Abstract
Intercellular transport of auxin is mediated by influx and efflux carriers in the plasma membrane and subjected to developmental and environmental regulation. Here, using the auxin-sensitive Arabidopsis thaliana root hair cell system and the tobacco (Nicotiana tabacum) suspension cell system, we demonstrate that the protein kinase PINOID (PID) positively regulates auxin efflux. Overexpression of PID (PIDox) or the auxin efflux carrier component PINFORMED3 (PIN3, PIN3ox), specifically in the root hair cell, greatly suppressed root hair growth. In both PIDox and PIN3ox transformants, root hair growth was nearly restored to wild-type levels by the addition of auxin, protein kinase inhibitors, or auxin efflux inhibitors. Localization of PID or PIN3 at the cell boundary was disrupted by brefeldin A and staurosporine. A mutation in the kinase domain abrogated the ability of PID to localize at the cell boundary and to inhibit root hair growth. These results suggest that PIDox- or PIN3ox-enhanced auxin efflux results in a shortage of intracellular auxin and a subsequent inhibition of root hair growth. In an auxin efflux assay using transgenic tobacco suspension cells, PIDox or PIN3ox also enhanced auxin efflux. Collectively, these results suggest that PID positively regulates cellular auxin efflux, most likely by modulating the trafficking of PIN and/or some other molecular partners involved in auxin efflux.
INTRODUCTION
Auxin moves from cell to cell with a polarity. In shoots, auxin moves basipetally from the shoot apex to the basal region. In the root, auxin movement is both basipetal through the central tissues and acropetal through the epidermis (Lomax et al., 1995). This vectorial auxin movement has long been implicated in various aspects of plant growth and development, such as apical dominance, vascular development, tropisms, and pattern formation during embryogenesis (Lomax et al., 1995; Morris et al., 2004).
Polar cell-to-cell auxin transport is thought to be mediated by carrier proteins that are asymmetrically localized in the plasma membrane (for the latest reviews, see Morris et al., 2004; Paponov et al., 2005). ALTERED RESPONSE TO AUXIN AND GRAVITY1 (AUX1) and LIKE AUX are the auxin influx carrier components, and PINFORMED (PIN) proteins are a group of auxin efflux carrier components. In addition, p-glycoproteins (PGPs) also are involved in auxin transport (Geisler and Murphy, 2006). Eight PIN family genes have been identified in Arabidopsis thaliana, and the biological functions of five of them have been characterized (Morris et al., 2004; Paponov et al., 2005). There is a great deal of accumulating evidence that PINs are critical auxin efflux carrier components: the molecular topology of the PIN proteins, their ability to transport auxin and auxin analogues in yeast, their polar distribution patterns in plant cells, the disruption of auxin polar transport in pin mutants, and the phenocopies of PIN mutations by auxin transport inhibitors (Morris et al., 2004). The subcellular dynamics of PIN localization provide additional clues to their biological function and their finely tuned regulation during plant development. Initiation of leaf primordia depends on the asymmetrical localization of PIN1 in shoot apical meristem cells (Reinhardt et al., 2003), and the gravitropic stimulus rapidly changes the direction of PIN3 trafficking in root cap columella cells (Friml et al., 2002).
Protein phosphorylation and dephosphorylation have been implicated in the regulation of auxin transport. In tobacco (Nicotiana tabacum) suspension cells, the kinase inhibitors staurosporine (ST) and K252a inhibited auxin efflux, and a subset of phosphatase inhibitors had an inhibitory effect on both auxin efflux and influx (Delbarre et al., 1998). Arabidopsis protein phosphatase 2A was shown to have a negative effect on auxin transport (Rashotte et al., 2001). Several lines of evidence suggest that the Ser/Thr protein kinase PINOID (PID) acts as a regulator of polar auxin transport in Arabidopsis: (1) the phenotype of the pid loss-of-function mutant resembles that of pin1, and auxin transport is decreased in the stems of pid plants (Bennett et al., 1995; Benjamins et al., 2001); (2) the biological activity of PID is sensitive to auxin transport inhibitors (Benjamins et al., 2001); (3) PID expression and its functional effect do not spatially overlap (Benjamins et al., 2001); and (4) PID regulates the subcellular polarity of PIN (Friml et al., 2004). The first two observations are indicative of PID's role as a positive regulator of auxin transport. On the other hand, the last observation suggests that PID acts as a binary switch to regulate the direction of auxin flow. It has also been proposed that PID functions as a component of auxin signaling pathways. This notion was based on the observation that exogenous application of auxin was unable to rescue the root phenotype caused by PID overexpression (using the cauliflower mosaic virus 35S promoter, P35S:PID) (Christensen et al., 2000; DeLong et al., 2002). Thus, at least three potential roles of PID have been suggested to date: as a positive regulator of auxin transport, a binary switch for PIN polarity, and a negative regulator of auxin signaling. All of these putative functions were derived from studies of the same P35S:PID Arabidopsis transgenic system.
The purpose of the current study was to characterize the function of PID using a simple biological system, the root hair cell system. The root hair is the tubular outgrowth of a root epidermal cell. Development of the root hair cell can be divided largely into three stages: fate determination to become a hair or nonhair cell, hair initiation, and hair elongation (Schiefelbein, 2000). Auxin has been shown to be a potent stimulator of hair growth (i.e., initiation and elongation stages) without affecting the cell fate–determining process (Okada and Shimura, 1994; Schiefelbein, 2000). Alteration of auxin-related events, such as cellular auxin influx (AUX1), expression of auxin-responsive genes (Aux/IAAs), and proteolysis signaling (Auxin-Resistant1 [AXR1]), influences root hair growth, providing indirect evidence for a positive role of auxin in this process (Masucci and Schiefelbein, 1994, 1996; Okada and Shimura, 1994; Leyser et al., 1996; Pitts et al., 1998; Reed, 2001). A recent study also showed that PGP4-mediated auxin transport could be implicated in root hair growth (Santelia et al., 2005). Exogenous auxin rescued hair growth in the Arabidopsis hairless rhd6 mutant, restoring it to near wild-type levels (Masucci and Schiefelbein, 1994, 1996) (Figures 1A and 1B), and enhanced hair elongation of wild-type roots (Pitts et al., 1998). These studies of auxin mutants and exogenous auxin treatment suggest that the intracellular level of auxin in the hair cell is critical to properly execute the morphogenesis and growth of the root hair.
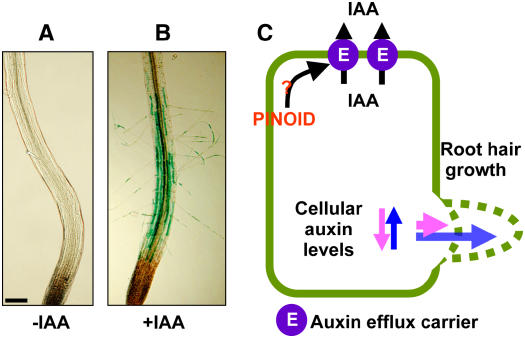
Figure 1.
The Auxin-Sensitive Root Hair System and the Working Hypothesis of This Study.
(A) and (B) Roots from hairless rhd6 mutant Arabidopsis harboring the PE7:GUS transgene. Seedlings were untreated (A) or treated with 30 nM IAA (B). IAA induces root hairs and hair cell–specific (PE7:GUS) expression in the transgenic mutant root. Bar = 100 μm.
(C) Scheme of the regulation of intracellular auxin levels. Higher auxin efflux in the hair cell decreases cellular auxin levels and shortens the root hair. By contrast, less or normal auxin efflux activity maintains enough auxin inside the hair cell to sustain hair elongation. PID as a positive regulator of auxin efflux would inhibit root hair elongation if overexpressed.
Taking advantage of the auxin-sensitive single cell root hair system, we demonstrated that the protein kinase PID functions as a positive regulator of cellular auxin efflux. The expression of PID or PIN was driven by a root hair cell–specific promoter, which made it possible to confine related functions of overexpressed PID or PIN to the hair cell. Our results indicated that (1) increased auxin efflux (i.e., by overexpression of PID or PIN) decreases the level of auxin in the hair cell, (2) low levels of auxin result in the inhibition of root hair growth, and (3) antagonists against PID activity or auxin efflux restore root hair growth (Figure 1C). We also directly showed the activities of PID and PIN for cellular auxin efflux in a second, independent system, the transgenic tobacco suspension cell system.
