Abstract
Plastids contain tetradecameric Clp protease core complexes, with five ClpP Ser-type proteases, four nonproteolytic ClpR, and two associated ClpS proteins. Accumulation of total ClpPRS complex decreased twofold to threefold in an Arabidopsis thaliana T-DNA insertion mutant in CLPR2 designated clpr2-1. Differential stable isotope labeling of the ClpPRS complex with iTRAQ revealed a fivefold reduction in assembled ClpR2 accumulation and twofold to fivefold reductions in the other subunits. A ClpR2:(his)6 fusion protein that incorporated into the chloroplast ClpPRS complex fully complemented clpr2-1. The reduced accumulation of the ClpPRS protease complex led to a pale-green phenotype with delayed shoot development, smaller chloroplasts, decreased thylakoid accumulation, and increased plastoglobule accumulation. Stromal ClpC1 and 2 were both recruited to the thylakoid surface in clpr2-1. The thylakoid membrane of clpr2-1 showed increased carotenoid content, partial inactivation of photosystem II, and upregulated thylakoid proteases and stromal chaperones, suggesting an imbalance in chloroplast protein homeostasis and a well-coordinated network of proteolysis and chaperone activities. Interestingly, a subpopulation of PsaF and several light-harvesting complex II proteins accumulated in the thylakoid with unprocessed chloroplast transit peptides. We conclude that ClpR2 cannot be functionally replaced by other ClpP/R homologues and that the ClpPRS complex is central to chloroplast biogenesis, thylakoid protein homeostasis, and plant development.
INTRODUCTION
Intracellular proteolysis is important for regulation of metabolic and signaling pathways as well as protein homeostasis and viability of cells and organelles. Proteolysis in chloroplasts and nonphotosynthetic plastids of plants is poorly understood but is important for plant development and plastid function. Several processing peptidases and proteases are present in plastids, including the stromal chloroplast processing peptidase SPP (Richter and Lamppa, 1998), the thylakoid processing peptidases cTPA (Oelmuller et al., 1996), TPP (Kirwin et al., 1987), and thylakoid/envelope SPase I (Inoue et al., 2005), stromal and thylakoid members of the Deg, FtsH, and Clp families (Adam et al., 2001; Sokolenko et al., 2002; Peltier et al., 2004b), and thylakoid-bound proteases SppA (Lensch et al., 2001) and Egy1 (Chen et al., 2005).
The Clp proteins form the largest plastid-localized protease family, with five Ser-type ClpP (P1,P3-6) proteases, four ClpR (R1-4) proteins, three Clp AAA+ chaperones (C1, C2, and D), and several additional members (ClpS1, S2, and T) with unknown functions (Adam and Clarke, 2002; Peltier et al., 2004b; Adam et al., 2006). The ClpR proteins lack the three catalytic amino acid residues that are conserved across ClpP proteins (Peltier et al., 2001). All members of the plastid-localized Clp proteolytic system have been identified at the protein level by mass spectrometry both in chloroplasts of Arabidopsis thaliana leaves and in nongreen plastids in Brassica rapa roots and Brassica oleracea petals, with the exception of a potential regulator, ClpT (Peltier et al., 2004b). Transcript analysis of most CLP genes showed constitutive expression in roots and leaves of Arabidopsis with only minor changes in gene expression under specific stress conditions or during senescence (Shanklin et al., 1995; Nakabayashi et al., 1999; Zheng et al., 2002).
The functional importance and substrates of the Clp family in plant growth, development, and protein homeostasis in nongreen plastids and chloroplasts is largely unknown. One ClpP subunit, ClpP1, is plastid encoded and was shown to be essential for shoot development in tobacco (Nicotiana tabacum) (Shikanai et al., 2001; Kuroda and Maliga, 2003). Downregulation of the plastid-encoded CLPP1 gene in the green algae Chlamydomonas reinhardtii suggested that ClpP1 is involved in the degradation of the thylakoid-bound subunits of cytochrome b6f and photosystem II (PSII) complex (Majeran et al., 2000, 2001). Loss of expression of the CLPC1 chaperone resulted into reduced plant growth and chloroplast development, but homozygous plants are autotrophic and seeds are viable (Constan et al., 2004; Sjogren et al., 2004; Kovacheva et al., 2005). This partial redundancy of ClpC1 is likely due to the expression of the ClpC2 homologue (Kovacheva et al., 2005). Although ClpC accumulates predominantly in the stroma, ClpC1 also associated with the chloroplasts protein translocation machinery in the inner envelope, interacting in particular with Tic110 and Tic40 (Akita et al., 1997; Nielsen et al., 1997). Consistently, loss of ClpC1 leads to lower protein translocation rates in isolated chloroplasts (Constan et al., 2004; Kovacheva et al., 2005). The Clp machinery in the photosynthetic bacterium Synechococcus sp PCC 7942 consists of three CLPP genes, one CLPR gene, and one CLPX gene. CLPP1,2 are dispensable, but CLPP3 and CLPR are essential (Schelin et al., 2002; Barker-Astrom et al., 2005).
The Clp machinery is simpler and best understood in the prokaryote Escherichia coli, where it initially was discovered (Hwang et al., 1987; Katayama-Fujimura et al., 1987). The Clp protease forms a bipartite structure of a Ser-type protease complex and an associated ATPase complex that serves in substrate recognition and feeding (Sauer et al., 2004). The catalytic chamber of the Clp protease core is enclosed in a barrel-shaped structure formed by the juxtaposition of two homoheptameric rings of ClpP peptidases, all encoded by a single gene. The diameter of the cavity is 50 Å, and the diameter of entrances at either end of the complex was reported as 10 Å (Wang et al., 1997). This suggested that only peptides and proteins in an unfolded state can enter the cavity, which is in agreement with biochemical data showing that ATPase and ATP hydrolysis are required for the degradation of folded proteins (Maurizi et al., 1994). We suggested that conserved (putative) pores at the interface of the two heptameric rings were a preferable route for release of proteolytic products (Peltier et al., 2004b). Recently, structural loosening in this region was shown to be important for product release (Gribun et al., 2005; Kang et al., 2005; Sprangers et al., 2005). The functional role of Clp complexes in E. coli and other bacteria, such as Bacillus subtilis, involves (1) degradation of misfolded proteins, (2) stress responses, and (3) gene regulation by proteolysis of transcription factors. Recognition of proteins by the ClpXP machinery in bacteria occurs via N- or C-terminal tags or via internal domains (Flynn et al., 2003; Chandu and Nandi, 2004).
To understand the function of the Clp gene family in Arabidopsis, it is important to understand how the different Clp proteins interact with each other and if there is any functional redundancy within the Clp family. Three-dimensional (3D) homology modeling showed that the ClpP/R proteins fit well together in a tetradecameric complex and also suggested unique contributions for each protein (Peltier et al., 2004b). The 3D modeling suggests that ClpS1,2 fit well on the axial sites of the ClpPR cores. In contrast with plastids, plant mitochondria contained a single ∼320-kD homotetradecameric ClpP2 complex, without association of ClpR or ClpS proteins (Peltier et al., 2004b). It is surprising that the Clp core composition appears identical in chloroplasts and nongreen plastids, despite the remarkable differences in plastid proteome composition. This suggests that regulation of plastid proteolysis by the Clp machinery is not through differential regulation of CLPP,R,S gene expression, but rather through substrate recognition mechanisms and regulated interaction with chaperone-like molecules (ClpS1,2 and others) to the ClpPR core. Still, it is not clear why the plastid Clp complex has evolved to such complexity, and little is known about its substrates.
In this study, we provide evidence for a functional role of one of the noncatalytic ClpR proteins, ClpR2. ClpR2 has 22 to 54% sequence similarity to the other ClpR proteins and lacks the L1 insertion loop of 10– to 12–amino acid residues present in the other ClpR subunits (Peltier et al., 2004a). We isolated, functionally complemented, and characterized a T-DNA insertion mutant in the 5′ untranslated region of the CLPR2 gene in Arabidopsis and named it clpr2-1. We conclude that ClpR2 is a nonredundant member of the ClpPRS protease complex and is needed for plant development, despite its high sequence similarity to other expressed plastid ClpP/R proteins and the absence of the conserved catalytic residues.
RESULTS
Downregulation of CLPR2 Expression Leads to a Yellow/Pale-Green Phenotype and Reduced Growth
The accumulation of noncatalytic ClpR components in the plastid-localized ClpPR core is puzzling and is unique to photosynthetic organisms. The Arabidopsis genome contains four CLPR genes. Protein products of all four genes target to chloroplasts and nongreen plastids and assemble into a 325- to 350-kD ClpPRS protease complex (Peltier et al., 2004b). The processed CLPR gene products (after removal of the predicted chloroplast transit peptides [cTPs]) have 27 to 39% sequence similarity with each other and between 24 and 39% sequence similarity to ClpP1,3-6 proteins. ClpR2 (At1g12410) has the highest sequence similarity to these ClpP proteins (between 32 and 38%) compared with the other ClpR proteins. ClpR2 is different from the other ClpR proteins in that it lacks a 10– to 12–amino acid insertion domain unique to ClpR proteins. Our 3D models suggested that this domain is protruding into the tunnel of the ClpPR core and possibly affects substrate presentation to the catalytic sites of the ClpPR core (Peltier et al., 2004b).
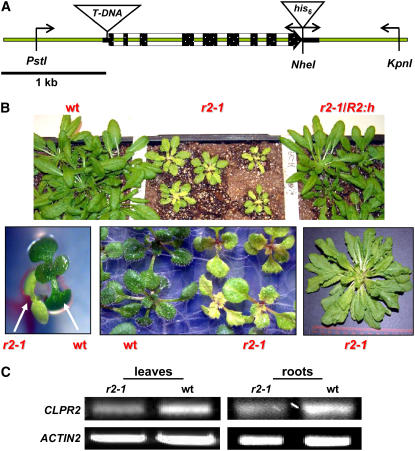
To understand the functional significance of ClpR2 in Arabidopsis, we screened the T-DNA insertion collection established by the SALK Institute (http://signal.salk.edu/cgi-bin/tdnaexpress) (Alonso et al., 2003). BLAST searching of the genomic sequence for CLPR2 against this T-DNA insertion collection (in Columbia-0 ecotype) identified one potentially tagged line, SALK_046378 (Figure 1A). Germination of the seeds of this line resulted in a segregating population of wild type–like plants and smaller seedlings with serrated, yellow leaves that developed into pale-green plants (Figure 1B). PCR-based genotyping and sequencing (data not shown) confirmed that the pale-green plants were homozygous for a T-DNA insertion 7 bp upstream of the translational start codon of CLPR2 (Figure 1A). We named this mutant clpr2-1. The yellow/pale-green phenotype of the F2 progeny of a clpr2-1 backcross (to the wild type) segregated in a 1:3 ratio (data not shown).
Figure 1.
Isolation and Complementation of a T-DNA Insertional Mutant for CLPR2.
(A) The T-DNA insertion site is 7 bp upstream of the translational start codon of CLPR2. Exons are indicated in black. The fragment used for complementation is shown through primer annealing sites (arrows) as well as the introduced restriction sites. The T-DNA (clpr2-1) and (his)6 (R2:h) tags are not drawn to scale. Bar = 1 kb.
(B) Homozygous clpr2-1 shows delayed development and a yellow/pale-green phenotype but can germinate and be maintained on soil. Younger leaves are paler with serrated leaves. The complemented line (clpr2-1/R2:h) shows a wild-type phenotype.
(C) Semiquantitative RT-PCR profile for CLPR2 (top panels) and ACTIN2 (bottom panels) in leaves and roots of clpr2-1 and the wild type.
