Abstract
In plants, many gene transcripts are very unstable, which is important for the tight control of their temporal and spatial expression patterns. To identify cellular factors controlling the stability of unstable mRNAs in plants, we used luciferase imaging in Arabidopsis thaliana to isolate a recessive mutant, stabilized1-1 (sta1-1), with enhanced stability of the normally unstable luciferase transcript. The sta1-1 mutation also causes the stabilization of some endogenous gene transcripts and has a range of developmental and stress response phenotypes. STA1 encodes a nuclear protein similar to the human U5 small ribonucleoprotein–associated 102-kD protein and to the yeast pre-mRNA splicing factors Prp1p and Prp6p. STA1 expression is upregulated by cold stress, and the sta1-1 mutant is defective in the splicing of the cold-induced COR15A gene. Our results show that STA1 is a pre-mRNA splicing factor required not only for splicing but also for the turnover of unstable transcripts and that it has an important role in plant responses to abiotic stresses.
INTRODUCTION
Gene expression is controlled at the transcriptional and posttranscriptional levels. The instability of mRNAs facilitates the tight control of specific temporal and spatial expression patterns. In higher plants, the control of mRNA stability has been associated with growth, development, and response to hormones as well as biotic and abiotic stresses (Abler and Green, 1996; Carrington and Ambros, 2003; Kuhn and Schroeder, 2003; Shi et al., 2003). Much effort has been made to understand the RNA silencing pathway for the degradation of mRNAs containing sequences complementary to short regulatory RNAs, such as microRNAs (miRNAs) and small interfering RNAs (siRNAs) (Voinnet, 2002; Bartel and Bartel, 2003; Carrington and Ambros, 2003). miRNAs and siRNAs assemble in endonuclease-containing complexes termed RISC and miRNP, respectively, and can target homologous RNA sequences for endonucleolytic cleavage (Hamilton and Baulcombe, 1999; Hammond et al., 2000; Zamore et al., 2000; Hutvagner and Zamore, 2002). Factors involved in miRNA or siRNA biogenesis or actions are important determinants of the abundance of target mRNAs.
Many endogenous mRNAs with a high turnover rate are not targeted by miRNAs or siRNAs. Some of these unstable mRNAs in plants contain, as instability determinants, multiple overlapping AUUUA sequences or downstream element sequences that are not AU-rich (Ohme-Takagi et al., 1993; Johnson et al., 2000). However, the primary or secondary sequence features conferring instability to most of the unstable mRNAs are not known. The cellular machinery important for the degradation of the unstable mRNAs is expected to consist of RNases, RNase inhibitors, RNA binding proteins, and, potentially, other cellular factors. To identify the cellular factors regulating RNA stability, two Arabidopsis thaliana mutants defective in downstream element–mediated mRNA decay were isolated (Johnson et al., 2000). However, the genes responsible for the mutant phenotypes have not been identified (Johnson et al., 2000). In contrast with the paucity of genetic studies of mRNA stability control in multicellular organisms, including plants, extensive genetic analysis has been conducted in yeast and has elucidated general mRNA decay mechanisms. The main pathway for the turnover of both unstable and stable transcripts in yeast is the deadenylation-dependent decapping pathway (Caponigro and Parker, 1996). In addition, yeast has mRNA surveillance systems that detect and degrade aberrant mRNAs (Hilleren and Parker, 1999), which include malprocessed transcripts and transcripts with premature nonsense codons. Nonsense-mediated decay also occurs in plants, but whether the mechanisms are the same as in yeast is unclear.
Because of their sessile nature, plants have evolved sophisticated mechanisms to cope with environmental challenges (Zhu, 2002). Recently, RNA metabolism was shown to be important in plant responses to abiotic stresses (Forment et al., 2002; Gong et al., 2002b; Kuhn and Schroeder, 2003). The expression of the RS domain of an SR-like splicing protein, SRL1, conferred salt tolerance to Arabidopsis, suggesting an important role of pre-mRNA splicing in salt tolerance (Forment et al., 2002). los4, an Arabidopsis mutant defective in a DEAD box–RNA helicase similar to the yeast RNA export factor Dbp5p, showed impaired chilling and freezing tolerance (Gong et al., 2002b). At least five genes involved in RNA metabolism have been implicated in plant responses to drought and the stress hormone abscisic acid (ABA) (Kuhn and Schroeder, 2003; Razem et al., 2006). The ABA-hypersensitive hyl1 Arabidopsis mutant is defective in a double-stranded RNA binding protein (Lu and Fedoroff, 2000). HYL1 appears to affect the production of some miRNAs that in turn regulate the expression levels of their target genes (Han et al., 2004). ABH1 and SAD1 from Arabidopsis and AKIP1 from Vicia faba appear to be directly involved in RNA processing, which somehow affects ABA responses. ABH1 encodes a large subunit of a dimeric mRNA cap binding complex (Hugouvieux et al., 2001), whereas SAD1 encodes an Lsm small ribonucleoprotein (snRNP) similar to the yeast Lsm5p (Xiong et al., 2001a). AKIP1 is a single-stranded RNA binding protein homologous with hnRNP A/B (Li et al., 2002). A very recent study revealed that FCA, an RNA binding protein that controls flowering time, is an ABA receptor important for ABA regulation of flowering (Razem et al., 2006).
We have used the firefly luciferase reporter gene driven by the stress-responsive RD29A promoter to facilitate genetic dissection of plant responses to abiotic stresses (Chinnusamy et al., 2002; Lee et al., 2002). The reporter gene system has allowed for the identification of a number of signaling components important for the transcriptional regulation of stress-responsive genes (Lee et al., 2001; Xiong et al., 2001a, 2001b, 2001c). In addition, this reporter system has led to the isolation of a DNA glycosylase that is essential for preventing transcriptional gene silencing (Gong et al., 2002a). We noticed that the luciferase transcript used in our studies is very unstable in Arabidopsis (Ishitani et al., 1998). Although the instability sequence in the luciferase reporter gene is not known, this unstable reporter has permitted us to isolate several Arabidopsis mutants with altered regulation of mRNA stability. Here, we present the characterization of one such mutant, stabilized1 (sta1), the cloning of the STA1 gene, and the surprising finding that STA1 is required for both pre-mRNA splicing and the degradation of some transcripts. In addition, STA1 is upregulated by cold stress, and sta1-1 mutant plants show altered responses to various abiotic stresses.
RESULTS
The sta1-1 Mutation Enhances the Stability of the Luciferase Transcript
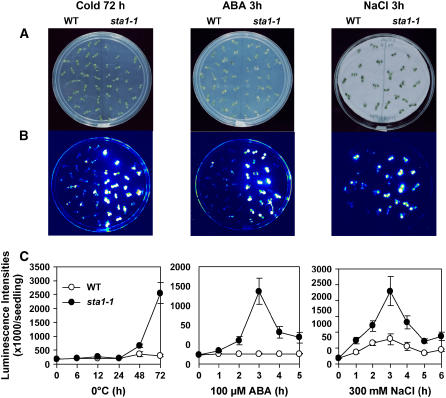
We previously described a mutant screening strategy that uses a low-light luminescence imaging system and transgenic Arabidopsis expressing the firefly luciferase reporter driven by the stress-inducible RD29A promoter (Chinnusamy et al., 2002; Lee et al., 2002). Using this system, we isolated many mutants that show altered luminescence responses under stress conditions. One such mutant, recovered from a population of ecotype Columbia plants expressing the RD29A-luciferase transgene (hereafter called the wild type) mutated with the use of ethyl methanesulfonate, showed higher luminescence than the wild type after cold, ABA, or NaCl treatment. The mutant, named sta1-1, was characterized in this study after several backcrosses were performed.
RD29A promoter–driven luciferase (RD29A-LUC) activity was tested with seedlings grown on Murashige and Skoog (MS) agar medium supplemented with 3% sucrose. Under the tested stress conditions, sta1-1 mutant seedlings showed higher luminescence than did the wild type (Figure 1). Under cold conditions, 72 h of treatment enhanced the luminescence intensity in the sta1-1 mutant much more than in the wild type (Figure 1C). ABA or NaCl treatment also induced higher luminescence in sta1-1 than in the wild type, with a peak at 3 h of treatment.
