Abstract
Cereal seed development depends on the intimate interaction of filial and maternal tissues, ensuring nourishment of the new generation. The gene jekyll, which was identified in barley (Hordeum vulgare), is preferentially expressed in the nurse tissues. JEKYLL shares partial similarity with the scorpion Cn4 toxin and is toxic when ectopically expressed in Escherichia coli and tobacco (Nicotiana tabacum). In barley, jekyll is upregulated in cells destined for autolysis. The gene generates a gradient of expression in the nucellar projection, which mediates the maternal–filial interaction during seed filling. Downregulation of jekyll by the RNA interference technique in barley decelerates autolysis and cell differentiation within the nurse tissues. Flower development and seed filling are thereby extended, and the nucellar projection no longer functions as the main transport route for assimilates. A slowing down in the proliferation of endosperm nuclei and a severely impaired ability to accumulate starch in the endosperm leads to the formation of irregular and small-sized seeds at maturity. Overall, JEKYLL plays a decisive role in the differentiation of the nucellar projection and drives the programmed cell death necessary for its proper function. We further suggest that cell autolysis during the differentiation of the nucellar projection allows the optimal provision of basic nutrients for biosynthesis in endosperm and embryo.
INTRODUCTION
In contrast with the situation in animals, the tight interaction between the mother plant and its developing offspring during embryogenesis does not involve a vascular connection. Rather, intercellular exchange is effected via the apoplasm (Wang et al., 1994; Patrick and Offler, 2001). Thus, specific maternal plant tissues (so-called nurse tissues) act as a bridge between the two generations and provide the environment for the developing zygote and filial tissue. The space necessary for the growth of the new organism is generated by programmed cell death (PCD) of nurse tissues, and the contents of dying cells are remobilized to feed the new growing organism as described for some monocots and dicots (Smart, 1994; Wu and Cheung, 2000; Greenwood et al., 2005). The need for this source of nourishment is particularly critical during periods when the external environment destabilizes steady state source/sink relationships. In extreme situations, these can directly affect seed set, with the immediate postfertilization period being particularly sensitive to metabolic disturbance (Westgate and Boyer, 1986). Metabolic stress during this time depresses endosperm cell division and endoreduplication and inhibits other cellular events that precede the synthesis of storage products (Ober et al., 1991; Cheikh and Jones, 1994; Artlip et al., 1995). While the mechanics of these adjustments vary, the maternal nurse function is universally important.
The molecular basis of the establishment and function of nurse tissues is poorly understood (Lopes and Larkins, 1993; Lam, 2004). Its key role in plant sexual reproduction, however, has been confirmed repeatedly in studies of several female-sterile (Robinson-Beers et al., 1992; Reiser and Fischer, 1993) and male-sterile (Chaudhury, 1993) mutants. The interdependence between, and the developmentally orchestrated PCD of nurse tissues, depends on both the identity of the gametophyte and the developmental stage of the filial organism (Norstog, 1974; Thorne, 1985; Dominguez et al., 2001). The genes involved in these processes clearly play a role in determining crop productivity and therefore represent potential targets for intervention (Goetz et al., 1999; Wang et al., 2003). Poaceae species provide the majority of carbohydrates for the human diet, but only a small number of genes active in the nurse tissues of cereal seeds have been identified to date (Doan et al., 1996; Chen and Foolad, 1997; Sturaro et al., 1998; Drea et al., 2005; Greenwood et al., 2005). The role of these in the establishment of nurse tissues, and hence their potential strategic significance for crop improvement, although likely, has yet to be experimentally proven.
Recently, we used expression macroarray expression analysis to identify genes active during barley (Hordeum vulgare) seed development and have isolated a cDNA sequence that is highly expressed during early development (Sreenivasulu et al., 2002). The gene is abundant in the nurse tissues of both the male and female sporophyte, reaching an expression maximum during the differentiation of the nucellar projection. This tissue provides the main conduit of solutes to zygotic tissues and is therefore critical for the process of seed filling in barley and wheat (Triticum aestivum) (Duffus and Cochrane, 1993; Wang et al., 1994). The conflicting role of this gene in the life and death of reproductive nurse tissues inspired the name Jekyll, in reference to the schizophrenic character of the Robert Louis Stevenson novel.
RESULTS
Jekyll Is a Single-Copy Gene in Barley, Encoding a Novel Class of Small Proteins
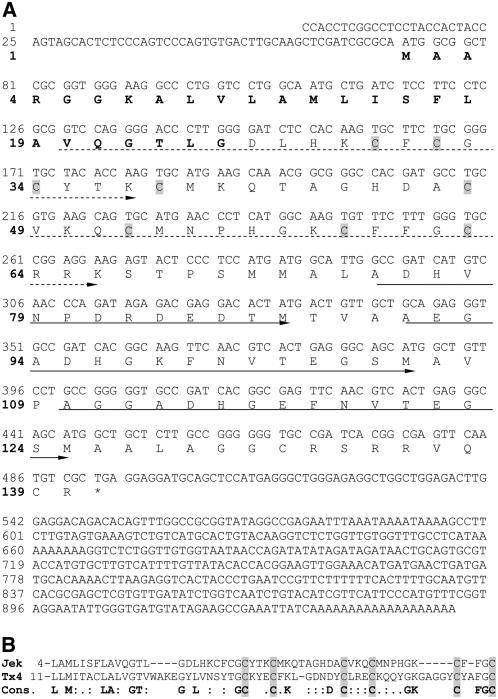
To reveal whether the jekyll EST clone HY09L21 represents a full-length mRNA, a phage cDNA library generated from developing barley caryopses (Weschke et al., 2000) was used to isolate nine clones, each with the same sequence and length as HY09L21. From a homology search of a large barley EST set (Kunne et al., 2005), 78 entries, all identical to the previously cloned sequences, were identified. By genome walking, a fragment 1335 bp upstream of the translation start codon with a TATA-box (GCTATAA) 39 to 46 bp upstream of the putative transcription start was isolated. Jekyll cDNA encodes a predicted cationic and amphipathic 140-residue protein (Figure 1A) with a secondary structure composed of two α-chains and one β-sheet followed by three α-chains (ααβααα-fold). Neither the cDNA nor the protein sequence share any significant similarity to any known sequences deposited in the public domain. The protein has the following features: a 25–amino acid putative signal peptide at the N terminus, three almost perfect direct repeats in the C-terminal region, and a Cys-rich domain (including 8 of the 10 Cys residues present in the complete sequence) in the central part of the protein. The predicted translation product is also rich in Ala (18 residues) and Gly (15 residues). PSI-BLAST analysis uncovered a sequence with partial similarity in the signal peptide and Cys-rich region (Figure 1B); this encodes toxin 4 from the scorpion Centruroides noxius (Vazquez et al., 1993).
Figure 1.
Structure of jekyll cDNA and Its Deduced Protein.
(A) Nucleotide sequence of jekyll cDNA and its deduced amino acid sequence. The putative signal peptide is shown in bold. Cys residues in the Cys-rich domain are shaded in gray. Continuous arrows mark nearly perfect repeats, and dashed arrows label less-conserved repeats. The stop codon is indicated by an asterisk.
(B) Amino acid alignment of similar regions of JEKYLL (Jek) and Toxin 4 (Tx4) from the scorpion C. noxius. The consensus sequence (Cons.) is shown below in bold. Matched Cys residues are shaded in gray.
DNA gel blot analysis showed that jekyll is a single-copy gene in barley (data not shown). Homologous sequences are present in the close relatives wheat and rye but not in other plants (see Supplemental Figures 1A and 1B online). Four wheat ESTs with strong similarity to jekyll were identified in the public database, and these were used to construct two contigs, the derived protein sequences of which show 66.3% similarity to one another and 52.3 and 55.1% similarity to barley JEKYLL. The domain structure of the predicted wheat proteins is similar to that of barley; in particular, both possess a nearly identical 25-residue signal peptide sequence at the N terminus and a highly conserved C terminus. The central portions of the proteins are variable, but the positions of the Cys residues are well conserved (see Supplemental Figure 1C online).