Homozygous clpr2-1 plants are viable on soil but have delayed development with a doubled flowering time compared with the wild type when grown under a 10-h light/dark cycle (Figure 1B). Nevertheless, clpr2-1 plants do set seeds that germinate on soil. No visual phenotype was observed in roots of homozygous clpr2-1 (data not shown). The phenotype of clpr2-1 plants grown on agar plates was unaffected by supplementation with sucrose (1 to 2%) (data not shown). RT-PCR in the clpr2-1 plants showed that the transcript encoding for the full-length cDNA was present both in leaves and in roots. However, mRNA levels were reduced more than twofold in the mutant compared with the wild type (Figure 1C). This suggested a correlation between reduced CLPR2 expression and the yellow/pale-green phenotype.
Functional Complementation of clpr2-1 with a (His)6-Tagged Genomic CLPR2 Transgene
To unambiguously determine if the yellow phenotype was truly linked to the T-DNA insertion in CLPR2, we functionally complemented clpr2-1 with a tagged genomic construct. A 3481-bp genomic fragment containing At1g12410 (CLPR2) was selected. This fragment included the endogenous promoter region (678 bp) as well as terminator (944 bp) (Figure 1A). The genomic DNA was PCR amplified in two separate fragments to introduce a C-terminal hexa-histidine (his6) tag to aid in recognition of the expression of transgenic ClpR2 protein. Homology modeling of ClpR2 predicted that the C terminus is exposed to the outer surface of the Clp core complex (Peltier et al., 2004b). We therefore chose to create a C-terminal fusion, as this is less likely to interfere with ClpR2 function. The reconstituted transgene (CLPR2:h) was inserted into the homozygous clpr2-1 genome through Agrobacterium tumefaciens–mediated transformation. Hygromycin-resistant plants were genotyped by PCR and DNA sequencing, and homozygotes for clpr2-1 with inserted CLPR2:h were selected. Importantly, these plants display a wild-type phenotype (Figure 1B), establishing that the yellow/pale-green phenotype of clpr2-1 is indeed due to reduced expression of CLPR2.
(His)6-Tagged ClpR2 Incorporates into the ClpPRS Complex in the Complemented clpr2-1 Line
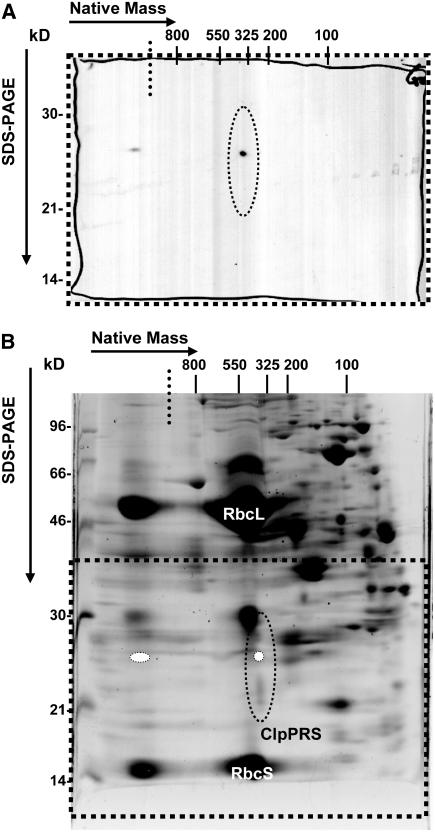
To determine if the transgenic (his)6-tagged ClpR2 in the complemented clpr2-1 line incorporated into the ClpPRS complex, stroma was collected from purified chloroplasts of clpr2-1/R2:h and analyzed on a native gel system in triplicate. Duplicate gels were blotted to a polyvinylidene difluoride (PVDF) membrane and detected using monoclonal (his)6 antiserum (Figure 2A), and another gel was stained with Sypro Ruby (Figure 2B). The protein gel blots gave a signal in the stacking gel and at ∼325 kD (in the native dimension) and 26 kD in the second denaturing dimension (Figure 2A). Comparing the stained gel with the protein gel blot showed that the spot in the higher mass complex at >800 kD corresponded with the stack of the gel (i.e., where protein aggregates run in the gel), and the ∼325-kD spot (native dimension) corresponded to the ClpPRS complex. This shows that full complementation was also achieved at the protein (assembly) level. The amount of ClpR2:(his)6 in the 325-kD ClpPRS complex was quantified by comparing the signal intensity to commercial purified (his)6:ubiquitin fusion protein (data not shown). This suggested that transgenic ClpR2:(his)6 fusion protein represented ∼0.01% of the total stromal protein.
Figure 2.
Incorporation of Transgenic ClpR2:(his)6 into the 325-kD ClpPRS Complex.
The incorporation of transgenic ClpR2:(his)6 into the 325-kD ClpPRS complex in the complemented clpr2-1 line was analyzed. Native stroma was obtained from purified intact chloroplasts and separated in triplicate first on CN-PAGE gels, followed by SDS-PAGE in the second dimension. Two 2D gels were blotted to PVDF membranes and probed with anti-polyHis serum (A), and the third gel was stained with Sypro Ruby (B).
(A) Immunoblot of the bottom part (dotted square) of a CN-PAGE separation of stromal complexes from the complemented line clpr2-1/R2:h. The region where the Clp core complex subunits migrate is enclosed by a dotted ellipse.
(B) CN-PAGE as in (A) but stained with Sypro Ruby. Dotted square and ellipse are as in (A); relative position of the anti-polyHis signal is represented by white ovals. The dotted lines indicate the transition from stacking to resolving gel in the first dimension.
The Stromal Proteome of clpr2-1 and Accumulation of the ClpPRS Complex
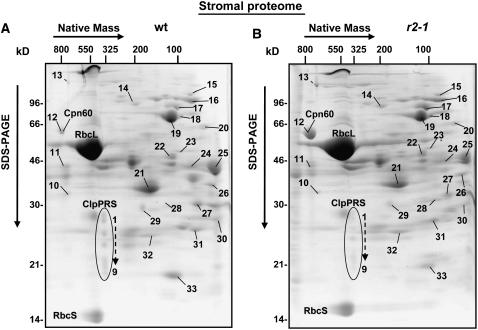
To investigate the stromal proteome and the ClpPRS complex in clpr2-1, native stroma was obtained from intact clpr2-1 and wild-type chloroplasts and separated by two-dimensional (2D) gels using colorless native PAGE (CN-PAGE) in the first dimension (Figures 3A and 3B). Three sets of gels with 1 mg of protein were obtained from independent stromal biological replicates and stained with Sypro Ruby. Gels were then scanned and spots were quantified. Spot volumes were normalized to the total spot volume on each gel. Spots were matched to proteins identified in an earlier study on the oligomeric stromal proteome of the wild type (Peltier et al., 2006), and several were confirmed by nano-liquid chromatography–electrospray tandem mass spectrometry (nanoLC-ESI-MS/MS) analysis. We were in particular interested to determine the expression levels of the stromal proteases and chaperones in clpr2-1. Therefore, we experimentally verified the identities of matched protein spots expected to contain such proteins (see Supplemental Table 1 online).
Figure 3.
Native Stromal Proteome in clpr2-1.
CN-PAGE separation and Sypro Ruby staining of 1 mg of stromal proteins from wild-type (A) and clpr2-1 (B) chloroplasts from fully developed rosettes. The identified proteins are indicated with spot numbers. Protein identities were determined by nanoLC-ESI-MS/MS in this study or for the wild type in a parallel and independent study (Peltier et al., 2006). For the complete data set, see Supplemental Table 1 online. Spot numbers 1 to 9 correspond to the ClpPRS complex. Spot numbers 10 to 31 correspond to the following proteins: 10, uridylyltransferase-related; 11, glyceraldehyde-3-phosphate dehydrogenase A-1,2 (GAPA-1,2); 12, Cpn60-α,β-1,2; 13, ferredoxin-dependent Glu synthase (Fd-GOGAT 1); 14, ClpC1,C2,D; 15, metalloproteases (M16) (AtPreP1,2); 16, lipoxygenase AtLOX2; 17, HSP90, elongation factor Tu-G (EF-G), and starch branching enzyme class II (SBEII); 18, cpHSP70-1,2 (DnaK homologues); 19, Transketolase-1 (TKL-1); 20, plastid phosphoglucomutase (PGM1); 21, fructose-bisphosphate aldolase-1,2 (SFBA-1,2); 22, fructose-bisphosphatase (FBPA); 23, monodehydroascorbate reductase (MDHAR); 24, phosphoglycerate kinase-2 (PGK-2); 25, phosphoglycerate kinase-1 (PGK-1); 26, annexin (AnnAt1); 27, sedoheptulose-bisphosphatase (SBPase); 28, GrpE-1; 29, thiazole biosynthetic enzyme (THI1); 30, 3-β-hydroxy-δ5-steroid dehydrogenase; 31, triosephosphate isomerase-1 (TPI-1); 32, Cpn21 (also Cpn20), carbonic anhydrase-1 (CA1); 33, peptidylprolyl isomerase ROC4.
We identified members of the GroES/GroEL system (Cpn60/Cpn21), the DnaK/DnaJ system (Hsp70 and GrpE), cpHsp90, and Hsp100 (ClpC1, C2, and D), and two metalloproteases of the M16 family (ZnMP). We did not detect any obvious mitochondrial, cytosolic, or other nonchloroplast proteins. The most striking difference between clpr2-1 and the wild type was the approximately threefold upregulation in clpr2-1 of the CPN60 chaperone complex at ∼800 kD (spot 12; Figure 3). Otherwise, the overall spot pattern (native masses and expression levels) was very similar for the wild type and clpr2-1. For instance, ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) and other abundant metabolic enzymes accumulated at wild-type levels. Spots corresponding to EF-Tu/Hsp90 (spot 17), Cpn21 (spot 32), and two Zn metalloproteases (spot 15) showed very little changes. A detailed quantitative comparative proteome analysis is in progress using stable isotopic labeling with cleavable isotope-coded affinity tagging.
The ClpPRS core complex in the stroma of clpr2-1 and the wild type resolved into a complex of 325 to 350 kD with spots between 20 and 30 kD (Figures 3 and 4B), as reported earlier for wild-type plants (Peltier et al., 2004b). Detailed mass spectrometric analysis showed that all 11 plastid-localized ClpP/R/S subunits were present in the stromal Clp core of clpr2-1 (see Supplemental Table 1 online). The sum of all Clp spot intensities, normalized against the total spot intensity, was 2.5- to 3-fold lower in clpr2-1 compared with the wild type.
Figure 4.
Quantitative Comparative Analysis of ClpPRS Complex in the Wild Type and clpr2-1 Using Differential Stable Isotope Labeling with iTRAQ.
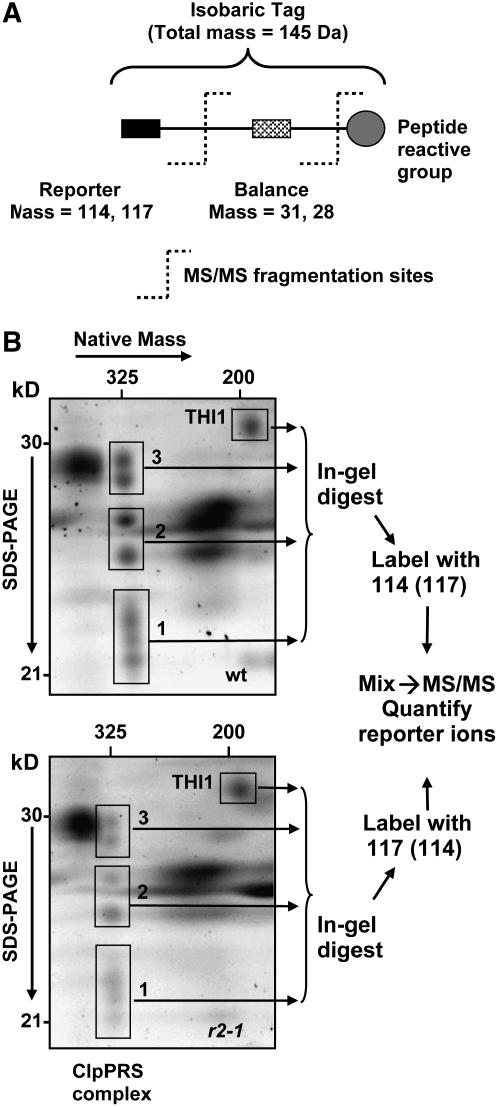
(A) Schematic representation of the isobaric iTRAQ tags used for the differential labeling. The tag consists of a balance group (28 or 31 D) and a reporter group (114 or 117 D), which is released upon fragmentation in the mass spectrometer.