Figure 1.
Comparison of Luminescence Images and Intensity between the Wild Type and sta1-1 under Stress.
(A) Wild-type and sta1-1 seedlings on MS agar plates.
(B) Luminescence images corresponding to plates in (A).
(C) Quantification of luminescence intensities over the time periods indicated (n = 20 for cold stress and ABA, n = 10 for NaCl treatment; error bars indicate sd).
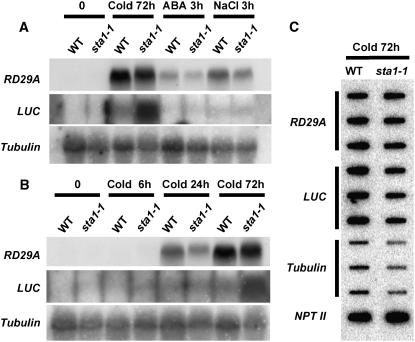
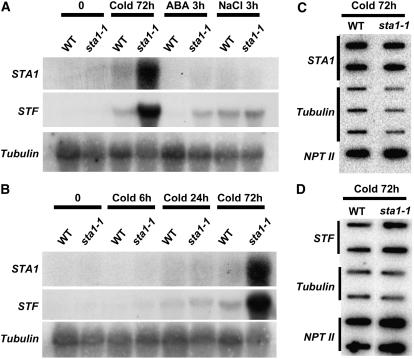
The steady state levels of the luciferase transcript and the endogenous RD29A transcript were examined by RNA gel blot analysis in both the wild type and the sta1-1 mutant. Because of its very unstable nature, the luciferase transcript was virtually undetectable in wild-type plants, even after cold, ABA, or NaCl treatment (Figure 2). However, a high level of luciferase mRNA was detected in sta1-1 after 72 h of cold treatment (Figure 2). This result also revealed that the endogenous RD29A transcript level was not higher in the sta1-1 mutant than in the wild type under any of the conditions tested (Figures 2A and 2B). Therefore, it is unlikely that the sta1-1 mutation caused increased transcription from the RD29A promoter, because both the transgene and the endogenous RD29A gene have the same promoter.
Figure 2.
Comparison of Expression Levels between the Wild Type and sta1-1 of Endogenous RD29A and the RD29A-LUC Transgene by RNA Hybridization and Nuclear Run-On Analysis.
(A) RNA hybridization with total RNA (20 μg) from samples treated with cold (72 h), ABA (3 h), or NaCl (3 h).
(B) RNA hybridization with total RNA (20 μg) from samples treated with cold (0, 6, 24, or 72 h).
(C) Nuclear run-on analysis with 72-h cold-treated samples.
To further examine whether the enhanced luciferase transcript level in sta1-1 is attributable to increased transcription or posttranscriptional changes in stability, nuclear run-on assays were performed. We used samples collected after 72 h of cold treatment, when the dramatic difference between the wild type and sta1-1 in luciferase transcript abundance was observed (Figures 2A and 2B). The nuclear run-on results showed that the sta1-1 mutant did not have higher transcription rates for either the endogenous RD29A or the luciferase transgene than the wild type (Figure 2C). Together, these results suggest that the higher level of luciferase transcript in sta1-1 is the result of enhanced luciferase transcript stability.
Physiological and Developmental Phenotypes of the sta1-1 Mutant
Because of the potential role of mRNA stability control in plant stress responses and development, the sta1-1 mutant was examined for possible stress tolerance and developmental phenotypes.
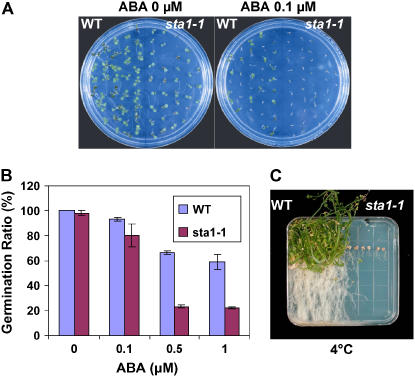
ABA is a stress hormone with inhibitory functions in seed germination and root growth. For the germination test on ABA, we considered germination as the emergence of cotyledons rather than of radicles. In the absence of exogenous ABA, the wild type and sta1-1 germinated completely on MS agar medium with 3% sucrose at 4 d after imbibition. However, when 0.1 μM ABA was added to the medium, the germination rate of sta1-1 seeds at 4 d was reduced to virtually zero (i.e., no cotyledon emergence), whereas ∼60% of the wild-type seeds could still germinate (Figure 3A). Three days later, the wild-type seeds overcame the inhibitory effect of 0.1 μM ABA to reach a germination rate of ∼92%. At that time, the germination rate of sta1-1 seeds was also improved to ∼90% in the presence of 0.1 μM ABA (Figure 3B). sta1-1 seeds, however, still showed a lower germination rate than did wild-type seeds at 0.5 and 1.0 μM ABA even at 7 d after imbibition (Figure 3B).
Figure 3.
sta1-1 Germination and Chilling Sensitivity.
(A) Germination test of the wild type and sta1-1 on MS agar medium with ABA (0 or 0.1 μM). Photographs were taken at 4 d after imbibition.
(B) Germination rates of the wild type and sta1-1 on MS medium with ABA (0, 0.1, 0.5, or 1 μM). Rates were scored at 7 d after imbibition. Data from three replicate experiments are shown. Error bars indicate se.
(C) Chilling sensitivity test of the wild type and sta1-1 at 4°C. Four-day-old seedlings were transferred to 4°C, and the photograph was taken ∼6 months later.
Low-temperature responses of the sta1-1 mutant were tested at 4°C. Four-day-old seedlings grown under normal conditions on a vertical plate were transferred to 4°C under light. Under this cold condition, sta1-1 was clearly damaged and all seedlings were eventually killed, whereas the wild-type seedlings were alive and growing (Figure 3C). These results showed that the sta1-1 mutant is chilling-sensitive.
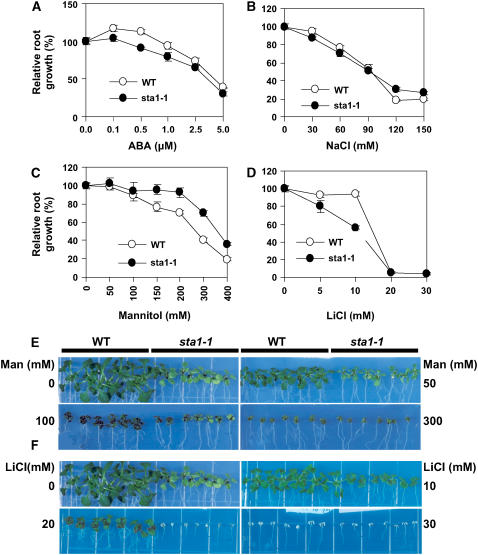
Root growth is affected by various stress conditions and has often been used as an index for stress sensitivity. The root growth of the wild type and the sta1-1 mutant was compared on MS agar medium supplemented with ABA, NaCl, mannitol, or LiCl. With ABA, sta1-1 showed lower relative root growth than the wild type, although the difference became smaller at higher ABA concentrations (Figure 4A). ABA at low concentrations is known to have a stimulatory rather than an inhibitory effect on root growth (Davies and Zhang, 1991). In our analysis, this root growth–promoting effect of ABA was observed at 0.1 to 0.5 μM ABA (Figure 4A). Although both the wild type and sta1-1 responded positively in root growth to 0.1 μM ABA, the positive ABA effect was much smaller in sta1-1 than in the wild type. At 0.5 μM ABA, the root growth of the wild-type seedlings was still promoted, but sta1-1 root growth was inhibited (Figure 4A). This result suggests that sta1-1 mutant seedlings are hypersensitive to ABA inhibition of root growth.
Figure 4.
sta1-1 Sensitivity to Various Salt and Osmotic Stress Conditions.
(A) to (D) Comparisons of the wild type and sta1-1 in root growth on MS agar medium with ABA (A), NaCl (B), mannitol (C), and LiCl (D). Root growth was measured relative to controls. At least eight seedling roots were measured for each data point. Error bars indicate se.
