Abstract
Vitamin B6 represents a highly important group of compounds ubiquitous in all living organisms. It has been demonstrated to alleviate oxidative stress and in its phosphorylated form participates as a cofactor in >100 biochemical reactions. By means of a genetic approach, we have identified a novel mutant, rsr4-1 (for reduced sugar response), with aberrant root and leaf growth that requires supplementation of vitamin B6 for normal development. Cloning of the mutated gene revealed that rsr4-1 carries a point mutation in a member of the PDX1/SOR1/SNZ (for Pyridoxine biosynthesis protein 1/Singlet oxygen resistant 1/Snooze) family that leads to reduced vitamin B6 content. Consequently, metabolism is broadly altered, mainly affecting amino acid, raffinose, and shikimate contents and trichloroacetic acid cycle constituents. Yeast two-hybrid and pull-down analyses showed that Arabidopsis thaliana PDX1 proteins can form oligomers. Interestingly, the mutant form of PDX1 has severely reduced capability to oligomerize, potentially suggesting that oligomerization is important for function. In summary, our results demonstrate the critical function of the PDX1 protein family for metabolism, whole-plant development, and vitamin B6 biosynthesis in higher plants.
INTRODUCTION
Vitamins are crucial compounds that are required for a broad variety of biochemical reactions. For some organisms, including humans, vitamin B6 is not internally synthesized and as such must be part of the nutrient uptake of these organisms. Vitamin B6 vitamers differ in a variable group present at the 4′ position that can constitute either a hydroxyl (pyridoxol [PN]), an aldehyde (pyridoxal [PL]), or an amino (pyridoxamine [PM]) group. Also, vitamin B6 must be phosphorylated (pyridoxal-5′-phosphate [PLP] or pyridoxamine-5′-phosphate) to be active as a cofactor. PLP plays a decisive role in amino acid metabolism, cocatalyzing transamination, decarboxylation, and α,β-elimination reactions of amino acid metabolism (Drewke and Leistner, 2001). Besides these roles, PLP also represents an important cofactor for the degradation of lipids and storage carbohydrates, such as glycogen (Friedrich, 1988; Isler et al., 1988; Combs, 1998). Apart from its function as a cofactor, vitamin B6 is also thought to act as a protective agent against reactive oxygen species, such as singlet oxygen (Bilski et al., 2000; Chen and Xiong, 2005).
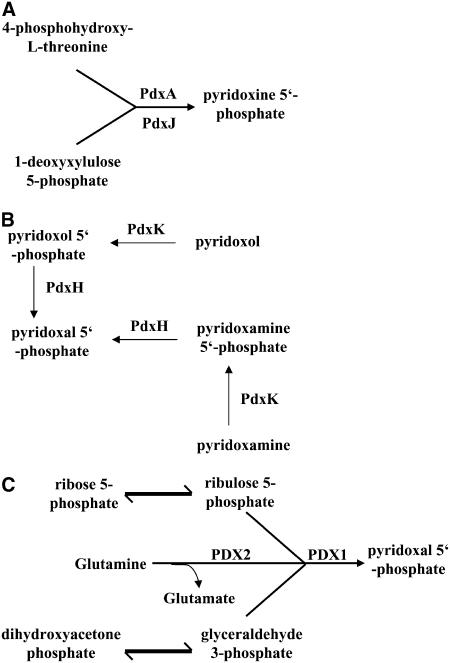
Three pathways have been described for vitamin B6 biosynthesis. First, a de novo deoxyxylulose 5-phosphate (DXP)–dependent pathway that is present in eubacteria, such as Escherichia coli. In this pathway, 4-phosphohydroxy-l-threonine and DXP are used as precursors for the synthesis of pyridoxine, which is catalyzed by two pyridoxine synthase proteins, PdxA and PdxJ (Drewke et al., 1993, 1996; Laber et al., 1999) (Figure 1A). Secondly, the salvage pathway, which has been described in bacteria, fungi, and plants and requires activity of two enzymes, pdxH and pdxK, a vitamin B6 oxidase, and a vitamin B6 kinase, respectively (Yang et al., 1996; Mittenhuber, 2001) (Figure 1B). In plants, the Arabidopsis thaliana salt-sensitive mutant salt overly sensitive4 (sos4) has been described that carries a mutation in a pdxK homolog (Shi et al., 2002; Shi and Zhu, 2002). Loss of SOS4 leads to altered root hair development and increased sensitivities toward Na+, K+, and Li+ ions, which correlate with an altered cellular ion content (Shi et al., 2002; Shi and Zhu, 2002). The third possible route of pyridoxine biosynthesis is a de novo DXP-independent pathway, such as those found in bacteria, archaea, and eukarya (Mittenhuber, 2001; Tambasco-Studart et al., 2005) (Figure 1C). The occurrence of this pathway was recently demonstrated in plants, Plasmodium falciparum, and Bacillus subtilis and involves two proteins, PDX1/SOR1/SNZ (for Pyridoxine biosynthesis protein 1/Singlet oxygen resistant 1/Snooze) and PDX2/SNO (further denoted as PDX1 and PDX2, respectively) (Burns et al., 2005; Tambasco-Studart et al., 2005; Gengenbacher et al., 2006). PDX1 and PDX2 act as a PLP synthase using Gln, ribose-5-phosphate (or ribulose-5-phosphate), and glyceraldehyde-3-phosphate (or dihydroxyacetone phosphate) to synthesize PLP (Burns et al., 2005; Gengenbacher et al., 2006; Tambasco-Studart et al., 2005). The PDX1 and PDX2 genes were independently described in different organisms and are often in close proximity in the genome (Tanaka et al., 2005). The Arabidopsis genome contains three PDX1 proteins, located in the cytosol, which are named according to Tambasco-Studart et al. (2005) as PDX1.1, PDX1.2, and PDX1.3. In addition, a PDX2 homolog is found in Arabidopsis (Chen and Xiong, 2005; Tambasco-Studart et al., 2005). It is interesting to note that only PDX1.1 and PDX1.3 function in PLP biosynthesis, whereas PDX1.2 does not (Tambasco-Studart et al., 2005). To date, the knowledge of the in planta function of PDX1 and PDX2 proteins comes exclusively from genetic approaches. Loss of PDX1.3 leads to shorter, pyridoxine-dependent root growth and hypersensitivity toward oxidative stress (Chen and Xiong, 2005), and pdx2 null mutants display an embryo-lethal phenotype (Tambasco-Studart et al., 2005).
Figure 1.
Schematic Drawing of Pyridoxine Biosynthesis.
(A) DXP-dependent pathway.
(B) Salvage pathway.
(C) DXP-independent pathway.
In this study, we describe a mutant, rsr4-1, that was originally identified in an ethyl methanesulfonate–based screen to generate reduced sugar response (rsr) mutants showing a reduced activation of a sugar-responsive patatin class I promoter (Martin et al., 1997). The mutant showed a surprising requirement for vitamin B6 for normal development. Cloning of the mutated gene revealed that rsr4-1 carries a point mutation in a member of the PDX1 family. The mutation caused reduced PL content and broadly affected amino and organic acid metabolism. Furthermore, our work describes expression patterns of all PDX1 genes and demonstrates that members of the family assemble into homodimers and heterodimers with other PDX1 proteins and with PDX2, a process that is strongly affected in the mutated rsr4-1 protein.
RESULTS
rsr4-1 Is Broadly Affected in Plant Development
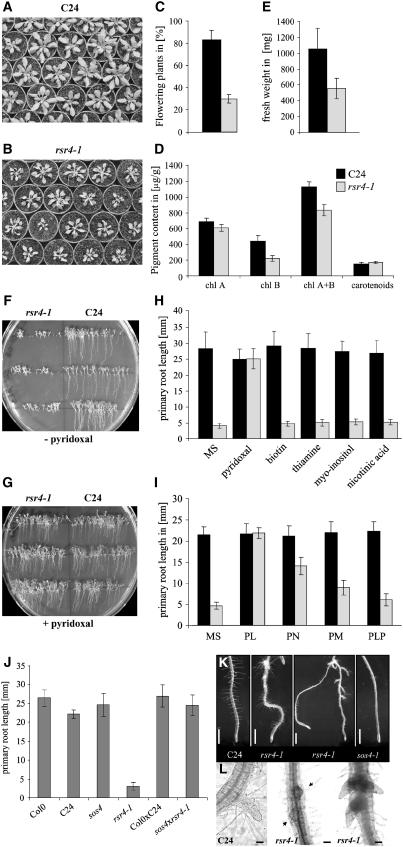
rsr4-1 plants display several prominent aerial and root phenotypes. Flowering time under long-day conditions (16 h light) was delayed, in comparison with wild-type C24 plants (Figure 2C), but mutant plants displayed no change in leaf number at the time of flowering, with an average of 22 rosette leaves (data not shown). However, leaf size was reduced in the mutant, resulting in decreased rosette growth and reduced fresh weight (Figure 2E). rsr4-1 develops yellowish leaves due to an ∼25% reduction of chlorophyll content (Figure 2D). The most striking change in rsr4-1 mutants was a strong reduction in primary root growth in sterile culture on minimal medium (Figures 2F to 2H). Surprisingly, addition of different vitamins to the medium revealed that this phenotype was fully complemented by the provision of pyridoxine to the mutant. This effect was specific to pyridoxine since all other vitamins tested were ineffective in complementing the mutant's root growth (Figure 2H). Likewise, additions of sugar (100 mM glucose and 60 mM sucrose) were also ineffective in reverting the phenotype (data not shown). Interestingly, addition of the different vitamin B6 vitamers to the growth medium revealed that PL was most effective in normalizing rsr4-1 root growth, whereas PLP was ineffective (Figure 2I). The presence of PL in the medium also complemented the aerial phenotypes of reduced leaf growth and bleaching (data not shown).