Jekyll mRNA Is Abundant in Short-Lived Nurse Tissues in the Reproductive Organs of Barley
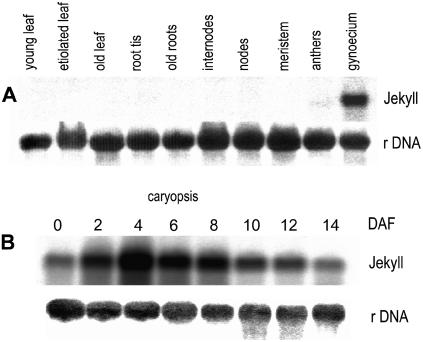
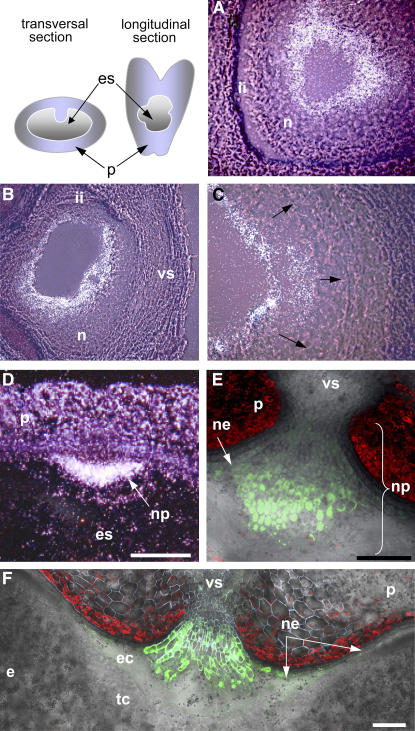
RNA gel blot hybridization experiments showed that jekyll in barley is represented by a single ∼900-bp product, consistent with the length of the cloned cDNA. The sequence was expressed in developing flowers (very weak in anthers and high in gynoecium) and throughout all stages of caryopsis development (peaking at 4 d after flowering [DAF]; Figure 2A) but not in any other plant organs (Figure 2B) or in germinating seeds (data not shown). In early caryopses, the level of expression was highest in the inner cell layers of the nucellus (Figure 3A). At later stages of seed development, jekyll expression spread outwards and was followed by the autolysis of highly vacuolated cells. The disintegration of lysed cells provides space for the enlarging endosperm cavity (Norstog, 1974), and the process continued in the dorsal and lateral sides of the seed until 4 DAF (Figures 3B and 3C). In the crease region, the nucellus remained intact, where it forms the nucellar projection (Figure 3D). The peak of jekyll expression (4 to 6 DAF) coincided temporally and spatially both with the differentiation of parenchyma tissues within the nucellar projection and with the vacuolization of the nucellar epidermis (Linnestad et al., 1998). Expression levels remained low in the undifferentiated cells adjacent to the vascular tissues and in the epidermis within the ventral region of the caryopsis (Figures 3E and 3F). Over time, jekyll expression decreased, but the spatial pattern of expression was preserved (data not shown).
Figure 2.
jekyll Transcripts Are Present in Flowers and throughout Caryopsis Development.
(A) Total RNA (10 μg per line) from caryopses at various developmental stages was hybridized with the jekyll probe (above) and with 25S rDNA (below) as a quantitative control.
(B) Total RNA (10 μg per line) from various barley tissues was hybridized with jekyll (above) and 25S rDNA probe (below).
Figure 3.
Localization of jekyll Expression in Developing Caryopses.
(A) and (B) jekyll expression patterns in the nucellar tissues shortly after pollination (A) and 2 DAF (B). Hybridization sites are visualized as a white signal.
(C) Expansion of expression toward the integuments (arrows).
(D) The nucellar projection at 4 DAF.
(E) and (F) A gradient of jekyll expression within the nucellar projection 6 and 8 DAF; pjekyll:GFP (see Methods).
e, endosperm; ec, endospermal cavity; es, embryo sac; ii, integuments; n, several layers of nucellar cells; ne, nucellar epidermis; np, nucellar projection; p, pericarp; tc, transfer cells, vs, vascular tissues. Bars = 120 μm.
Accumulation of JEKYLL Is Coupled with the mDifferentiation of Nucellar Tissue
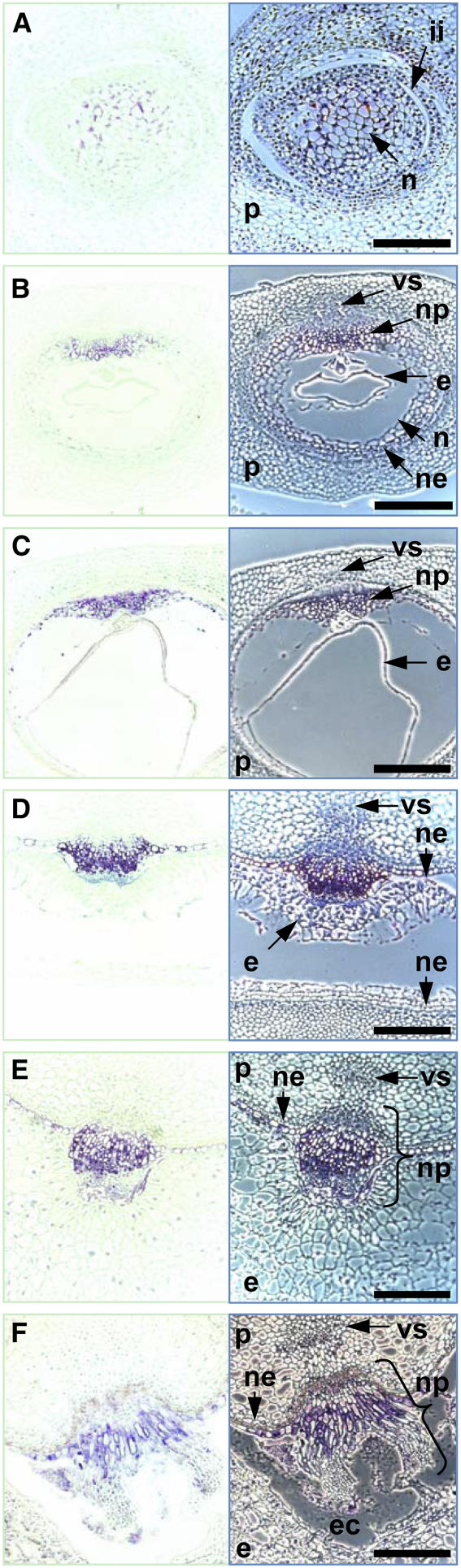
Immunodetection showed that JEKYLL was present in nucellar tissues before anthesis. In particular, it was concentrated in the fully expanded cells in the central part of the ovule (Figure 4A). At 2 to 4 DAF, its level increased dramatically in the cells of the nucellar projection (Figures 4B and 4C), and beyond this stage, its accumulation became progressively restricted to the nucellar projection tissue (Figure 4D). The appearance of JEKYLL in the nucellar epidermis coincided with the vacuolization of the epidermal cells in the crease. The protein was barely detectable either in the distal part or in the small mitotic basal cells of the nucellar projection but was present in expanding cells, reaching a maximal concentration in highly vacuolated cells with the morphology of transfer cells (Figures 4D and 4E). Small concentrations were present in the partially autolyzed tissues at the margins of the nucellar projection, but the protein was absent in the attached endosperm (Figures 4E and 4F). Thus, levels of JEKYLL coincided both temporally and spatially with transcript levels. The protein accumulated in concert with the differentiation of nucellar tissues, with maximal accumulation in expanding cells being rapidly followed by cell autolysis.
Figure 4.
Immunodetection of JEKYLL in Longitudinal Sections of the Gynoecium and the Transverse Sections of the Developing Caryopsis.
The target is present in the gynoecium (A) and developing caryopsis ([B] to [F]), as visualized by blue staining (left panel) of the tissue section (right panels; phase contrast microscopy). Abbreviations are as in Figure 3. Bars = 300 μm.