RESULTS
Root Hair Cell–Specific Overexpression of PID Inhibits Root Hair Growth
To characterize the function of the PID protein kinase in auxin efflux at the cellular level, we overexpressed PID specifically in the developing root hair cell. To determine the subcellular localization of PID in the hair cell, PID was fused to the gene encoding green fluorescent protein (GFP), and PID-GFP was expressed using the root hair cell–specific promoter of ARABIDOPSIS EXPANSIN A7 (ATEXPA7) (Cho and Cosgrove, 2002). The ATEXPA7 promoter (PE7) begins to operate immediately before root hair morphogenesis begins. There were two reasons for using PE7 in this study. First, the activity of PE7 corresponds precisely, both spatially and temporally, to the site of auxin action during root hair development. This made it possible to localize PID function in vivo to the time and place where auxin functions in root hair morphogenesis. Second, the use of a root hair cell–specific promoter confined the phenotypic effect of PID only to the hair cell, excluding complicating pleiotropic effects that might result from the use of a universal promoter.
Overexpression of PID in the root hair cell (PIDox; PE7:PID-GFP) greatly suppressed root hair development (Figures 2B, 2C, and 3). The inhibitory effect of PIDox was more severe on root hair elongation than on hair initiation (Figure 3). Most of the 74 T1 transformants exhibited the apparent root hair phenotype. Progeny of four randomly selected independent T1 lines were used for further analyses. The defect in root hair growth of the PIDox transformants was fully rescued by the addition of exogenous indole-3-acetic acid (IAA) (Figure 2D). This result suggested that PID positively regulates auxin efflux, decreasing intracellular auxin concentration below a certain threshold required for root hair growth. Steady auxin supply through an exogenous source was able to restore intracellular auxin concentration to levels that support hair growth. The recovery of root hairs by auxin treatment in the PIDox root indicates that PID acts upstream of auxin signaling.
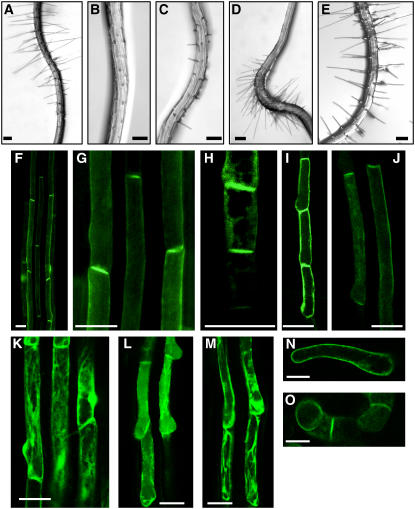
Figure 2.
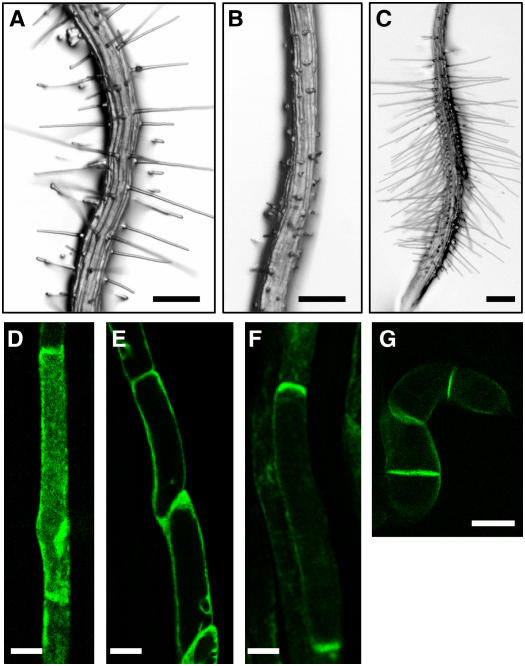
Root Hair Cell–Specific Overexpression of PID Suppresses Root Hair Growth.
(A) The control transformant (PE7:GFP) expressing only GFP in the hair cell, driven by the root hair cell–specific ATEXPA7 promoter (PE7).
(B) to (D) PE7:PID-GFP transformants expressing PID-GFP in the hair cell, untreated ([B], severe phenotype transformant; [C], moderate phenotype transformant) or treated with 30 nM IAA (D).
(E) PE7:mPID-GFP transformant expressing a mutated form of PID (mPID)–GFP with a point mutation in the ATP binding domain.
(F) to (J) Confocal microscopy images of the roots of PE7:PID-GFP transformants. Images of fully elongated (or mature; [F], [G], [I], and [J]) and elongating (H) hair cells are shown.
(K) A confocal microscopy image of the root of the control PE7:GFP transformant.
(L) and (M) Confocal microscopy images of the roots of PE7:mPID-GFP transformants, expressing mPID-GFP.
(N) and (O) Confocal microscopy images of tobacco BY-2 cells harboring PTA:PID-GFP. Cells were treated with dexamethasone (Dex) to induce expression, as described in Methods.
Bars = 100 μm for (A) to (E) and 30 μm for (F) to (O).
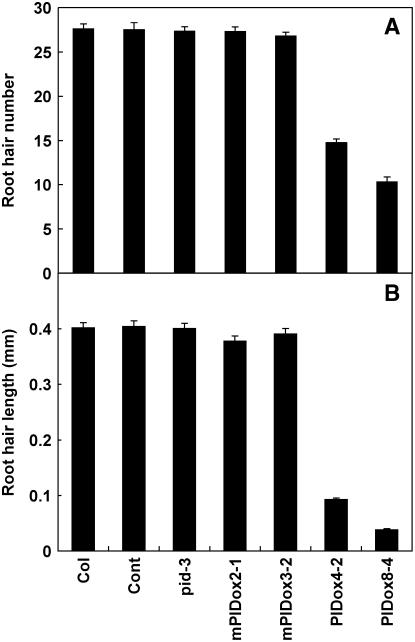
Figure 3.
Root Hair Development in PID Transformants.
Root hair number (A) and root hair length (B) in control (Cont; PE7:GFP), loss-of-function pid-3 mutant, wild-type PID-overexpressing transformants (PE7:PID-GFP; PIDox), and mutant PID-overexpressing transformants (PE7:mPID-GFP; mPIDox). Two independent homozygous lines for each transformant were analyzed. Data represent means ± se for each line (n = 240 for root hair length, n = 30 for root hair number).
By contrast, overexpression of a mutated form of PID (mPIDox), which was defective in ATP binding activity as a result of a single point mutation (Christensen et al., 2000), had almost no inhibitory effect on root hair growth (Figures 2E and 3), indicating that the protein kinase activity of PID was critical for the inhibition of root hair growth and probably for enhancing auxin efflux as well. We examined 60 independent T1 mPIDox lines and found all of them to have normal root hair growth.
PID Is Targeted to the Cell Periphery through Its Kinase Activity
The roots of PE7:PID-GFP transgenic plants, which expressed the biologically active PID-GFP fusion protein, exhibited obvious hair cell–specific expression of the fusion protein (Figures 2F to 2J). Although the distribution of PID-GFP was somewhat diffuse throughout the cytoplasm, PID-GFP appeared to be concentrated at the cell periphery, at both the apical and basal ends of the hair cell, rather than asymmetrically at one end only. This was evident when transgene expression was discontinued in the hair cell file (see the apical and basal ends of the left hair cell in Figure 2J). The distribution pattern of PID in Arabidopsis root hair cells was mimicked in tobacco cv Bright Yellow 2 (BY-2) cells when we expressed PID-GFP in the tobacco cell using a Dex-inducible promoter system (Aoyama and Chua, 1997). PID-GFP localized along the cell periphery, and there were high concentrations of protein in the boundary between two consecutive cells (Figures 2N and 2O). In contrast with PID-GFP, GFP alone exhibited mostly dispersed localization throughout the cytoplasm (Figure 2K).
When we examined the mutated form of PID (mPID-GFP), we observed a largely dispersed cytoplasmic distribution (Figures 2L and 2M), very much like that seen with GFP alone. This dispersed distribution pattern of mPID-GFP was most likely not attributable to levels of mPID-GFP expression, because we found that both weak and strong expression levels gave rise to similar distribution patterns (see Supplemental Figure 4 online). These results suggested that the kinase activity of PID was critical for the trafficking of PID to the cell periphery.