Figure 2.
Phenotypic Changes Exhibited by rsr4-1.
(A) and (B) Overview of C24 (A) and rsr4-1 (B).
(C) Flowering time is delayed in the mutant. Approximately 85% of C24 wild-type plants flowered after 38 d in comparison with only 30% of rsr4-1.
(D) rsr4-1 has reduced chlorophyll content.
(E) Fresh weight is reduced in the mutant. Values given are averages from n = 60 complete rosettes of C24 and rsr4-1.
(F) and (G) rsr4-1 develops a short root on Murashige and Skoog (MS) medium (F), which can be ameliorated by the addition of PL (n = 30) (G).
(H) and (I) Specificity of PL-dependent complementation of root growth. Roots were measured 10 d after plating (n = 30).
(J) Crossing of the salt-sensitive mutant sos4 into rsr4-1 revealed that in F1 progeny (n = 30), rsr4-1 root growth is complemented.
(K) sos4 has no or very few root hairs; by contrast, rsr4-1 does not have an obvious root hair phenotype. Bars = 1 mm.
(L) rsr4-1 develops a short primary root that has lateral roots budding in close proximity to each other in the absence of PL. Bars = 100 μM. Error bars in this and subsequent figures represent se.
The short root phenotype of rsr4-1 was accompanied by early development of lateral roots, often at the base of the hypocotyl (Figure 2K). In the absence of vitamin B6, we also observed that lateral roots frequently initiated closer to each other in comparison with the wild type (Figures 2K and 2L). Another pyridoxine mutant that was recently described is sos4, which is affected in a pyridoxine kinase (Shi et al., 2002). Like rsr4-1, the sos4 mutation is recessive and leads to reduced root elongation (Shi and Zhu, 2002). However, crossing of rsr4-1 with sos4 resulted in complementation of the root elongation growth in the F1 generation, demonstrating that the mutants are affected at different loci (Figure 2J). sos4 mutants also displayed defects in root hair development (Shi and Zhu, 2002). Comparison of rsr4-1 with sos4 showed that rsr4-1 had no obvious defects in root hair development, further underscoring the difference between the two mutants (Figure 2K). In addition, sos4 is hypersensitive to salt stress (Shi et al., 2002), whereas we could not observe changed salt sensitivities for rsr4-1 mutants (data not shown).
rsr4-1 Carries a Single Recessive Point Mutation on the Upper Arm of Chromosome 5
To analyze the nature and location of the mutated locus within the Arabidopsis genome, a cross between rsr4-1 and Columbia-0 (Col-0) was performed. In the F1 generation, 50 tested plants showed a clear loss of the vitamin B6 dependence, with all plants developing long roots in the absence of the vitamin (data not shown). The short root phenotype segregated in the F2 generation in a 3:1 pattern. From 249 plants, 183 developed a normal root compared with 66 plants with a short root (χ2 = 0.301; χ2 test with one degree of freedom), thus indicating that the pyridoxine-dependent phenotype of rsr4-1 is recessive. To identify the mutated locus in rsr4-1, 55 F2 plants with short roots were selected for a rough mapping approach using cleaved-amplified polymorphic sequence (CAPS) and simple sequence length polymorphic (SSLP) markers as described by Martin et al. (1997). The mutated locus was mapped to the upper arm of chromosome 5 with closest coupling to SSLP markers CTR1,2 (979,763 bp) and nga158 (1,698,614 bp) (Table 1). The use of a new CAPS marker located 506,908 bp below the top of chromosome 5 showed that the number of recombinants was lower at this position in comparison with nga158 (Table 1). Based on Kosambi's map function (Kosambi, 1944), we calculated the position of RSR4 to be within an area of ∼8 centimorgans from the very top of chromosome 5.
Table 1.
Mapping Data for RSR4
| Marker | C24/C24 | C24/Col-0 | Col-0/Col-0 |
|---|---|---|---|
| CTR1,1 | 42 | 9 | 4 |
| nga158 | 45 | 8 | 2 |
| 506908 | 47 | 8 | 0 |
DNA of 55 F2 plants derived from rsr4-1 × Col-0 were used for CAPS and SSLP mapping. Segregation data on specific fragment patterns are given in the order C24/C24, C24/Col-0, and Col-0/Col-0.
rsr4-1 Is Affected in PDX1.3
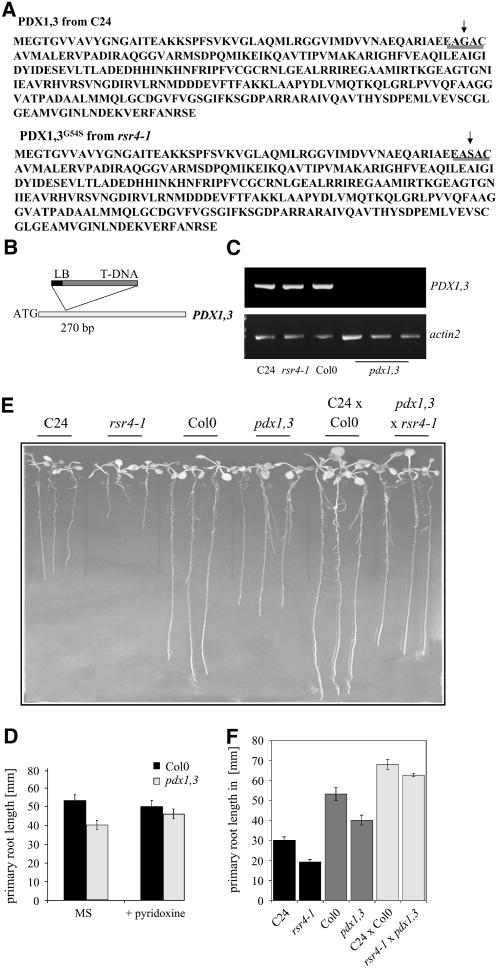
Since rsr4-1 requires pyridoxine for normal growth, it appears to be highly likely that the mutant is affected in biosynthesis of this vitamin. One of the PDX1 genes, PDX1.3, is located on the upper arm of chromosome 5 within the area we identified in our rough mapping approach. Sequencing of the corresponding PDX1.3 open reading frames from C24 wild type and rsr4-1 led to the identification of a point mutation at position 162 from G to A in rsr4-1. This mutation caused an amino acid exchange from Gly to Ser (PDX1.3G54S) (Figure 3A). Unfortunately, it remains unclear what the effect of the mutation is on the protein conformation since structural data from members of the PDX1 family are not available. Using the NNPREDICT program (Kneller et al., 1990), an α-helix is predicted in this area, but no obvious changes appeared to be induced by the mutation.
Figure 3.
rsr4-1 Is Affected in a Member of the PDX1 Family.
(A) A single point mutation at position 54 led to an amino acid exchange from Gly in C24 (top sequence) to Ser in the mutant (bottom sequence). The position of the mutation is indicated with an arrow and underlined.
(B) Scheme of the T-DNA insertion in PDX1.3/RSR4. LB, left border.
(C) RT-PCR analysis of C24, rsr4-1, Col-0, and three independent pdx1.3 plants that were homozygous for a T-DNA insertion.
(D) PDX1.3/RSR4 plants show a slightly shorter root in comparison with Col-0 wild-type plants that can be complemented by supplying PL to the plants.
(E) Phenotype of 12-d-old seedlings from C24, rsr4-1, Col-0, PDX1.3/RSR4, and F1 plants derived from crossings with C24 × Col-0 and pdx1.3 × rsr4-1.
(F) Quantification of primary root length growth 12 d after plating for 20 parental and F1 plants.