Jekyll Expression Inhibits the Growth of Escherichia coli and Promotes Cell Autolysis in Transgenic Tobacco Root Epidermis
In JEKYLL overexpressing E. coli, cell growth stopped following isopropyl-1-thio-β-d-galactopyranoside (IPTG) induction of protein synthesis. To define the peptide domain(s) responsible for this growth inhibition, constructs consisting of the Cys-rich region either alone or in combination with other structural domains were prepared (see Supplemental Figure 2 online, inset) and transformed into DL21 E. coli cells. A drastic inhibition of growth was associated with the expression, even on its own, of the Cys-rich domain (see Supplemental Figure 2 online). Since other domains were ineffective, the Cys-rich domain must itself be responsible for cell death.
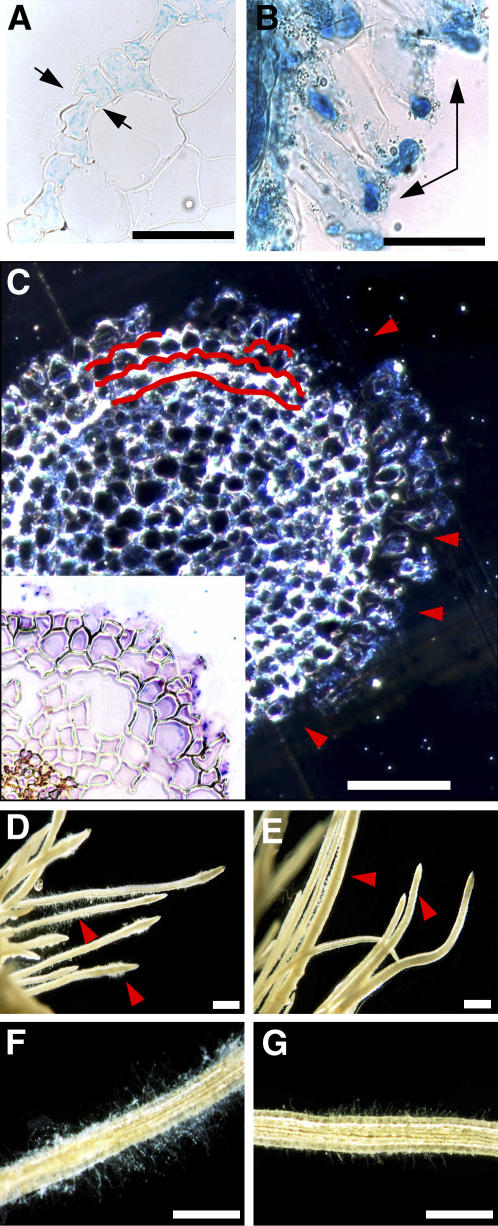
When JEKYLL cDNA was expressed ectopically in tobacco (Nicotiana tabacum) plants, driven by an ethanol-inducible promoter, plants wilted within 24 h of induction and later died, whereas control plants (the wild type and plants expressing β-glucuronidase (GUS) under the same promoter) continued to grow normally (see Supplemental Figure 3 online). The activation of GUS expression by ethanol occurred first in the epidermal cell layer of the root elongation region and in the root hairs. Later, the expression spread to the cortex and finally moved toward the shoot (Figures 5A and 5B). This pattern mirrors the route of solute uptake and transport, which is performed by epidermal cells and root hairs specialized for this function (Gilroy and Jones, 2000). Likewise, JEKYLL was expressed first in the epidermal cells and in the root cortex of plants (Figure 5C, inset). Cells accumulating the protein underwent rapid expansion, following which they self-destructed (Figure 5C). The loss of root hairs was the first recognizable phenotypic effect of jekyll expression (Figures 5D to 5G). Following the disruption of root function, the stem and leaves started to wilt. After ∼48 h, roots were completely macerated. Finally, death of transgenic plants was observed in all four independently transformed transgenic tobacco lines (12 to 20 plants per line).
Figure 5.
Alcohol-Induced jekyll Expression in Transformed Tobacco Roots.
(A) and (B) Spatial patterns of promoter activity after induction with ethanol in roots of tobacco transformed with pAlc-GUS grown under hydroponics. GUS staining in the epidermis ([A]; arrows) and root hairs ([B]; arrows) as primary sites of promoter activity. Bars = 40 μm.
(C) Disintegration of the outer cell layers in roots of transgenics after the ethanol induction coincides spatially with JEKYLL immunodetected with specific antibodies (shown in inset). Extensive cell damage within two or three outer cell layers of the root are indicated in red. Bar = 150 μm.
(D) to (G) Root hairs of wild-type plants ([D] and [F]) and severe reduction of root hair growth in transgenics expressing JEKYLL ([E] and [G]). Bars = 1 mm.
Root hair and whole plant growth were restored when jekyll expression was turned off by removing plants to an inducer-free medium. Thus, the primary effect of jekyll expression appears to be to damage the epidermis and root hairs, and the subsequent loss of turgor and eventual plant death are the result of failure in root function. The expression of jekyll evidently prevents epidermal cell development (root hair building and/or growth) and dramatically undermines cell integrity.
RNA Interference–Mediated Downregulation of Jekyll Impairs Flower and Seed Development
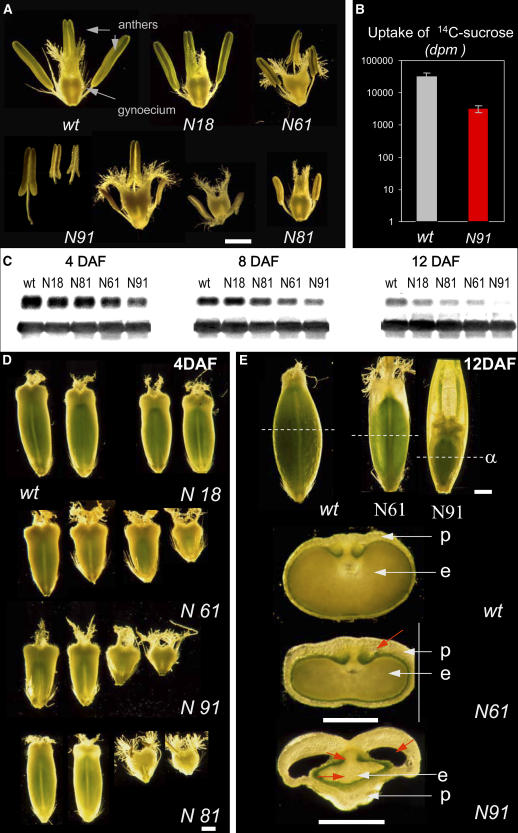
A group of 46 independent barley transformants were generated using Agrobacterium tumefaciens harboring pVECNpass construct to achieve RNA interference (RNAi)–mediated jekyll repression. No phenotypic differences between transgenic and wild-type plants were observed until flowering. Later on, the extent of phenotypic alteration was dependent on the degree of jekyll downregulation. Four T2 lines exhibiting a weak (N18), moderate (N61 and N81), and strong (N91) phenotype were chosen for detailed investigation. Although the number of flowers, as well as their morphology, remained unchanged (Figure 6A), the development of the transgenic flowers took at least 6 to 7 d longer than the wild-type ones, and both gynoecia and anthers exhibited size irregularities. This phenotype was strongly expressed in N91 and N81 and to a lesser extent in N18 and N61. To assess possible effects on carbohydrate uptake of the flowers, 14C-sucrose was fed to the stem of both transgenic and wild-type plants. Wild-type and transgenic flowers differed in isotope incorporation after 48 h by >10-fold (Figure 6B), indicating a significant decrease in the sink strength of jekyll downregulated flowers. The expression of jekyll, as measured by RNA gel blot hybridization, varied between transgenic lines from almost 100 to 20% of the wild-type mRNA level (Figure 6C). The extent of mRNA repression was consistent with the severity of phenotype (Table 1, Figures 6D and 6E). Developing caryopses were characterized by a reduction in their dorso-ventral diameter (Figure 6E). However, despite showing retarded growth, transgenic endosperms were histologically normal (Duffus and Cochrane, 1993). The dry weight of mature seeds was reduced in all transgenics (Table 1); in addition, pericarp structure was subtly changed. In extreme cases, additional ear cavities were observed, reflecting an imbalance in the growth of the pericarp and endosperm (Figure 6E, arrow in N91). Strongly affected caryopses died during the prestorage or early storage phases. Lastly, the number of seeds reaching maturity was lower in all transgenics compared with the wild type, varying from ∼90% of the wild type in N18, 50% in N61, to only 20% in line N91. Overall, it appears that jekyll downregulation leads to impaired growth of the reproductive organs and to a decrease in final seed yield.