NPA Restores Root Hair Development in PIDox Transformants
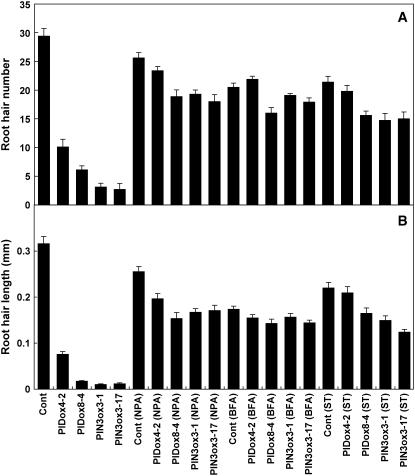
If PID inhibits root hair development through the enhancement of auxin efflux, inhibitors of auxin efflux carriers should be able to block the effect of PIDox on root hair growth. We tested this hypothesis using the auxin efflux inhibitor naphthylphthalamic acid (NPA) with two or four randomly chosen independent PIDox lines (Figures 4C and 5; see Supplemental Figures 2A and 2B online). Both root hair number and length of the root in PIDox transgenic lines increased with increasing concentrations of NPA, recovering nearly to control levels in the presence of 10 μM NPA (see Supplemental Figures 2A and 2B online). These results strongly support the idea that auxin efflux is involved in PIDox-induced root hair inhibition. It is likely that NPA inhibited the auxin efflux that was enhanced by PID, resulting in a restoration of auxin levels inside the PIDox root hair cell and a restoration of hair growth.
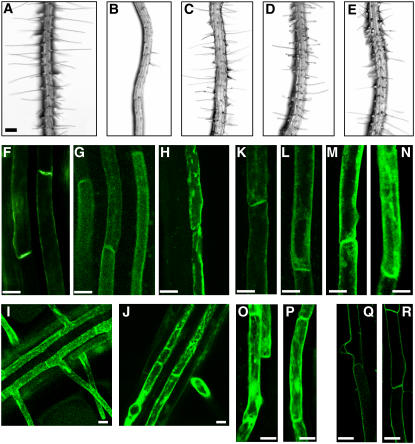
Figure 4.
Effects of NPA, BFA, and ST on Root Hair Growth of PIDox Roots, and the Subcellular Localization of PID-GFP.
(A) Control (PE7:GFP) transformant, untreated. Bar = 100 μm for (A) to (E).
(B) to (E) Roots of PIDox (PE7:PID-GFP) transformants either untreated (B) or treated with 1 μM NPA (C), 5 μM BFA (D), or 0.25 μM ST (E).
(F) to (H) Confocal microscopy images of roots from PE7:PID-GFP transformants treated with 0 μM (F), 10 μM (G), or 50 μM (H) BFA. Bars = 10 μm for (F) to (R).
(I) and (J) Confocal microscopy images of roots of PE7:GFP transformants treated with 0 μM (I) or 50 μM (J) BFA.
(K) to (N) Confocal microscopy images of root hair cells of PE7:PID-GFP transformants treated with 0 μM (K), 2 μM (L), or 5 μM ([M] and [N]) ST.
(O) and (P) Confocal microscopy images of root hair cells of PE7:GFP transformants treated with 0 μM (O) or 5 μM (P) ST.
(Q) and (R) Confocal microscopy images of root hair cells of PE7:AHA2-GFP transformants treated with 0 μM (Q) or 5 μM (R) ST.
Figure 5.
NPA, BFA, and ST Restore Root Hair Growth of PIDox and PIN3ox Transformants.
Root hair number (A) and root hair length (B) in control (Cont; PE7:GFP) transformants, PIDox transformants (PE7:PID-GFP), and PIN3ox transformants (PE7:PIN3-GFP) treated with 1 μM NPA, 5 μM BFA, or 0.25 μM ST. Two independent homozygous lines for each transformant were observed. Data represent means ± se for each transformant (n = 64 for root hair length, n = 8 for root hair number).
Brefeldin A Internalizes PID Proteins and Restores Root Hair from PIDox Roots
Brefeldin A (BFA) blocks PIN trafficking to the plasma membrane (Geldner et al., 2001, 2003), which in turn causes a decrease in auxin efflux from the cell (Delbarre et al., 1998). We were interested in whether blocking the trafficking of PINs in the PIDox root hair cell by BFA would affect the root hair growth phenotype caused by the overexpression of PID. When seedlings were treated with BFA for 24 h, root hair number and length in the control plants (PE7:GFP) decreased with increasing concentrations of BFA. By contrast, in PIDox plants, the root hair parameters were somewhat restored by BFA, with the optimum effect in the ∼5 to 10 μM range (Figures 4D and 5; see Supplemental Figures 2C and 2D online). These data support our hypothesis that BFA would affect the ability of PIDox to increase auxin efflux, resulting in the restoration of auxin levels in the cell and subsequent hair cell growth.
Because BFA treatment has been shown to result in the internalization of PIN proteins (Geldner et al., 2001, 2003) and the cellular localization of PID was similar to the known distribution of PIN proteins (Figures 2F to 2J, 2N, and 2O; see Figures 7E to 7G below), we were interested in whether BFA interfered with the localization of PID. PID-GFP was distributed diffusely throughout the cytoplasm after a 2-h incubation in liquid medium containing 10 μM BFA (Figure 4G), which was clearly distinguished from the nontreated root cell (Figure 4F). Internalized compartments of PID-GFP were evident upon treatment with 50 μM BFA (Figure 4H). This effect of BFA on PID localization implied that the similar molecules targeted by BFA in PIN trafficking could be involved in modulating PID trafficking as well.
Figure 7.
Hair Cell–Specific Overexpression of PIN3 Inhibits Root Hair Growth.
(A) to (C) Roots from PE7:GFP (control; [A]) and PIN3ox (PE7:PIN3-GFP; [B] and [C]) transformants. Treatment with IAA (30 nM) restored root hairs in PIN3ox transformants (C). Bars = 100 μm.
(D) to (F) Confocal microscopy images of the root hair cells from PE7:GFP (D) and PE7:PIN3-GFP ([E] and [F]) transformants. Bars = 10 μm.
(G) A confocal microscopy image of transgenic tobacco BY-2 cells expressing PTA:PIN3-GFP. Bar = 30 μm.
Inhibition of Kinase Activity Restores Root Hairs in PIDox Roots and Interferes with PID Trafficking
PID is a Ser/Thr protein kinase, and its kinase activity was shown to be required for its biological function (Christensen et al., 2000). We hypothesized that inhibition of Ser/Thr kinase activity by ST would abolish the inhibitory effect of PIDox on root hair growth. Root hair number and length in control plants (PE7:GFP) decreased in the presence of increasing concentrations of ST, whereas the roots in PIDox lines showed significant recovery, in particular in the range of ∼0.1 to 1.0 μM ST (Figure 4E; see Supplemental Figures 2E and 2F online). One of the PIDox lines showed root hair recovery at 0.25 μM ST, to the level of growth of the control plant treated with the same concentration of ST (Figure 5). We did the same experiment with a hairless rhd6 mutant that shared several phenotypic features with PIDox lines, including restoration of root hair growth by ethylene and water stress as well as auxin (Cho and Cosgrove, 2002; our unpublished observation). In contrast with PIDox transformants, root hairs were not restored in the rhd6 mutant by ST treatment (see Supplemental Figure 2E online). This finding suggested that the ST effect is unique to PIDox and thus probably specific to PID or its regulatory kinases (Zegzouti et al., 2006). Although ST treatment restored root hairs in the PIDox transformants, primary root growth was decreased significantly by the inhibitor. This suggests that ST had both a selective effect on root hair growth and a general toxic effect on primary root growth. We also examined the effect of PD98059, an inhibitor of mitogen-activated protein kinase kinase (Reiners et al., 1998). However, in the concentration range tested (∼10 to 250 μM), PD98059 failed to restore root hairs in the PIDox root (data not shown), providing additional evidence that the phenotype of PIDox transformants was mediated by a specific Ser/Thr kinase. It was surprising that, despite its toxic properties, ST specifically restored root hair growth in PIDox transformants. In a certain concentration range, ST may act primarily on very selective targets. Because ST restored root hair growth from PIDox hair cells, we assumed that PID-mediated protein phosphorylation was one of those targets.
Treatment with ST also disrupted the normal cellular localization of PID-GFP. Internalization of PID-GFP was observed upon treatment with 2 μM ST (or with 0.25 μM for 24 h on the solid medium) (Figure 4L), and the effect was even greater at 5 μM ST (Figures 4M and 4N). By contrast, treatment with 5 μM ST did not affect the localization of GFP, a cytosolic protein marker (Figure 4P), or of Arabidopsis plasma membrane H+-ATPase 2 (AHA2-GFP), a plasma membrane protein marker (Figure 4R), in the root hair cell, indicating that the ST effect was specific for PID.