To confirm that the mutation found in PDX1.3 is responsible for the rsr4-1 mutant phenotype, we first tried to ectopically express a PDX1.3 cDNA in the rsr4-1 background. Unfortunately, despite several attempts, we obtained only very low transformation efficiency of the mutant, and among the few plants we did obtain, we did not isolate complemented mutants. Alternatively, to corroborate that the PDX1.3G54S mutation is responsible for the rsr4-1 mutant phenotype, an allelism test was performed in which rsr4-1 was crossed into a pdx1.3 T-DNA insertion mutant. For this cross, a T-DNA insertion mutant from the SALK center annotated as SALK_086418 was used (Alonso et al., 2003; Chen and Xiong, 2005). In this mutant, the insertion was located 270 bp downstream of the ATG and led to a complete loss in PDX1.3 expression (Figures 3B and 3C). RT-PCR analysis on C24 and rsr4-1 revealed that the mutation had no impact on PDX1.3G54S expression since expression was invariant in the mutant and wild type. The pdx1.3 mutant showed a weak bleaching of the leaves (data not shown) and, like rsr4-1, a shorter, pyridoxine-dependent root growth (Figure 3D). For allelism testing, the short root phenotype of both mutants was employed. To exclude heterotic effects, the parental ecotypes (Col-0 and C24) were additionally crossed in parallel. Analyses of the F1 generation revealed that the mutant cross displayed shorter roots than the control C24 × Col-0 wild-type cross, and this difference is statistically significant (P < 0.05 for the mutant compared with the wild-type cross; Student's t test) (Figures 3E and 3F), demonstrating that the mutants were allelic to one another. These results confirm that the mutation in PDX1.3 (further denoted as PDX1.3/RSR4) is indeed responsible for the rsr4-1 mutant phenotype.
rsr4-1 Has Reduced Pyridoxine Content Correlating with Major Metabolic Changes
To further analyze changes in rsr4-1, contents of the different vitamin B6 vitamers were measured in rosette leaves from mutant and wild-type plants. PN content in both wild-type and mutant plants was very low and only detectable in one of the three samples analyzed (Table 2, first column). The second derivative, PM, was also low in content, but unlike PN, clearly detectable. Although the amount of PM varied between the different samples (Table 2, second column), no significant changes were detectable between the wild type and rsr4-1. Determination of PL content revealed this to be the most abundant of the three vitamin B6 derivatives. Interestingly, PL content was significantly altered in the mutant (Table 2, third column), being reduced in rsr4-1 to 37% of the level found in C24. This finding is important since it demonstrates that the mutation in rsr4-1 negatively affected vitamin B6 biosynthesis.
Table 2.
Measurements of PN, PM, and PL Contents in Rosette Leaves from 3-Week-Old Soil-Grown Plants
| Experiment | Genotype | PN (μmol/g FW) | PM (μmol/g FW) | PL (μmol/g FW) |
|---|---|---|---|---|
| 1 | C24 | 3.4 | 9.80 | 80.30 |
| 2 | C24 | ND | 2.70 | 48.20 |
| 3 | C24 | ND | 12.60 | 79.80 |
| Mean value (se) | C24 | 3.4 | 8.70 (4.17) | 69.43 (15.02) |
| 1 | rsr4-1 | 1.9 | 14.50 | 30.70 |
| 2 | rsr4-1 | ND | 12.80 | 21.10 |
| 3 | rsr4-1 | ND | 5.40 | 23.60 |
| Mean value (se) | rsr4-1 | 1.9 | 10.90 (3.96) | 25.13 (4.07) |
Differences in PL contents between C24 and rsr4-1 are statistically significant (Student's t test results in P < 0.05). FW, fresh weight; ND, not detectable.
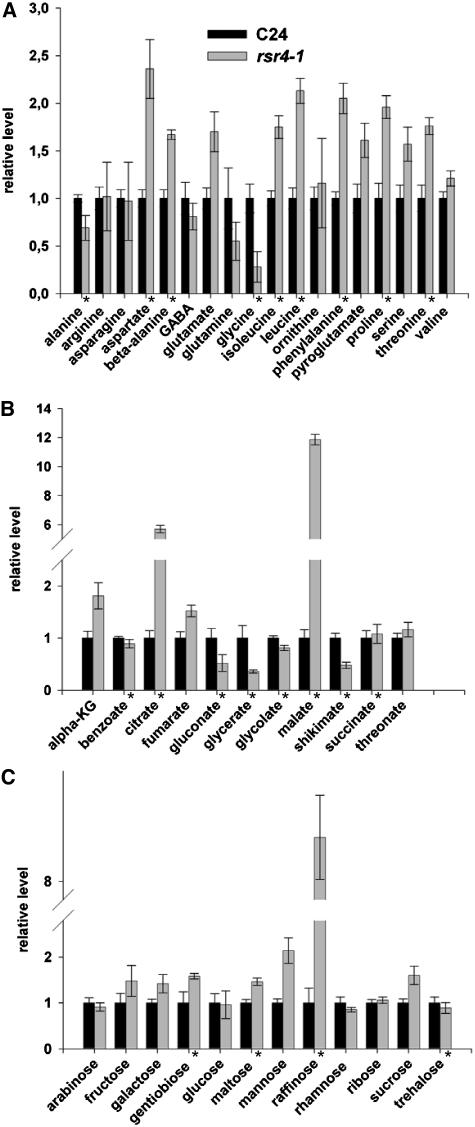
PL is the precursor for PLP, which is a key compound for several steps in amino acid metabolism. To investigate whether the reduced PL content in rsr4-1 negatively affected metabolism, we performed metabolite analysis paying special attention to amino acid biosynthesis in rosette leaves from wild-type and mutant plants. The levels for many of the amino acids detected were increased in rsr4-1, even those that require PLP-dependent enzyme activities for their biosynthesis, such as Ile and Asp (Figure 4A; see Supplemental Table 1 online). It is also noteworthy that constituents of the TCA cycle, such as malate, citrate, and fumarate, were highly upregulated (Figure 4B) and that shikimate, a key compound in indole-3-acetic acid (IAA) biosynthesis, was significantly reduced. Finally, relatively few changes in carbohydrate content were observed in the mutant, with the most marked change being the increased levels of the trisaccharide raffinose in the mutant (Figure 4C). Taken together, these metabolite data do not support the hypothesis that reduced vitamin B6 content correlates with reduction in amino acid biosynthesis. They do, however, reveal that changes in pyridoxine homeostasis have a broad effect on the plant metabolism and underscore the importance of vitamin B6 for metabolism in general.
Figure 4.
Metabolite Contents in Rosette Leaves of rsr4-1 Expressed Relative to C24 Levels.
(A) Contents of amino acids in C24 and rsr4-1. GABA, γ-aminobutyrate.
(B) Contents of organic acids. alpha-KG, α-ketoglutarate.
(C) Contents of sugars. Data have been normalized as described in Methods. Values given are averages from six independent samples. Asterisks mark significant differences (P < 0.05) between C24 and rsr4-1. Quantitative data are provided in Supplemental Table 1 online.
Complementation Analysis of an E. coli pdxJ19 Mutant
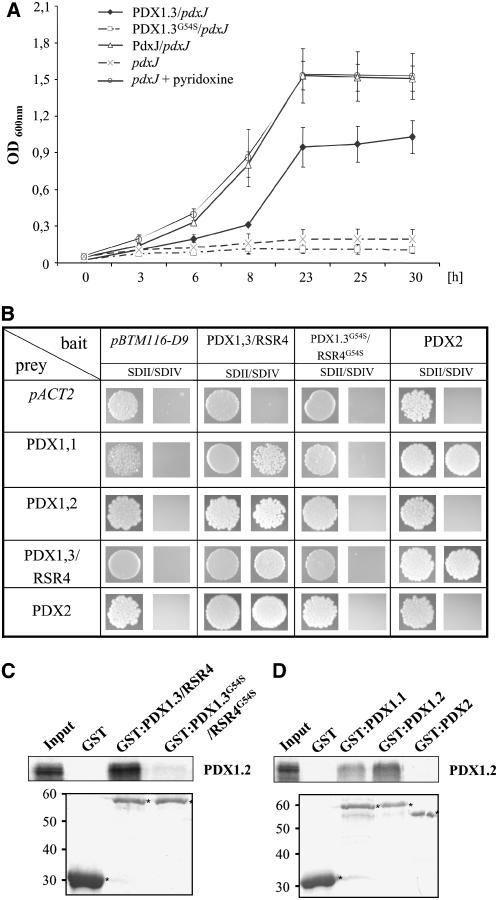
Wetzel et al. (2004) showed earlier that a PDX1 homolog from Cercospora nicotianae can functionally complement a pyridoxine auxotroph pdxJ E. coli mutant. A similar approach was followed to determine enzymatic functioning of PDX1.3/RSR4 and its mutated form from Arabidopsis. Here, both PDX1.3/RSR4 and PDX1.3G54S/RSR4G54S cDNAs were cloned into the isopropylthio-β-galactoside (IPTG)–inducible E. coli expression vector pCRT7/N and used for complementation analysis of a pdxJ19 E. coli mutant. Growth of the pdxJ19 mutant was optically measured at λ = 600 nm for 30 h and compared with the growth of a pdxJ19 mutant transformed with PDX1.3/RSR4, PDX1.3G54S/RSR4G54S, and a pdxJ expression construct in the presence or absence of PL. Figure 5A shows that the pdxJ19 mutant only grew in medium supplemented with PL. By contrast, IPTG-induced expression of both PdxJ and PDX1.3/RSR4 allowed growth of the pdxJ19 strain in PL-free medium, demonstrating complementation of the pdxJ19 mutant and participation of PDX1.3/RSR4 in vitamin B6 biosynthesis. In addition, these results confirm the ability of members of the PDX1 family to functionally complement mutations in a gene of unrelated sequence involved in pyridoxine synthesis through a different pathway. In comparison, the mutated PDX1.3G54S/RSR4G54S protein was not capable of complementing the pdxJ19 strain, which indicates that PDX1.3G54S/RSR4G54S is a nonfunctional protein with respect to vitamin B6 biosynthesis (Figure 5A), and this finding is consistent with the defects observed in the rsr4-1 mutant.