(E) and (F) Comparisons between the wild type and sta1-1 in seedling growth on MS agar medium with mannitol (Man) (E) or LiCl (F). Photographs were taken at 13 d after seedling transfer onto the treatment medium.
All experiments were performed three times except for LiCl treatment (two times) with different seeds lots, and each time nearly identical results were obtained.
The relative root growth rates of the wild type and sta1-1 were not significantly different on NaCl-containing plates, although it appeared that wild-type root growth became more sensitive to high concentrations (120 and 150 mM) than sta1-1 root growth (Figure 4B). Interestingly, the sta1-1 mutant showed an apparently higher level of tolerance in relative root growth to osmotic stress caused by mannitol (Figure 4C). The osmotic stress tolerance of sta1-1 can also be observed at the whole seedling level. The size of the wild-type seedlings on mannitol-containing plates was reduced greatly as the concentration of mannitol increased. By contrast, the size decrease in sta1-1 seedlings by mannitol was relatively small (Figure 4E). Indeed, the appearances of the wild-type and sta1-1 seedlings were very similar at 300 mM mannitol, whereas sta1-1 seedlings were much smaller without mannitol.
Differences between the wild type and sta1-1 were observed under LiCl treatment (Figures 4D and 4F). Approximately 100% of the relative root growth in the wild type was maintained with up to 10 mM LiCl, whereas sta1-1 displayed only ∼55% relative root growth at 10 mM LiCl (Figure 4D). The sta1-1 seedlings were all killed at 20 mM LiCl, whereas the wild-type seedlings were all alive under this condition (Figure 4F).
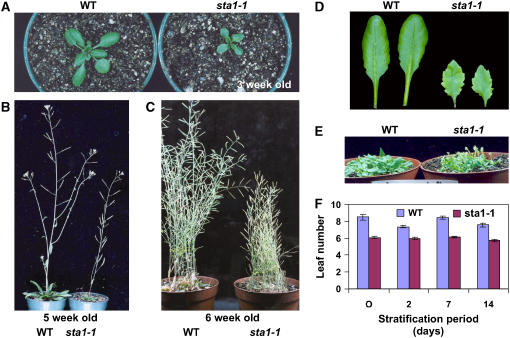
The sta1-1 plants were smaller in size and height than were wild-type plants (Figures 5A to 5C). sta1-1 mutant leaves were also smaller than wild-type leaves. In addition, the mutant leaves were more serrated and had a pointed leaf tip (Figure 5D). Although sta1-1 completed its life cycle at a similar rate as the wild type, the inflorescence of sta1-1 started bolting earlier than that of the wild type (Figures 5E and 5F). sta1-1 plants generally bolted at a leaf number of six, whereas the wild type started to bolt at a leaf number of approximately eight.
Figure 5.
Developmental Phenotypes of sta1-1.
(A) to (C) Morphology of wild-type and sta1-1 plants: 3 weeks old (A), 5 weeks old (B), and 6 weeks old (C).
(D) Comparison of leaf morphology between the wild type and sta1-1.
(E) Early-bolting phenotype of sta1-1.
(F) Leaf number comparison between the wild type and sta1-1 upon bolting after different stratification periods.
STA1 Encodes a Pre-mRNA Splicing Factor
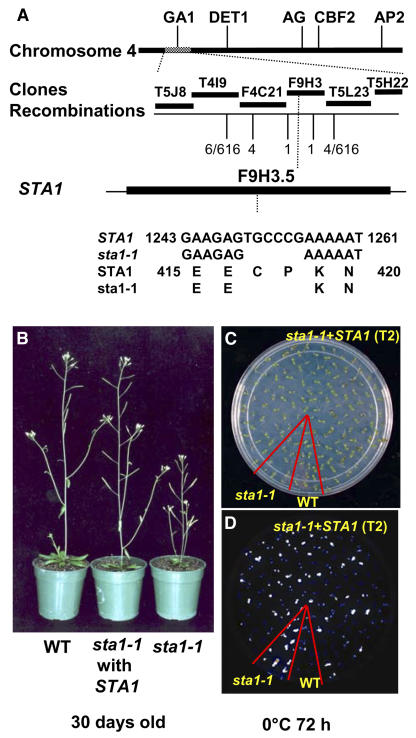
None of the 31 F1 plants derived from a cross between the wild type and sta1-1 showed a mutant phenotype in luminescence or development (see Supplemental Table 1 online). In the successive F2 generation, 23% of the progeny displayed mutant levels of luminescence (intensities of >1 × 106 counts per seedling after 72 h of cold treatment). These results suggest that a single recessive nuclear mutation is responsible for the mutant phenotypes conferred by sta1-1. To clone the mutation, F2 seeds from a cross between wild-type plants in the Landsberg erecta ecotype and sta1-1 were used as a mapping population. Seedlings with high luminescence after 72 h of cold treatment were mapped with known simple sequence length polymorphism (SSLP) markers as well as SSLP markers that were developed in this study. The mutation was limited to a 50-kb region between positions 30 and 80 kb on BAC clone F9H3. Through sequencing of genomic DNA in this region in the sta1-1 mutant, a mutation was found in the F9H3.5 gene (At4g03430) that had a computer annotation of “putative pre-mRNA splicing factor.” In the sta1-1 mutant, 6 bp (1249 to 1254 bp from the translation initiation site) were deleted from At4g03430, which resulted in two amino acid deletions in-frame in the open reading frame (ORF) (Figure 6A).
Figure 6.
Molecular Cloning of STA1 and Functional Complementation.
(A) Positional cloning of STA1. Numbers of recombinations are from 308 F2 progeny seedlings with the phenotype conferred by sta1-1. Markers used at the recombination positions were, from left, T4I9-29K, F4C21-27K, F9H3-80K, F9H3-32K, and F9H3-3K.
(B) Molecular complementation of the sta1-1 developmental defect with the wild-type STA1 gene.
(C) and (D) Molecular complementation of the RD29A-LUC expression defect with the wild type STA1 gene. Shown are seedlings on an MS agar plate (C) and the corresponding luminescence image (D).
To confirm that the correct gene was cloned, a genomic fragment containing 1513 bp upstream of the start codon, the 3090-bp ORF, and 253 bp downstream of the stop codon was introduced into the sta1-1 mutant through Agrobacterium tumefaciens–mediated transformation. Transgenic plants (T1) expressing this genomic fragment did not display sta1-1 developmental phenotypes (Figure 6B), and the subsequent T2 seedlings showed a 3:1 segregation ratio between seedlings with normal luminescence intensity and high luminescence intensity after cold treatment (Figure 6C). These results confirm that At4g03430 is the correct gene responsible for the phenotypes conferred by sta1-1.
The At4g03430 gene does not contain any intron, and its ORF has been confirmed by Yamada et al. (2003). The STA1 ORF is predicted to encode a polypeptide of 1029 amino acids with a molecular mass of ∼115.6 kD. The deduced amino acid sequence of STA1 exhibits a significant degree of overall similarity with human U5 snRNP-associated 102-kD protein (accession number O94906; 53% identity and 69% similarity). STA1 protein is also similar to the fission yeast pre-mRNA splicing factor PRP1p (accession number Q12381) and the budding yeast pre-mRNA splicing factor Prp6p (accession number P19735), with identities of 42 and 31% and similarities of 61 and 48%, respectively. STA1 exists as a single-copy gene in the Arabidopsis genome. We attempted to isolate a homozygous T-DNA knockout allele of sta1-1. The SALK line SALK_009304 contains a T-DNA insertion at 286 bp downstream from the translation initiation site, which likely represents a null allele. However, genotyping of 36 plants identified only wild-type and heterozygous alleles but not homozygous T-DNA mutants. This suggests that the homozygous T-DNA mutant is lethal.