(B) Spots containing Clp subunits and the reference spot THI1 were sliced as four gel pieces as indicated (1 to 4) from 2D gels (CN-PAGE followed by SDS-PAGE) of the stromal proteome of the wild type and clpr2-1. Gel pieces were washed and digested with trypsin. Peptides were extracted with formic acid and differentially labeled with the iTRAQ reagents (i.e., 114 for wild-type and 117 for clpr2-1 samples). After pairwise (pairs of identical spot numbers) mixing of samples from the wild type and clpr2-1, the samples were cleaned up using C18 microcolumns. The eluates for each pair were analyzed by reverse-phase nanoLC-ESI-MS/MS. After mass spectral data processing, sequence information was used for protein identification, and the reporter ions (114 and 117) were used for peptide quantification. The experiment was performed with two independent biological replicates, with a label switch between the replicates to avoid a possible systematic bias due to the label. Within one biological replicate, duplicate gels for the wild type and for clpr2-1 were used to reduce variability. Each sample was analyzed twice by MS/MS.
Quantification of the Individual ClpP/R/S Proteins in the Clp Core Complex Using iTRAQ
Since most of the 11 subunits of the Clp core complex migrate in overlapping spots on the 2D gels, we determined the relative accumulation levels using a combination of the 2D gels and differential stable isotope labeling with the amine-reactive isobaric tagging reagent iTRAQ (Figure 4A). This novel technique has been used with success in studies of human, yeast, and microbes (Ross et al., 2004; DeSouza et al., 2005; Hardt et al., 2005), but not yet in plants.
The gel spots containing the ClpP/R/S subunits were excised from the 2D native gels of chloroplast stroma from the wild type and clpr2-1 as three gel slices per gel (1, 2, and 3 in Figure 4B). In addition, we selected a well-resolved protein spot containing thiazole biosynthetic enzyme THI1 (At5g54770) as a reference (THI1 is in spot 29 in Figures 3A and 3B). We selected THI1 as a reference since it was very reproducible and well resolved with a similar molecular mass range (33 kD) and relative abundance as the ClpP/R/S subunits. Gel slices from the wild type and clpr2-1 were first digested with trypsin, followed by quantitative peptide extraction and differential labeling with either the heavy or light iTRAQ reagents (reporter of 114 or 117 D, respectively) and then analyzed by MS/MS as depicted in Figure 4B. The experiment was performed in two biological replicates and included a label switch to avoid a possible bias. To reduce variation due to 2D gel separation and peptide extractions, equivalent gel slices from two separate gels were pooled.
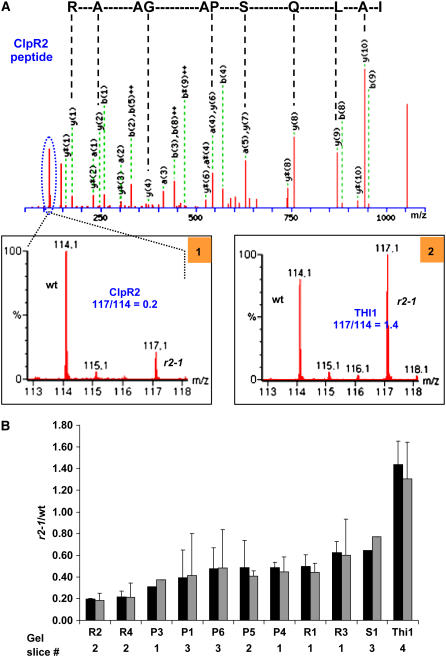
Figure 5A shows an example of the MS/MS spectrum of a double charged peptide (mass-to-charge ratio [m/z] = 599.84) belonging to the ClpR2 protein and the reporter ion pair at 114 and 117 generated in the collision cell of the mass spectrometer. Peptides from clpr2-1 were labeled with the heavier isotope (117 D) and the wild type with the light isotope (114 D). Inset 1 of Figure 5A shows a close-up of the reporter, with a calculated ratio between the areas under the 114 (wild type) and 117 (clpr2-1) reporter ion peaks of 0.2. This shows that accumulation of ClpR2 in the mutant is fivefold lower than in the wild type (normalized to total stromal protein). Inset 2 shows the reported region for a peptide belonging to the THI1 protein. In this case, the 117:114 ratio is 1.4, indicating that accumulation of THI1 is 40% higher in the mutant than in the wild type. High-quality MS/MS spectra and 114 to 117 reporter ratios were obtained for 85 peptides belonging to the ClpP/R/S and THI1 proteins. A complete list of these peptides and stable isotope ratios is provided in Supplemental Table 2 online.
Figure 5.
Quantitative Comparative Analysis of ClpPRS Complex in the Wild Type and clpr2-1 Using Differential Stable Isotope Labeling with iTRAQ.
(A) An example of the MS/MS spectrum of a double-charged peptide (m/z = 599.84; IALQSPAGAAR; the y-ion series is indicated) belonging to the ClpR2 protein and the reporter ion pair at 114 and 117 generated in the collision cell of the mass spectrometer. Peptides from clpr2-1 were labeled with the heavy tag (117) and the wild type with the light tag (114). Inset 1 shows a close-up of the 114 to 117 region of this MS/MS spectrum, with a calculated ratio between the areas under the 114 and 117 peaks of 0.2. This shows that accumulation of ClpR2 in the mutant is fivefold lower than in the wild type (normalized to total stromal protein). Inset 2 shows a close-up of the 114 to 117 area for a peptide belonging to the THI1 protein in gel slice 4. In this case, the 114:117 ratio is 1.4, indicating that THI1 is 40% higher in the mutant than in the wild type (normalized on basis of total stromal protein). High-quality MS/MS spectra and 114:117 reporter ratios were obtained for 85 peptides belonging to the ClpP/R/S and THI1 proteins. A complete list of these peptides and reporter ion ratios is listed in Supplemental Table 2 online.
(B) Summary of quantification of the different ClpP/R/S proteins accumulating in the ClpPRS complex displayed as ratios between clpr2-1 and the wild type. Standard deviations for the biological replicates are indicated. The black bars show uncorrected average ratios, and the gray bars show average ratios corrected for differences in protein loading, as determine by total spot volume per gel. Details for the peptides used for quantification are listed in Supplemental Table 2 online.
The graph in Figure 5B summarizes the quantification of each of the chloroplast-localized ClpP and ClpR proteins, ClpS1, and THI1 based on the peptides listed in Supplemental Table 2 online. The black bars show the averaged ratios from the two biological replicates without any corrections. The gray bars show very similar averaged ratios after corrections for small loading differences on the gels (for details, see legend of Figure 5). THI1 was 1.4-fold upregulated (1.2-fold after correction for total spot volume). Each of the ClpP/R subunits and ClpS1 accumulated in the complex at lower levels than the wild type. ClpR2 and ClpR4 were each reduced fivefold, while accumulation of the other ClpP/R subunits was reduced ∼2- to 2.5-fold, indicating a change in overall ClpP/R stoichiometry.
Overview of clpr2-1 Mesophyll Cells and Chloroplasts
To investigate the effect of reduced ClpR2 and ClpPRS complex accumulation on cell and chloroplast development, we analyzed the leaf anatomy and chloroplast size and numbers by light microscopy and laser scanning confocal microscopy (Figures 6A to 6F). The chloroplast ultrastructure was analyzed by transmission electron microscopy (TEM) (Figures 6G to 6L).
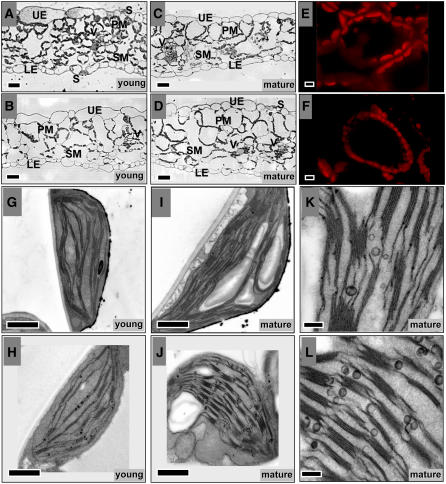
Figure 6.
Light, Confocal, and Transmission Electron Microscopy Analysis of Leaves of clpr2-1 and the Wild Type.
(A) to (D) Light microscopy of transverse sections from young, expanding leaf blades of the wild type (A) and clpr2-1 (B) and of mature leaves of the wild type (C) and clpr2-1 (D).
(E) and (F) Laser scanning confocal microscopy images of mesophyll cells of the wild type (E) and clpr2-1 (F). Only the red channel for chlorophyll autofluorescence is shown, collected at the same excitation intensities.
(G) to (L) TEM analysis of the ultrastructure of chloroplasts from young expanding leaves of wild-type (G) and clpr2-1 plants (H) and from mature leaves of wild-type (I) and clpr2-1 (J) leaves. Close-ups of plastoglobules in chloroplasts of mature leaves of the wild type (K) and clpr2-1 (L).
Bars = 25 μm in (A) to (D), 5 μm in (E) and (F), 1 μm in (G) to (J), and 0.2 μm in (K) and (L). UE, upper epidermis; LE, lower epidermis; PM, palisade mesophyll; SM, sponge mesophyll; S, stomata; V, vasculature.
Young emerging leaves of clpr2-1 contained much larger but fewer cells and were vacuolated compared with young emerging wild-type leaves of the same age (Figures 6A and 6B), while cell size in adult leaves was quite similar for clpr2-1 and the wild type (Figures 6C and 6D). Confocal microscopy of fully expanded leaves showed that chloroplasts in clpr2-1 are smaller and with lower fluorescence yield than in the wild type but are present in higher numbers per cell (Figures 6E and 6F). The increased number of chloroplasts per cell in clpr2-1 appears to compensate for the loss of chloroplast size.
TEM of thin sections of young, developing leaves showed that thylakoid accumulation was reduced in clpr2-1 with a higher number of plastoglobules (visible as electron dense particles) than the wild type (Figures 6G and 6H). During maturation of the leaf, thylakoid accumulation increased in clpr2-1, thus becoming more similar to the wild type (Figures 6I and 6J), although total thylakoid per chloroplast and per cell remained lower all through development, as will also be demonstrated at the biochemical level below. These observations are in agreement with a partial recovery of the phenotype in aging clpr2-1 plants from yellow to pale green. Comparisons of older, fully expanded leaves of the wild type and clpr2-1 showed that chloroplasts in clpr2-1 remain smaller than in the wild type, with higher numbers of thylakoid-associated plastoglobules and possibly an increased ratio of grana to stromal lamellae (Figures 6G to 6L).
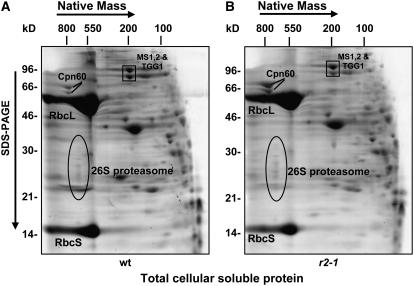
To estimate if the ratio of stromal chloroplast proteome per total soluble cellular proteome was much altered in clpr2-1, we compared the total soluble leaf proteome extracts from wild-type and clpr2-1 leaves using native 2D gels (Figure 7). Taking Rubisco and other abundant metabolic enzymes as a measure of the soluble chloroplast proteome and the proteasome, cytosolic Met synthase (MS1,2), and vacuolar thioglucoside glucohydrolase 1 (TGG1) (identified by MS/MS) as indicators of the rest of the cellular proteome, we showed that the total chloroplast protein/total cell protein ratio is not much changed in clpr2-1, in agreement with microscopic analysis described above.