Figure 5.
Functional Analysis of the PDX1 Family.
(A) PDX1.3/RSR4 complements a pdxJ19 E. coli mutant, whereas PDX1.3G54S/RSR4G54S completely failed.
(B) Yeast two-hybrid analysis on the PDX1 family and PDX2. SDII, selection medium for transformation with bait (pBTM112-D9) and prey (pACT2) plasmids supplemented with uracil and His; SDIV, selection medium for interaction studies without uracil and His supplements. Images were taken from single spots.
(C) and (D) Pull-down assays confirm results from the yeast assays for PDX1.1, PDX1.2, PDX1.3, PDX2, and PDX1.3G54S/RSR4G54S. Top panels show pull-down results; bottom panels show amounts of GST and GST fusion proteins used for pull-down assays. First lane of each assay shows 1 μL of [35S]-Met–labeled protein used for pull downs.
Assembly of PDX1 Proteins Is Disrupted by the rsr4-1 Mutation
It was previously described that the yeast PDX1 homolog Snz1p from Saccharomyces cerevisiae assembles into dimers and higher-order protein complexes (Padilla et al., 1998). For this reason, we tested interaction of all Arabidopsis PDX1 proteins and the mutated PDX1.3G54S/RSR4G54S. The different PDX1 genes were cloned in yeast two-hybrid vectors for interaction analysis. As shown in Figure 5B, PDX1.3/RSR4 assembled with a second PDX1.3 protein but was also able to interact with PDX1.1 and PDX1.2 in yeast two-hybrid assays. Comparable interaction patterns we observed for PDX1.1 and PDX1.2 in yeast (data not shown). PDX1.1 and PDX1.3/RSR4 could also assemble with PDX2, the second subunit of PLP synthase. Here, it is noteworthy that PDX1.2 did not assemble with PDX2. This finding is critical since Tambasco-Studart et al. (2005) reported that PDX1.1 and PDX1.3 but not PDX1.2 possess PLP biosynthetic activity. Finally, PDX1.3G54S/RSR4G54S completely failed to interact in the yeast assays with PDX1 and PDX2 proteins (Figure 5B).
To consolidate the data from the yeast two-hybrid experiments, pull-down assays were performed with glutathione S-transferase (GST) fusion proteins and in vitro–translated and [35S]-Met–labeled proteins. As a first approach, we tested in vitro–translated PDX1.1, PDX1.2, and PDX1.3G54S/RSR4G54S for interaction with GST fusion proteins. However, all proteins except for PDX1.2 bound unspecifically to GST and glutathione beads (data not shown). We therefore decided to focus on in vitro–translated PDX1.2 and to combine this PDX1 protein with affinity-purified GST and GST fusion proteins from E. coli. As shown in Figures 5C and 5D, GST:PDX1.3/RSR4 interacted with in vitro–translated PDX1.2, whereas the GST control did not. By contrast, hardly any interaction between PDX1.3G54S/RSR4G54S and PDX1.2 was detectable. Finally, both GST:PDX1.1 and GST:PDX1.2 assembled with in vitro–translated PDX1.2, but GST:PDX2 did not. In summary, these results indicate that PDX1 proteins function in variable protein complexes and demonstrate that replacement of a single residue in PDX1.3 has a profound effect on complex formation.
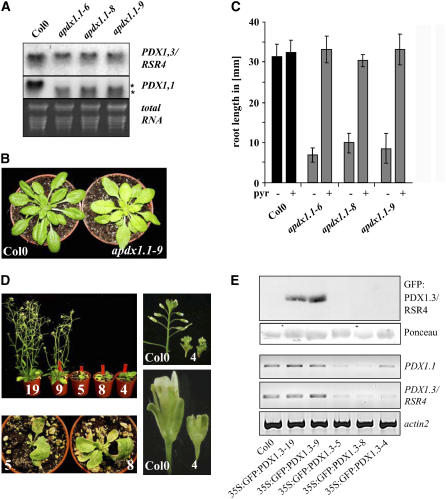
Reduced PDX1.1 Expression Leads to rsr4-1–Related Phenotypes
Based on the high homology between PDX1.1 and PDX1.3/RSR4, their ability to dimerize with each other and with PDX2, and their participation in PLP biosynthesis (Tambasco-Studart et al., 2005), we next asked whether PDX1.1 and PDX1.3/RSR4 are functionally redundant in planta. We used an antisense approach against PDX1.1 to address this question. For this purpose, a 464-bp C-terminal fragment of PDX1.1 was cloned in antisense orientation into the binary vector pCB302-3 (Xiang et al., 1999). Introduction of the construct into Col-0 wild-type plants resulted in 20 out of 86 of the primary transformants displaying yellowish leaves and shorter roots (Figure 6A), with this phenotype correlating reduced PDX1.1 gene expression (Figure 6B). Most importantly, PDX1.1 antisense lines generated a reduced and pyridoxine-dependent root growth comparable to rsr4-1 (Figure 6C). Intriguingly, the antisense effect was largely specific to PDX1.1 since PDX1.3/RSR4 expression was only slightly reduced in the antisense plants (Figure 6B). The pyridoxine-dependent root growth and appearance of yellowish leaves of the PDX1.1 antisense plants demonstrate overlapping functions of PDX1.1 and PDX1.3/RSR4 and are in keeping with the results of Tambasco-Studart et al. (2005), in suggesting that both proteins participate in vitamin B6 biosynthesis in planta.
Figure 6.
Reduced PDX1.1 Expression Leads to rsr4-1-Related Phenotypes, Indicating Functional Redundancy of the Two PDX1 Proteins.
(A) RNA gel blot analysis of three independent transgenic lines showing the expression of a partial cDNA of 466 bp in length from the C-terminal part of PDX1.1 in antisense orientation (here referred to as apdx1.1). All three plants strongly expressed the antisense construct (bottom band and marked by an asterisk). Hardly any expression was detectable for the endogenous PDX1.1.
(B) and (C) In comparison with Col-0 wild-type plants, apdx1.1 plants developed yellowish leaves (B) and shorter primary roots ([C]; n = 30). As such, they display a similar phenotype to rsr4-1. Root growth was recovered to wild-type levels by supplementation with PL. pyr, PL.
(D) Introduction of a 35S:GFP:PDX1.3 construct into Col-0 led to dramatic developmental changes in 20% of generated plants. Numbers indicate different transgenic lines.
(E) Protein gel blot (top panels) and RT-PCR analysis (bottom panels). Transgenic plants with wild type–like development expressed GFP:PDX1.3. By contrast, plants with strong mutant phenotypes did not express GFP:PDX1.3/RSR4 and had strongly reduced expression levels of both PDX1.1 and PDX1.3/RSR4 genes. To specifically detect endogenous gene expression, RT-PCR was performed using primers amplifying 5′- and 3′-untranslated regions.
Cosuppression of Both PDX1.1 and PDX1.3 Leads to Highly Abnormal Leaf and Flower Development
To investigate the effect of PDX1.3/RSR4 overexpression on plant development, we introduced a GFP:PDX1.3/RSR4 expression construct under the control of a 35S promoter (further referred to as 35S:GFP:PDX1.3) into Col-0 wild-type plants. Most of the resultant transformants displayed normal wild type–like growth. In these wild type–like plants, expression of GFP:PDX1.3/RSR4 was easily detectable (Figure 6E, 35S:GFP:PDX1.3-9 and 35S:GFP:PDX1.3-19), suggesting that overexpression of PDX1.3/RSR4 has no major impact on the plant. However, ∼20% of transformants were dramatically affected in growth: they had only very few, yellowish, and distorted rosette leaves (Figure 6D) and either died prior to the development of inflorescences (Figure 6D, 35S:GFP:PDX1.3-5 and -8) or generated small flowers that produced no seeds (Figure 6D, 35S:GFP:PDX1.3-4). It is important to note that the phenotypes were much stronger than the ones we found for rsr4-1 and apdx1.3 plants. Since we did not detect GFP:PDX1.3/RSR4 in these plants (Figure 6E, 35S:GFP:PDX1.3-5, -8, and -4), we assumed that introduction of the 35S:GFP:PDX1.3 construct led to cosuppression of both PDX1.1 and PDX1.3/RSR4. As shown in Figure 6E, RT-PCR analysis on wild-type and transgenic plants revealed that the aberrant growth clearly correlated with a reduced expression of both PDX1 genes, whereas no change was detectable for the plants expressing GFP:PDX1.3/RSR4 protein.