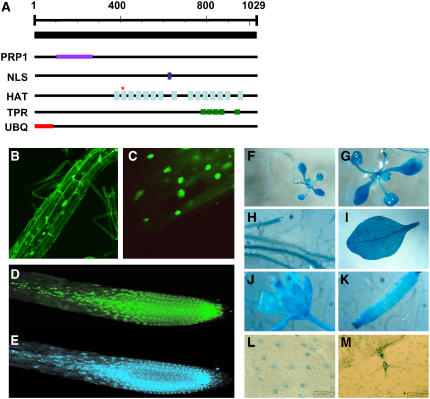
Domain analysis predicted that the STA1 protein has 15 HAT (for half a tetratricopeptide repeat [TPR]) helix domains and 5 TPR domains as well as a PRP1 splicing factor N-terminal domain and a bipartite nuclear localization signal (Figure 7A; see Supplemental Table 2 online). The HAT domain has a sequence and structure similar to the TPR domain and is found in many RNA processing proteins (Preker and Keller, 1998). HAT domains are present in multiple repeats, and it is believed that intramolecular HAT–HAT interaction provides a protein–protein interaction surface. The sta1-1 mutation took place in one of the HAT domains (Figure 7A). Therefore, it is likely that the protein interaction surface provided by the HAT domain is affected by the mutation in STA1. The TPR domain is a degenerate 34–amino acid repeat forming two α-helices and is often arranged in tandem. Proteins with TPR repeats are involved in many cellular events, such as cell cycle control, splicing, transcription, protein folding, and protein transport (Blatch and Lassle, 1999). TPR participates in these activities by mediating protein–protein interactions. Interestingly, the N-terminal 85 amino acid residues in STA1 are predicted to contain a ubiquitin domain by the PROSITE Database of Protein Families and Domains (http://www.expasy.org/prosite) (Figure 7A). This ubiquitin domain was also found in the STA1 homolog in rice (Oryza sativa), but not in humans or yeast, which suggests an evolutionary divergence of the plant protein.
Figure 7.
Characterization of STA1.
(A) Predicted domain in the STA1 protein. The asterisk represents the mutation site in sta1-1. PRP1, PRP1 splicing factor N-terminal domain; NLS, nuclear localization signal; HAT, half a TPR; TPR, tetratricopeptide repeat; UBQ, ubiquitin.
(B) to (D) Confocal microscopic images of an Arabidopsis root expressing the GFP-STA1 fusion protein.
(E) A 4′,6-diamidino-2-phenylindole–stained root corresponding to the root in (D).
(F) to (M) Expression of STA1 promoter–GUS in Arabidopsis. Expression in whole seedlings ([F] and [G]), root (H), leaf (I), flower (J), silique (K), guard cells (L), and trichome (M). For observation of guard cells and trichomes, the epidermal layer was peeled from leaves.
To investigate the subcellular localization of STA1, we generated transgenic Arabidopsis expressing a green fluorescent protein (GFP)–STA1 fusion protein. The green fluorescence was detected in nuclei, which suggests a nuclear localization of the STA1 protein (Figures 7B to 7E). This observation is consistent with the presence of a nuclear localization signal in the STA1 amino acid sequence (Figure 7A; see Supplemental Table 2 online).
To determine the tissue distribution of STA1 expression, an ∼1.5-kb sequence upstream of the STA1 initiation codon was amplified by PCR and used to drive the expression of the β-d-glucuronidase (GUS) reporter gene. STA1 promoter–GUS transgenic Arabidopsis plants were assayed to detect GUS expression. The GUS reporter gene was expressed in all tissues tested, although leaf epidermal cells did not seem to have strong expression. In leaf epidermis, GUS staining was detected preferentially in guard cells and trichomes. The STA1 promoter–GUS expression results suggest a largely ubiquitous expression pattern of STA1 (Figures 7F to 7M).
STA1 Is Stress-Inducible and Required for Both Pre-mRNA Splicing and mRNA Turnover
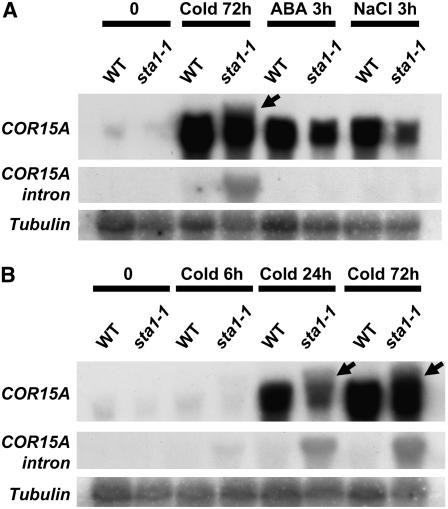
The notion that STA1 is a pre-mRNA splicing factor and that the sta1-1 mutant may be defective in pre-mRNA splicing is supported by experimental evidence. Results of RNA gel blot analysis revealed an additional, slightly higher band when the COR15A gene was used as a probe (Figures 8A and 8B). This higher band was present only in cold stress–treated sta1-1 plants. The size of the higher band appeared to be the same as that of the unspliced COR15A transcript. To test this notion, we PCR-amplified the intron present in the COR15A ORF and labeled this fragment as a probe for RNA gel blot analysis. As expected, the intron probe detected a signal only in cold stress–treated sta1-1 plants, and the size of the signal was the same as that of the upper band detected by the COR15A cDNA (Figures 8A and 8B). These results demonstrate that the sta1-1 mutant is indeed defective in pre-mRNA splicing.
Figure 8.
COR15A Expression in sta1-1.
Total RNA (20 μg) from 14-d-old seedlings after cold, ABA, or NaCl treatment (A) or different cold stress durations (B) was subjected to RNA hybridization with the probes shown. Arrows indicate COR15A nonspliced transcript.
It is interesting that the COR15A splicing defect occurred only under cold stress conditions, even though COR15A was also induced by ABA or NaCl (Figure 8A). The preferential splicing defect under cold stress and the increased cold sensitivity of the sta1-1 mutant prompted us to test whether STA1 might be preferentially needed under cold stress and thus that its expression might be upregulated by cold. Indeed, we found that the STA1 transcript level is upregulated by cold stress but not by ABA or NaCl (Figures 9A and 9B). Surprisingly, we found that the cold-induced STA1 transcript level was substantially higher in sta1-1 than in the wild type (Figures 9A and 9B). Results of nuclear run-on assays revealed no substantial difference in STA1 transcription rates between wild-type and sta1-1 plants (Figure 9C). Thus, the STA1 transcript is highly unstable, because it was not detectable without cold stress by RNA gel blot analysis, even though the STA1 promoter has strong constitutive activities (Figure 7). Therefore, the sta1-1 mutation causes the stabilization of the normally unstable STA1 transcript. It should be noted that our STA1 promoter–driven GUS expression construct does not contain a 21-bp sequence immediately upstream of the start codon or the 3′ untranslated region of STA1, which might be important in posttranscriptional regulation of STA1 expression. Thus, it is possible that STA1 gene expression may not be constitutive or ubiquitous.
Figure 9.
Expression of STF and STA1 in the Wild Type and sta1-1.
(A) and (B) RNA gel blot analysis of wild-type and sta1-1 total RNA (20 μg) with STA1 and STF probes after different treatments (A) or different cold durations (B).
(C) and (D) Nuclear run-on analysis with samples after 72 h of cold treatment. STA1 (C) and STF (D) were analyzed.
To identify other endogenous genes with enhanced transcript stability in the sta1-1 mutant, full genome microarray analysis was performed with the use of Affymetrix 24K GeneChips. Total RNA extracted from 14-d-old seedlings of the wild type and the sta1-1 mutant grown under normal conditions was used for the transcript profiling. After statistical analysis, we found that the transcript levels of 71 genes were significantly (P ≤ 0.05) higher by at least twofold in sta1-1 than in the wild type (see Supplemental Table 3 online). The STA1 gene itself was not included in our list of 71 genes, probably because our microarray analysis was performed with seedlings not under cold treatment. However, the microarray result still indicated that the level of the SAT1 transcript was ∼1.97 times higher, with a P value of 0.016, in sta1-1 than in the wild type grown under normal conditions. One of the 71 genes, steroid sulfotransferase (STF; At2g03760), which was determined to have a high transcript level in sta1-1 by the microarray assay, was tested by RNA gel blot analysis. STF was found to be strongly upregulated by cold and NaCl stress and slightly upregulated by ABA (Figures 9A and 9B). Consistent with the microarray result, RNA gel blot analysis showed that the STF transcript level was higher in sta1-1 than in the wild type, particularly after 72 h of cold stress (Figures 9A and 9B). It is noteworthy that under NaCl stress, the STF transcript level was only slightly higher in sta1-1 than in the wild type (Figure 9A). This finding is consistent with the enhanced requirement for STA1 in facilitating transcript turnover under cold stress. To investigate whether the higher STF transcript level under cold stress is also attributable to transcript stabilization, we performed nuclear run-on assays, which revealed no substantial difference in the transcription rate for STF between the wild type and the sta1-1 mutant (Figure 9D). Therefore, the higher level of STF transcript in the mutant appears to be also caused by enhanced transcript stability.