Figure 6.
Flower and Seed Phenotypes Associated with Repression of Jekyll.
(A) Reduction of gynoecium and anther size in transgenics.
(B) Uptake of sucrose (in dots per minute [dpm]) by developing flowers in the wild type and line N91 (averaged from 15 independent measurements and represented in a logarithmic scale; means ± sd are shown).
(C) Total RNA from the wild type and transgenics at 4, 8, and 12 DAF hybridized with jekyll probe (above) and 25S rDNA (below).
(D) Changes in size and form of 4-DAF caryopses of the wild type and transgenics.
(E) Decrease in size and change in morphology of 12-DAF wild-type and transgenic caryopses. Orientations of anatomical cross sections are marked by dashed lines (α). Changes in seed anatomy are indicated by red arrows. e, endosperm; p, pericarp. Bars = 1 mm.
Table 1.
Phenotypic Characteristics of Selected Lines of T2 Transgenic Barley Plants Transformed with the Construct pVECNpass
| Plant Line | Level of jekyll Expression (%)a | Plant Height (cm) | Number of Spikes per Plant | Number of Corns per Spike | 100 Corn Weight (g) | Starch Content (mg/g) |
|---|---|---|---|---|---|---|
| Wild type | 100.0 | 56.4 ± 2.3a | 18.0 ± 2.4a | 21.6 ± 3.8a | 4.42 ± 0.67a | 455 ± 46a |
| N18 | 74.8 | 52.0 ± 2.0a | 17.7 ± 2.4a | 20.2 ± 4.9a | 3.04 ± 0.05b | 365 ± 26b |
| N61 | 21.3 | 49.3 ± 3.3b | 12.8 ± 4.8a | 13.4 ± 2.8b | 2.10 ± 0.36c | 340 ± 18b |
| N81 | 20.8 | 52.4 ± 3.2a | 15.0 ± 4.0a | 6.8 ± 4.6c | 2.14 ± 0.23c | ND |
| N91 | 25.3 | 51.4 ± 7.7a | 15.2 ± 5.0a | 5.2 ± 3.3c | 2.02 ± 0.38c | 361 ± 63b |
The values are given as means ± sd. Values followed by the same letter within a column are not significantly different at P < 0.05. ND, not detected.
Levels of jekyll mRNA expression were estimated by RNA gel blot analysis at 4 DAF. Hybridization signals were quantified as described (Weschke et al., 2000) and given in relative units as a percentage of expression to the wild type estimated as 100%.
The Differentiation Gradient Is Altered in the Nucellar Projection of Transgenic Barley Seeds
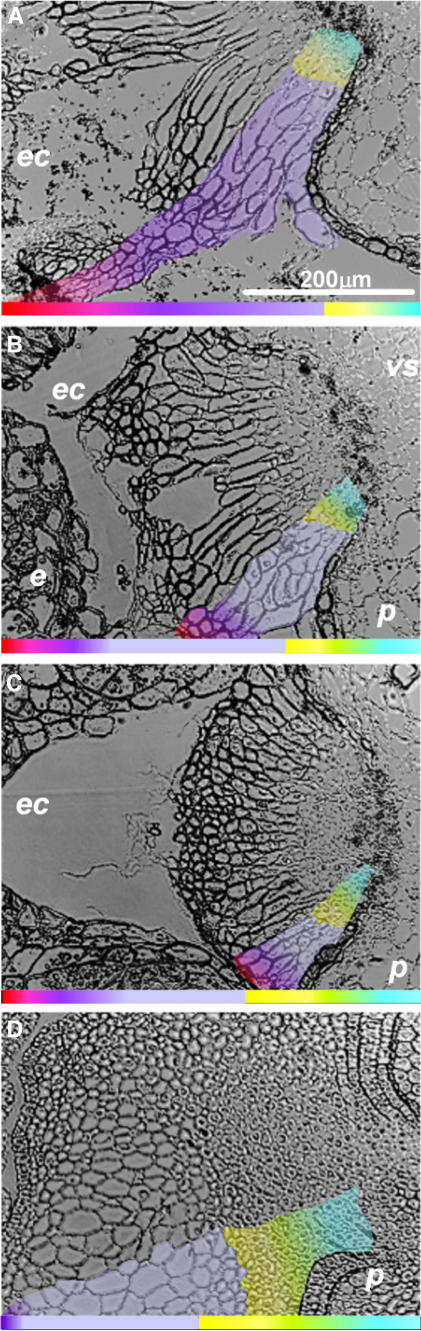
We performed comparative analysis of tissue structures in wild-type and transgenic plant lines with respect to the different levels of jekyll repression (Figure 7). Four tissue types along the radial axis in wild-type plants have been defined (Norstog, 1974; Duffus and Cochrane, 1993) and coded in colors (Figure 7A): (1) the basal region facing the main vascular bundle; (2) the middle zone of elongating cells; (3) transfer cells; and (4) the cell debris region, adjacent to the endosperm transfer cells. In transgenic N18, there was little change in zonation pattern (cf. Figures 7A and 7B). However, the basal region adjacent to the vascular tissue, which is populated by small isodiametric cells, constitutes an additional cell layer. The extent of radial cell expansion in the middle and the transport cell regions was decreased and the amount of cell debris reduced. These tendencies were exaggerated in line N61, in which the basal region featured an additional three cell layers. Cell elongation was dramatically reduced in the central part of the nucellar projection, and the cell debris region was barely recognizable (Figure 7C). Structural changes were expressed even more noticeably in line N91 (Figure 7D), where the nucellar projection was formed mostly by small cells (at least 10 additional cell layers) and irregularly expanded cells, which did not show transfer cell morphology. The region of cell debris was absent, and the nucellar projection had lost its regular tissue structure, instead being transformed into a massive callus-like organ. In this extreme case, the endosperm barely reached the cellularization stage, and further development of the caryopsis failed. Thus, the downregulation of jekyll has its primary impact on cell fate in the nucellar projection by affecting cell differentiation/expansion and preventing autolysis.
Figure 7.
Changes in the Cellular Structure of the Nucellar Projection Is Associated with Repression of jekyll.
(A) Cross section of the nucellar projection of a 12-DAF wild-type caryopsis. The basal region adheres to the vascular tissue and is visible as one layer of small dense cytoplasmic cells (type I) traced in blue. One to two layers of elongated cells (type II) are in yellow. Transfer cells (type III), distinguished by maximal expansion along the radial axis, are in turquoise. Autolysing cells (type IV) and the cell debris region occupy the endosperm cavity and are shown in violet/red.
(B) to (D) The nucellar projection of transgenic lines N18, N61, and N91 at 12 DAF. The relative distribution of the corresponding regions of transgenic caryopses is represented by color bars (below). Bar = 200 μm.
e, endosperm; ec, endospermal cavity; ne, nucellar epidermis; p, pericarp, vs, vascular tissues.