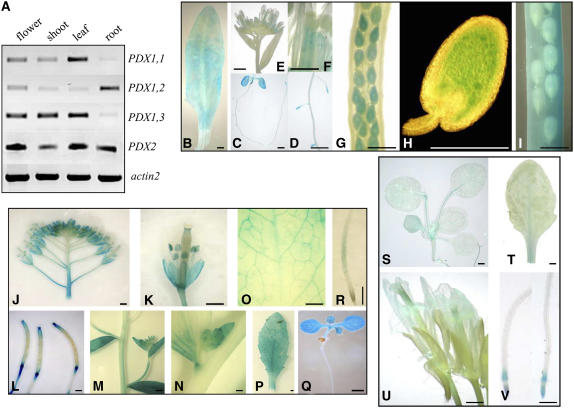
Expression Analysis of the Arabidopsis PDX1 Family and PDX2
To examine in which tissues the different members of the PDX1 family and PDX2 are expressed, RT-PCR and PDX1 promoter:GUS analysis were performed. As shown in Figure 7A, PDX1.3/RSR4 is expressed in flowers, shoots, and leaves but only very weakly in root tissue. This was surprising since the most striking phenotype of rsr4-1 was root related. A similar expression pattern was observed for PDX1.1, whereas PDX1.2 showed low expression in shoot and leaf, with clearly stronger expression in flower and root. In comparison, PDX2 was strongly expressed in all tissues examined.
Figure 7.
Expression Analysis of PDX1 and PDX2 Genes.
(A) RT-PCR analysis of different plant tissues (flower, shoot, leaf, and root) for the PDX1 gene family and PDX2.
(B) to (I) PDX1.1 promoter:GUS analysis of leaf (B), 4-d-old seedling (C), lateral roots (D), flowers ([E] and [F]), siliques ∼5 d after fertilization (G) and corresponding seed (H), and differentiated siliques (I).
(J) to (Q) PDX1.3/RSR4 promoter:GUS analysis of flower tissue ([J] and [K]), siliques (L), shoot and cauline leaves (M), budding shoots (N), rosette leaves ([O] and [P]), 4-d-old seedling (Q), and root (R).
(S) to (V) PDX1.2 promoter:GUS analysis of 10-d-old seedling (S), leaf (T), flower (U), and root tips (V).
Bars = 1 mm.
For more detailed description of the PDX1 expression patterns, the different promoters of PDX1.1 (1511 bp), PDX1.2 (1808 bp), and PDX1.3/RSR4 (1136 bp) were cloned in front of a GUS reporter gene. As expected, promoter:GUS lines from PDX1.1 and PDX1.3/RSR4 (further referred to as proPDX1.1:GUS and proPDX1.3:GUS, respectively) had overlapping expression patterns being highly active in leaves and only weakly so in roots. However, proPDX1.1:GUS lines showed staining in tips of primary and lateral roots (Figure 7D), a pattern that was not observed in the proPDX1.3:GUS lines (Figure 7R). Another difference in expression was present in flowers and siliques: proPDX1.1:GUS lines showed a strong dot-like staining in pistils (Figure 7F). Furthermore, at young silique stages (∼5 d after pollination), strong staining in the endosperm was detectable in proPDX1.1:GUS lines, with no staining present in the embryo (Figure 7H). At later stages when seeds were fully developed, staining was restricted to the surrounding mantle tissue of the siliques (Figure 7I). In comparison, proPDX1.3:GUS lines showed strong staining in anthers and sepals (Figures 7J and 7K). Furthermore, proPDX1.3:GUS lines showed GUS expression only at the top and base of differentiated siliques, with no staining detectable in the seeds at all stages (Figure 7L). In proPDX1.2:GUS seedlings, a weak expression was present in the leaf vascular tissue (Figure 7S). Hardly any staining was detectable in differentiated leaves where GUS staining only appeared in the central vein and similarly weak; in flowers, we observed GUS expression only in the pistil (Figure 7U). Most interestingly, in the proPDX1.2:GUS lines was a root tip–specific expression pattern: in emerging root buds, only the very tip showed GUS expression (data not shown), whereas in older roots, a second area appeared, shortly after the root tip, that showed strong GUS expression (Figure 7V).
DISCUSSION
Vitamin B6 is known to protect against oxidative stress and in its phosphorylated form represents an important cofactor for a variety of biochemical processes (Ehrenshaft et al., 1999; Drewke and Leistner, 2001). Despite these facts, the effect of vitamin B6 on plant development and the mechanisms controlling biosynthesis of the vitamin are still poorly understood. The work presented here shows that rsr4-1 plants are directly affected in PDX1.3/RSR4. This was demonstrated on the one hand by a map-based cloning approach and on the other hand by allelism testing against a pdx1.3 null mutant. At this point, we cannot fully account for the observed low transformation efficiency and the lack of PDX1.3 transgene expression in and thereby complementation of rsr4-1. However, we suspect that the presence of the mutated PDX1.3G54S/RSR4G54S protein interfered with both ectopic expression of a PDX1.3/RSR4 cDNA and Agrobacterium tumefaciens–mediated transformation of the mutant. While preparing this article, two T-DNA insertion mutants in PDX1.3/RSR4 exhibiting the same phenotypic changes in root and shoot development that we observed for rsr4-1 were described (Chen and Xiong, 2005). We have subsequently confirmed that one of these T-DNA lines (SALK_086418) is allelic to rsr4-1. However, with respect to stress tolerance, there is a clear difference between rsr4-1 and pdx1.3; for example, Chen and Xiong (2005) described pdx1.3 null mutants as salt sensitive similar to sos4. We failed to observe similar behavior in rsr4-1; however, it remains possible that this is related to the enhanced Pro and raffinose contents of this mutant. Pro is a known compatible compound that protects against a variety of stress conditions, such as drought and salt stress (Nanjo et al., 1999; Hellmann et al., 2000; Yonamine et al., 2004). A similar role is described for raffinose in the context of the amelioration during various abiotic stress factors (Taji et al., 2002; Zuther et al., 2004). This difference between rsr4-1 and pdx1.3 indicates that in contrast with pdx1.3, PDX1.3G54S/RSR4G54S is not a simple loss-of-function mutation since the presence of the mutated protein appears to exhibit an additional metabolic impact. It will be interesting in the future to assess whether similar metabolic changes occur in the pdx1.3 null mutant to those detected here for rsr4-1. Despite the fact that the mutants displayed some differences, many of the results presented here are in keeping with those described previously. For example, our in planta measurements of reduced PL levels in rsr4-1 that demonstrate the requirement of PDX1.3 for vitamin B6 biosynthesis further support the recently published in vitro data of Tambasco-Studart et al. (2005), who showed activity in PLP biosynthesis for PDX1.1 and PDX1.3/RSR4. In addition, our complementation analyses of the E. coli pdxJ19 mutant with PDX1.3/RSR4 and PDX1.3G54S/RSR4G54S confirm the results from Wetzel et al. (2004) and further underscore that the G54S point mutation disrupted biosynthetic activity of the protein.
In B. subtilis, Burns et al. (2005) described that dimerization of PDX1 and PDX2 produces an active PLP synthase, and a similar situation is postulated for the Arabidopsis PLP synthase (Tambasco-Studart et al., 2005). In addition, Padilla et al. (1998) demonstrated that S. cerevisiae PDX1 proteins interact with PDX2 and other PDX1 family members and might even hypothetically participate in larger protein complexes. Our findings in both yeast two-hybrid and pull-down experiments provide strong support for the idea of dimerization-induced activity: first, the assembly of Arabidopsis PDX1.1 and PDX1.3/RSR4 into both homodimers and heterodimers failed when paired with the PDX1.3G54S/RSR4G54S mutant; second, we were able to detect interaction of PDX1.1 and PDX1.3 with PDX2. Interestingly, PDX1.2, which is not active as a PLP synthase (Tambasco-Studart et al., 2005), also assembled with PDX1.1 and PDX1.3 but failed to interact with PDX2. In this context, it is also noteworthy that PDX1.2 did not complement the pdxJ19 E. coli mutant (data not shown). Hence, these findings imply that in planta PLP synthase activity is coupled with the ability of PDX1 proteins to interact with PDX2.
It is surprising that rsr4-1 or reduced PDX1.1 expression alone had such dramatic effects on Arabidopsis development. Given their similar activity, location, and expression patterns, PDX1.1 and PDX1.3/RSR4 would have been expected to be functionally equivalent and thus capable of complementing for the loss of one another. However, the related phenotypes of rsr4-1/pdx1.3 and PDX1.1 antisense lines clearly demonstrated that this is not the case. Rather, it indicates a situation in which both are required to act synergistically to establish vitamin B6 homeostasis. The dissection of the specific functions of the two PDX1 proteins will be a central focus of our future work on the PDX1 protein family.