Figure 7.
Oligomeric Soluble Cellular Proteomes of the Wild Type and clpr2-1.
CN-PAGE separation and Sypro Ruby staining of 2 mg of total soluble proteins from fully developed rosettes of wild-type (A) and clpr2-1 (B) plants. The predominant chloroplast stromal components are the Rubisco large and small subunits (RbcL and RbcS, respectively). The extrachloroplastic 26S proteasome complex (ovals) and cytosolic Met synthase 1,2 (MS1, At5g17920; MS2, At3g03780) and vacuolar TGG1 (At5g20980) are indicated.
Thylakoid Pigment Accumulation, PSII Damage, and Light Sensitivity of clpr2-1
The leaf color and microscopy analyses showed that biogenesis of the thylakoid membrane system is delayed and partially disrupted. Analysis of the pigment and protein content of purified thylakoid membranes from the wild type and clpr2-1 showed that the ratio of chlorophyll (a+b) to thylakoid protein was only 3% lower in clpr2-1 than in the wild type but that thylakoids of clpr2-1 accumulated 18% more carotenoids per amount of thylakoid protein than wild-type plants. The chlorophyll a/b ratio was 10% lower in clpr2-1 (Table 1). This was also reflected in the 25% increase in carotenoid/chlorophyll (a+b) ratio in clpr2-1. The carotenoid/chlorophyll (a+b) ratio in clpr2-1/R2:h was similar to the wild type, indicative of full complementation (Table 1). The amount of thylakoid protein per total soluble leaf protein was only 25 to 30% of the wild type, supporting the microscopy observations of the reduced amount of thylakoid membrane structures in clpr2-1 leaf.
Table 1.
Physiological Parameters for the Wild Type, clpr2-1, and clpr2-1/R2:h Grown at 100 μmol·m−2·s−1 followed by a Shift to 1000 μmol·m−2·s−1 for Two or Four Light Cycles
| Light Intensity | Parameter | Wild Type | r2-1 | r2-1/R2:h |
|---|---|---|---|---|
| 100 μE/m2s | Ca/Cb | 2.26 ± 0.033 | 2.05 ± 0.078a | 2.18 ± 0.051b |
| Ca+b/Cx+c | 5.87 ± 0.293 | 4.67 ± 0.484a | 5.73 ± 0.374 | |
| Fv/Fm | 0.84 ± 0.003 | 0.66 ± 0.036a | 0.83 ± 0.002a | |
| Thyl-Prot/Ca+b | 7.3 | 7.1 | ND | |
| Thyl-Prot/Cx+c | 53.4 | 45.3 | ND | |
| 1000 μE/m2s | Ca/Cb1 | 2.22 ± 0.079 | 2.28 ± 0.216 | 2.50 ± 0.571 |
| Ca+b/Cx+c1 | 5.78 ± 0.391 | 3.76 ± 0.667a,c | 5.68 ± 1.161 | |
| Fv/Fm2 | 0.76 ± 0.014d | 0.66 ± 0.010a | 0.79 ± 0.014b,d |
Measurements are means ± sd (n = 8). Student's t test probability for comparisons against the wild type (a and b) or against normal light conditions (c and d) are as follows: a and d, P < 0.001; b and c, 0.001 < P < 0.01. Ca, Cb, Ca+b, and Cx+c, concentration of chlorophylls a and/or b and xanthophylls + carotene; Fv/Fm, ratio of variable fluorescence versus maximum fluorescence of PSII; Thyl-Prot, thylakoid proteins. Light stress exposure: 1, four light cycles; 2, two light cycles. ND, not determined.
To investigate the functional status of the thylakoid-bound photosynthetic apparatus of clpr2-1 and the complemented line clpr2-1/R2:h, we measured the ratio between the variable chlorophyll fluorescence (Fv) to maximum chlorophyll fluorescence (Fm) (Fv/Fm) of plants grown at 100 μmol·m−2·s−1. This Fv/Fm ratio is a measure of PSII efficiency (Krause and Weis, 1991). Fv/Fm of clpr2-1 leaves was 22% lower than that of the wild type, most likely indicating a significant inactive PSII population. Importantly, the Fv/Fm ratio of the complemented line was similar to the wild type. We also subjected wild-type, clpr2-1, and clpr2-1/R2:h plants to 2 d (two light cycles) of high light treatment (1000 μmol·m−2·s−1). The high light treatment resulted in a 10% decline of Fv/Fm for both wild-type and complemented lines, whereas Fv/Fm of clpr2-1 leaves did not decrease any further (Table 1).
Starch Accumulation during the Day and Night Cycle
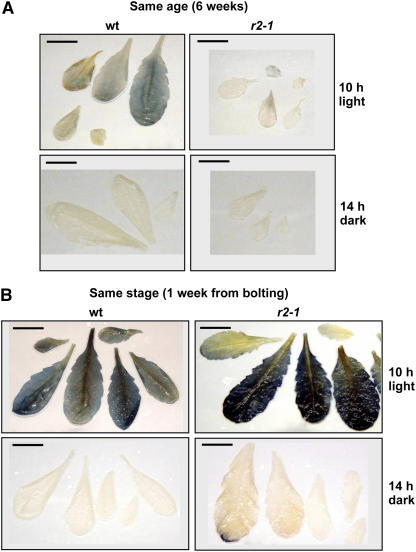
The microscopy and pigment analyses suggested a delayed and reduced accumulation of the thylakoid membrane system and its photosynthetic electron transport components. To evaluate if this was reflected in accumulation of reduced carbohydrates, we measured starch accumulation during the day and night cycle. Leaves from the wild type and clpr2-1 either from plants of identical age (6 weeks after sowing) or identical growth stage (1 week prior to bolting) were stained with iodine for the presence of starch at the end the light period and at the end of the dark period. This showed that starch accumulation levels at the end of the light cycle were reduced in clpr2-1 compared with same aged wild-type plants (Figure 8A). By contrast, when comparing wild-type and clpr2-1 rosettes of the same developmental stage, starch accumulation was comparable or even higher in clpr2-1 than in the wild type, with the exception of very young developing leaves of clpr2-1 (Figure 8B). These young leaves did not accumulate much starch, probably reflecting their delayed chloroplast development.
Figure 8.
Starch Accumulation and Starch Breakdown in Wild-Type and clpr2-1 Leaves.
To determine the net balance of reduced carbohydrate production (supply) and consumption (demand), leaves of the wild type and clpr2-1were stained (by iodine) for starch accumulation at the end of the light or dark periods. (A) shows starch staining of the wild type and clpr2-1 of 6-week-old plants, and (B) shows starch staining of the wild type and clpr2-1 ∼1 week prior to bolting.
Mass Spectrometry Analysis of Thylakoid Proteins
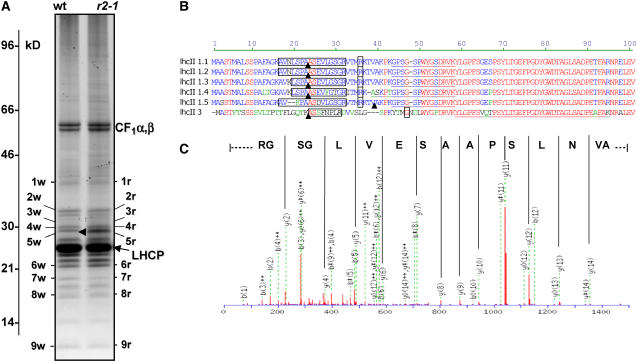
Protein profiles of purified thylakoids from the wild type and clpr2-1 were compared on one-dimensional (1D) gels loaded on an equal thylakoid protein basis and stained with the fluorescent dye Sypro Ruby. This allowed visualization of the most abundant thylakoid proteins (Figure 9A). At this level of resolution, the protein profiles were very similar, with the exception of one band at 29 kD (4) (Figure 9A). This gel band in clpr2-1 and the wild type, as well as an additional eight bands in clpr2-1 and equivalent bands in the wild type, were excised, and proteins were identified by nanoLC-ESI-MS/MS (all data are available in Supplemental Table 3 online). As expected, we predominantly identified the abundant proteins of the photosynthetic electron transport chain. These proteins all seem to migrate at expected apparent molecular masses and collectively indicated the presence of a normal electron transport chain in clpr2-1.
Figure 9.
Thylakoid Protein Profiles of Chloroplasts from Wild-Type and clpr2-1 Leaves and Identification by Mass Spectrometry.
(A) Thylakoid proteins were purified from leaves of wild-type and clpr2-1 plants and were separated by tricine SDS-PAGE and stained with Sypro Ruby, and 100-μg proteins were loaded in each lane. Molecular markers are indicated. Bands that are numbered were excised and digested with trypsin, and peptides were extracted and analyzed by nanoLC-ESI-MS/MS. A complete listing of all identified proteins is given in Supplemental Table 3 online.
(B) Sequence alignment of the N termini of LHCII1.1 and 2 (At1g29910 and At1g29920), LHCII1.3 (At1g29930), LHCII1.4 (At2g34430), LHCII1.5 (At2g34420), and LHCII.3 (At5g54270). Boxed residues indicate the peptides identified by MS/MS in clpr2-1, containing the predicted cTP cleavage site indicated by arrowheads. The most N-terminal peptides identified by MS/MS previously in extensive thylakoid proteome analyses of wild-type plants are underlined (Friso et al., 2004; Peltier et al., 2004a). N termini determined by Edman sequencing reported in the literature and in SWISS-PROT are indicated as single boxed residues (see Gomez et al., 2003).
(C) MS/MS fragmentation of doubly charged precursor ion with m/z of 764.40. This is a 1526.79-D peptide AVNLSPAASEVLGSGR matching to LHCII-1.1 and 2 (At1g29910 and At1g29920) with a Mowse score of 75.
However, in the clpr2-1 lane, but not in the wild-type lane, we consistently found peptides matching against several (25 to 27 kD) light-harvesting complex II (LHCII) proteins in bands 8r (at ∼17 kD) and 9r (10 kD). This strongly suggests accumulation of LHCII breakdown products in clpr2-1 but not in the wild type.
Surprisingly, the mass spectrometric analysis of bands 3r, 4r, and 5r between 28 and 30 kD in clpr2-1, but not in the wild type, reproducibly identified different and unique peptides matching to the predicted cTPs of LHCII-1.1, LHCII-1.2, LHCII-1.3, LHCII-1.4, LHCII-1.5, and LHCII-3 (Figure 9B). The interpreted MS/MS spectrum for one of those peptides (1526.79 D) matching against LHCII-1.1 and -2 is shown in Figure 9C. These peptides were not found below the main LHCII band at 25 to 27 kD. This showed that these five members of the LHCII family accumulated in part in their precursor form in the clpr2-1 thylakoids.