DISCUSSION
In this study, we used a genetic approach to identify a novel factor important in mRNA turnover. The recessive sta1-1 mutation causes the stabilization of not only the firefly luciferase reporter gene transcript but also transcripts from at least two endogenous genes (STA1 itself and STF). Interestingly, STA1 encodes a pre-mRNA splicing factor. Indeed, sta1-1 mutant plants are defective in the splicing of the COR15A gene. Our work thus identifies a cellular factor required for both transcript turnover and RNA splicing. Furthermore, we found that STA1 expression is upregulated by cold stress, and the gene appears to be essential under cold stress conditions.
Pre-mRNA splicing is an indispensable process for removing introns from pre-mRNA for proper gene expression in eukaryotic cells and is performed by the spliceosome, a multicomponent complex of small nuclear RNAs and many protein factors (Jurica and Moore, 2003). Small nuclear RNAs include U1, U2, U4, U5, and U6, and each constitutes a snRNP with several protein subunits. Although the spliceosome is made up of ∼300 polypeptides that include many other proteins (Rappsilber et al., 2002; Zhou et al., 2002), these snRNPs form the core. The remaining non-snRNP protein factors are known to participate in recruiting the core splicing machinery and/or connecting splicing to other processes, such as transcription, 5′ end capping, and 3′ end cleavage/polyadenylation (Proudfoot et al., 2002). During the splicing processes, the structures and components of the snRNP complexes change dynamically. For example, during the spliceosome activation process, U5 snRNP undergoes a dramatic remodeling by tightly associating with SKIP (for Ski Oncogene–Interacting Protein), the Prp19 complex, and other factors in exchange for other U5 snRNP–associated proteins, such as 15- and 100-kD proteins (Makarov et al., 2002). The newly remodeled 35S U5 snRNPs persist throughout the splicing catalytic processes until dissociation from mRNA. After the dissociation, 35S U5 snRNP is converted into 20S U5 snRNP, an abundant form of U5 snRNP.
The sequence of STA1 suggests that it is a U5 snRNP–associated protein. It has high similarities to human U5 snRNP–associated 102-kD protein (accession number O94906), fission yeast pre-mRNA splicing factor Prp1p (accession number Q12381), and budding yeast pre-mRNA splicing factor Prp6p (accession number P19735). In budding yeast, Prp6p mediates the interactions between U4/U6 snRNP and U5 snRNP. U4/U6 snRNP and U5 snRNP form a tri-snRNP before being integrated into the spliceosome. Galisson and Legrain (1993) showed that the U4/U6-U5 tri-snRNP did not accumulate in the prp6 mutant. Instead, the U4/U6 snRNP and U5 snRNP were present separately in the mutant, which suggests a Prp6p requirement in U4/U6-U5 tri-snRNP formation. The U5 snRNP–associated 102-kD protein, human Prp6p homolog, is also thought to bridge U4/U6 snRNP and U5 snRNP through specific interaction with the U4/U6 snRNP–associated 61-kD protein (Makarov et al., 2000; Makarova et al., 2002). Thus, it is likely that the STA1 gene product has a similar function in plant U4/U6-U5 tri-snRNP formation. However, we found that STA1 was not capable of complementing the temperature-sensitive growth phenotype of the prp6-1 yeast mutant Se1 (Urushiyama et al., 1997) (data not shown). Nevertheless, the in vivo splicing defect in sta1-1, together with the strong sequence similarity between STA1 and the human and yeast proteins, support STA1 as a bona fide splicing factor. The inability of STA1 to complement the yeast mutant is likely attributable to sequence and possibly functional divergence of the plant protein. The N-terminal region of STA1 has a ubiquitin domain not observed in nonplant proteins. Interestingly, the PatchCalling database, which provides data on global protein–protein interactions by use of industrialized, high-throughput yeast two-hybrid technology (Uetz et al., 2000; http://portal.curagen.com/pathcalling_portal/index.htm), contained an interaction between Prp6p and Ubc9p, a ubiquitin-conjugating enzyme in budding yeast. This suggests that ubiquitin conjugation may also be required for the proper function of STA1 homologs in nonplant systems. STA1 might have acquired the ubiquitin moiety as an intramolecular domain during evolution.
It is interesting to speculate on how the sta1-1 mutation enhances transcript stability or how STA1 normally promotes transcript turnover. Evidence suggests that posttranscriptional processes such as RNA processing, export, translational regulation, and degradation are interconnected. In addition, RNA processing events, including splicing, can be coupled to transcription in higher eukaryotes (Proudfoot et al., 2002; Jensen et al., 2003). As RNA emerges from transcription, it is packaged into a messenger ribonucleoprotein complex. Failure to form proper messenger ribonucleoprotein may lead to retention by a nuclear surveillance system, resulting in mRNA degradation. We suggest that STA1 and possibly the entire spliceosome may be part of the nuclear surveillance system that recognizes and degrades certain transcripts before they exit the nucleus. It is not known what primary or secondary sequence features are recognized by this surveillance system. It is interesting that all three transcripts (luciferase, STA1, and STF) confirmed to be stabilized in sta1-1 do not have introns. However, many of the 71 genes showing enhanced transcript levels in the mutant do have introns. We do not know whether the intronless genes are directly or indirectly affected by the sta1-1 mutation.
Among the 71 genes with high expression levels in the sta1-1 mutant are two spliceosomal proteins. They include proteins that are homologous with U5 snRNP–associated 200-kD protein (At2g42270) and U4/U6 snRNP–associated 90-kD Prp3 protein (At1g28060). Results of many studies showed these proteins to be present in the spliceosome, along with U5 snRNP–associated 102-kD protein, the human counterpart of STA1 (Anthony et al., 1997; Jurica et al., 2002; Makarov et al., 2002; Rappsilber et al., 2002; Zhou et al., 2002). Therefore, the higher expression of one U5 snRNP–associated protein gene and one U4/U6 snRNP–associated protein gene in the sta1-1 mutant might imply a cellular mechanism to compensate for the defect in STA1. Complementation of defects in the splicing machinery by overexpressing other components is not surprising. The addition of a high amount of SR proteins, auxiliary splicing factors rich in Arg-Ser (RS) dipeptides, compensated for the loss of U1 snRNP function or U2AF depletion in an in vitro splicing system (Crispino et al., 1994; Tarn and Steitz, 1994; MacMillan et al., 1997). A cellular compensation system for sta1-1 defects could possibly affect RNA processing, which in turn may have resulted in the enhanced transcript stability of certain genes in the sta1-1 mutant. Recently, UBP1, a U-rich intron and 3′ untranslated region binding hnRNP, was cloned and characterized in tobacco (Nicotiana tabacum) (Lambermon et al., 2000). UBP1 overexpression led to the enhanced stability of certain transcripts. UBP1 can bind the 3′ untranslated region of mRNAs and thus may protect the mRNAs from exonucleolytic degradation (Lambermon et al., 2000). Interestingly, UBP1-enhanced transcript stability was observed only for intronless gene transcripts or less efficiently spliced intron-containing transcripts (Lambermon et al., 2000).