rsr4-1 mutant plants are evidently affected in whole-plant development. This becomes obvious by the dramatic changes in pyridoxine-dependent root growth and late flowering behavior. One potential explanation for the yellowish leaf phenotype of both pdx1 mutants could be that they harbor defects in heme and consequently chlorophyll biosynthesis, which requires activity of the PLP-dependent δ-aminolevulinate synthase (Papenbrock and Grimm, 2001). However, we cannot currently formally exclude the possibility that this phenotype may merely reflect the considerable metabolic changes displayed by the mutant. The low expression of both PDX1.1 and PDX1.3/RSR4 in root tissue is also striking especially in context of the clear reduction in primary root growth in the absence of PL supplementation. One potential explanation for the aberrant root development could be that the mutant displays a restricted supply of photosynthate to the root. However, the metabolite profiles clearly demonstrated that content of the major transported amino acids and sugars, such as asparagine and sucrose, were comparable to the wild type or even higher in the mutant, and the aerial phenotype in the mutant is not consistent with a reduced transport function (Riesmeier et al., 1993). When taken together, these arguments strongly imply that the shorter root is not merely a consequence of reduced supply of these metabolites. Alternatively, the aberrant root growth of rsr4-1 can be connected to reduced shikimate levels in the mutant. This compound represents a key precursor for biosynthesis of the phytohormone IAA, which is a critical determinant for normal root development (Knaggs, 2001; Poupart et al., 2005). Our observation of weak expression of PDX1.3/RSR4 within the root elongation zone also suggests that it is likely that a local supply of PLP is required to support root elongation. However, it is also possible that the roots requirement for pyridoxine is mainly met by supply from the photosynthesizing tissues of the plant, since these organs show strongest expression of PDX1.1 and PDX1.3/RSR4. Such a situation would require that plants encode for vitamin B6 transporters, such as those previously described for yeast Transport of pyridoxine protein1 (Tpn1p) (Stolz and Vielreicher, 2003). Although our search of the Arabidopsis databases for related Tpn1p proteins did not yield any suitable candidates, this does not necessarily mean that the vitamin is not transported within the plant. For example, Tpn1p belongs to the purine-cytosine permease family, and permeases with similar substrate specificities are present in Arabidopsis (Bürkle et al., 2003; Wormit et al., 2004). In this context, it is interesting to mention that we observed complementation of the aerial rsr4-1 phenotype by feeding B6 vitamers through the root, which indicates transport capacities of the plant for the vitamin. Hence, it will be interesting in the future to test whether these Arabidopsis proteins possess transport affinities for PN, PL, or PM, to determine contents of pyridoxine and its derivates in different Arabidopsis organs, and to perform isotope tracer studies at the whole-plant level.
Originally rsr4-1 was found in a screen for mutants that showed aberrant sugar-dependent regulation of a patatin class I promoter from potato (Solanum tuberosum) displaying a reduced response to exogenously supplied sucrose and glucose (Martin et al., 1997). These authors also clearly demonstrated that basic mechanisms controlling patatin promoter activity are conserved between potato and Arabidopsis. As for the pyridoxine-deficient phenotype, a rough mapping approach localized the mutation responsible for reduced GUS activity to the upper arm of chromosome 5 (Martin et al., 1997). The clear genetic coupling of both phenotypes, reduced GUS expression and reduced root elongation, makes it likely that they are both caused by the same point mutation in PDX1.3/RSR4. An influence of PDX1.3/RSR4 on patatin promoter activity appears to be likely since the promoter is regulated by both sugar and Gln; in this context, the metabolic flux from source to sink organs appears to be the crucial regulatory factor (Peña-Cortéz et al., 1992; Martin et al., 1997). The dramatic changes in amino acid content and in some carbohydrates, which can be taken as an indication of changes in transport or metabolism of these compounds in the mutant, could conceivably have resulted in a reduced patatin promoter activity. The suggestion of altered IAA metabolism (Chen and Xiong, 2005; this study) also presents a possible explanation for the fact that a sugar sensing screen resulted in an isolate affected in pyridoxine metabolism. Many ABA mutants were isolated in a similar fashion (Arenas-Huertero et al., 2000; Laby et al., 2000), and recent studies indicate similar links between sugar and auxin metabolism (Moore et al., 2003; Loreti et al., 2005). Another attractive alternative is that transcription factors specific for cis-regulatory elements within the patatin promoter are affected by the rsr4-1 mutation. For example, the patatin promoter contains a consensus element called the B-box. This element is proposed to be responsible for sugar-inducible and tuber-specific expression of the patatin gene in potato (Zourelidou et al., 2002). As for the potato tuber, the root is a typical sink tissue, and the patatin promoter is mainly active in these organs (Peña-Cortéz et al., 1992; Martin et al., 1997). Recently, Zourelidou et al. (2002) showed that the B-box is the binding site for a new class of transcription factor that was named Storekeeper. Interestingly, several Storekeeper-related proteins are encoded in Arabidopsis (e.g., At4g00270), which might specifically bind to the B-box element like their homologs from potato. Thus, it is feasible to assume that activity or expression of these transcription factors can be negatively affected in pdx1-3/rsr4-1, leading to reduced patatin promoter activity. However, establishment of the interconnectivity of PDX1.3/RSR4 activity and patatin promoter-dependent gene expression will constitute part of our future work on the Arabidopsis PDX1 family.
Conclusions
This work demonstrates that PDX1.3/RSR4 is required for vitamin B6 biosynthesis in planta. Mutation in rsr4-1 strongly affected both development and metabolism of the mutant, and this is most likely the result of the reduced PDX1.3G54S/RSR4G54S ability to interact with other members of the PDX1 family and PDX2. These findings are of general consequence since they give novel insights in mechanisms controlling vitamin B6 biosynthesis in higher plants and emphasize the significance of this compound for plant development and metabolism. In the future, it will be vital to dissect the individual roles of each member of the PDX1 family. It will also be of interest what kind of substrates are used in planta by PDX1 proteins and what kind of additional PDX1 interacting proteins are required to achieve efficient biosynthesis of vitamin B6.
METHODS
Tissue Culture, Plant Growth, and Transformation
Arabidopsis thaliana ecotypes C24 and Col-0 were grown in a greenhouse under standard conditions and in tissue culture on solid MS medium (Murashige and Skoog, 1962). For root elongation growth assays, vitamins were supplemented to the medium (2.43 μM PN, PM, PL, or PLP, 2.5 μM biotine, 0.3 μM thiamine, 560 μM myo-inositol, and 4.06 μM nicotinic acid; Sigma-Aldrich) as recommended by Murashige and Skoog (1962). rsr4-1 was backcrossed twice into C24 to eliminate secondary mutations and to outcross the Pat(B33)-GUS transgene prior to performance of phenotypical and physiological analysis. Plants were transformed using an Agrobacterium tumfaciens–mediated transfer protocol according to Clough and Bent (1998).
Mapping of RSR4
For map-based cloning of RSR4, F2 plants derived from a cross between rsr4-1 and Arabidopsis Col-0 were selected for reduced root growth on MS medium without PL. Genomic DNA was isolated by standard techniques and used for SSLP- and CAPS-based mapping of the mutated loci as described earlier (Martin et al., 1997). PCR products for the CAPS marker were directly digested with KpnI to detect polymorphism. For primer sequences used in this and following sections, see Supplemental Table 2 online.
Generation of DNA Constructs
PDX1.3/RSR4 promoter and full-length PDX1 open reading frames were amplified from genomic DNA. The PDX1.3/RSR4 promoter (1136 bp upstream from PDX1.3/RSR4 ATG) was cloned into the BamHI-XbaI sites of the binary vector pCB308 (Xiang et al., 1999). For antisense constructs, PDX1.3/RSR4 open reading frames in the suitable orientation were cut out with SacI-EcoRV and cloned into the SacI-SmaI sites of the binary vector pCB302-3 (Xiang et al., 1999). For expression in Escherichia coli, PDX1.3/RSR4 and PDX1.3/RSR4G54S were cloned into pCRT7/N (Invitrogen). PdxJ from E. coli was cloned into a modified pACS1 vector (Clontech). For GST fusion constructs, full-length cDNAs were cloned into the GATEWAY-compatible pDEST15 expression vector. GFP construct was generated in binary vector pK7FWG2 (Karimi et al., 2002). All clones were fully sequenced and controlled for correct sequence and orientation before use in the different assays.
Complementation Analysis
For complementation analysis, an E. coli pdxJ19 mutant (http://cgsc.biology.yale.edu:80/cgi-bin/sybgw/cgsc/Strain/11585; Taylor, 1972) was used. For assays, the pdxJ19 mutant was transformed with either wild-type pdxJ from E. coli or PDX1.3/RSR4 and PDX1.3/RSR4G54S from Arabidopsis. The different strains were grown overnight in 100 mL of Luria-Bertani medium supplemented with PL (0.5 μM) and thiamine (1 μg/mL). One milliliter of the overnight cultures was added to 100 mL of M9 minimal medium (Sambrook and Russell, 2001) supplemented with 0.3 mM IPTG (Sigma-Aldrich) and thiamine (1 μg/mL). Only in the case of untransformed pdxJ19 mutant was M9 culture medium supplemented with PL (0.5 μM) to control growth in parallel to a pdxJ19 culture without PL. Growth at 37°C over 30 h was optically quantified at λ = 600 nm with a Kontron Uvicon 860 photometer.
Quantitation of Vitamin B6
Extraction of vitamin B6 was performed following a modified procedure described by Bognar and Ollilainen (1997) using 25 g of rosette leaves from 21-d-old soil-grown C24 and rsr4-1 plants. The content of B6 vitamers was analyzed according to a modified HPLC procedure described earlier (Arenz et al., 1996). Each of the three samples was measured twice to assure for correct quantification of B6 vitamers.