These results, together with the result from the analysis of mPID (PE7:mPID-GFP), suggested that PID kinase activity was critical for the inhibition of root hair growth and also for maintaining the proper subcellular localization of PID.
PID and PINs Expressed in the Arabidopsis Root Hair Cell
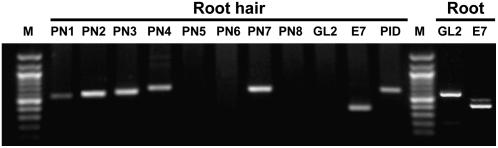
To determine whether PID or any of the PIN proteins were expressed in the growing root hair cell, RT-PCR was performed using hair cell–specific RNA as the template and gene-specific primers for PIN1 to PIN8 and PID. Gene-specific primers were designed to include intron(s), so that the PCR products amplified from cDNAs could be distinguished from PCR products from genomic DNA (see Supplemental Table 1 online for the expected sizes of the products from cDNAs and genomic DNA for each gene). The PCR-derived fragments examined in Figure 6 correspond to products amplified from cDNAs. Root hair cell–specific RNA was purified from 2-d-old growing root hairs as described in Methods. Forty-five cycles of amplification gave rise to DNA fragments corresponding to PID, PIN1, PIN2, PIN3, PIN4, and PIN7 (Figure 6). As controls, the root hair cell–specific ATEXPA7 (PE7) transcript was also amplified, but the non-hair cell–specific GLABRA2 (GL2) transcript was not amplified from root hair–specific RNA, indicating that the RNA template was specific for the root hair cell. GL2 was amplified when whole root RNA was used as a template. These results suggest that PID and many of the PIN proteins function in the root hair cell.
Figure 6.
PID and PINs Are Expressed in the Root Hair Cell.
RT-PCR analysis of RNA from Arabidopsis root hair cells (Root hair) or whole root tissues (Root). E7 (ATEXPA7) was used as a positive control for hair cell–specific amplification; GL2 (GLABRA2) was used as a control for non-hair cell–specific amplification. Gene-specific primer sets were designed to include intron(s) to discriminate amplification from cDNAs versus genomic DNA.
Overexpression of PIN in the Root Hair Cell Inhibits Root Hair Growth
PIN refers to a group of auxin efflux carrier components, and it has been shown that heterologous expression of PIN proteins in yeast facilitated the efflux of auxin and auxin analogues from the cell (Chen et al., 1998; Luschnig et al., 1998). We hypothesized that overexpression of a PIN protein in root hair cells would decrease cellular auxin levels and inhibit root hair growth. To test this hypothesis, we introduced a transgene encoding PIN3 fused to GFP, PE7:PIN3-GFP (PIN3ox), into Arabidopsis. All 52 T1 PIN3ox transformants exhibited reduced length and number of root hairs (Figure 7B). Two homozygous PIN3ox lines maintained only ∼10 and 5% of control levels of root hair number and length, as seen by comparison with PE7:GFP transformants (Figure 5). The addition of IAA fully restored root hairs in the PIN3ox root (Figure 7C). These results confirm that enhanced efflux of auxin from the root hair cell inhibits root hair growth.
PIN3-GFP localized to the cellular boundary in both Arabidopsis root hair cells and tobacco BY-2 cells (Figures 7E to 7G), as would be expected based on its properties as a membrane protein. In general, PIN3-GFP was detectable along the entire hair cell boundary in most of the T1 lines (Figure 7E). However, in ∼30% of them, there was a more focused localization at the longitudinal end of the cell (Figure 7F). This end-localization of PIN3 occurred on both sides of the hair cell, rather than on one particular end, similar to the localization of PID-GFP (Figures 2F to 2J), indicating a correlation in the spatial distribution of PID and PIN in the cell. In tobacco cells as well, PIN3-GFP was more concentrated at the boundary between two neighboring cells (Figure 7G).
Inhibition of Auxin Efflux, PIN Trafficking, or Protein Kinase Activity Restores Root Hairs in PIN3ox Transformants
To determine whether the inhibition of auxin efflux activity (and the subsequent restoration of normal levels of auxin in the hair cell) restored root hair growth in PIN3ox transformants, seedlings were treated with varying concentrations of NPA. At concentrations as low as 0.1 μM, NPA increased root hair number and length of PIN3ox roots, and treatment with 10 μM NPA restored root hair development almost to the level of the control seedlings (Figures 5 and 8A to 8D; see Supplemental Figures 3A and 3B online). PIN3ox transformants were also treated with BFA to test the effect of the disruption of PIN trafficking on auxin efflux and hair growth. Root hair number and length were restored in PIN3ox roots upon BFA treatment, with an optimal effect at 1 to 5 μM BFA, whereas root hair growth in control (PE7:GFP) plants decreased with increasing concentrations of BFA (Figures 5, 8E, and 8F; see Supplemental Figures 3C and 3D online). Our observations are consistent with the previous results from tobacco cells, in which BFA decreased the efficiency of auxin efflux carriers and enhanced the cellular accumulation of auxin in a certain concentration range (Delbarre et al., 1998; Petrášek et al., 2003). To determine whether PIN3ox-mediated stimulation of auxin efflux (and the subsequent inhibition of root hair growth) depended on protein kinase activity, PIN3ox transformant seedlings were treated with ST. The protein kinase inhibitor had a dramatic effect on root hair growth in the PIN3ox root (Figures 5, 8G, and 8H; see Supplemental Figures 3E and 3F online), restoring hair number and length. Along with the results of ST treatment of PIDox seedlings, this result was very intriguing, as the biochemical processes for root hair morphogenesis were restored by a broad-spectrum kinase inhibitor. This strongly supported the notion that ST acts on specific kinase targets involved in the regulation of auxin efflux components, including PIN proteins.
Figure 8.
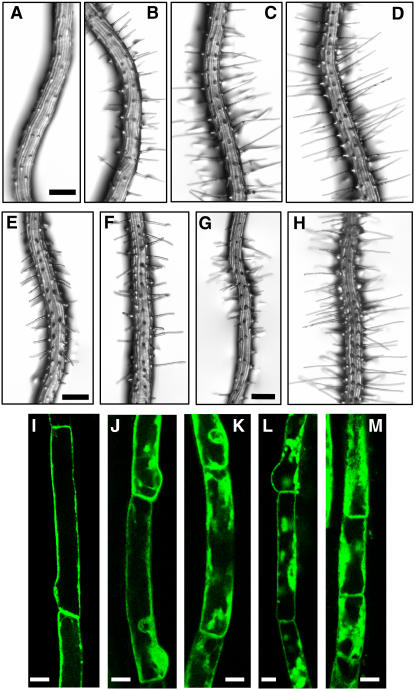
Effects of NPA, BFA, and ST on Root Hair Growth of PIN3ox Roots, and Subcellular Localization of PIN3-GFP.
(A) to (H) Roots of PIN3ox (PE7:PIN3-GFP) transformants treated with 0 μM (A), 0.1 μM (B), 0.5 μM (C), or 1.0 μM (D) NPA; 1.0 μM (E) or 5.0 μM (F) BFA; or 0.1 μM (G) or 0.5 μM (H) ST. Bars = 100 μm.
(I) to (M) Confocal microscopy images of root hair cells from PE7:PIN3-GFP transformants. The seedlings were treated with 0 μM (I), 2 μM (J), or 5 μM (K) ST or 10 μM (L) or 50 μM (M) BFA. Bars = 10 μm.
Subcellular Localization of PIN3 in the Hair Cell Is Disrupted by the Protein Kinase Inhibitor
To determine whether kinase activity was required for the correct trafficking of PIN3, we examined the effect of ST on the subcellular localization of PIN3-GFP in PE7:PIN3-GFP transformants. When transgenic seedlings were treated with 2 to 5 μM ST for 2 h, PIN3-GFP internalized into discrete compartments, suggesting that ST interfered with the normal trafficking of PIN3 to the plasma membrane (Figures 8J and 8K). A similar result was observed when newly formed hair cells were treated with 0.5 μM ST for 24 h. The presence of intracellular PIN3-GFP vesicles induced by treatment with ST resembled the internalization pattern induced by BFA treatment (Figures 8L and 8M), suggesting that protein phosphorylation plays a role in PIN trafficking to the plasma membrane.
The overall effects of NPA, BFA, and ST on PIN3ox transformants were similar to their effects on PIDox transformants, suggesting that PID and PIN function in the same biochemical pathway to regulate auxin efflux.