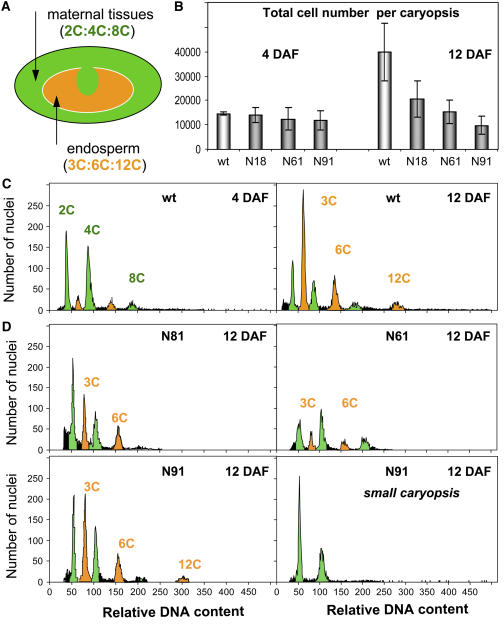
Proliferation of Endosperm Nuclei Is Delayed in Transgenic Caryopses with an Aberrant Nucellar Projection
The decrease in size and fresh weight of transgenic caryopses (Table 1) suggested perturbations in endosperm cell number and/or in the endoreduplication levels (Brunori et al., 1993). Both comparative flow cytometry and a conventional histological study of wild-type and transgenic caryopses showed that, at 4 DAF, cell number per caryopsis did not differ significantly from one another. This suggested that variation in cell expansion was responsible for the variable caryopsis size of transgenics at the early postfertilization stage. Within 7 d, the cell population of wild-type caryopses was doubled; by contrast, neither transgenic line achieved this level of cell multiplication (Figures 8A and 8B). Thus, cell proliferation in transgenic caryopses was impaired starting after the endosperm cellularization stage. The maternal tissue is targeted by jekyll downregulation, and the filial tissues are thereby indirectly affected. Flow cytometry (Arumuganathan and Earle, 1991) was exploited to distinguish nuclei from the diploid pericarp from those from triploid endosperm cells. Up to 4 DAF, the ratio cell numbers in the pericarp and endosperm were identical in wild-type and transgenic plants. The predominant nuclei were 2C and 4C, since maternal tissue occupies the majority of the caryopsis (Figure 8C, left panel). From 12 DAF, the number of endosperm nuclei (3C to 12C) exceeded the number of pericarp cells in wild-type plants, reaching 60% of the total cell population (Figure 8C, right panel). By contrast, in all the transgenics, the number of endosperm nuclei was much lower (Figure 8D). In the small caryopses of one transgenic line, the number was reduced by up to 99%, but in larger ones, the effect was less extreme (27 to 41% reduction; Figure 8D, bottom panels). Thus, jekyll downregulation affects the proliferation of endosperm (filial) but not pericarp (maternal) cells.
Figure 8.
Ploidy Level and Total Cell Numbers in Developing Caryopses of Wild-Type and Transgenic Plants.
(A) The various ploidy levels present in the developing caryopsis.
(B) Total cell number per caryopsis in the wild type and transgenics 4 and 12 DAF. Values given as means ± sd (n = 5).
(C) Flow cytometric analysis of nuclei from wild-type caryopses 4 (left) and 12 DAF (right).
(D) Flow cytometric analysis of nuclei from transgenic caryopses 12 DAF.
Wild-type endosperm cells were present as 3C, 6C, and 12C in the proportion 4:2:1 (Figure 8C). Cells with 3C and 6C nuclei are likely mitotic, while 12C nuclei are characteristic of cells entering endopolyploidization (Engelen-Eigles et al., 2001). In transgenic plants, the proportion of 12C cells was substantially reduced compared with the wild type (Figure 8D).
The retardation of endosperm development, therefore, is a major effect of jekyll downregulation, while proper JEKYLL function in the nucellar projection ensures the developmentally predetermined proliferation of endosperm.
Storage Patterns in Transgenic Caryopses Reveal Defects in Flux Exchange between Pericarp and Endosperm
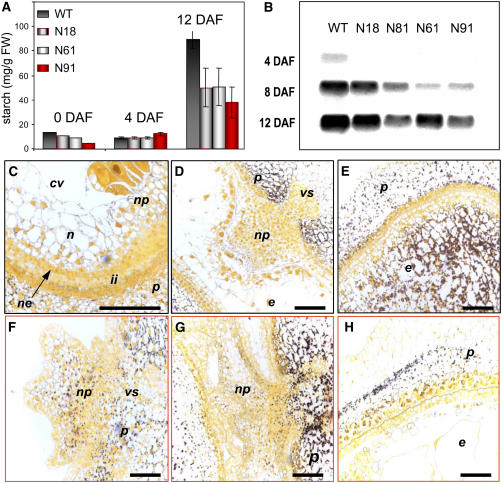
The cellular parameters of the endosperm and its carbohydrate uptake ability determine the starch storage capacity of the caryopsis (Zinselmeier et al., 1995; Smidansky et al., 2002). Starch accumulation in transgenic and wild-type plants was analyzed by a combination of biochemical assay (Figure 9A), expression of the endosperm-specific small subunit of AGPase, a key enzyme of starch synthesis (Figure 9B), and histochemical tissue staining (Figures 9C to 9H). In the wild type, transient accumulation of starch began in the maternal tissues of the young caryopsis (nucellar tissues and pericarp) but was utilized during further development (Figures 9D and 9E). The main starch storage is typically initiated in the endosperm (Simmonds and O'Brien, 1981; Wobus et al., 2004). The opposite trend was apparent in jekyll downregulated plants, where an increased density of starch grains was observed within the pericarp and integuments and some starch remained in the nucellus until 12 DAF. In line N91, starch content was actually increased, accompanied by a callus-like growth of the nucellar projection (Figures 9F to 9H). The prolonged deposition of starch in the nucellus of transgenic plants was associated with a delay in carbohydrate accumulation within the maternal tissues. In these cases, the accumulation of starch in the endosperm was negligible compared with that achieved in wild-type plants.
Figure 9.
Effect of Jekyll Repression on Starch Content and Starch Distribution during Seed Development.
(A) Starch content in developing caryopses 0, 4, and 12 DAF. Error bars indicate sd. FW, fresh weight.
(B) Hybridization of total RNA from wild-type and transgenic caryopses 4, 8, and 12 DAF with the endosperm-specific small subunit of ADP-glucose pyrophosphorylase probe.
(C) to (E) Iodine staining of cross sections of wild-type seeds. Transient starch accumulation is visible in the nucellus 2 DAF (C) and pericarp 4 DAF (D), followed by starch accumulation in endosperm at 8 DAF (E). Note the absence of starch in the nucellar projection.
(F) to (H) Similar staining of caryopses of transgenic N91 shows starch deposition within the nucellar projection at 4 DAF (F). The area of starch deposition increases with time, caryopsis at 6 DAF (G). At 12 DAF, starch is deposited primarily in the pericarp of the transgenic caryopsis (H) but not in the endosperm. Abbreviations are as in Figure 3. Bars = 300 μm.
The storage of starch in the endosperm is associated with the upregulation, typically by 4 DAF, of AGPase (Tomlinson and Denyer, 2003). In all transgenics, the expression of the endosperm-specific small subunit of AGPase was delayed, and its level was lowest in lines N61 and N91 (Figure 9B). During the major storage phase, starch accumulation rate in these lines was twofold to threefold lower than in wild-type plants. By 12 DAF, the starch content in the transgenics was only half that in the wild type (Figure 9A); as a result, the starch content of transgenic seeds was reduced at maturity (Table 1), despite the longer period (3 to 5 weeks) required for maturation.
Starch accumulation in the endosperm relies on assimilate delivery from the pericarp via the nucellar projection (Wang et al., 1994; Tomlinson and Denyer, 2003; Wobus et al., 2004). Therefore, any reduction in nutrient transport into the embryo sac limits resources for endosperm growth and storage product accumulation, especially during the immediate postfertilization period (Schussler and Westgate, 1995). In this study, the main storage process in transgenic endosperms was slowed, but at the same time, there was evidence for prolonged transient storage in the transgenic pericarp. We infer that this reflects damage to the bridge between the sites of metabolite provision (pericarp) and uptake (endosperm). Overall, the normal functioning of the nucellar projection, which is the route for metabolite delivery and the site of jekyll expression, is compromised in jekyll downregulated seeds.