Electron Transport Chain Protein Complexes Are Slightly Downregulated
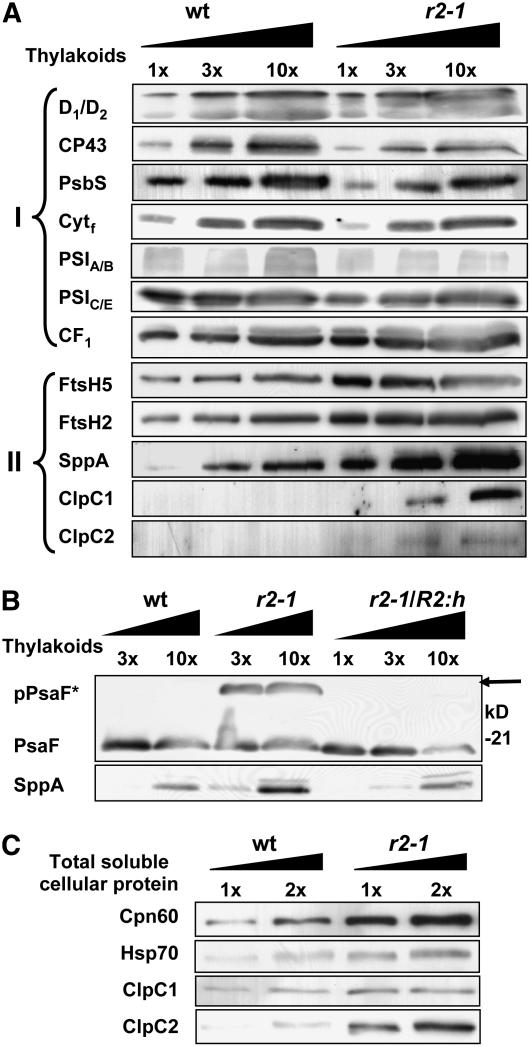
To determine levels of the photosynthetic electron transport complexes (PSI, PSII, and the cytochrome b6f complex) and the ATP synthase, a protein gel blot analysis was performed on thylakoid membranes purified from full rosettes of clpr2-1 and the wild type and loaded on an equal thylakoid protein basis (Figure 10A, Group I). Accumulation of representative protein subunits of each of these complexes was analyzed. The thylakoid purification and complete protein gel blotting analysis for proteins of Groups I and II were done at least twice from independent sets of wild-type and clpr2-1 plants. Standard errors are indicated in Figure 10A. To ensure accurate quantification, a titration of both wild-type and clpr2-1 thylakoid proteins (1x-3x-10x) was loaded on the gels (Figure 10A).
Figure 10.
Immunoblot Analyses of Chloroplast Protein Populations.
To evaluate the effect of reduced ClpPRS protease complex accumulation on the major thylakoid protein complexes, as well as proteases and chaperones in stroma and thylakoid, a protein gel blot analysis was performed. Titrations (indicated as 1x, 2x, 3x, and 10x) of thylakoid proteins ([A] and [B]) or total soluble protein extracts (C) from the wild type, clpr2-1, and the complemented line were separated by SDS-PAGE and blotted onto PVDF membranes.
(A) Membranes were probed with antibodies generated against different proteins of PSI and II, the cytochrome b6f complex, F-type ATP-synthase (Group I), and different chloroplast chaperones and proteases (Group II). Group I proteins are as follows: D1/2, the D1 and D2 proteins of PSII; PsbS, a unique antenna protein in PSII; Cytf, the 32-kD heme binding cytochrome f protein in the cytochrome b6f complex; PSIA/B, the reaction center A and B proteins of PSI; PSIC/E, PsaC and PsaE of PSI; CF1, the peripheral α- and β-subunits of the thylakoid ATP synthase complex. The protein ratios in clpr2-1/wild type were determined by two or three independent experiments and were consistently close to one. Group II proteins are as follows: FtsH2 and 5, thylakoid members of the FtsH Zn metalloprotease family; SppA, an ATP-independent light-induced Ser-type thylakoid protease; ClpC1,2, Hsp100 chaperones in the Clp family. Protein ratios in clpr2-1/wild type were quantified in two to three independent experiments and were on average 2.7 (FtsH5), 2 (FtsH2), 10 (SppA), and >10 (ClpC1 and C2).
(B) Protein gel blot of purified thylakoid membrane proteomes of the wild type, clpr2-1, and the complemented line clpr2-1/R2:h, probed with anti-PsaF. A strong cross-reacting band at ∼25 kD can be seen in clpr2-1 but not in the wild type or clpr2-1/R2:h. This band is the unprocessed PsaF protein (marked as pPsaF and indicated with a solid arrow). The bottom panel shows the response of antiserum directed against thylakoid protease SppA in the wild type, clpr2-1, and clpr2-1/R2:h. Lanes were loaded with 10, 30, and 100 μg of thylakoid protein (1x, 3x, and 10x, respectively).
(C) Protein gel blot of total soluble leaf proteomes of the wild type and clpr2-1. Equal protein concentrations (1x and 2x) were loaded for the wild type and clpr2-1 and probed with antisera directed against Cpn60, Hsp70, ClpC1, and ClpC2. Protein ratios in clpr2-1/wild type were quantified in one to three independent experiments and were on average 6 (Cpn60), 2 (HSP70), 2 (ClpC1), and 7 (ClpC2).
The protein gel blots showed that accumulation of PSII core (as measured by D1/D2 and CP43) and PsbS, the PSI core (as measured by PsaA/B and PsaC/E), the ATP synthase complex (as measured by the CF1 α- and β-subunits), and the cytochrome b6f complex (as measured by cytochrome f) were not significantly altered when loaded on a total thylakoid protein basis. As expected, when probing cytochrome f on the basis of total cellular protein extracts (not just thylakoid proteins), accumulation was reduced severalfold (data not shown), indicative of reduced accumulation of photosynthetic membranes per cell and confirming the observations from microscopy in Figure 6. We also probed with PsaF, an ∼18-kD integral PSI protein with a cleavable lumenal transit peptide (Figure 10B). The PsaF level was not significantly altered in clp2-1, in agreement with the immunoblot data for PsaA/B and PsaC/E. Interestingly, a strong signal was present several kilodaltons above the PsaF mature protein in clpr2-1 but not in the wild type. The apparent mass corresponds well with the calculated mass of the precursor of PsaF (24 kD). Thus, processing of the cTP is not only affected in several members of the LHCII family but likely also for PsaF. The clpr2-1/R2:h line did not show the unprocessed PsaF protein, demonstrating that the processing defect was indeed related to the reduced CLPR2 expression (Figure 10B).
Thylakoid Proteases and Stromal Chaperones Are Differentially Upregulated in clpr2-1
To determine if reduced accumulation of the ClpPRS complex resulted in a change in expression of thylakoid proteases, we probed the thylakoid proteome from the wild type and clpr2-1 for the two most prominent thylakoid-localized members of the Zn metalloprotease FtsH family, FtsH5 and FtsH2 (also named Var1 and Var2; Sakamoto et al., 2003; Yu et al., 2004). On equal thylakoid protein basis, both FtsH homologues were twofold upregulated. We also probed for thylakoid protease SppA (Lensch et al., 2001), which was more than fivefold upregulated in clpr2-1 but not in the complemented line (Figures 10A and 10B). Preliminary protein gel blot analysis with antiserum against thylakoid protease EGY1 (Chen et al., 2005) showed nearly twofold increased accumulation in the mutant (data not shown). ClpC1,2 chaperones (predominantly accumulating in the stroma) were below detection on the protein gel blots of thylakoids from the wild type. By contrast, both are present at significant levels in thylakoids of clpr2-1, at least 10-fold higher levels than in the wild type (Figure 10A). Protein gel blot analysis of the total soluble leaf proteome of clpr2-1 and the wild type showed that ClpC1 upregulated approximately twofold, and ClpC2 upregulated approximately sevenfold (Figure 10C). Stromal chaperones Cpn60 and Hsp70 showed a sixfold and twofold increase, respectively, in protein accumulation in clpr2-1 on a total soluble leaf proteome basis (Figure 10C).
DISCUSSION
Proteases are involved in numerous aspects of biogenesis and maintenance of chloroplasts, including degradation of cTPs, damaged proteins, and partially assembled complexes, as well as adaptation to environmental conditions and breakdown of chloroplasts during senescence. Given the evolutionary origin of chloroplasts, it is not surprising that all plastid proteases identified so far are homologues of bacterial proteases (reviewed in Adam et al., 2001; Sokolenko et al., 2002; Sakamoto, 2006).
Plant plastids in roots, petals, and leaves contain tetradecameric ClpPR protease core complexes with products of nine different genes and plant-specific ClpS subunits with unknown function (Peltier et al., 2004b). It is unclear why the plastid Clp core complex has evolved to such complexity compared with nonphotosynthetic organisms. Plastid-encoded ClpP1 is essential for shoot development (Shikanai et al., 2001; Kuroda and Maliga, 2003), but it is unknown if the nuclear-encoded components of the Clp core complex (ClpP3-6, ClpR1-4, and ClpS1,2) have any functional redundancy. Functions of the E. coli Clp machinery include degradation of specific regulatory proteins as well as general removal of aggregated and misfolded proteins, including N-end rule substrates (Erbse et al., 2006) and nascent peptide chains stalled on cytosolic ribosomes (for a review, see Sauer et al., 2004). Given its abundance and complexity, the plastid Clp system seems well suited to play a central function in plastid protein homeostasis and biogenesis, reminiscent of the central role of the nuclear/cytosolic proteosome in plant development (Moon et al., 2004; Smalle and Vierstra, 2004).
Downregulation of CLPR2 Gene Expression Results in Reduced ClpR2 and ClpPRS Complex Accumulation
clpr2-1 showed a fivefold reduced accumulation of assembled ClpR2 protein in the chloroplast, based on differential stable isotope labeling with iTRAQ and mass spectrometry. Since ClpR2 has no known conserved catalytic residues and it is part of an oligomeric ClpPRS protease core complex, it was important to determine the accumulation and composition of the ClpPRS complex in clpr2-1. The comparative analysis of the oligomeric chloroplast stromal proteome with 2D gels showed that relative accumulation of the ClpPRS complex was 2.5- to 3-fold reduced.
Employing the relatively new iTRAQ method, we demonstrated that reduced CLPR2 expression resulted in a differential reduction in accumulation of ClpP/R proteins; ClpR2 and ClpR4 were each reduced fivefold, while the other subunits were on average reduced by a factor of 2.5. The ClpPR complex is a double ring structure with seven ClpP/R subunits in each ring. Thus, the complex contains 14 ClpP/R subunits that are normally filled with products of nine different CLPP and CLPR genes. In our previous study, we showed that ClpP4 and ClpP5 are the most abundant proteins, with three copies each, followed by ClpR4, with two copies per complex (Peltier et al., 2004b). The other ClpP/R subunits were estimated to be present in one copy each. Making a simple calculation, the iTRAQ data show an ∼2.5-fold reduction in complex accumulation [2(3 × 0.45) + 2 × 0.2 + 1 × 0.2 + 5(1 × 0.45) = 5.6 and 5.6/14 = 0.40] corresponding to the 2.5- to 3-fold reduction determined from the 2D gels. Although the standard deviations for some of the ClpP,R proteins were quite large, the fivefold downregulation of assembled ClpR2 and ClpR4 and ∼2.5-fold downregulation of most of the other assembled ClpPR subunits must logically result in a heterogeneous ClpPRS complex population. The simplest scenario is that all copies of ClpR2 accumulate in a wild type–like complex, whereas the additional copies of the other ClpP,R subunits assemble into one (or more) modified complex. Judging from the clpr2-1 phenotype, the differential downregulation is not an effective compensatory mechanism.
Reduced CLPRS Complex Accumulation Affects Chloroplast Size, Reduces Thylakoid Accumulation, and Delays Plant Development
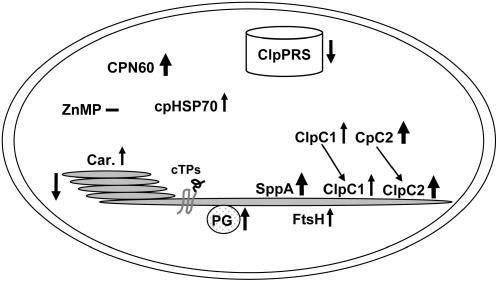
clpr2-1 showed delayed development and flowering and abnormal leaf anatomy and leaf morphology. Chloroplast size was reduced, but this loss of chloroplast volume was compensated for by a higher number of chloroplasts per cell. Correspondingly, the ratio between total chloroplast stromal proteome and total soluble cellular proteome was not changed in fully developed leaves of clpr2-1. By contrast, the amount of thylakoids and associated chlorophyll was twofold to threefold reduced in clpr2-1 both on a total cell basis and a chloroplast basis; consequently, leaves appeared pale green. Most of these phenotypic abnormalities gradually diminished with plant age. Complementation of clpr2-1 showed that each of these phenotypic parameters was linked to the T-DNA insertion in CLPR2. Figure 11 schematically summarizes the consequences of downregulation of CLPR2 for the chloroplast as observed in this study.