PIDox or PIN3ox Increases Auxin Retention inside the Cell
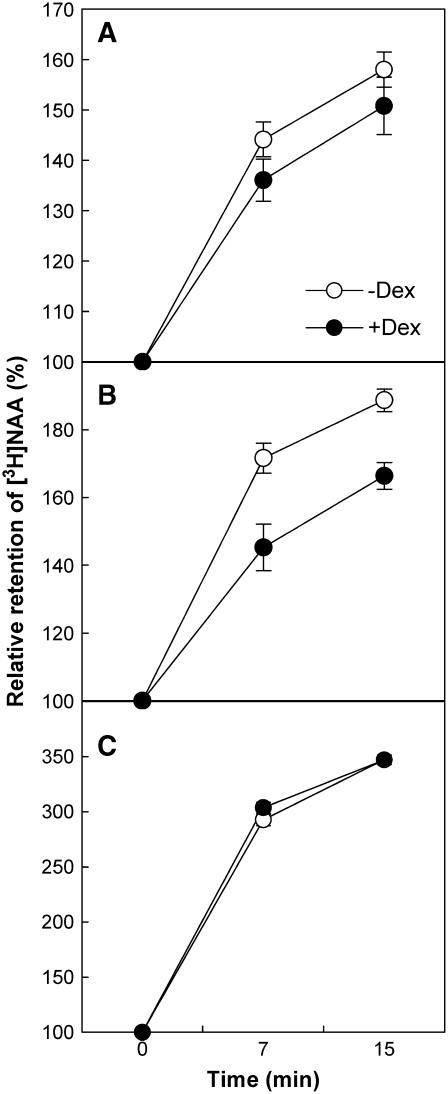
We also examined the function of PID and PIN in auxin efflux in a second, independent system, tobacco BY-2 cells. The Dex-inducible promoter system (Aoyama and Chua, 1997) was used to overexpress PID (PTA:PID-GFP) or PIN3 (PTA:PIN3-GFP) in tobacco cells, and the retention of [3H]naphthalene acetic acid (NAA) in the cell was measured. Dex treatment resulted in 18 and 37% decreases of [3H]NAA in PIDox and PIN3ox transgenic cells, respectively (Figures 9A and 9B). Dex treatment had no effect on auxin retention in control cells (Figure 9C). The difference in retention rates of PIDox and PIN3ox somehow reflects the inhibitory strength of root hair growth by two transgenes—that is, stronger root hair inhibition in PIN3ox than in PIDox. The molecular characteristics of PIN, the responsiveness of PID to NPA, and the fact that NAA is able to freely diffuse into cells suggest that the decreased retention of [3H]NAA in PIDox and PIN3ox transgenic tobacco cells was most likely attributable to enhanced auxin efflux from these cells.
Figure 9.
Retention of [3H]NAA in Tobacco Cells Is Decreased by PIDox or PIN3ox.
Relative retention of 3H-labeled NAA in PIDox (PTA:PID-GFP; [A]), PIN3ox (PTA:PIN3-GFP; [B]), and control (C) tobacco BY-2 cells. PID and PIN3 expression was driven by the Dex-inducible PTA promoter system for 24 h before measurement. Data represent means ± se from nine replicates. Averages for +Dex are significantly different from those for –Dex at P < 0.05 for (A) and P < 0.01 for (B).
DISCUSSION
The Root Hair System as a Model for Auxin Transport
In this study, we used the in planta single cell system, the root hair cell, as a biological marker to characterize the cellular function of the PID protein kinase. Numerous genetic and physiological studies have shown that auxin positively regulates root hair growth (see Introduction). Here, we provide additional evidence that the intracellular level of auxin in the hair cell is critical for root hair growth. Root hair cell–specific overexpression of PIN3 severely inhibited root hair growth (Figures 5 and 7B). In this experiment, PIN3 was expressed only in the root hair cell immediately before the root hair began to grow. Our results indicate that the inhibition of hair growth by PIN3 overexpression was attributable to excessive auxin efflux, resulting in decreased auxin levels inside the hair cell. Another experiment with the hair cell–specific transgenic system showed that the auxin-responsiveness of hair growth is regulated by the hair cell itself. In the dominant Arabidopsis axr2 mutant, there is a point mutation in the gene encoding AXR2/IAA7, such that AXR2/IAA7 is resistant to proteasome-mediated turnover. As a result, auxin-responsive genes are continually suppressed. The axr2 mutant has a pleiotropic phenotype, consisting of defects in auxin responses, lesions in apical dominance, gravitropism, stem and root elongation, and root hair development (Wilson et al., 1990). When axr2 was overexpressed using the root hair–specific ATEXPA7 promoter, root hair development was completely blocked (see Supplemental Figure 1 online), and there was no detectable effect on other organs and tissues. This result indicated that the root hair cell is where the auxin-responsive genes function in root hair growth.
The root hair system used in this study was a very robust and consistent system for exploring auxin transport mechanisms. ST can inhibit many different protein kinases involved in diverse aspects of cellular metabolism (Yamaki et al., 2002). It is striking that this broad-spectrum kinase inhibitor specifically rescued the biological process of root hair growth in both PIDox and PIN3ox roots (Figures 4E, 5, 8G, and 8H). BFA, a vesicle trafficking inhibitor, also has wide-ranging inhibitory effects on cellular processes, such as the elongation of stems and coleoptiles and pollen tube growth (Satiat-Jeunemaitre et al., 1996). This inhibitor also restored root hair growth in PIDox and PIN3ox transformants, almost to the level seen in control plants (Figures 4D, 5, 8E, and 8F). These robust yet seemingly specific properties of root hair development indicate that the root hair can serve as a unique in planta marker system for exploring the mechanism of auxin transport at the single cell level.
Is PID an Auxin Signaling Component or an Auxin Transport Regulator?
Overexpression of PID in Arabidopsis using the cauliflower mosaic virus 35S promoter (P35S:PID) resulted in several different phenotypes, such as reduced root elongation, delayed lateral root formation, reduced expression of auxin-responsive DR5:GUS, and collapse of the primary root (Christensen et al., 2000; Benjamins et al., 2001). However, application of auxins, either IAA or membrane-diffusible NAA, failed to rescue these phenotypes (Christensen et al., 2000; DeLong et al., 2002). These results provide evidence that PID is a negative regulator of auxin signaling. Another study using the same P35S:PID Arabidopsis transformant showed that NPA rescued the root phenotypes of the transformant, suggesting that PID functions in auxin transport (Benjamins et al., 2001).
The functional marker system used in these previous studies was principally the root apex, where the root meristem collapses and where DR5:GUS was expressed. In terms of auxin action, the root apex is a highly complicated region (Kramer, 2004). Auxin metabolism and multidirectional auxin flow through various cell types take place in the root apex (Morris et al., 2004; Ljung et al., 2005). When the expression of PID is driven by the 35S promoter, the protein will act with different levels of strength, depending on the promoter activity in different cell types, and will have multiple targets, making it difficult to yield reliable assessments of the phenotypic effects of overexpression.
In this study, because we limited PID expression to the root hair cell, we were able to obtain consistent results that suggested that PID is a positive regulator of auxin efflux. First, PIDox using the root hair–specific promoter PE7 resulted in a defective root hair phenotype, and this defect was fully rescued by the addition of IAA (Figure 2D). IAA-induced restoration of root hairs was mimicked in PIN3ox transformants (Figure 7C). The rescue of PIDox by exogenous IAA indicated that auxin influx occurred normally in the hair cell and that PID acts upstream of auxin signaling. Second, treatments that inhibited auxin efflux rescued the PIDox root hair phenotype. Inhibition of auxin efflux by NPA and inhibition of PIN trafficking by BFA restored root hair growth of the PIDox root (Figures 4C, 4D, and 5). The restoration of root hair growth by these inhibitors also was observed in the PIN3ox root (Figures 5 and 8). These results suggested that by affecting PINs and/or other efflux components, high levels of PID activate auxin efflux in the hair cell, resulting in cellular auxin levels below the threshold required for root hair growth. The decreased retention of radiolabeled NAA in PIDox tobacco cells (Figure 9) was additional direct evidence that PID is a positive regulator of auxin efflux.
Is PID a Binary Switch for the Subcellular Polarity of PIN Proteins or a Positive Regulator for Auxin Efflux Activity?