DISCUSSION
Jekyll Encodes a Toxic Protein Essential for Sexual Reproduction
We have described the isolation and functional characterization of jekyll, which is highly expressed in the developing barley caryopsis and not found in any other plant organs except flowers. Because of low molecular weight, JEKYLL belongs to a group of small proteins, widely represented in both plants and animals, involved in the regulation of various cellular functions (Waters et al., 1996). Like JEKYLL, many of these proteins are developmentally regulated (Coca et al., 1994; DeRocher and Vierling, 1994; Waters et al., 1996; Wehmeyer et al., 1996). A well-studied example is the defensins, which have a distinct function in either seed maturation or the defense response (Thomma et al., 2002). They are small cationic, amphypathic peptides with eight Cys residues and are structurally similar to the first antifungal peptide isolated from barley (Mendez et al., 1990; Thomma et al., 2002). However, JEKYLL shows no similarity to the defensins (or any other known plant proteins) at the level of either amino acid composition or secondary structure. Curiously, it shares some features with a scorpion toxin, the most conspicuous of which is position of the six Cys residues in the domain that has a toxic effect both on E. coli and tobacco. Whether the basis of this toxicity of JEKYLL is similar to that of the scorpion toxin (thought to be caused by the formation of structure interfering with sodium channel function; Rodríguez de la Vega and Possani, 2005) is unknown.
The expression of jekyll is associated with cell death in barley. The gene is upregulated only in deteriorating maternal tissues, which border and nurse the new generation in the male and female sporophytes. This tissue death ensures the survival of the newly growing organism and is precisely regulated. As occurs also when jekyll is suppressed, any interference in development (Mariani et al., 1990) or metabolism (Goetz et al., 1999) of the nurse tissue leads to growth arrest and/or male sterility. In the female sporophyte, jekyll is upregulated precisely in those nucellar cell layers that are attached to the gametophyte and programmed for subsequent autolysis. Nucellar PCD is critical for tissue homeostasis, defense, and sink establishment of ovules (Wu and Cheung, 2000).
The impact of JEKYLL on filial tissue development has been elucidated by specifically modulating its expression levels. Flowers of transgenic barley plants received less carbohydrate from the maternal tissue, needed longer to develop, and were smaller in size. Transgenic lines with a strong phenotype produced few mature seeds. These observations underline the crucial role of JEKYLL in flower development and, consequently, seed set.
Since the first description of nurse tissues of barley (Norstog, 1974), only three genes expressed in the nucellus have been identified: NUC1, with no information on its function (Doan et al., 1996); nucellin, a homolog of the dicot vacuolar-processing protease (Chen and Foolad, 1997; Linnestad et al., 1998); and Hvex1, a putative extensin (Sturaro et al., 1998). Whether these genes are expressed in the male sporophyte is as yet unknown, as is their contribution to nurse function and/or sexual reproduction. In this work, the expression of jekyll in nurse tissues and its functional involvement in the sexual reproduction of barley has been experimentally proven.
JEKYLL Governs Cell Differentiation in the Nucellus and Is Required for the Establishment of the Nucellar Projection
What is the role of JEKYLL in the establishment of nurse tissues? jekyll expression was followed in the nucellus of wild-type caryopses (developmentally controlled expression), in roots of transgenic tobacco plants (induced expression), and in the nucellus of transgenic caryopses (RNAi-mediated downregulated expression). In the nucellus, the accumulation of JEKYLL was associated with structural changes in the nucellar projection: first in cells entering expansion, then in differentiating transfer cells, and finally reaching a maximum in elongated cells shortly before their autolysis. The autolysis of cells was also observed in tobacco epidermal cells where jekyll expression was induced: these cells stopped dividing, underwent uncontrolled cell expansion, and finally died, instead of the normal process of acquiring the developmentally programmed root hair morphology (Gilroy and Jones, 2000). Thus, jekyll switched epidermal cell fate to autolysis.
The opposite effect occurred in transgenic barley seeds with reduced jekyll expression, where the larger number of small cells present in the nucellar projection suggested a prolongation of mitosis and/or a defect in cell expansion/autolysis. In transgenics showing a slightly reduced level of jekyll expression (e.g., line N18), normal levels of mitotic activity and cell expansion are present in the nucellar projection, but autolysis is hampered. Where jekyll is more repressed (line N61) there was a clear decrease in cell expansion and a slower autolysis, while mitotic activity appeared to be controlled. Finally, when jekyll expression was severely downregulated (line N91), mitotic cells could not be switched into differentiation and expansion. Instead, they generated a callus-like growth and neither differentiated nor deteriorated.
We suggest that jekyll expression is sufficient to terminate cell proliferation, but higher amounts of the protein are necessary to release/facilitate expansion and autolysis. Plants do not appear able to compensate for jekyll downregulation, which suggests an important function for JEKYLL. Overall, JEKYLL seems to be necessary for the establishment of the nucellus structure in the barley caryopsis, presumably via its involvement in the termination of mitotic activity and the facilitation of cell expansion, followed by cell autolysis.
JEKYLL Controls Seed Filling by Promoting the Nourishment of the Endosperm
In wild-type plants, the high metabolic activity of short-lived nurse tissues first attracts a flow of metabolite (Andersen et al., 2002; Sun et al., 2004) and later ensures that sufficient nutrient is available for the gametophytic and filial tissues by releasing cell contents through autolysis (Wu and Cheung, 2000; Wang et al., 2003). The architecture of the nucellar projection reflects these abilities in that proliferating cells are positioned in the upper part (solute uptake), maximally elongated transport cells in the middle part (solute translocation), and autolysing cells in the lower part, adjacent to the endosperm cavity (solute release). Both this differentiation gradient (Wang et al., 1994) and the gradient of jekyll expression are arranged along the metabolite transport route (Duffus and Cochrane, 1993) and act to guarantee metabolite flow and release toward proliferating filial tissues.
The critical role of jekyll in the functional establishment of the nucellar projection and its significant impact on barley seed development and assimilate storage have been elucidated by the transgenic approach applied in this study. We have noted that (1) a decelerated/failed autolysis of nurse tissues leads to reduced amounts of cell debris being released into the apoplast, thus diminishing nutrient availability for reutilization by filial tissues. The same is true for secretion products delivered by the Golgi apparatus and the endoplasmic reticulum from degenerating cells, which retain their integrity even after the autolytic digestion of cell walls (Dominguez et al., 2001). (2) Proper transmission and diffusion within the nucellar projection are compromised because the cell elongation and differentiation steps essential for adapting transfer cell function (Patrick and Offler, 2001) were defective. (3) Cell proliferation and starch accumulation in the nucellus confirm its own biosynthetic activity dominating over degradation processes, which commonly contribute to nourishment (Norstog, 1974). In terms of the maternal/filial interaction, these changes suggest a functional shift from provision of nutrition to competition for nutrition and thus are critical for the survival of the new generation, especially during sink/source adjustment episodes. Thus, when JEKYLL control over cell fate and autolysis/death in nucellar tissues is lost, structural changes in the nucellar projection are induced that interfere with its function as the main assimilate transport route.
The delivery of nutrients is known to have a major impact on the proliferation and onset of endopolyploidization in the endosperm (Westgate and Boyer, 1986; Zinselmeier et al., 1995). Moreover, many nutritional compounds perform regulatory or signal functions (Wobus and Weber, 1999; Williams et al., 2000; Rolland et al., 2002; Koch, 2004), and hormone transport commonly occurs via the same transport route (Kamboj et al., 1998; Yang et al., 2002). In this context, an operational nucellar projection is required for endosperm development. In our experiments, all caryopses with defective jekyll function developed a lower sink and a reduced storage capacity during the major assimilate storage period. They contained less starch at maturity, were of small size, and the plants yielded less harvestable seeds than the wild type. Hence, we propose that jekyll represents a further mechanism for maternal control (Jarvi and Eslick, 1975; Lynch and Walsh, 1998; Alleman and Doctor, 2000) during seed development.