Metabolic and Pigment Quantification
For determination of metabolite content, an established gas chromatography–mass spectrometry procedure (Roessner et al., 2001; Roessner-Tunali et al., 2003) that had been adopted for Arabidopsis tissues (Giege et al., 2003) was used. For this purpose, rosette leaves (100 mg fresh weight) were harvested from 21-d-old soil-grown Arabidopsis plants. Having confirmed that the absolute values determined by this method were comparable with those previously recorded in Arabidopsis using other published methods (Carrari et al., 2005), it was decided to present data as relative values with respect to the wild-type control as described by Roessner et al. (2001). For chlorophyll quantification, rosette leaves (200 mg fresh weight) from 30-d-old plants cultured in soil were ground in liquid nitrogen, and carotenoids and chlorophyll a/b were extracted twice with 80% acetone. Pigment content was immediately photometrically measured under dimmed light conditions and quantified according to Lichtenthaler (1987).
RNA Isolation and Expression Analysis
For tissue-specific expression analysis of PDX1.3/RSR4G54S and PDX1.3/RSR4 null mutants, 4-week-old, flowering, soil-grown plants were used. For RT-PCR analysis, RNA was isolated using a plant RNA isolation kit (Macherey and Nagel). RT-PCR was done using gene-specific primers (see Supplemental Table 2 online) and a one-step RT-PCR kit (Qiagen). For RNA gel blot analysis, RNA isolation, hybridization, and signal detection were done using standard techniques. The 5′- and 3′-untranslated areas of PDX1.3/RSR4 and PDX1.1 were used alongside the primers mentioned above for RT-PCR analysis to allow specific hybridization.
Yeast Two-Hybrid Assays
A lexA-based yeast two-hybrid system was used as described by Weber et al. (2005) with pBTM116-D9 (kindly provided by Erich Wanker) as bait plasmid and pACT2 (Clontech; GenBank accession no. U29899) as prey plasmid. Both vectors were modified with a GATEWAY cassette (Invitrogen). cDNAs of PDX1 and PDX2 genes were cloned into pDONR221 (Invitrogen) and subsequently introduced into pBTM116-D9 and pACT2. For yeast two-hybrid assays, yeast cells were transformed with bait and prey plasmid constructs as described before (Weber et al., 2005). Cells were grown on synthetic dextrose (SD) minimal medium (Sambrook and Russell, 2001) supplemented with Leu and His (SDII). Selected colonies were diluted 1:2000 in autoclaved distilled water before transfer to SD minimal medium without supplements (SDIV) for control of interaction. Photographs of single drops from diluted colonies were taken 3 d after transfer.
Pull-Down Assays and Protein Gel Blot Analysis
In vitro–translated PDX1.2 and PDX2 were synthesized with the TNT reticulocyte lysate system (Promega) using l-[35S] trans-labeled Met (Amersham). For pull-down assays from reticulolysate, PDX1.2 and PDX2 were incubated at 4°C for 2 h with GST, GST:PDX1.1, GST:PDX1.2, GST:PDX1.3, GST:PDX1.3G54S, and GST:PDX2. Binding assays and washings were done in standard buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, and 0.2% Nonidet P-40) and as described before (Hellmann et al., 2003). Proteins were resolved on SDS-PAGE. Products were detected by autoradiography. Protein extraction and GFP immunodetection were done as described before (Hellmann et al., 2003) using a GFP antibody from Santa Cruz Biotechnology (catalog no. sc-8334).
GUS Staining and Microscopy
For detection of GUS expression, seedlings and different tissues were incubated up to 24 h in GUS staining solution at 37°C according to Jefferson et al. (1989). Pictures were taken with an Olympus C-4040 zoom digital camera mounted on a SZX12 stereomicroscope (Olympus) or an Axioskop 2 plus microscope (Zeiss).
Vitamin B6 Quantification and pdxJ19 Bioassays
Determination of vitamin B6 was performed following a modified procedure described by Bognar and Ollilainen (1997). In detail, 25 g (fresh weight) of rosette material of 3-week-old soil-grown Arabidopsis C24 wild type and rsr4-1 mutant plants were powdered in liquid nitrogen and suspended in 45 mL of 0.1 M HCl. The suspension was boiled for 30 min at 120°C in a pressure cooker. After centrifugation of the suspension for 30 min at 15,000g, the supernatant was filled up to 50 mL with 0.1 M HCl. For quantitative analysis of the three vitamin B6 derivatives, the pH of this extract was adjusted to 4.8, and a volume of 12.5 mL was further subjected to enzymatic hydrolysis by adding 2 mg of acid phosphatase and 15 mg of β-glucosidase (Sigma-Aldrich) solved in a total volume of 2 mL. This solution was gently shaken at 37°C in a water bath for 18 h. The pH was then adjusted to 3.0, and the solution was boiled for 15 min at 100°C to stop the enzymatic reaction. After centrifugation at 15,000g for 30 min, the supernatant was subjected to HPLC analysis or to a qualitative bioassay as described below. Control reactions with standards of the phosphorylated B6 vitamers showed that under the applied conditions the enzymatic hydrolysis was quantitative. Note that the weight of the starting material of Arabidopsis wild-type and mutant rosettes was exactly equal and that the extraction procedure was performed in parallel in exactly the same manner. The content of B6 vitamers PN, PL, and PM was determined according to a modified procedure described earlier (Arenz et al., 1996). As eluents, solvents A (13 mM trifluoric acid), B (0.5 M potassium phosphate buffer, pH 5.0), and C (0.5 sodium phosphate buffer, pH 5.6) were used running a ternary gradient as follows: elution was started with 100% solvent A (0 to 15 min), increasing buffer B from 0 to 100% (15 to 20 min), followed by an increase of buffer C from 0 to 100% (25 to 30 min). After elution, the column was reconditioned with 100% A (35 to 45 min). Pyridoxine-hydrochloride, pyridoxal-hydrochloride, and PM dihydrochloride (Fluka) served as standards.
Accession Numbers
Sequence from this article can be found in the GenBank/EMBL data libraries under accession numbers PDX1.1 (At2g38230), PDX1.2 (At3g16050), PDX1.3 (At5g01410), and PDX2 (At5g60540).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Metabolite Contents of C24 and rsr4-1.
Supplemental Table 2. Primers Used for the Different Methodical Approaches.
Supplementary Material
Acknowledgments
We thank Sutton Mooney for critical reading, Jiang-Kang Zhu for donating sos4 seeds, Erich Wanker for pBTM116-D9 vector donation, and Alexandra Meier, Verena Schade, and the gardeners for excellent technical help and care of the plants. This work was supported in part by the Deutsche Forschungsgemeinschaft (HE3224/6-1 to H.H. and J.E.L.) and by the Department of Energy DOE (DE-FG02-04ER15542 to W.B.F.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hanjo Hellmann (hellmann@zedat.fu-berlin.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.036269.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero, F., Arroyo, A., Zhou, L., Sheen, J., and Leon, P. (2000). Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 14 2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Arenz, A., Klein, M., Fiehe, K., Groß, J., Drewke, C., Hemscheidt, T., and Leistner, E. (1996). Occurrence of neurotoxic 4-O-methylpyridoxine in ginkgo biloba leaves, ginkgo medications and Japanese ginkgo food. Planta Med. 62 548–551. [DOI] [PubMed] [Google Scholar]
- Bilski, P., Li, M.Y., Ehrenshaft, M., Daub, M.E., and Chignell, C.F. (2000). Vitamin B6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem. Photobiol. 71 129–134. [DOI] [PubMed] [Google Scholar]
- Bognar, A., and Ollilainen, V. (1997). Influence of extraction on the determination of vitamin B6 in food by HPLC. Z. Lebensm. Unters. Forsch. 204 327–335. [Google Scholar]
- Bürkle, L., Cedzich, A., Dopke, C., Stransky, H., Okumoto, S., Gillissen, B., Kuhn, C., and Frommer, W.B. (2003). Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J. 34 13–26. [DOI] [PubMed] [Google Scholar]
- Burns, K.E., Xiang, Y., Kinsland, C.L., McLafferty, F.W., and Begley, T.P. (2005). Reconstitution and biochemical characterization of a new pyridoxal-5′-phosphate biosynthetic pathway. J. Am. Chem. Soc. 127 3682–3683. [DOI] [PubMed] [Google Scholar]
- Carrari, F., Coll-Garcia, D., Schauer, N., Lytovchenko, A., Palacios-Rojas, N., Balbo, I., Rosso, M., and Fernie, A.R. (2005). Deficiency of a plastidial adenylate kinase in Arabidopsis results in elevated photosynthetic amino acid biosynthesis and enhanced root growth. Plant Physiol. 137 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., and Xiong, L. (2005). Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stresses. Plant J. 44 396–408. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Combs, G.F., Jr. (1998). The Vitamins, Fundamental Aspects in Nutrition and Health. (San Diego, CA: Academic Press).