Figure 11.
Summary of the Analysis of clpr2-1.
Simplified summary of the consequences of reduced CLPR2 expression for chloroplast protein homeostasis. Arrows indicate the differential response, with thickness of the arrow correlating to the fold change. Car., carotenoid/thylakoid protein ratio; PG, plastoglobule; ZnMP, Zn2+ metalloprotease.
Reduced ClpPRS Complex Accumulation Triggers Upregulation of Plastid Chaperones
Extensive analysis of the homooligomeric ClpP protease complex in bacteria has shown that the core complex needs ATP-dependent ClpA/X chaperones of the HSP100 family to recruit and unfold its substrates. Without these chaperones, the Clp complex can only degrade small peptides (reviewed in Sauer et al., 2004). Chloroplasts contain three Clp chaperones (named ClpC1, ClpC2, and ClpD), and they are homologues to ClpA. Although ClpC1 accumulates predominantly in the stroma, it also associates with the protein import channel located in the inner chloroplast envelope, where it is involved in import of nuclear-encoded chloroplast proteins (Akita et al., 1997; Nielsen et al., 1997). Analysis of null mutants for CLPC1 and CLPC2 have recently been described (Constan et al., 2004; Park and Rodermel, 2004; Sjogren et al., 2004; Kovacheva et al., 2005). The clpc1 null mutant has a pale-green phenotype that appears milder than clpr2-1, while clpc2 mutants have no obvious phenotype. Protein gel blot analysis of total soluble leaf proteins in clpr2-1 (this study) showed that ClpC1 and in particular ClpC2 were severalfold upregulated in clpr2-1 and surprisingly also accumulated at the thylakoid surface (Figure 11). This suggests that the Clp system is actively involved in unfolding and/or degradation of thylakoid proteins or thylakoid protein fragments. This is consistent with observations in the green algae C. reinhardtii, showing that reduced expression of the plastid-encoded CLPP1 gene results in slower degradation rates of the thylakoid-bound subunits of cytochrome b6f and PSII complexes (Majeran et al., 2000, 2001).
Chloroplasts also contain other chaperone systems that do not deliver substrates to the ClpPRS core complex. These are the GroES/GroEL or Cpn60/Cpn21/Cpn10 system, the DnaK/DnaJ or Hsp70/GrpE system, Hsp90, and the Hsp100 protein ClpB3 (Cao et al., 2003). These chaperone systems are constitutively expressed and are involved in several aspects of plastid biogenesis and protein homeostasis, including protein sorting, protein (un)folding, and assembly (Jackson-Constan et al., 2001; van Wijk, 2001). We observed that in particular the 800-kD Cpn60 complex was strongly upregulated in clpr2-1. This Cpn60α,β complex is a tetradecameric barrel-like structure with two stacked rings of seven subunits and a central pore in which proteins can be (re)folded in an ATP-dependent manner. An Arabidopsis null mutant in Cpn60α was seedling lethal (Apuya et al., 2001). The upregulation of Cpn60 in clpr2-1 suggests an increased need for protein (re)folding in the chloroplast.
Unexpected Defects in Thylakoid Biogenesis and Upregulation of Thylakoid Proteases
The carotenoid levels in the thylakoids of clpr2-1 were significantly increased, both when normalized to thylakoid protein and when normalized to total chlorophyll, and are most likely an indication of increased levels of oxidative stress. The reduced efficiency of PSII (or accumulation of inactive PSII complexes) and the increased accumulation of plastoglobules are consistent with increased oxidative and/or other stresses. These plastoglobules are lipid-rich, thylakoid-associated particles, and recent plastoglobule proteome analyses clearly showed that these particles isolated from clp2-1 and the wild type both contain similar sets of enzymes involved in various tocopherol and other isoprenoid-related pathways as well as fatty acid/lipid metabolism (Vidi et al., 2006; Ytterberg et al., 2006).
When we compared the purified thylakoids isolated from developed leaf rosettes of clpr2-1 and the wild type, we found very little change in accumulation of the subunits of the photosynthetic electron transport chain. Surprisingly, thylakoids of clpr2-1 accumulated low levels of several unprocessed chlorophyll a/b binding proteins of PSII (LHCII-1,1,2,3,4,5 and LHCII-3) and unprocessed PsaF, an integral membrane protein of PSI with a cleavable lumenal transit peptide. Processing of PsaF was restored by complementation with CLPR2:h.
Classical in vitro studies have shown that unprocessed LHCII (pLHCII) proteins can integrate into the thylakoid membrane (Chitnis et al., 1986; Cline, 1986). However, under optimal conditions, pLHCII is processed rapidly by the general stromal processing peptidase (SPP) in the stroma, followed by targeting and insertion into the thylakoid membrane (reviewed in Keegstra and Cline, 1999). It is highly unlikely that the Clp protease complex is directly involved in processing of the cTPs. The delayed and reduced accumulation of the thylakoid membrane system in clpr2-1 and this accumulation of unprocessed thylakoid membrane proteins suggest that the removal of the cTP is coupled more tightly with thylakoid biogenesis than suggested from in vitro studies. Alternatively, processing by SPP (Richter and Lamppa, 1998) is occurring at suboptimal rates due to a general imbalance of chloroplast protein homeostasis, possibly engaging most chaperones in removal of protein aggregates rather than refolding and facilitating SPP-dependent processing.
We also found accumulation of LHCII breakdown products in clpr2-1 but not in the wild type. Interestingly, clpr2-1 thylakoids showed increased expression levels of two ATP-dependent metalloproteases, FtsH2,5, and the thylakoid-bound ATP-independent Ser-type SppA protease (Lensch et al., 2001). In particular, SppA was dramatically upregulated (>10-fold) in clpr2-1 but was normal in the complemented line. Substrates for SppA are currently not known nor has an Arabidopsis mutant for SppA been analyzed. A large number of studies have analyzed the thylakoid FtsH proteases and showed that FtsH1,2,5,8 form thylakoid complexes that degrade unassembled thylakoid proteins and are involved in the D1 repair cycle of PSII (Yu et al., 2005; Zaltsman et al., 2005; Sakamoto, 2006). Thus, the upregulation of these thylakoid proteases in clpr2-1 suggests an accelerated turnover of thylakoid proteins; these could be unassembled proteins that have accumulated in excess or proteins that need removal because of (oxidative) damage. We note that while the FtsH mutants show variegated phenotypes, clpr2-1 is not variegated. Interestingly, a ClpC2 mutant was able to suppress variegation in FtsH2 mutants through an unknown mechanism, excluding a direct physical interaction or activation/deactivation (Park and Rodermel, 2004). We favor the interpretation that ClpC2 is needed for thylakoid biogenesis and homeostasis. When ClpC2 levels are low, thylakoid biogenesis is slowed down, removing the need for a threshold concentration of FtsH2 (for a brief discussion, see Adam et al., 2006).
Why Does Reduced ClpPRS Complex Accumulation Lead to an Imbalance in Thylakoid Protein Homeostasis?
The differential upregulation of thylakoid proteases and the accumulation of low levels of LHCII breakdown products and unprocessed LHCII and PsaF proteins suggest an imbalance in thylakoid protein homeostasis (Figure 11). The major photosynthetic complexes in the thylakoid membrane each contain a mixture of nuclear- and chloroplast-encoded proteins that assemble in defined stoichiometries; balanced chloroplast protein homeostasis thus requires coordinated expression of the nuclear and chloroplast genomes (reviewed in Rochaix, 2004). Given its abundance and complexity, the plastid Clp system seems well suited to play a central role in plastid protein homeostasis and biogenesis. The ClpPRS complex might (1) regulate plastid gene expression via turnover of nuclear-encoded proteins that control plastid transcription, mRNA processing, or translation, (2) degrade unassembled and/or damaged proteins and breakdown products generated by thylakoid proteases, and (3) degrade misfolded and aggregated stromal and thylakoid proteins.
Several scenarios are conceivable that explain the surprising imbalanced thylakoid protein homeostasis in clpr2-1. One explanation is that the ClpPRS complex is involved in the turnover of (at least) some of the hundreds of nuclear-encoded proteins that control different aspects of plastid gene expression. Since many (∼30) of the plastid-encoded proteins are part of thylakoid-localized photosynthetic complexes, this would lead to imbalance between nuclear- and plastid-encoded proteins and accumulation of unassembled proteins. Increased rates of proteolysis are needed to remove these unassembled proteins; indeed, upregulation of thylakoid proteases (FtsH and SppA) was observed in this study. The Clp system likely also acts as a secondary protease that degrades breakdown products generated by the (primary) thylakoid proteases, as suggested from accumulation of such products in clpr2-1. It is likely that the ClpPRS complex is also involved in degradation of misfolded and aggregated stromal and thylakoid proteins. During and after import of nuclear-encoded proteins, chaperones are needed for refolding and possibly for the presentation of imported precursor proteins to SPP. Increased engagement of the ClpC,D chaperones in removal of aggregates might compete with this protein import and cTP processing, thus leaving a subfraction of chloroplast proteins unprocessed, as observed in this study.
ClpR2 Is a Key Component of the ClpPRS Core Complex
The strong phenotype of clpr2-1 demonstrates that CLPR2 is not redundant, despite the presence of three other expressed CLPR homologues. The absence of the three conserved catalytic residues makes it highly unlikely that ClpR2 has peptidase activity itself. Thus, our data suggest that ClpR2 is (1) needed as a structural component of the ClpPRS core and/or (2) provides a unique functional contribution to the core complex. The differential response of the other ClpP,R proteins did not provide an effective compensatory mechanism. Future studies will be needed to further unravel structural and functional relationships and contributions of the ClpP/R subunits.
METHODS
Plant Growth, Mutant Isolation, and Genotyping
JP82_1C09 (later renamed SALK_046378) and wild-type Arabidopsis thaliana (Columbia-0) seeds were surface sterilized, vernalized, and germinated on Murashige and Skoog medium (Sigma-Aldrich) or directly on soil under continuous 100 μmol·m−2·s−1 light at 23°C in controlled growth chambers (Percival). Antibiotic resistance was tested by adding 50 mg/L kanamycin to the medium and showed that the SALK_046378 line had lost kanamycin resistance, which is quite common (see http://signal.salk.edu/tdna_FAQs.html). Genomic DNA was extracted by grinding leaf tissue in extraction buffer (0.2 M Tris-HCl, pH 9.0, 0.4 M LiCl, 25 mM EDTA, and 1% [w/v] SDS) followed by isopropanol precipitation 1:1 (v/v) (adapted from Krysan et al., 1996). PCR was performed using Platinum Taq DNA polymerase (Invitrogen), and primers used were as follows: CLPR2-specific, 5′-CTTCATTGAGATCGTGAGATTGGTT-3′ and 5′-GGCCTGTATTCTTGGCTAGTTCATT-3′; T-DNA–specific (left border), 5′-GGCAATCAGCTGTTGCCCGTCTCACTGGTG-3′, 5′-CACCAGTGAGACGGGCAACAGCTGATTGCC-3′, and 5′-CCCAGGCTTTACACTTTATGCTTCCGGCTC-3′ (Integrated DNA Technologies). For RT-PCR, total RNA was isolated with an RNeasy plant mini kit (Qiagen). First strand was synthesized from equal amounts of total RNA with Superscript III RT-RNaseH (Invitrogen), and primers used were as follows: CLPR2 (At1g12410) 5′-AGATCTGGCGGTCTCGTTTAATACA-3′ and 5′-GCTAGCCCCTGCGCTTTCGTCTTGGC-3′ (nested primers 5′-CATATGTCTGGGACTGGGAAAGCGAGCAGG-3′ and 5′-GAATTCCTAGCCTAGCCCTGCGCTTTCGTC-3′) and ACTIN2 (At3g18780) 5′-GCAACTGGGATGATATGGAAAAGA-3′ and 5′-CAAACGAGGGCTGGAACAAGACT-3′. Twenty cycles were used for ACTIN2 and 20 to 30 for CLPR2. Sequence verification and analyses were performed using VectorNTI Suite 9.0 (Invitrogen).