Jekyll-like genes are restricted to species, such as barley, wheat, and rye (Secale cereale), which form a massive nucellar projection during the onset of seed filling. Similar sequences were not found in plants that, although possessing a nucellus, do not build a nucellar projection. Endogenous upregulation of jekyll is developmentally controlled and coincides with the massive proliferation of endosperm nuclei. During the later developmental stages, when the endosperm becomes the major sink organ and determines carbohydrate uptake (Smidansky et al., 2002; Borisjuk et al., 2004), the need for its nourishment is superfluous, and jekyll expression is reduced. The size of the mature seed is however predetermined at an early developmental stage (Brunori et al., 1993; Engelen-Eigles et al., 2001). In conclusion, we suggest that jekyll is a part of a regulatory system governing cell fate in nurse tissues and especially in the nucellar projection of developing caryopsis. Its involvement in, and its crucial impact on, the maternal/filial interaction has been established during sexual reproduction and early seed filling in barley.
METHODS
Plant Material
Wild-type and transgenic barley (Hordeum vulgare) and tobacco (Nicotiana tabacum) plants were grown under standard greenhouse conditions at 18°C (for barley) or 24°C (for tobacco) with 16 h of light and a relative air humidity of 60%. Determination of developmental stages for developing barley seeds and tissue isolations were performed as described (Weschke et al., 2000).
Cloning of Jekyll and Sequence Analysis
The barley EST clone HY09L21 (EMBL accession number AL508766) was used to screen a seed-specific phage cDNA library (Weschke et al., 2000). Positive clones were selected, sequenced, and compared with the entire clone. Additionally, search of the EST collection from Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (http://gprc.ipk-gatersleben.de) resulted in a number of additional ESTs, identical to the previous clones. Promoter region was isolated using a GenomeWalker kit (BD Biosciences). EST clones from wheat (Triticum aestivum) were found in the EST database (accession numbers BJ237646, BJ246112, BJ235819, and BJ247336) (http://www.shigen.nig.ac.jp/wheat/komugi/top/top.jsp).
DNA and RNA Procedures
Standard DNA and RNA cloning procedures were performed as described by Sambrook et al. (1989). DNA gel blot analysis from plant total leaf DNA (10 μg per probe) digested with appropriate restriction enzymes was performed as previously described (Radchuk et al., 2005). The jekyll fragment derived in PCR with the oligos 5′-CGTGGATCCGATCTCCACAAGTGCTTCTG-3′ and 5′-GAAGAAGCTTAATTCTCGGCCTATACCG-3′ was used as a probe. Total RNA was extracted from tissue samples using a Purescript RNA isolation kit (Biozym) and used for RNA gel blot analyses as described (Radchuk et al., 2005).
Plasmid Constructions
The RNAi construction pVECNpass for barley stable transformation was generated and consisted of the jekyll promoter together with the 5′-upstream region (1332 nucleotides), sense fragment of jekyll (259 nucleotides), the first intron of GA20 oxidase from potato (Solanum tuberosum) (199 nucleotides), and antisense fragment of jekyll (259 nucleotides). The appropriate DNA fragments were PCR amplified and cloned into modified pUC19 vector using of the specific restriction sites. The used primers and restriction sites were as follows: for jekyll promoter and 5′-upstream region, 5′-TACTCGAGGGCACGCGTGGTCGACG-3′ (XhoI underlined, as are further restriction sites ) and 5′-GAGCCACTAGTGCGCGATCGAGCTTGC-3′ (SpeI); for jekyll sense fragment, 5′-GCACTAGTGGCTCGCGGTGGGAAGG-3′ (SpeI) and 5′-TGCAGCAACAGATCTAGTGTCCTCGTC-3′ (BglII); for jekyll antisense fragment 5′-GCTCTAGAGGCTCGCGGTGGGAAGG-3′ (XbaI) and 5′-TGCAGCAACGGATTCAGTGTCCTCGTC-3′ (BamHI). The whole cassette was cut out from pUC19 with PstI enzyme and cloned into the appropriate restriction site of intermediate vector pBluescript SK, and then, by cutting out with ApaI-NotI restriction enzymes, the cassette was introduced into corresponding sites of the binary vector pWVec8 (Wang et al., 2001).
Attempts to create a vector overexpressing jekyll gene driven by a constitutive 35S promoter were unsuccessful because transformed Escherichia coli cells did not grow after transformation with the resulted construct. Therefore, for stable overexpression of JEKYLL in plants, a construct was generated consisting of jekyll gene in an alcohol-inducible expression system (Caddick et al., 1998). For this, the coding part of jekyll was PCR amplified with the primers 5′-GCAAGGATCCATGGCGGCTCGCGGTGGGAA-3′ (BamHI) and 5′-GAATGGATCCTCAGCGACATTGAACTCGCCGTG-3′ (BamHI) and cloned into the BamHI restriction site of the vector pACN (Caddick et al., 1998) between the modified AlcA promoter and nos terminator. The sense orientation was checked by restriction analysis and sequencing. The whole cassette was then cut out by HindIII endonuclease and integrated into the appropriate site of binary alcohol inducible system binSRNA to generate the pAlc-jekyll construct used for tobacco plant transformation. The construct pAlc-GUS was kindly provided by U. Sonnewald.
For overexpression of jekyll in E. coli, the coding region of jekyll without signal peptide was PCR amplified using the primer pair 5′-CGTGGATCCGATCTCCACAAGTGCTTCTG-3′ (BamHI is underlined) and 5′-GAAGCTCGAGGCGACATTGAACTCGCCGTG-3′(XhoI is underlined) and cloned into the bacterial expression vector pET23a between BamHI-XhoI restriction sites, resulting in the vector pJ2.
For functional characterization of different JEKYLL protein domains, full-length and different truncated sequence fragments were cloned in frame into the pET23a vector between BamHI and XhoI insertion sites. The fragments were amplified by PCR using the following primers: for construct pJ1, 5′- GCAAGGATCCATGGCGGCTCGCGGTGGGAA-3′ (BamHI) and 5′-GAAGCTCGAGGCGACATTGAACTCGCCGTG-3′ (XhoI); for pJ3, 5′-GCAAGGATCCATGGCGGCTCGCGGTGGGAA-3′ (BamHI) and 5′-CGTGCTCGAGTCACTCTGCAGCAAC-3′ (XhoI); for pJ4, 5′-GATGGATCCAAGTGCTTCTGCGGGT-3′ (BamHI) and 5′-GAGTACTCTCGAGCCGGCACCCAAAG-3′ (XhoI); for pJ5, 5′-CCTGGATCCATGTGTTTCTTTGGGTGC (BamHI) and 5′-ACTTGCCGTGCTCGAGACCCTCTGC-3′ (XhoI); and for pJ6, 5′-GATGGGATCCGCCGATCATGTCAA-3′ (BamHI) and 5′-CAGGCTCGAGCATGCTGCCCTCAG-3′ (XhoI).
To create the pjekyll:green fluorescent protein (GFP) construct consisting of GFP driven by the jekyll promoter, the sequence of the jekyll promoter was inserted using blunt ends in front of the gfp sequence. The whole cassette was then cut out from the plasmid with the SfiI restriction enzyme and cloned into the same restriction site of the p6U vector (DNA Cloning Service), resulting in the pjekyll:GFP construct.
Plant Transformation Procedures
RNAi transgenic barley lines were generated by Agrobacterium tumefaciens–mediated transformation of immature embryos derived from H. vulgare cv Golden Promise essentially as described (Wang et al., 2001). Genetic transformation of barley with the construct pJekyll:GFP was performed as described by Kumlehn et al. (2006).
Tobacco plants (ecotype Havana) were transformed with the construct pAlc-jekyll by A. tumefaciens strain EHA105 as previously described (Bäumlein et al., 1991). Transgenic tobacco plants carrying the GUS gene under alcohol inducible promoter was kindly supplied by U. Sonnewald. T2 transgenic tobacco plants with one copy of the transgene were used for analyses. To establish hydroponic plant cultures, the seedlings were planted in sterile sand and adopted to the greenhouse conditions. The plants were removed, cut out from the roots, and placed in darkened glass vessels containing liquid sterile Murashige and Skoog medium (Murashige and Skoog, 1962) without sugars and phytohormones. Plants built new roots in 7 to 10 d and were used for induction of transgene expression. To activate the alcohol-inducible promoter, tobacco plants were placed in new Murashige and Skoog medium supplemented with 2% ethanol and left for up to 7 d.