The function of STA1 appears to be essential in Arabidopsis, because sta1 null mutants are lethal. Yeast prp6 null mutants also appear to be lethal, as indicated in the Saccharomyces Genome Database (http://www.yeastgenome.org/) (Giaever et al., 2002). It is likely that the in-frame deletion of two amino acids in sta1-1 represents a weak mutant allele. Promoter-GUS analyses suggest that STA1 expression may be constitutive. Indeed, the sta1-1 mutant exhibits a phenotypic defect in the absence of cold or other stress, which also suggests that sta1-1 is not strictly a temperature-sensitive allele. However, the mutant defect is most severe under cold stress, as indicated by the malsplicing of COR15A in the cold and the dramatic chilling sensitivity of the mutant plants. The cold stress phenotypes of sta1-1 plants suggest an important requirement of STA1 under cold stress. This requirement is reflected in the cold stress upregulation of STA1 expression. Nevertheless, it is still possible that the sta1-1 allele may be sensitive to cold and thus that cold temperature may exacerbate the mutant defect by causing a more severe defect in the mutant protein.
sta1-1 plants are also altered in their responses to ABA and salt stress. The mutant is hypersensitive to ABA in germination and root growth, as are the phenotypes of abh1 and sad1, both of which affect RNA metabolism (Hugouvieux et al., 2001; Xiong et al., 2001a). In addition, sta1-1 plants show a serrated leaf phenotype that is also observed in the abh1, although sta1-1 but not abh1 is smaller and bolts early (Hugouvieux et al., 2002). However, the ABA hypersensitivity in sta1-1 is not as strong as that in abh1 or sad1. Unlike the abh1 or sad1 mutant, sta1-1 does not have a detectable phenotype in stomatal opening or closing (data not shown), even though the STA1 promoter–GUS is expressed in guard cells. The weak existence or absence of certain ABA phenotypes in sta1-1 plants may be because the mutation (an in-frame deletion of two amino acid residues) causes only a partial loss of function under these conditions. sta1-1 plants are also hypersensitive to LiCl (Figures 4D and 4F). This phenotype is consistent with the defect of sta1-1 in pre-mRNA splicing, as LiCl is well known for its inhibitory effect on RNA-processing enzymes (Dichtl et al., 1997). A recently identified ABA receptor, FCA, is a nuclear RNA binding protein that regulates flowering in response to ABA (Razem et al., 2006). Associated with FY, FCA autoregulates its own mRNA by promoting premature cleavage and polyadenylation (Macknight et al., 2002; Quesada et al., 2003; Simpson et al., 2003). Thus, future investigations of the potential involvement of STA1 in FCA-mediated ABA signaling would be interesting.
METHODS
Plant Materials and Growth Conditions
RD29A-LUC–expressing Arabidopsis thaliana ecotype Columbia gl1 (referred to here as the wild type) plants were mutated by ethyl methanesulfonate to generate M2 seeds. Surface-sterilized M2 seeds were plated on MS (Murashige and Skoog salt base; JRH Biosciences) agar (0.6%) plates supplemented with 3% sucrose and placed at room temperature (22 ± 1°C) under continuous light after 2 to 3 d of cold stratification. Seven-day-old seedlings were used to screen for altered LUC expression in response to low temperature, ABA, or NaCl treatment with the use of a video-imaging system consisting of a charge-coupled device camera (CCD-512SB; Princeton Instruments), a controller (Princeton Instruments), and a computer with WinView image-processing software, as described previously (Chinnusamy et al., 2002; Lee et al., 2002). When necessary, seedlings were transferred to soil pots and allowed to grow in a growth chamber with cycles of 16 h of light at 22°C and 8 h of dark at 18°C.
Physiological Characterization
Stresses were applied to 1-week-old wild-type and mutant seedlings grown on the same MS agar plate. For cold treatment, the plates were placed at 0°C in the dark for the designated times. For ABA treatment, 100 μM ABA [(±)-cis,trans-ABA; Sigma-Aldrich] dissolved in sterile water was sprayed uniformly on the leaves of the seedlings. ABA-treated plates were kept at room temperature (22 ± 1°C) under cool-white light for the designated times. For NaCl treatment, seedlings were transferred to a filter paper saturated with 300 mM NaCl in MS solution. The seedlings were then incubated under light at room temperature for the designated times.
For germination tests, surface-sterilized seeds were placed on MS agar (0.6%) plates supplemented with ABA at the designated concentrations. The plates were cold-treated for 2 d at 4°C to promote uniform germination. Seven days later, germination was scored. Cotyledon appearance was considered to be germination.
For growth analysis, 4-d-old seedlings grown vertically on MS agar (1.2%) square plates were transferred onto MS agar (1.2%) supplemented with various salts. Root length was measured 4 d later, and seedling growth phenotypes were examined 13 d later. For chilling tests, 4-d-old seedlings grown at room temperature on MS agar (1.2%) square plates were transferred to 4°C under continuous light conditions. Phenotypes were then monitored.
Gene Expression Analysis
Nine-day-old seedlings grown on MS agar plates were used for RNA gel blot analysis. After stress treatments as described in the text, total RNA was extracted and analyzed as described previously (Liu and Zhu, 1997).
For nuclear run-on analysis, nuclei were isolated from 2-week-old seedlings treated with cold (0°C) for 72 h. The nuclei isolation and in vitro transcription reactions were performed as described (Dorweiler et al., 2000). Comparable amounts of labeled RNA for treated and untreated samples were used for filter hybridization. Slot blots on nitrocellulose filter membranes were prepared with 100 ng of denatured purified gene fragments or an equivalent amount of denatured linearized plasmid per slot. For comparison, two to three slots were used for each probe. Prehybridization and hybridization were performed as described by Dorweiler et al. (2000). After hybridization, the strips were washed for 15 min with 5× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS at 42°C and then with 2× SSC and 0.1% SDS for 15 min at room temperature. The strips were visualized with the use of a STORM 860 PhosphorImager (Molecular Dynamics), and signals were quantified with ImageQuant software (Molecular Dynamics). Background was subtracted from each signal before normalizing the probe to the signals for control.
Probes used for both RNA hybridization analysis and nuclear run-on analysis were as follows: the RD29A gene–specific probe from the 3′ noncoding region (Liu and Zhu, 1997); the COR15A cDNA (Gilmour et al., 1992; Lin and Thomashow, 1992), kindly provided by M.F. Thomashow; and the COR15A intron fragment amplified from Arabidopsis genomic DNA by use of the primer pair COR15AI-F (5′-AAGGATCTTAGCAGGCAATGTT-3′) and COR15AI-R (5′-CAAAGGTTTCAAAACACATATCCA-3′). Full-length coding regions of both luciferase and STA1 were used to detect each transcript signal. STF (At2g03760), NPT II, and tubulin probe fragments were amplified from plasmid construct, cDNA clone, or genomic DNA by PCR with the following primer pairs: STF-F (5′-TGAAGCTAAAGATTCCGACATTATC-3′) and STF-R (5′-AGTATCTCTCCATCCTCCAATCTCT-3′); NPT II-F (5′-ATGACTGGGCACAACAGACA-3′) and NPT II-R (5′-AATATCACGGGTAGCCAACG-3′); and Tubulin-F (5′-CGTGGATCACAGCAATACAGAGCC-3′) and Tubulin-R (5′-CCTCCTGCACTTCCACTTCGTCTTC-3′).
For Affymetrix GeneChip array analysis, 20 μg of total RNA from wild-type and sta1-1 seedlings grown for 14 d at 22°C with a cycle of 16 h of light and 8 h of darkness was extracted by use of the RNeasy plant mini kit (Qiagen) and used to make biotin-labeled complementary RNA targets. Affymetrix Arabidopsis ATH1 genome array GeneChips, which contain >22,500 probe sets representing ∼24,000 genes, were used, and hybridization, washing, and staining were performed at the Genetic Analysis and Technology Core Facility at the University of Arizona. The microarray assay included data sets from five biological replicates of wild-type plants and two biological replicates of the sta1-1 mutant, and these seven data sets were used for statistical analysis to determine genes with higher transcript levels in sta1-1. Expression measures from Affymetrix cell intensity files were background-corrected, normalized, and summarized using the robust multiarray average algorithm from the affy package of Bioconductor (Irizarry et al., 2003; Gentleman et al., 2004). Differentially expressed genes were identified by statistical analysis implemented in the LIMMA package of Bioconductor (Gentleman et al., 2004; Smyth, 2004). Through this analysis, the P values were obtained from the distribution of the moderated t statistic, the ratio of the log2(fold change between the wild type and sta1-1) to its se, and were corrected for multiple testing according to Benjamini and Hochberg (1995). Genes with at least a twofold higher transcript level than the wild type and a false discovery rate–adjusted P value of <0.05 were considered to be genes with significantly higher transcript levels in sta1-1.
