. Real-time, sequence-specific detection of nucleic acids during strand displacement amplification. Anal. Chem. 1999;276:177–187. doi: 10.1006/abio.1999.4350. [DOI] [PubMed] [Google Scholar]
- 22.Heid C.A., Stevens J., Livak K.J., Williams P.M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 23.Solinas A., Brown L.J., McKeen C., Mellor J.M., Nicol J.T.G., Thelwell N., Brown T. Duplex scorpion primers in SNP analysis and FRED amplications. Nucleic Acids Res. 2001;29:e96. doi: 10.1093/nar/29.20.e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saiki R.K., Gelfand D.H., Stoffel S., Scharf S.J., Higuchi R., Horn G.T., Mullis K.B., Erlich H.A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1998;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 25.Saha B.K., Tian B., Bucy R.P. Quantitation of HIV-1 by real-time PCR with a unique fluorogenic probe. J. Virol. Methods. 2001;93:33–42. doi: 10.1016/s0166-0934(00)00288-3. [DOI] [PubMed] [Google Scholar]
- 26.Leone G., van Schijndel H., van Gemen B., Kramer F.R., Schoen C.D. Molecular Beacon Probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res. 1998;26:2150–2155. doi: 10.1093/nar/26.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker G.T., Fraiser M.S., Schram J.L., Little M.C., Nadeau J.G., Malinowski D.P. Strand displacement—an isothermal in vitro DNA amplification technique. Nucleic Acids Res. 1992;20:1691–1696. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]