Complementation
Genomic DNA fragments were amplified using Taq polymerase and primers 5′-CAACTGCAGTTTGCAGCGATATTCCGATGAC-3′, 5′-AGAGGCTAGCTAGTGGTGGTGGTGGTGGTGCCCTGCGCTTTCGTCTTGGCGA-3′, 5′-CTAGGCTAGCTCTTTTTGTTTGTTT-3′, and 5′-CCGGGTACCAAGCTTTTGGTGGGGTGATTC-3′ and subcloned into pGEM-T easy (Promega). The terminator sequence was subsequently inserted downstream of the promoter-CDS sequence as an NheI-KpnI fragment. The reconstituted CLPR2:h gene was then cloned into pCAMBIA-1300 (www.cambia.org) as a PstI-KpnI fragment. Restriction enzymes were from Invitrogen or New England Biolabs. The resulting vector was electroporated into Agrobacterium tumefaciens strain GV3101 (a kind gift of J. Hua) and used for floral dipping according to Clough (2005). Transgenic plants were selected based on hypocotyl length after dark germination under selective pressure for hygromycin resistance (20 mg/L), adapted from Lahser and Wright-Minogue (2003).
Isolation of Intact Chloroplasts and Protein Analysis by 1D and 2D Gels
All samples for protein analysis were from plants grown on soil under a 10-h-light/14-h-dark cycle at 23°/19°C (light/dark) and 100 μmol·m−2·s−1. Intact chloroplasts were purified from leaf rosettes, and thylakoids and stroma were collected as described by Peltier et al. (2004b). Native stromal proteomes of the wild type, clpr2-1, and clpr2-1/R2:h were separated by CN-PAGE as described originally by Schägger et al. (1994) and Peltier et al. (2001, 2004b), followed by SDS-PAGE. Approximately 1 mg of stromal protein was loaded in each lane (for the first dimension). The resulting 2D gels were stained with Sypro Ruby (Invitrogen), and gels were scanned using a CCD camera (FluorS; Bio-Rad). Spot volumes were determined, corrected for background, and normalized to total spot volume using Phoretix software (Genomic Solutions).
Purified thylakoid proteins were separated on 1D tricine gels and stained with Sypro Ruby as by Peltier et al. (2004a) or on 1D Laemmli gels and transferred to a PVDF membrane for protein gel blot analysis with different polyclonal or monoclonal antisera. Protein gel blot analysis was performed using standard procedures using chemiluminescence for detection. Protein loading was between 1 and 100 μg and was optimized for each antiserum. Antisera used in this study were generous gifts from numerous colleagues, namely, anti-SppA from A. Sokolenko, anti-FstH1 and anti-FtsH5 from W. Sakamoto, anti-ClpC1 and anti-ClpC2 from S. Rodermel, anti-PsbS from K.K. Niyogi, and anti-Egy1 from N. Li. Antiserum directed against PsaF and against purified PSI core, cross-reacting with PsaA/B, PsaC, and PsaE/C, were from H. Scheller. Anti-(His)6 was obtained from Sigma-Aldrich. All mass spectrometry–based protein identification was performed after in-gel digestion with trypsin (Promega). Extracted peptides were then analyzed by reverse-phase nanoLC-ESI-MS/MS (Q-TOF 1.5; Waters), with parameters as described by Peltier et al. (2004a). Mass spectral data were submitted to an in-house Mascot 2.0 search engine (www.matrixscience.com) and searched against The Arabidopsis Information Resource Protein Data Bank version 5.0. The Mascot output was automatically processed by in-house software (Q. Sun and K.J. van Wijk, unpublished data) followed by manual verification.
Differential Stable Isotope Labeling with iTRAQ
Stained spots were excised from 2D native gels (wild type and clpr2-1), cut into 1-mm3 cubes, and washed with 50 mM triethylammonium bicarbonate, pH 8.0, and 100% acetonitrile (ACN) (1:1). After trypsin digestion, the peptides were extracted four times with 5% formic acid (FA) and 100% ACN (1:1) followed by dehydration with 100% ACN. After drying in a vacuum concentrator, each extract was resuspended in dissolution buffer containing 0.5 M triethylammonium bicarbonate and labeled with the iTRAQ reagents according to the manufacturer's instructions (Applied Biosystems). The samples were then incubated at room temperature for 60 min. After labeling, the pairs of wild-type and clpr2-1 samples were combined (e.g., gel band 1 wild type with gel band 1 from the mutant), dried down, and reconstituted in 5% FA. The samples were then desalted and concentrated using zip tips (C18; Millipore), with repeated passage of the flow-through over the zip tip to maximize peptide recovery. The eluates from the zip tips were collected in one tube (total 100 μL), dried, and resuspended in 15 μL of 5% FA. The labeled peptide mixtures were separated by online reverse-phase nanoLC-ESI-MS/MS using 150-min gradients with 95% water, 5% ACN, and 0.1% FA (solvent A) and 95% ACN, 5% water, and 0.1% FA (solvent B) at a flow rate of 200 nL/min. Data acquisition and processing of MS/MS spectra were performed as described above, except that the collision energy values were increased by 15%. The area of the light and heavy isotope tags were calculated using Mascot Distiller after applying the correction factors as indicated by the manufacturer, using in-house written software (Q. Sun and K.J. van Wijk, unpublished data).
Microscopy
The fixation protocol was modified from Gluaert (1975). Briefly, the leaf materials were excised and placed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate, pH 6.8, for 0.5 h at room temperature, 10 min of which was under vacuum. The samples were then transferred to 4°C for 1.5 h. Following fixation, the samples were washed with 0.1 M sodium cacodylate buffer, pH 6.8, for three 10-min incubations at 4°C. After washing, the samples were placed at 4°C overnight in 1% osmium tetroxide. After three buffer washes, the samples were dehydrated through a graded series of ethanol. After dehydration, the samples were infiltrated through a graduated series of ethanol and Spurr's resin and then embedded in 100% Spurr's resin with accelerator curing at 70°C overnight. Images were taken on an FEI TECNAI 12 BioTwin microscope (Philips). For confocal microscopy, fresh mature leaves were mounted in water. Imaging was performed on a Leica TCS SP2 laser scanning confocal microscope.
Miscellaneous
As a measure of the efficiency of PSII, Fv/Fm values from 10-min dark-adapted leaves were measured on the wild type, clpr2-1, and clpr2-1/R2:h using a chlorophyll fluorescence instrument (FMS2; HansaTech). For starch staining, leaves were decolorized in hot 80% (v/v) ethanol, stained with iodine solution, and washed several times with water (Zeeman et al., 1998). Chlorophyll concentrations were determined spectrometrically in 80% acetone (Lichtenthaler, 1987), and protein determinations were done with the bicinchoninic acid assay (Smith et al., 1985) and the Bradford assay (Bradford, 1976).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: ClpP1, ATCG00670; ClpP3, At1g66670; ClpP4, At5g45390; ClpP5, At1g02560; ClpP6, At1g11750; ClpR1, At1g49970; ClpR2, At1g12410; ClpR3, At1g09130; ClpR4, At4g17040; ClpS1, At4g25370; ClpS2, At4g12060; ClpC1, At5g50920; ClpC2, At3g48870; SppA, At1g73990; CPN60α1, At2g28000; CPN60-β1, At3g13470; CPN60β2, At1g55490; ZnMP-1, At3g19170; ZnMP-2, At1g49630; FtsH5, At5g42270; FtsH2, At2g30950; cpHSP70-1, At4g24280; cpHSP70-2, At5g49910; LHCII-1.1, At1g29910; LHCII-1.2, At1g29920; LHCII-1.3, At1g29930; LHCII-1.4, At2g34430; LHCII-1.5, At2g34420; LHCII-3, At5g54270; and PsaF, At1g31330.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Proteins Identified from a Two-Dimensional CN-PAGE and SDS-PAGE Gel with Stroma from Purified Intact Chloroplasts of the Wild Type and clpr2-1.
Supplemental Table 2. iTRAQ-Labeled Peptides That Were Used for Quantification of the Subunits of the ClpPRS Complex and the Reference Protein THI1.
Supplemental Table 3. Proteins Identified from a One-Dimensional Gel with Purified Thylakoids from the Wild Type and clpr2-1.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation (MCB 0343444) and the U.S. Department of Energy (DE-FG02-04ER15560) to K.J.v.W. We thank all members of the van Wijk laboratory, Jian Hua, and Thomas Owens for their generous help and suggestions. We also thank Steven Rodermel, Zach Adam, Anna Sokolenko, Krishna K. Niyogi, Wataru Sakamoto, Hendrik Scheller, and Ning Li for generous gifts of various antisera and Mandayam Parthasarathy and Anita Aluisio of the electron microscopy facility at Cornell University for the TEM analysis.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Klaas J. van Wijk (kv35@cornell.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.042861.
References
- Adam, Z., Adamska, I., Nakabayashi, K., Ostersetzer, O., Haussuhl, K., Manuell, A., Zheng, B., Vallon, O., Rodermel, S.R., Shinozaki, K., and Clarke, A.K. (2001). Chloroplast and mitochondrial proteases in Arabidopsis. A proposed nomenclature. Plant Physiol. 125 1912–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam, Z., and Clarke, A.K. (2002). Cutting edge of chloroplast proteolysis. Trends Plant Sci. 7 451–456. [DOI] [PubMed] [Google Scholar]
- Adam, Z., Rudella, A., and van Wijk, K.J. (2006). Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol. 9 234–240. [DOI] [PubMed] [Google Scholar]
- Akita, M., Nielsen, E., and Keegstra, K. (1997). Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 136 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Apuya, N.R., Yadegari, R., Fischer, R.L., Harada, J.J., Zimmerman, J.L., and Goldberg, R.B. (2001). The Arabidopsis embryo mutant schlepperless has a defect in the chaperonin-60alpha gene. Plant Physiol. 126 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker-Astrom, K., Schelin, J., Gustafsson, P., Clarke, A.K., and Campbell, D.A. (2005). Chlorosis during nitrogen starvation is altered by carbon dioxide and temperature status and is mediated by the ClpP1 protease in Synechococcus elongatus. Arch. Microbiol. 183 66–69. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid a sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Cao, D., Froehlich, J.E., Zhang, H., and Cheng, C.L. (2003). The chlorate-resistant and photomorphogenesis-defective mutant cr88 encodes a chloroplast-targeted HSP90. Plant J. 33 107–118. [DOI] [PubMed] [Google Scholar]
- Chandu, D., and Nandi, D. (2004). Comparative genomics and functional roles of the ATP-dependent proteases Lon and Clp during cytosolic protein degradation. Res. Microbiol. 155 710–719. [DOI] [PubMed] [Google Scholar]
- Chen, G., Bi, Y.R., and Li, N. (2005). EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J. 41 364–375. [DOI] [PubMed] [Google Scholar]
- Chitnis, P.R., Harel, E., Kohorn, B.D., Tobin, E.M., and Thornber, J.P. (1986). Assembly of the precursor and processed light-harvesting chlorophyll a/b protein of Lemna into the light-harvesting complex II of barley etiochloroplasts. J. Cell Biol. 102 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, K. (1986). Import of proteins into chloroplasts. Membrane integration of a thylakoid precursor protein reconstituted in chloroplast lysates. J. Biol. Chem. 261 14804–14810. [PubMed] [Google Scholar]
- Clough, S.J. (2005). Floral dip: Agrobacterium-mediated germ line transformation. Methods Mol. Biol. 286 91–102. [DOI] [PubMed] [Google Scholar]
- Constan, D., Froehlich, J.E., Rangarajan, S., and Keegstra, K. (2004). A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiol. 136 3605–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSouza, L., Diehl, G., Rodrigues, M.J., Guo, J., Romaschin, A.D., Colgan, T.J., and Siu, K.W. (2005). Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and cICAT with multidimensional liquid chromatography and tandem mass spectrometry. J. Proteome Res. 4 377–386. [DOI] [PubMed] [Google Scholar]
- Erbse, A., Schmidt, R., Bornemann, T., Schneider-Mergener, J., Mogk, A., Zahn, R., Dougan, D.A., and Bukau, B. (2006). ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439 753–756. [DOI] [PubMed] [Google Scholar]
- Flynn, J.M., Neher, S.B., Kim, Y.I., Sauer, R.T., and Baker, T.A. (2003). Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11 671–683. [DOI] [PubMed] [Google Scholar]
- Friso, G., Giacomelli, L., Ytterberg, A.J., Peltier, J.B., Rudella, A., Sun, Q., and Wijk, K.J. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell 16 478–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluaert, A.M. (1975). Fixation, Dehydration and Embedding of Biological Specimens. (New York: North-Holland/American Elsevier).