Positional Cloning
For genetic mapping of the sta1-1 mutation, sta1-1 in the Columbia ecotype was crossed with the wild type in ecotype Landsberg erecta. The resulting F1 plants were allowed to self, and the F2 seeds were collected. Homozygous sta1-1 mutants in the segregating F2 population were selected on the basis of their high luminescence under stress conditions. Mapping of the mutation was performed with SSLP markers (Bell and Ecker, 1994). For fine mapping, new SSLP markers were developed with the use of the Cereon Arabidopsis polymorphism collection at http://www.Arabidopsis.org/Cereon/index.html. Primers for SSLP markers were as follows: T4I9-29K (5′-TTGATCGATCGTCTCGTATTTC-3′ and 5′-TTGGCCATTACTTTGGATCA-3′), F4C21-27K (5′-GCTCGTGACGTGGCTATCTT-3′ and 5′-TGGGGGTCAAAACTCAAAAC-3′), F9H3-80K (5′-GATCGGAAAACCAGAAACGA-3′ and 5′-TTTCCGGCAAAATTGTAACAG-3′), F9H3-32K (5′-CCGTTACACATAATAAAGGGTTTTC-3′ and 5′-CGTTACTAATGGATTTAGAGTGAGTGA-3′), and F9H3-3K (5′-GTAGGTCCCCAGCCTTGATT-3′ and 5′-TTGAAAACTGCTGACGGAGA-3′).
Plasmid Construction and Plant Transformation
The F9H3 BAC clone was obtained from the ABRC and used as a PCR template. For sta1-1 complementation, the 4954-bp genomic DNA fragment of STA1 covering from 1513 bp upstream of the start codon to 253 bp downstream of the stop codon was amplified with LA Taq polymerase (Takara Shuzo), with F9H3 BAC DNA used as a template with the following primers: F9H3.5gKpnI-F (5′-TGTTGGTACCCTTATTGTAGCAATACTTGTTCTTA-3′) and F9H3.5gSalI-R (5′-TACAGTCGACAAAAGAAGTTTAATAGCTGAACA-3′). The resulting fragment was cloned into pCAMBIA1200 between the KpnI and SalI sites, generating pCAM1200-HC15.
For the STA1 promoter–driven GUS construct, a 1493-bp fragment spanning from −1513 to ∼−21 bp upstream of the STA1 ORF was amplified by PCR, with F9H3 BAC DNA used as a template and the primer pair F9H3.5pSalI-F (5′-GTTGGTCGACTTATTGTAGCAATACTTGTTCTTA-3′) and F9H3.5pNcoI-R (5′-CCGGTCCATGGAACCAAACTATAAAAATCTCT-3′). The STA1 promoter fragment was then cloned into pCAMBIA1381 between the SalI and NcoI sites, resulting in pCAM1381-HC15-GUS.
For the construct for the GFP-STA1 fusion protein, the STA1 ORF was amplified by PCR with F9H3 BAC DNA used as a template and the primer pair F9H3.5gSalI-F (5′-GATTAGGTCGACATGGTGTTTCTCTCGATTCCAAAC-3′) and F9H3.5gXmaI-R (5′-ATTGATCCCGGGCAGCAGAATTCTCTTCCTTGCTCAA-3′). The amplified STA1 ORF was subcloned into pEZTNL between the XhoI and XmaI sites, resulting in pEZTNL-HC15-GFP.
pCAM1200-HC15 and pCAM1381-HC15-GUS were transferred to Agrobacterium tumefaciens GV3101 (pMP90), and pEZTNL-HC15-GFP was transferred to A. tumefaciens LBA4404, by electroporation at 1250 V with capacitance of 25 μF and resistance of 400 Ω. After appropriate antibiotic selection and PCR confirmation, selected agrobacteria were grown at 28°C in Luria-Bertani broth (1% [w/v] bacto-tryptone, 0.5% [w/v] bacto-yeast extract, and 1% [w/v] NaCl, pH 7.0) overnight and then used for in planta floral infiltration.
GUS Staining
Hygromycin-resistant, STA1 promoter–GUS transgenic Arabidopsis seedlings and plant parts (T1 generation) were stained in GUS assay buffer (3 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, 0.1 M Na-phosphate, pH 7, 0.1% Triton X-100, and 8 mM β-mercaptoethanol) for 12 h at 37°C, followed by incubation in 70% ethanol to remove chlorophyll.
Microscopy
Glufosinate-ammonium–resistant GFP-STA1 transgenic seedlings selected in soil by spraying 30 mg/L Finale (AgrEvo Environmental Health) were mounted on glass slides, and green fluorescence images were taken with use of a Bio-Rad MRC1024 confocal laser-scanning microscope with a 488-nm excitation laser and a 522/DF35 emission filter.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative data library under accession numbers At2g03760 (STF), At2G42540 (COR15A), At4g03430 (STA1), and At5G52310 (RD29A) and in the GenBank/EMBL data libraries under accession numbers O94906 (human U5 snRNP–associated 102-kD protein), Q12381 (fission yeast pre-mRNA splicing factor Prp1p), and P19735 (budding yeast pre-mRNA splicing factor Prp6p). Microarray data are available from the Gene Expression Omnibus data repository (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE4662.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Genetic Analysis of the sta1-1 Mutant.
Supplemental Table 2. Predicted Domains in the STA1 Protein.
Supplemental Table 3. List of Genes with Expression Levels Significantly Higher by at Least Twofold in sta1-1 Than in the Wild Type.
Supplementary Material
Acknowledgments
We thank Tokio Tani for the gift of the prp6-1 yeast mutant, Sohail Khan for microarray statistical analysis, and Rebecca Stevenson for excellent technical assistance. This work was supported by National Institutes of Health Grant R01GM-0707501, U.S. Department of Agriculture National Research Initiative Grant 2003-00751, and National Science Foundation Grants IBN-0212346 and MCB-0241450 to J.-K.Z.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jian-Kang Zhu (jian-kang.zhu@ucr.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.042184.