It was recently shown that PID protein kinase regulates the polarity of the subcellular distribution of PIN proteins in various Arabidopsis cell types (Friml et al., 2004). In root tissues, a low dose of PID (in wild-type cells) resulted in the localization of PIN1, PIN2 (in the cortex), and PIN4 at the apical membrane of the cell (toward the root apex), whereas a high dose of PID (in P35S:PID) shifted the localization of these PINs to the basal end of the cell (toward the root base). The authors concluded that PID acted as a binary switch for PIN polarity. It was suggested that this apical-to-basal shift of PIN polarity induced by a high dose of PID caused a change of auxin flux from acropetal to basipetal, resulting in a depletion of auxin in the root tip tissues and, consequently, a collapse of the root meristem.
In this study, when expressed in the root hair cell, PIN3 did not exhibit a particular polarity in the plasma membrane. PIN3-GFP was observed along the entire hair cell membrane and was concentrated at both apical and basal ends (Figures 7E and 7F). The distribution pattern of PIN1-GFP in the hair cell was identical to that of PIN3-GFP—that is, it had no particular polarity (data not shown). These results suggest that PIN polarities may occur selectively in certain cell types that are needed for auxin polar transport and redistribution.
Considering the nonpolarity of PIN distribution in the root hair cell, the suppression of hair growth by PIDox was most likely attributable to the activation of PIN proteins (or other efflux components) rather than to a change of PIN polarity. The polarity change of PINs, if any, would not influence auxin levels in the hair cell. Potential mechanisms for activating auxin efflux are more likely to be gene or catalytic activation of the efflux carrier components or enhancement of trafficking of those components to the plasma membrane.
Both pharmacological and genetic approaches indicated that PID-mediated protein phosphorylation is involved in facilitating auxin efflux in Arabidopsis root hair cells. The protein kinase inhibitor ST interfered with the trafficking of PIN3 to the plasma membrane (Figures 8J and 8K) and restored root hair growth in PIN3ox transformants (Figures 5, 8G, and 8H). These results suggested that PID-mediated protein phosphorylation enhances cellular auxin efflux by facilitating the trafficking of PIN proteins or other efflux components to the plasma membrane. In light of the apparent cell type–dependent polarity of different PINs, PID might function in the polar trafficking of PINs in those cells and tissues involved in auxin conduction, whereas in other cell types, such as developing root hair cells, PID may simply facilitate PIN trafficking to the plasma membrane.
PID Might Enhance Cellular Auxin Efflux, Probably by Working in the Proximity of PINs
Numerous studies involving immunohistochemical analysis (Morris et al., 2004) and several studies using reporter fusion proteins (Benková et al., 2003; Friml et al., 2004; Blilou et al., 2005) have shown that PINs localize to the plasma membrane. It has also been shown that BFA is effective at disrupting the localization of PIN proteins (Geldner et al., 2001, 2003). In this study, it was shown that PIN3 localized along the entire cell boundary, or sometimes to both the apical and basal ends of Arabidopsis hair cells and tobacco BY-2 cells (Figures 7E to 7G). Interestingly, the location of PID in Arabidopsis hair cells and tobacco cells was very similar to that of PIN3 (Figures 2F to 2J, 2N, and 2O), and treatment with BFA or with the kinase inhibitor ST caused the internalization of both proteins (Figures 4G, 4H, 4L to 4N, and 8J to 8M).
The amino acid sequence of PID does not include any predicted membrane association motifs (Christensen et al., 2000). In this study, we show a certain similarity in the subcellular dynamics of PID and PIN3, suggesting that PID probably acts in the proximity of PIN proteins and that the molecular machinery modulating one protein most likely modulates the cellular behavior of the other as well. It was shown previously that PID functions as a molecular switch for PIN polarity, implying the molecular function of PID in the process of protein trafficking (Friml et al., 2004). Our results, by showing the spatial and functional overlap between PID and PIN3, support the idea that PID may be one of the essential components modulating PIN trafficking in its vicinity.
The subcellular polarity among PINs is basically governed by cell type–specific mechanisms (Paponov et al., 2005). However, the apical–basal localization of PIN1 is opposite that of the influx component AUX1 in the same root protophloem cell (Swarup et al., 2001), indicating that the cues for polar trafficking of PINs could also be derived from their molecular structure. If, in this mechanism, PID conferred molecule specificity for trafficking of the PIN proteins, PID would likely have to interact with specific interfacial molecules for each PIN or with a specific motif in the PIN protein. The similarity of the internalization patterns of PIN3 either by ST or BFA treatment (Figures 8J to 8M) implies that a PID-mediated protein phosphorylation process may be coupled with PIN trafficking between the plasma membrane and intracellular compartments. However, PID's phosphorylation targets in the PIN trafficking machinery remain to be elucidated.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana was the model plant in this study; the Columbia ecotype was the wild type. rhd6 seeds were obtained from J.W. Schiefelbein (University of Michigan), and pid-3 and axr2-1 seeds were from the ABRC. The seeds were sowed on agarose plates containing 4.3 g/L Murashige and Skoog nutrient mix (Sigma-Aldrich), 1% sucrose, 0.5 g/L MES, pH 5.7, with KOH, and 0.8% agarose. Three-day-old treated seeds were germinated at 23°C under 16-h-light/8-h-dark photoperiods. For pharmacological experiments, 3-d-old seedlings were transferred to new plates containing the indicated chemicals and incubated for an additional 1 d, after which root hairs were examined. For observation of subcellular localization of PID-GFP and PIN3-GFP after BFA or ST treatment, seedlings were incubated either in solid medium containing the chemicals for 24 h or in liquid medium for 2 h. Control agarose plates and liquid medium included the same concentrations of DMSO that were included in the chemical (BFA and ST) treatments. Transformed plants were selected on hygromycin-containing plates (10 μg/mL). For observation of root hairs, homozygous transformants were planted on antibiotic-free medium, and T1 and T2 lines were selected on hygromycin-containing medium. Hygromycin did not particularly interfere with root hair development, as shown with the control PE7:GFP transformants in each experiment.
Transgene Constructs
To express genes specifically in the root hair cells, the ATEXPA7 promoter PE7 (Cho and Cosgrove, 2002) was used. The binary vector pGPTV-HYG (Becker et al., 1992) was the basic cloning vector. PE7:GFP was as described by Cho and Cosgrove (2002). For the PE7:PID-GFP construct, a genomic fragment of PID was obtained by PCR using primers 5′-CCATTTCTAGATTTAACTTCGATTTTC-3′ (containing an XbaI site) and 5′-CGTAGCCCGGGTAACAAAGTAATCGAACGCCGCT-3′ (containing an XmaI site) using Arabidopsis genomic DNA as the template. The PCR product was cloned into XbaI-XmaI sites upstream of the GFP gene, producing a PID-GFP fusion protein. For the PE7:mPID-GFP construct, the point-mutated mPID fragment (Asp-205 to Ala) (Christensen et al., 2000) was produced by the megaprimer PCR method (Ke and Madison, 1997). The first PCR was performed using primers 5′-CCATTTCTAGATTTAACTTCGATTTTC-3′ (containing an XbaI site) and 5′-TTTCAGGCTTCAGAGCTCTGTAGATGATA-3′ (containing the point mutation) to produce the megaprimer. The second PCR was done using the megaprimer and 5′-CGTAGCCCGGGTAACAAAGTAATCGAACGCCGCT-3′ (containing an XmaI site) to produce mPID. PE7:mPID-GFP was generated from PE7:PID-GFP by replacing PID with mPID. For PE7:axr2 (see Supplemental Figure 1 online), the coding region of the axr2 dominant mutant gene was obtained by PCR from genomic DNA of the mutant plant and inserted upstream of GFP in the PE7:GFP vector.
For the PE7:PIN3-GFP construct, PIN3-GFP was generated by insertion of GFP into the PIN3 genomic fragment (–27 to 2726 bp, relative to the start codon) at position 1487. The 5′ and 3′ genomic fragments of PIN3 and the GFP coding sequence were obtained by PCR using the following primer sets: 5′-CGTTGGTCGACGGTTTTCCATTTTTG-3′ (containing a SalI site) and 5′-GGAGGCCCGGGTTTTCGTTGACTTGC-3′ (containing an XmaI site) for the 5′ fragment of PIN3; 5′-GTCAACCCGGGAATATGCCTCCGGCGA-3′ (containing an XmaI site) and 5′-GACTCGAGCTCGGGGCTTTCATAAC-3′ (containing a SacI site) for the 3′ fragment of PIN3; and 5′-TCGCCCCCGGGGTGAGCAAGGGCG-3′ (containing an XmaI site) and 5′-CGGCCCCCGGGCTTGTACAGCTCGTCC-3′ (containing an XmaI site) for GFP. These PCR products and the PE7 fragment, which was amplified by PCR using primers 5′-TGAATAAGCTTTTGGTTCTAATG-3′ (containing a HindIII site) and 5′-ACCCAGTCGACCCTCTTTTTCTTTAT-3′ (containing a SalI site), were cloned into the binary vector pCAMBIA1300-NOS (Hyg+).