- Gomez, S.M., Bil, K.Y., Aguilera, R., Nishio, J.N., Faull, K.F., and Whitelegge, J.P. (2003). Transit peptide cleavage sites of integral thylakoid membrane proteins. Mol. Cell. Proteomics 2 1068–1085. [DOI] [PubMed] [Google Scholar]
- Gribun, A., Kimber, M.S., Ching, R., Sprangers, R., Fiebig, K.M., and Houry, W.A. (2005). The ClpP double ring tetradecameric protease exhibits plastic ring-ring interactions, and the N termini of its subunits form flexible loops that are essential for ClpXP and ClpAP complex formation. J. Biol. Chem. 280 16185–16196. [DOI] [PubMed] [Google Scholar]
- Hardt, M., Witkowska, H.E., Webb, S., Thomas, L.R., Dixon, S.E., Hall, S.C., and Fisher, S.J. (2005). Assessing the effects of diurnal variation on the composition of human parotid saliva: Quantitative analysis of native peptides using iTRAQ reagents. Anal. Chem. 77 4947–4954. [DOI] [PubMed] [Google Scholar]
- Hwang, B.J., Park, W.J., Chung, C.H., and Goldberg, A.L. (1987). Escherichia coli contains a soluble ATP-dependent protease (Ti) distinct from protease La. Proc. Natl. Acad. Sci. USA 84 5550–5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, K., Baldwin, A.J., Shipman, R.L., Matsui, K., Theg, S.M., and Ohme-Takagi, M. (2005). Complete maturation of the plastid protein translocation channel requires a type I signal peptidase. J. Cell Biol. 171 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Constan, D., Akita, M., and Keegstra, K. (2001). Molecular chaperones involved in chloroplast protein import. Biochim. Biophys. Acta 1541 102–113. [DOI] [PubMed] [Google Scholar]
- Kang, S.G., Dimitrova, M.N., Ortega, J., Ginsburg, A., and Maurizi, M.R. (2005). Human mitochondrial ClpP is a stable heptamer that assembles into a tetradecamer in the presence of ClpX. J. Biol. Chem. 280 35424–35432. [DOI] [PubMed] [Google Scholar]
- Katayama-Fujimura, Y., Gottesman, S., and Maurizi, M.R. (1987). A multiple-component, ATP-dependent protease from Escherichia coli. J. Biol. Chem. 262 4477–4485. [PubMed] [Google Scholar]
- Keegstra, K., and Cline, K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwin, P.M., Elderfield, P.D., and Robinson, C. (1987). Transport of proteins into chloroplasts. Partial purification of a thylakoidal processing peptidase involved in plastocyanin biogenesis. J. Biol. Chem. 262 16386–16390. [PubMed] [Google Scholar]
- Kovacheva, S., Bedard, J., Patel, R., Dudley, P., Twell, D., Rios, G., Koncz, C., and Jarvis, P. (2005). In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 41 412–428. [DOI] [PubMed] [Google Scholar]
- Krause, G.H., and Weis, E. (1991). Chlorophyll fluorescence and photosynthesis: The basics. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42 313–349. [Google Scholar]
- Krysan, P.J., Young, J.C., Tax, F., and Sussman, M.R. (1996). Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc. Natl. Acad. Sci. USA 93 8145–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda, H., and Maliga, P. (2003). The plastid clpP1 protease gene is essential for plant development. Nature 425 86–89. [DOI] [PubMed] [Google Scholar]
- Lahser, F.C., and Wright-Minogue, J. (2003). Selection of hygromycin-resistant Arabidopsis seedlings. Biotechniques 34 28–30. [DOI] [PubMed] [Google Scholar]
- Lensch, M., Herrmann, R.G., and Sokolenko, A. (2001). Identification and characterization of SppA, a novel light-inducible chloroplast protease complex associated with thylakoid membranes. J. Biol. Chem. 276 33645–33651. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 148 350–382. [Google Scholar]
- Majeran, W., Olive, J., Drapier, D., Vallon, O., and Wollman, F.A. (2001). The light sensitivity of ATP synthase mutants of Chlamydomonas reinhardtii. Plant Physiol. 126 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran, W., Wollman, F.A., and Vallon, O. (2000). Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b(6)f complex. Plant Cell 12 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizi, M.R., Thompson, M.W., Singh, S.K., and Kim, S.H. (1994). Endopeptidase Clp: ATP-dependent Clp protease from Escherichia coli. Methods Enzymol. 244 314–331. [DOI] [PubMed] [Google Scholar]
- Moon, J., Parry, G., and Estelle, M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi, K., Ito, M., Kiyosue, T., Shinozaki, K., and Watanabe, A. (1999). Identification of clp genes expressed in senescing Arabidopsis leaves. Plant Cell Physiol. 40 504–514. [DOI] [PubMed] [Google Scholar]
- Nielsen, E., Akita, M., Davila-Aponte, J., and Keegstra, K. (1997). Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J 16 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelmuller, R., Herrmann, R.G., and Pakrasi, H.B. (1996). Molecular studies of CtpA, the carboxyl-terminal processing protease for the D1 protein of the photosystem II reaction center in higher plants. J. Biol. Chem. 271 21848–21852. [DOI] [PubMed] [Google Scholar]
- Park, S., and Rodermel, S.R. (2004). Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis. Proc. Natl. Acad. Sci. USA 101 12765–12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier, J.B., Cai, Y., Sun, Q., Zabrouskov, V., Giacomelli, L., Rudella, A., Ytterberg, A.J., Rutschow, H., and van Wijk, K.J. (2006). The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell. Proteomics 5 114–133. [DOI] [PubMed] [Google Scholar]
- Peltier, J.B., Ripoll, D.R., Friso, G., Rudella, A., Cai, Y., Ytterberg, J., Giacomelli, L., Pillardy, J., and Van Wijk, K.J. (2004. b). Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J. Biol. Chem. 279 4768–4781. [DOI] [PubMed] [Google Scholar]
- Peltier, J.B., Ytterberg, J., Liberles, D.A., Roepstorff, P., and van Wijk, K.J. (2001). Identification of a 350-kDa ClpP protease complex with 10 different Clp isoforms in chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 276 16318–16327. [DOI] [PubMed] [Google Scholar]
- Peltier, J.B., Ytterberg, A.J., Sun, Q., and van Wijk, K.J. (2004. a). New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J. Biol. Chem. 279 49367–49383. [DOI] [PubMed] [Google Scholar]
- Richter, S., and Lamppa, G.K. (1998). A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc. Natl. Acad. Sci. USA 95 7463–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix, J.D. (2004). Genetics of the biogenesis and dynamics of the photosynthetic machinery in eukaryotes. Plant Cell 16 1650–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, P.L., et al. (2004). Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3 1154–1169. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W. (2006). Protein degradation machineries in plastids. Annu. Rev. Plant Biol. 57 599–621. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W., Zaltsman, A., Adam, Z., and Takahashi, Y. (2003). Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15 2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, R.T., et al. (2004). Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell 119 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger, H., Cramer, W.A., and von Jagow, G. (1994). Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217 220–230. [DOI] [PubMed] [Google Scholar]
- Schelin, J., Lindmark, F., and Clarke, A.K. (2002). The clpP multigene family for the ATP-dependent Clp protease in the cyanobacterium Synechococcus. Microbiol. 148 2255–2265. [DOI] [PubMed] [Google Scholar]
- Shanklin, J., DeWitt, N.D., and Flanagan, J.M. (1995). The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: An archetypal two-component ATP-dependent protease. Plant Cell 7 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai, T., Shimizu, K., Ueda, K., Nishimura, Y., Kuroiwa, T., and Hashimoto, T. (2001). The Chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42 264–273. [DOI] [PubMed] [Google Scholar]
- Sjogren, L.L., MacDonald, T.M., Sutinen, S., and Clarke, A.K. (2004). Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol. 136 4114–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., and Vierstra, R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55 555–590. [DOI] [PubMed] [Google Scholar]
- Smith, P.K., Krohn, R.I., Hermanson, G.T., Mallia, A.K., Gartner, F.H., Provenzano, M.D., Fujimoto, E.K., Goeke, N.M., Olson, B.J., and Klenk, D.C. (1985). Measurement of protein using bicinchoninic acid. Anal. Biochem. 150 76–85. [DOI] [PubMed] [Google Scholar]
- Sokolenko, A., Pojidaeva, E., Zinchenko, V., Panichkin, V., Glaser, V.M., Herrmann, R.G., and Shestakov, S.V. (2002). The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr. Genet. 41 291–310. [DOI] [PubMed] [Google Scholar]
- Sprangers, R., Gribun, A., Hwang, P.M., Houry, W.A., and Kay, L.E. (2005). Quantitative NMR spectroscopy of supramolecular complexes: Dynamic side pores in ClpP are important for product release. Proc. Natl. Acad. Sci. USA 102 16678–16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk, K.J. (2001). Proteins involved in biogenesis of the thylakoid membrane. In Regulatory Aspects of Photosynthesis. Advances in Photosynthesis, B. Andersson and E.-M. Aro, eds (Dordrecht, The Netherlands: Kluwer Academic Press), pp. 153–175.
- Vidi, P.A., Kanwischer, M., Baginsky, S., Austin, J.R., Csucs, G., Dörmann, P., Kessler, F., and Bréhélin, C. (2006). Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J. Biol. Chem. 281 11225–11234. [DOI] [PubMed] [Google Scholar]
- Wang, J., Hartling, J.A., and Flanagan, J.M. (1997). The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 91 447–456. [DOI] [PubMed] [Google Scholar]
- Ytterberg, A.J., Peltier, J.B., and van Wijk, K.J. (2006). Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 140 984–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F., Park, S., and Rodermel, S.R. (2004). The Arabidopsis FtsH metalloprotease gene family: Interchangeability of subunits in chloroplast oligomeric complexes. Plant J. 37 864–876. [DOI] [PubMed] [Google Scholar]
- Yu, F., Park, S., and Rodermel, S.R. (2005). Functional redundancy of AtFtsH metalloproteases in thylakoid membrane complexes. Plant Physiol. 138 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman, A., Ori, N., and Adam, Z. (2005). Two types of FtsH protease subunits are required for chloroplast biogenesis and photosystem II repair in Arabidopsis. Plant Cell 17 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman, S.C., Umemoto, T., Lue, W.L., Au-Yeung, P., Martin, C., Smith, A.M., and Chen, J. (1998). A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell 10 1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, B., Halperin, T., Hruskova-Heidingsfeldova, O., Adam, Z., and Clarke, A.K. (2002). Characterization of chloroplast Clp proteins in Arabidopsis: Localization, tissue specificity and stress responses. Physiol. Plant. 114 92–101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.