References
- Abler, M.L., and Green, P.J. (1996). Control of mRNA stability in higher plants. Plant Mol. Biol. 32 63–78. [DOI] [PubMed] [Google Scholar]
- Anthony, J.G., Weidenhammer, E.M., and Woolford, J.L. (1997). The yeast Prp3 protein is a U4/U6 snRNP protein necessary for integrity of the U4/U6 snRNP and the U4/U6.U5 tri-snRNP. RNA 3 1143–1152. [PMC free article] [PubMed] [Google Scholar]
- Bartel, B., and Bartel, D.P. (2003). MicroRNAs: At the root of plant development? Plant Physiol. 132 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57 289–300. [Google Scholar]
- Blatch, G.L., and Lassle, M. (1999). The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. Bioessays 21 932–939. [DOI] [PubMed] [Google Scholar]
- Caponigro, G., and Parker, R. (1996). Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev. 60 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301 336–338. [DOI] [PubMed] [Google Scholar]
- Chinnusamy, V., Stevenson, B., Lee, B.-h., and Zhu, J.-K. (2002). Screening for gene regulation mutants by bioluminescence imaging. Science's STKE, http://stke.sciencemag.org/cgi/content/full/sigtrans;2002/140/pl10. [DOI] [PubMed]
- Crispino, J.D., Blencowe, B.J., and Sharp, P.A. (1994). Complementation by SR proteins of pre-messenger RNA splicing reactions depleted of U1 snRNP. Science 265 1866–1869. [DOI] [PubMed] [Google Scholar]
- Davies, W.J., and Zhang, J.H. (1991). Root signals and the regulation of growth and development of plants in drying soil. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42 55–76. [Google Scholar]
- Dichtl, B., Stevens, A., and Tollervey, D. (1997). Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 16 7184–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorweiler, J.E., Carey, C.C., Kubo, K.M., Hollick, J.B., Kermicle, J.L., and Chandler, V.L. (2000). mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12 2101–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forment, J., Naranjo, M.A., Roldan, M., Serrano, R., and Vicente, O. (2002). Expression of Arabidopsis SR-like splicing proteins confers salt tolerance to yeast and transgenic plants. Plant J. 30 511–519. [DOI] [PubMed] [Google Scholar]
- Galisson, F., and Legrain, P. (1993). The biochemical defects of prp4-1 and prp6-1 yeast splicing mutants reveal that the PRP6 protein is required for the accumulation of the U4/U6.U5 tri-snRNP. Nucleic Acids Res. 21 1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman, R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever, G., et al. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 387–391. [DOI] [PubMed] [Google Scholar]
- Gilmour, S.J., Artus, N.N., and Thomashow, M.F. (1992). cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol. Biol. 18 13–21. [DOI] [PubMed] [Google Scholar]
- Gong, Z.H., Morales-Ruiz, T., Ariza, R.R., Roldan-Arjona, T., David, L., and Zhu, J.-K. (2002. a). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111 803–814. [DOI] [PubMed] [Google Scholar]
- Gong, Z.Z., Lee, H., Xiong, L., Jagendorf, A., Stevenson, B., and Zhu, J.-K. (2002. b). RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. USA 99 11507–11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404 293–296. [DOI] [PubMed] [Google Scholar]
- Han, M.-H., Goud, S., Song, L., and Fedoroff, N. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 101 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren, P., and Parker, R. (1999). mRNA surveillance in eukaryotes: Kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA 5 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux, V., Kwak, J.M., and Schroeder, J.I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106 477–487. [DOI] [PubMed] [Google Scholar]
- Hugouvieux, V., Murata, Y., Young, J.J., Kwak, J.M., Mackesy, D.Z., and Schroeder, J.I. (2002). Localization, ion channel regulation, and genetic interactions during abscisic acid signaling of the nuclear mRNA cap-binding protein, ABH1. Plant Physiol. 130 1276–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner, G., and Zamore, P.D. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science 297 2056–2060. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., Hobbs, B., Collin, F., Beazer-Barclay, Y.D., Antonellis, K.J., Scherf, U., and Speed, T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264. [DOI] [PubMed] [Google Scholar]
- Ishitani, M., Xiong, L., Lee, H., Stevenson, B., and Zhu, J.-K. (1998). HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell 10 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, T.H., Dower, K., Libri, D., and Rosbash, M. (2003). Early formation of mRNP: License for export or quality control? Mol. Cell 11 1129–1138. [DOI] [PubMed] [Google Scholar]
- Johnson, M.A., Perez-Amador, M.A., Lidder, P., and Green, P.J. (2000). Mutants of Arabidopsis defective in a sequence-specific mRNA degradation pathway. Proc. Natl. Acad. Sci. USA 97 13991–13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica, M.S., Licklider, L.J., Gygi, S.R., Grigorieff, N., and Moore, M.J. (2002). Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica, M.S., and Moore, M.J. (2003). Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell 12 5–14. [DOI] [PubMed] [Google Scholar]
- Kuhn, J.M., and Schroeder, J.I. (2003). Impacts of altered RNA metabolism on abscisic acid signaling. Curr. Opin. Plant Biol. 6 463–469. [DOI] [PubMed] [Google Scholar]
- Lambermon, M.H.L., Simpson, G.G., Kirk, D.A.W., Hemmings-Mieszczak, M., Klahre, U., and Filipowicz, W. (2000). UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J. 19 1638–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.-h., Stevenson, B., and Zhu, J.-K. (2002). High-throughput screening of Arabidopsis mutants with deregulated stress-responsive luciferase gene expression using a CCD camera. In Luminescence Biotechnology: Instruments and Applications, K. Van Dyke, C. Van Dyke, and K. Woodfork, eds (Boca Raton, FL: CRC Press), pp. 557–564.
- Lee, H., Xiong, L., Gong, Z.Z., Ishitani, M., Stevenson, B., and Zhu, J.-K. (2001). The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 15 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.X., Kinoshita, T., Pandey, S., Ng, C.K.Y., Gygi, S.P., Shimazaki, K., and Assmann, S.M. (2002). Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature 418 793–797. [DOI] [PubMed] [Google Scholar]
- Lin, C.T., and Thomashow, M.F. (1992). DNA sequence analysis of a complementary DNA for cold-regulated Arabidopsis gene Cor15 and characterization of the Cor15 polypeptide. Plant Physiol. 99 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and Zhu, J.-K. (1997). Proline accumulation and salt-stress-induced gene expression in a salt-hypersensitive mutant of Arabidopsis. Plant Physiol. 114 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., and Fedoroff, N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12 2351–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight, R., Duroux, M., Laurie, R., Dijkwel, P., Simpson, G., and Dean, C. (2002). Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan, A.M., McCaw, P.S., Crispino, J.D., and Sharp, P.A. (1997). SC35-mediated reconstitution of splicing in U2AF-depleted nuclear extract. Proc. Natl. Acad. Sci. USA 94 133–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov, E.M., Makarova, O.V., Achsel, T., and Luhrmann, R. (2000). The human homologue of the yeast splicing factor Prp6p contains multiple TPR elements and is stably associated with the U5 snRNP via protein-protein interactions. J. Mol. Biol. 298 567–575. [DOI] [PubMed] [Google Scholar]
- Makarov, E.M., Makarova, O.V., Urlaub, H., Gentzel, M., Will, C.L., Wilm, M., and Luhrmann, R. (2002). Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298 2205–2208. [DOI] [PubMed] [Google Scholar]
- Makarova, O.V., Makarov, E.M., Liu, S.B., Vornlocher, H.P., and Luhrmann, R. (2002). Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 21 1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi, M., Taylor, C.B., Newman, T.C., and Green, P.J. (1993). The effect of sequences with high AU content on mRNA stability in tobacco. Proc. Natl. Acad. Sci. USA 90 11811–11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker, P.J., and Keller, W. (1998). The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem. Sci. 23 15–16. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. (2002). Integrating mRNA processing with transcription. Cell 108 501–512. [DOI] [PubMed] [Google Scholar]
- Quesada, V., Macknight, R., Dean, C., and Simpson, G.G. (2003). Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J. 22 3142–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber, J., Ryder, U., Lamond, A.I., and Mann, M. (2002). Large-scale proteomic analysis of the human spliceosome. Genome Res. 12 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razem, F.A., El-Kereamy, A., Abrams, S.R., and Hill, R.D. (2006). The RNA-binding protein FCA is an abscisic acid receptor. Nature 439 290–294. [DOI] [PubMed] [Google Scholar]
- Shi, H.Z., Lee, B.-h., Wu, S.J., and Zhu, J.-K. (2003). Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 21 81–85. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., Dijkwel, P.P., Quesada, V., Henderson, I., and Dean, C. (2003). FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113 777–787. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 1–25. [DOI] [PubMed] [Google Scholar]
- Tarn, W.Y., and Steitz, J.A. (1994). SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 8 2704–2717. [DOI] [PubMed] [Google Scholar]
- Uetz, P., et al. (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403 623–627. [DOI] [PubMed] [Google Scholar]
- Urushiyama, S., Tani, T., and Ohshima, Y. (1997). The prp1(+) gene required for pre-mRNA splicing in Schizosaccharomyces pombe encodes a protein that contains TPR motifs and is similar to Prp6p of budding yeast. Genetics 147 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O. (2002). RNA silencing: Small RNAs as ubiquitous regulators of gene expression. Curr. Opin. Plant Biol. 5 444–451. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Gong, Z.Z., Rock, C.D., Subramanian, S., Guo, Y., Xu, W.Y., Galbraith, D., and Zhu, J.-K. (2001. a). Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell 1 771–781. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Ishitani, M., Lee, H., and Zhu, J.-K. (2001. b). The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13 2063–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Lee, B.-h., Ishitani, M., Lee, H., Zhang, C.Q., and Zhu, J.-K. (2001. c). FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 15 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K., et al. (2003). Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302 842–846. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. (2000). RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101 25–33. [DOI] [PubMed] [Google Scholar]
- Zhou, Z.L., Licklider, L.J., Gygi, S.P., and Reed, R. (2002). Comprehensive proteomic analysis of the human spliceosome. Nature 419 182–185. [DOI] [PubMed] [Google Scholar]
- Zhu, J.-K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.