For the PTA:PID-GFP construct, the PID-GFP fragment was prepared by PCR amplification using primers 5-TTAACCTCGAGTTTCCCGGCGATGTTACG-3′ (containing an XhoI site) and 5′-AGCTCACTAGTCGCGGCCGCTTTACTTG-3′ (containing an SpeI site) from the PE7:PID-GFP construct and was inserted into the Dex-inducible vector system pTA7001 (Aoyama and Chua, 1997). For the PTA:PIN3-GFP construct, the PIN3-GFP fragment (with SalI and XbaI sites) was digested from PE7:PIN3-GFP and cloned into XhoI-SpeI sites of pTA7001.
All of the constructs were confirmed by nucleotide sequencing and introduced into Arabidopsis plants or tobacco (Nicotiana tabacum) BY-2 cells using Agrobacterium tumefaciens strain C58C1 (pMP90) (Bechtold and Pelletier, 1998). Transgene insertion in the Arabidopsis transformants was checked by PCR analysis of the genomic DNA and nucleotide sequencing. Four randomly chosen PID-GFP or PIN3-GFP lines (PE7:PID1 to PE7:PID4, PE7:PIN3,1 to PE7:PIN3,4) and one strong expressing PE7:PID8 line were used for analysis. Segregation ratios for hygromycin sensitivity of the transformants were 3:1 for PE7:PID-GFP1, PE7:PID-GFP3, and PE7:PID-GFP4, 7:1 for PE7:PID-GFP2, and 6:1 for PE7:PID-GFP8. PE7:PIN3-GFP lines showed ratios between 3:1 and 6:1. Unless mentioned otherwise, T3 homozygous transformants were used for the experiments.
Culture and Transformation of Tobacco BY-2 Cells
Tobacco BY-2 cells (Nagata et al., 1992) were cultured in darkness at 26°C with shaking (150 rpm). The liquid culture medium included 3% (w/v) sucrose, 4.3 g/L Murashige and Skoog salts, 100 mg/L inositol, 1 mg/L thiamine, 0.2 mg/L 2,4-D, and 200 mg/L KH2PO4, pH 5.8. The suspension cells were subcultured weekly. Stock BY-2 calli were maintained on solid medium with 0.8% (w/v) agar and subcultured monthly. Transgenic cells and calli were maintained on the same medium supplemented with 150 μg/mL hygromycin.
For transformation of tobacco BY-2 cells, 50 mL of a 3-d-old culture was cocultivated with 10 mL of Agrobacterium tumefaciens harboring PTA:GFP, PTA:PID-GFP, or PTA:PIN3-GFP in a Petri dish in the dark for 3 d at 26°C. Inoculated cells were washed three times and transferred onto solid medium supplemented with 150 μg/mL hygromycin and 200 μg/mL cefotaxime. After 3 weeks, positive calli were transferred onto new medium. Individual calli were then maintained on solid medium or resuspended in the liquid medium to obtain cell suspensions.
Auxin Retention Assay in Tobacco BY-2 Cells
Measurement of auxin retention was performed according to the methods of Delbarre et al. (1996) and Petrášek et al. (2003) with modifications. Briefly, cells (150 to 200 mg), after incubation with or without 10 μM Dex for 24 h, were filtered, resuspended, and equilibrated in uptake buffer (20 mM MES, 40 mM sucrose, and 0.5 mM CaSO4, pH adjusted to 5.7 by KOH) for 45 min with continuous orbital shaking. Equilibrated cells were collected by filtration, resuspended in fresh uptake buffer supplemented with or without 10 μM Dex, and incubated in the orbital shaker for 1.5 h in darkness at 26°C. [3H]NAA was mixed with 10 mL of the cell suspension for a final concentration of 5 nM [3H]NAA, and 0.5-mL aliquots were rapidly filtered by vacuum pressure on GF/C glass fiber filters (Whatman) at different time intervals (0, 7, and 15 min). Cell cakes were washed in 5 mL of distilled water, transferred to scintillation vials, and extracted with 1 mL of ethanol for 30 min, and radioactivity was determined by liquid scintillation counting. Nine replicates were performed for each time point.
Measurement of Root Hair Parameters
Root hair number and length were measured as described by Cho and Cosgrove (2002) with modifications. All of the hairs shown in the same root range (0.78 mm from the hair maturation region) were counted. To measure hair length, four consecutive hairs protruding perpendicularly from each side of the root, for a total of eight hairs from both sides of the root, were measured. Hair bulges shorter than 14 μm were considered arrested at the early bulge stage and were not counted.
Observation of Reporter Gene Expression
For detection of GFP, seedling roots were observed using a confocal laser scanning microscope (LSM 510; Carl Zeiss). GFP was detected using a 488-nm excitation filter and a 543-nm emission filter. Fluorescence images were digitized with the Zeiss LSM image browser version 2.80.1123. For detection of GFP fluorescence from BY-2 cells harboring PTA:GFP, PTA:PID-GFP, or PTA:PIN3-GFP, the transgenic cells were pretreated with 10 μM Dex.
RT-PCR for Detection of PIN and PID Expression in the Root Hair Cell
Root hair cell–specific RNA was purified from 2-d-old Arabidopsis seedling roots. Actively growing young root hairs were pulled by fine forceps and immersed directly into the RNA extraction buffer. The RNeasy plant mini kit (Qiagen) was used to extract total RNA. Whole root RNA was prepared by grinding the seedling roots in a 1.5-mL plastic tube. The cDNA was synthesized by Moloney murine leukemia virus reverse transcriptase, and PCR was conducted using CDSIII oligo d(T)30 primers (Clontech) and the Advantage 2 polymerase kit (Clontech) according to the manufacturer's instructions. Forty-five cycles were performed with the root hair cell–specific RNA template, and 30 cycles with the whole root RNA template. The gene-specific primer sets for PCR were as follows: 5′-CAAAACGACGCAGGCTAAGGTG-3′ and 5′-AAGTCCCTGTGTTTTGGTAATATCTCT-3′ for PIN1; 5′-TGGGCAAAAAAGGTAGCGACGT-3′ and 5′-CACCTTTGGGTCGTATCGCCTT-3′ for PIN2; 5′-TAGGAGGAGCCGAAGCAAGTCA-3′ and 5′-TTTCCCTTTCTTCCTTCTCCCTCTA-3′ for PIN3; 5′-CAGGGCTGAATAAAATGGGGTCTA-3′ and 5′-CGAGAAATAAATAGCCCAATCTCCGA-3′ for PIN4; 5′-CTTAACGCCGCAAGGGCGTTAC-3′ and 5′-AATAATATGCCAAAGTTGTTGGTAAGGCA-3′ for PIN5; 5′-AGGAGCCGCCGGAAAAGACAC-3′ and 5′-ATTCGGCTTCAGTGTGTTGTGTGAATC-3′ for PIN6; 5′-CGGACTACACAAGCTTCGGTGTA-3′ and 5′-GCCAAGCAGAACTTCTCCATTGA -3′ for PIN7; 5′-CAAGTAGACAACATCAATATCGAAAGTGG-3′ and 5′-CGCTGTGCTTAGAACATCTGCAT-3′ for PIN8; 5′-CCATTTCTAGATTTAACTTCGATTTTC-3′ and 5′-TAGATGATACCCAACATGTGTAG-3′ for PID; 5′-CTTTAGAGATGAAGCTCGTCGGC-3′ and 5′-TGTGACTTGTCATCAGCAATCTTCGAT-3′ for GL2; and 5′-GTATCATCCCAGTTGCATACCGAA-3′ and 5′-GTTTGGTAGGGCAAAAGCCTAG-3′ for ATEXPA7. Gene-specific primer pairs were designed to include intron(s) so that cDNA PCR products could be distinguished from genomic PCR products.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are At1G12560 (ATEXPA7), At1G79840 (GL2), At3G23050 (AXR2/IAA7), At2G34650 (PID), At1G73590 (PIN1), At5G57090 (PIN2), At1G70940 (PIN3), At2G01420 (PIN4), At5G16530 (PIN5), At1G77110 (PIN6), At1G23080 (PIN7), and At5G15100 (PIN8).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Root Phenotype of The PE7:axr2 Transformant.