- Drewke, C., Klein, M., Clade, D., Arenz, A., Muller, R., and Leistner, E. (1996). 4-O-phosphoryl-L-threonine, a substrate of the pdxC(serC) gene product involved in vitamin B6 biosynthesis. FEBS Lett. 390 179–182. [DOI] [PubMed] [Google Scholar]
- Drewke, C., and Leistner, E. (2001). Biosynthesis of vitamin B6 and structurally related derivatives. Vitam. Horm. 61 121–155. [DOI] [PubMed] [Google Scholar]
- Drewke, C., Notheis, C., Hansen, U., Leistner, E., Hemscheidt, T., Hill, R.E., and Spenser, I.D. (1993). Growth response to 4-hydroxy-L-threonine of Escherichia coli mutants blocked in vitamin B6 biosynthesis. FEBS Lett. 318 125–128. [DOI] [PubMed] [Google Scholar]
- Ehrenshaft, M., Chung, K.R., Jenns, A.E., and Daub, M.E. (1999). Functional characterization of SOR1, a gene required for resistance to photosensitizing toxins in the fungus Cercospora nicotianae. Curr. Genet. 34 478–485. [DOI] [PubMed] [Google Scholar]
- Friedrich, W. (1988). Vitamins. (Berlin and New York: De Gruyter).
- Gengenbacher, M., Fitzpatrick, T.B., Raschle, T., Flicker, K., Sinning, I., Muller, S., Macheroux, P., Tews, I., and Kappes, B. (2006). Vitamin B6 biosynthesis by the malaria parasite Plasmodium falciparum: Biochemical and structural insights. J. Biol. Chem. 281 3633–3641. [DOI] [PubMed] [Google Scholar]
- Giege, P., Heazlewood, J.L., Roessner-Tunali, U., Millar, A.H., Fernie, A.R., Leaver, C.J., and Sweetlove, L.J. (2003). Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15 2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann, H., Funck, D., Rentsch, D., and Frommer, W.B. (2000). Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiol. 122 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann, H., Hobbie, L., Chapman, A., Dharmasiri, S., Dharmasiri, N., del Pozo, C., Reinhardt, D., and Estelle, M. (2003). Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis. EMBO J. 22 3314–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isler, O., Brubacher, G., Ghisla, S., and Kräutler, B. (1988). Vitamine II. (Stuttgart and New York: Thieme).
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1989). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Knaggs, A.R. (2001). The biosynthesis of shikimate metabolites. Nat. Prod. Rep. 18 334–355. [DOI] [PubMed] [Google Scholar]
- Kneller, D.G., Cohen, F.E., and Langridge, R. (1990). Improvements in protein secondary structure prediction by an enhanced neural network. J. Mol. Biol. 214 171–182. [DOI] [PubMed] [Google Scholar]
- Kosambi, D.D. (1944). The estimation of map distance from recombination values. Ann. Eugen. 12 172–175. [Google Scholar]
- Laber, B., Maurer, W., Scharf, S., Stepusin, K., and Schmidt, F.S. (1999). Vitamin B6 biosynthesis: Formation of pyridoxine 5′-phosphate from 4-(phosphohydroxy)-L-threonine and 1-deoxy-D-xylulose-5-phosphate by PdxA and PdxJ protein. FEBS Lett. 449 45–48. [DOI] [PubMed] [Google Scholar]
- Laby, R.J., Kincaid, M.S., Kim, D.G., and Gibson, S.I. (2000). The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 23 587–596. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. (1987). Chlorophylls and carotenoids, the pigments of photosynthetic biomembranes. Methods Enzymol. 148 350–382. [Google Scholar]
- Loreti, E., Poggi, A., Novi, G., Alpi, A., and Perata, P. (2005). A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol. 137 1130–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, T., Hellmann, H., Schmidt, R., Willmitzer, L., and Frommer, W.B. (1997). Identification of mutants in metabolically regulated gene expression. Plant J. 11 53–62. [DOI] [PubMed] [Google Scholar]
- Mittenhuber, G. (2001). Phylogenetic analyses and comparative genomics of vitamin B6 (pyridoxine) and pyridoxal phosphate biosynthesis pathways. J. Mol. Microbiol. Biotechnol. 3 1–20. [PubMed] [Google Scholar]
- Moore, B., Zhou, L., Rolland, F., Hall, Q., Cheng, W.H., Liu, Y.X., Hwang, I., Jones, T., and Sheen, J. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signalling. Science 300 332–336. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Nanjo, T., Kobayashi, M., Yoshiba, Y., Kakubari, Y., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Lett. 461 205–210. [DOI] [PubMed] [Google Scholar]
- Padilla, P.A., Fuge, E.K., Crawford, M.E., Errett, A., and Werner-Washburne, M. (1998). The highly conserved, coregulated SNO and SNZ gene families in Saccharomyces cerevisiae respond to nutrient limitation. J. Bacteriol. 180 5718–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenbrock, J., and Grimm, B. (2001). Regulatory network of tetrapyrrole biosynthesis studies of intracellular signalling involved in metabolic and developmental control of plastids. Planta 213 667–681. [DOI] [PubMed] [Google Scholar]
- Peña-Cortéz, H., Liu, X., Sanchez-Serrano, J., Schmid, R., and Willmitzer, L. (1992). Factors affecting expression of patatin and proteinase inhibitor tt gene families in detached potato leaves: implications for their co-expression in developing tubers. Planta 186 495–502. [DOI] [PubMed] [Google Scholar]
- Poupart, J., Rashotte, A.M., Muday, G.K., and Waddell, C.S. (2005). The rib1 mutant of Arabidopsis has alterations in indole-3-butyric acid transport, hypocotyl elongation, and root architecture. Plant Physiol. 139 1460–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier, J.W., Flugge, U.I., Schulz, B., Heineke, D., Heldt, H.W., Willmitzer, L., and Frommer, W.B. (1993). Antisense repression of the chloroplast triose phosphate translocator affects carbon partitioning in transgenic potato plants. Proc. Natl. Acad. Sci. USA 90 6160–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner, U., Luedemann, A., Brust, D., Fiehn, O., Willmitzer, L., and Fernie, A.R. (2001). Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13 11–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali, U., Hegemann, B., Lytovchenko, A., Carrari, F., Bruedigam, C., Granot, D., and Fernie, A.R. (2003). Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133 84–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, W.R. (2001). Molecular Cloning: A Laboratory Manual, 3rd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Shi, H., Xiong, L., Stevenson, B., Lu, T., and Zhu, J.K. (2002). The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell 14 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., and Zhu, J.K. (2002). SOS4, a pyridoxal kinase gene, is required for root hair development in Arabidopsis. Plant Physiol. 129 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz, J., and Vielreicher, M. (2003). Tpn1p, the plasma membrane vitamin B6 transporter of Saccharomyces cerevisiae. J. Biol. Chem. 278 18990–18996. [DOI] [PubMed] [Google Scholar]
- Taji, T., Ohsumi, C., Iuchi, S., Seki, M., Kasuga, M., Kobayashi, M., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2002). Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 29 417–426. [DOI] [PubMed] [Google Scholar]
- Tambasco-Studart, M., Titiz, O., Raschle, T., Forster, G., Amrhein, N., and Fitzpatrick, T.B. (2005). Vitamin B6 biosynthesis in higher plants. Proc. Natl. Acad. Sci. USA 102 13687–13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T., Tateno, Y., and Gojobori, T. (2005). Evolution of vitamin B6 (pyridoxine) metabolism by gain and loss of genes. Mol. Biol. Evol. 22 243–250. [DOI] [PubMed] [Google Scholar]
- Taylor, A.L. (1972). Experiments in Molecular Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Weber, H., Bernhardt, A., Dieterle, M., Hano, P., Mutlu, A., Estelle, M., Genschik, P., and Hellmann, H. (2005). Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Physiol. 137 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel, D.K., Ehrenshaft, M., Denslow, S.A., and Daub, M.E. (2004). Functional complementation between the PDX1 vitamin B6 biosynthetic gene of Cercospora nicotianae and pdxJ of Escherichia coli. FEBS Lett. 564 143–146. [DOI] [PubMed] [Google Scholar]
- Wormit, A., Traub, M., Florchinger, M., Neuhaus, H.E., and Mohlmann, T. (2004). Characterization of three novel members of the Arabidopsis thaliana equilibrative nucleoside transporter (ENT) family. Biochem. J. 383 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, C., Han, P., Lutziger, I., Wang, K., and Oliver, D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40 711–717. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Zhao, G., and Winkler, M.E. (1996). Identification of the pdxK gene that encodes pyridoxine (vitamin B6) kinase in Escherichia coli K-12. FEMS Microbiol. Lett. 141 89–95. [DOI] [PubMed] [Google Scholar]
- Yonamine, I., Yoshida, K., Kido, K., Nakagawa, A., Nakayama, H., and Shinmyo, A. (2004). Overexpression of NtHAL3 genes confers increased levels of proline biosynthesis and the enhancement of salt tolerance in cultured tobacco cells. J. Exp. Bot. 55 387–395. [DOI] [PubMed] [Google Scholar]
- Zourelidou, M., de Torres-Zabala, M., Smith, C., and Bevan, M.W. (2002). Storekeeper defines a new class of plant-specific DNA-binding proteins and is a putative regulator of patatin expression. Plant J. 30 489–497. [DOI] [PubMed] [Google Scholar]
- Zuther, E., Buchel, K., Hundertmark, M., Stitt, M., Hincha, D.K., and Heyer, A.G. (2004). The role of raffinose in the cold acclimation response of Arabidopsis thaliana. FEBS Lett. 576 169–173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.