Supplemental Figure 2. Effects of Chemicals on the Restoration of Root Hair Growth in PE7:PID-GFP Lines.
Supplemental Figure 3. Effects of Chemicals on the Restoration of Root Hair Growth in PE7:PIN3-GFP Lines.
Supplemental Figure 4. Expression Dosage Is Unlikely to Affect the Distribution Pattern of mPID.
Supplemental Table 1. Expected Sizes of PCR Products in RT-PCR for PINs and PID.
Supplementary Material
Acknowledgments
We thank Zee-Won Lee at the Korea Basic Science Institute for kindly helping with confocal microscopy imaging, Youngsook Lee at the Pohang University of Science and Technology and Jiří Friml at the Universität Tübingen for kindly providing PE7:AHA2-GFP and PPIN1:PIN1-GFP transgenic Arabidopsis seeds, respectively, and Remko Offringa at Leiden University and the PlantD members for helpful discussions. The pTA7001 vector was kindly provided by Nam-Hai Chua. We are also grateful to Mi Sook Cho in our laboratory and Suk-Yoon Kwon and Young Pyo Lee at the Korea Research Institute of Bioscience and Biotechnology for helping with the auxin retention assay. This study was supported by grants from the Korea Research Foundation (KRF-2003-041-C00349 and KRF-2004-041-C00366), the Plant Diversity Research Center of the 21st Century Frontier Research Program (PF0330506-00), the Korea Science and Engineering Foundation Environmental Biotechnology Research Center (R15-2003-012-02003-0), and partly by the research fund of Chungnam National University in 2004.
This article is dedicated to Hans Kende on the occasion of his retirement.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hyung-Taeg Cho (htcho@cnu.ac.kr).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.035972.
References
- Aoyama, T., and Chua, N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11 606–612. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Arabidopsis Protocols, J.M. Martinez-Zapater and J. Salinas, eds (Totowa, NJ: Humana), pp. 259–266. [DOI] [PubMed]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20 1195–1197. [DOI] [PubMed] [Google Scholar]
- Benjamins, R., Quint, A., Weijers, D., Hooykaas, P., and Offringa, R. (2001). The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128 4057–4067. [DOI] [PubMed] [Google Scholar]
- Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. [DOI] [PubMed] [Google Scholar]
- Bennett, S.R.M., Alvarez, J., Bossinger, G., and Smyth, D.R. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8 505–520. [Google Scholar]
- Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., Heidstra, R., Aida, M., Palme, K., and Scheres, B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433 39–44. [DOI] [PubMed] [Google Scholar]
- Chen, R., Hilson, P., Sedbrook, J., Rosen, E., Caspar, T., and Masson, P.H. (1998). The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. USA 95 15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H.-T., and Cosgrove, D.J. (2002). The regulation of Arabidopsis root hair initiation and expansin gene expression. Plant Cell 14 3237–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S.K., Dagenais, N., Chory, J., and Weigel, D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100 469–478. [DOI] [PubMed] [Google Scholar]
- Delbarre, A., Muller, P., and Guern, J. (1998). Short-lived and phosphorylated proteins contribute to carrier-mediated efflux. Plant Physiol. 116 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarre, A., Muller, P., Imhoff, V., and Guern, J. (1996). Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 198 532–541. [DOI] [PubMed] [Google Scholar]
- DeLong, A., Mockaitis, K., and Christensen, S. (2002). Protein phosphorylation in the delivery of and response to signals. Plant Mol. Biol. 49 285–303. [PubMed] [Google Scholar]
- Friml, J., Wisniewska, J., Benkova, E., Mendgen, K., and Palme, K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415 806–809. [DOI] [PubMed] [Google Scholar]
- Friml, J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 862–865. [DOI] [PubMed] [Google Scholar]
- Geisler, M., and Murphy, A.S. (2006). The ABC of auxin transport: The role of p-glycoproteins in plant development. FEBS Lett. 580 1094–1102. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A., and Jürgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112 219–230. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Friml, J., Stierhof, Y.-D., Jürgens, G., and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413 425–428. [DOI] [PubMed] [Google Scholar]
- Ke, S.-H., and Madison, E.L. (1997). Rapid and efficient site-directed mutagenesis by single-tube ‘megaprimer’ PCR method. Nucleic Acids Res. 25 3371–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, E.M. (2004). PIN and AUX/LAX proteins: Their role in auxin accumulation. Trends Plant Sci. 9 578–582. [DOI] [PubMed] [Google Scholar]
- Leyser, H.M.O., Pickett, F.B., Dharmasiri, S., and Estelle, M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10 403–413. [DOI] [PubMed] [Google Scholar]
- Ljung, K., Hull, A.K., Celenza, J., Yamada, M., Estelle, M., and Mormanly, J. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17 1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax, T.L., Muday, G.K., and Rubery, P.H. (1995). Auxin transport. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 509–530.
- Luschnig, C., Gaxiola, R.A., Grisafi, P., and Fink, G.R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci, J., and Schiefelbein, J.W. (1994). The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin and ethylene-associated process. Plant Physiol. 106 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci, J.D., and Schiefelbein, J.W. (1996). Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, D.V., Friml, J., and Zažímalová, E. (2004). The transport of auxin. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 437–470.
- Nagata, T., Nemoto, Y., and Hasezawa, S. (1992). Tobacco BY2 cell line as the ‘HeLa’ cell in the cell biology of higher plants. Int. Rev. Cytol. 13 1–30. [Google Scholar]
- Okada, K., and Shimura, Y. (1994). Modulation of root growth by physical stimuli. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 665–684.
- Paponov, I.A., Teale, W.D., Trebar, M., Blilou, K., and Palme, K. (2005). The PIN auxin efflux facilitators: Evolutionary and functional perspectives. Trends Plant Sci. 10 153–200. [DOI] [PubMed] [Google Scholar]
- Petrášek, J., Černá, A., Schwarzerová, K., Elčkner, M., Morris, D.A., and Zažímalová, E. (2003). Do phytotropins inhibit auxin efflux by impairing vesicle traffic? Plant Physiol. 131 254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts, R.J., Cernac, A., and Estelle, M. (1998). Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16 553–560. [DOI] [PubMed] [Google Scholar]
- Rashotte, A.M., DeLong, A., and Muday, G.K. (2001). Genetic and chemical reductions in protein phosphatase activity alter auxin transport. Plant Cell 13 1683–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6 420–425. [DOI] [PubMed] [Google Scholar]
- Reiners, J.J., Lee, J.-Y., Clift, R.E., Dudley, D.T., and Myrand, S.P. (1998). PD98059 is an equipotent antagonist of the aryl hydrocarbon receptor and inhibitor of mitogen-activated protein kinase kinase. Mol. Pharmacol. 53 438–445. [DOI] [PubMed] [Google Scholar]
- Reinhardt, D., Pesce, E.-R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260. [DOI] [PubMed] [Google Scholar]
- Santelia, D., Vincenzetti, V., Azzarello, E., Bovet, L., Fukao, Y., Düchtig, P., Mancuso, S., Martinoia, E., and Geisler, M. (2005). MDR-like ABC transporter AtPGP4 is involved in auxin-mediated lateral root and root hair development. FEBS Lett. 579 5399–5406. [DOI] [PubMed] [Google Scholar]
- Satiat-Jeunemaitre, B., Cole, L., Bourett, T., Howard, R., and Hawes, C. (1996). Brefeldin A effects in plant and fungal cells: Something new about vesicle trafficking? J. Microsc. 181 162–177. [DOI] [PubMed] [Google Scholar]
- Schiefelbein, J.W. (2000). Constructing a plant cell: The genetic control of root hair development. Plant Physiol. 124 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup, R., Friml, J., Marchant, A., Ljung, K., Sandberg, G., Palme, K., and Bennett, M. (2001). Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 15 2648–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, A.K., Pickett, F.B., Turner, J.C., and Estelle, M. (1990). A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 222 377–383. [DOI] [PubMed] [Google Scholar]
- Yamaki, K., Hong, J., Hiraizumi, K., Ahn, J.W., Zee, O., and Ohuchi, K. (2002). Participation of various kinases in staurosporine induced apoptosis of RAW 264.7 cells. J. Pharm. Pharmacol. 54 1535–1544. [DOI] [PubMed] [Google Scholar]
- Zegzouti, H., Anthony, R.G., Jahchan, N., Bögre, L., and Christensen, S.K. (2006). Phosphorylation and activation of PINOID by the phospholipid signaling kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 6404–6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.