Overexpression of JEKYLL in E. coli, Microsequencing, and Antibody Production
The plasmid pJ2 was introduced in E. coli strain BL21 (DE3) pLysS (Novagen). Expression of the recombinant protein was induced by incubation of transformed E. coli cells for 3 h at 37°C in Luria-Bertani medium supplemented with 0.5 mM IPTG and purified through one-step affinity chromatography on nickel-nitrilotriacetic acid agarose resins using attached His tag. Proper production of JEKYLL in E. coli cells was checked by partial protein sequencing of the first 10 amino acids of the isolated protein. The overexpressed protein was dissolved in 8 M urea, purified using Ni-NTA resins (Qiagen), eluted at pH 5.0 according to manufacturer's instructions, and dialyzed three times against PBS, pH 8.0. Two hundred micrograms of the purified protein was used to immunize rabbits intradermally and boosted after 2 weeks with the same antigen. Polyclonal anti-Jekyll antibodies were produced according to Harlow and Lane (1988) and tested by protein gel blot hybridization in a dilution of 1:1000. Specificity of binding was ascertained by competition with a 100-fold molar excess of respective antigens.
Expression of Different Protein Domains in E. coli Cells
To study toxic capacities of different domains of JEKYLL, the constructs pJ1 to pJ6 were transformed into the E. coli strain BL21 and grown at 37°C in Luria-Bertani medium supplemented with 100 μg/mL ampicillin and 100 μg/mL IPTG for 8 h. The rate of bacterial growth was estimated every hour by measurement of optical density at A600.
Histochemical Techniques
Caryopses were fixed in 2.5% glutaraldehyde and 50 mM sodium cacodylate, pH 7.0, or in 4% (w/v) paraformaldehyde and 50 mM potassium phosphate buffer, pH 7.0, under slight vacuum for 4 h at room temperature, rinsed in cacodylate buffer, dehydrated, and embedded in Paraplast Plus (Sherwood Medical). Sections were cut at 10 to 15 μm on a microtome, transferred on poly-l-lysine–treated slides (Sigma Diagnostics), and dried overnight at 45°C. As a general stain, toluidine blue was used according to Gerlach (1977) and Carson (1990). In situ hybridization was performed according to Borisjuk et al. (1995). The same cDNA fragment as for blot hybridizations was used as a probe after labeling with [33P]dCTP. The immunostaining and structural investigations were performed using microsections of seeds and roots embedded in butyl-methyl methacrylate (Baskin et al., 1992). Briefly, barley caryopses were sliced into small pieces and fixed 1.5 to 3 h in 4% paraformaldehyde in PBS with 10 mm DTT, washed in PBS with DTT for 2 h, and dehydrated in ethanol series. The embedding in butyl-methyl methacrylate was followed by polymerization at 20°C for 48 h under UV light. Sections of 3- to 5-μm thickness were cut on a microtome (RM 2165; Leica). After de-embedding with acetone and rehydration, the sections were stained with toluidine blue and/or used for immunodetection. The immunolocalization procedure was performed with an affinity-purified anti-JEKYLL polyclonal antibody using a corresponding VASTASTAIN ABC-AP kit (Alkaline Phosphatase Substrate Kit III). The histochemical detection of GUS activity was performed as described (Jefferson et al., 1987) and evaluated microscopically by an Axioscope (Carl Zeiss).
Flow Cytometry Analysis
Suspensions of nuclei were obtained by chopping plant tissues with a sharp razor blade in nuclei-stabilizing buffer (1.21 g Tris, 0.5 g NaCl, 0.47 g Triton X-100, and 107 g MgCl2 × 6H2O, pH 7.0). The 4′,6-diamidino-2-phenylindole (Sigma-Aldrich) at a final concentration of 50 μg mL−1 was added to the nuclei suspension 30 min before analysis. Nuclear DNA content was measured using a flow cytometer. The Foulgen staining was performed for localization of nuclei in situ within the tissue sections of the caryopses.
Biochemical Procedures
For measurement of 14C-sucrose uptake, stems of barley plants were cut ∼25 cm below spikes and put into 100 mL of solution containing 10 mM sucrose, 5 mM glutamine, 5 mM asparagine, 5 mM KH2PO4, 10 mM MES, pH 7.0, and 500 μL [U-14C] sucrose (7.4 MBq mL−1; Amersham-Buchler). After 24 h of incubation in the light (∼400 μmol m−2 s−1), the flowers of wild-type and transgenic plants were removed from the spikes and frozen in liquid nitrogen. Subsequently, the plant material was homogenized in 2 mL methanol (60% [v/v]). Radioactivity was determined by a liquid scintillation counter (Rotiszint). Counts were corrected for background and quenching by external standards. Starch determination was performed essentially as described (Rolletschek et al., 2005).
Computational Analyses
Searches of the National Center for Biotechnology Information databases were performed with BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). Alignments of amino acid sequences were performed using ClustalW (DNAstar). Sequence data were analyzed using Lasergene software (DNAstar).
Accession Number
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AM261729.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Genomic Organization of Jekyll in Barley and Other Plants.
Supplemental Figure 2. The Influence of JEKYLL Domains on Growth of E. coli Cells.
Supplemental Figure 3. Influence of Jekyll on Phenotype of Transgenic Tobacco Plants Seven Days after Induction of the Gene Expression.
Supplementary Material
Acknowledgments
This work was supported in part by the projects GABI-SEED (FKZ 0312282) and GABI-SEED 2 (FKZ 0313115) of the German Ministry for Education and Research. We thank R. Manteuffel for advice and help with antibody production, W. Weschke for advice during molecular biological work, M. Hajirezaei for technical support during the 14C-isotope studies, and T. Sharbel and H. Block for advice and help with flow cytometry. We appreciate the preliminary work of S. Gubatz on immunodetection and especially thank B. Claus and M. Melzer for confocal microscopy. We also thank U. Siebert, A. Stegmann, G. Einert, E. Fessel, and K. Blaschek for excellent technical assistance. Special thanks to S. Schulze and K. Davidsen (Max-Planck-Institut) for stable barley transformation and to U. Tiemann and K. Lipfert for excellent artwork.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instruction for Authors (www.plantcell.org) is: Ljudmilla Borisjuk (borysyuk@ipk-gatersleben.de).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.041335.
References
- Alleman, M., and Doctor, J. (2000). Genomic imprinting in plants: Observations and evolutionary implications. Plant Mol. Biol. 43 147–161. [DOI] [PubMed] [Google Scholar]
- Andersen, M.N., Asch, F., Wu, Y., Jensen, C.R., Naested, H., Mogensen, V.O., and Koch, K.E. (2002). Soluble invertase expression is an early target of drought stress during the critical, abortion-sensitive phase of young ovary development in maize. Plant Physiol. 130 591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artlip, T.S., Madison, J.T., and Setter, T.L. (1995). Water deficit in developing endosperm of maize: Cell division and nuclear DNA endoreduplication. Plant Cell Environ. 18 1034–1049. [Google Scholar]
- Arumuganathan, K., and Earle, E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9 208–218. [Google Scholar]
- Baskin, T., Busby, C.H., Fowke, L.C., Sammut, M., and Gubler, F. (1992). Improvements in immunostaining samples embedded in methacrylate: Localization of microtubules and other antigens throughout developing organs in plants of diverse taxa. Planta 187 405–413. [DOI] [PubMed] [Google Scholar]
- Bäumlein, H., Boerjan, W., Nagy, I., Bassüner, R., Van Montagu, M., Inze, D., and Wobus, U. (1991). A novel seed protein gene from Vicia faba is developmentally regulated in transgenic tobacco and Arabidopsis plants. Mol. Gen. Genet. 225 459–467. [DOI] [PubMed] [Google Scholar]
- Borisjuk, L., Rolletschek, H., Radchuk, R., Weschke, W., Wobus, U., and Weber, H. (2004). Seed development and differentiation: A role for metabolic regulation. Plant Biol. 6 375–386. [DOI] [PubMed] [Google Scholar]
- Borisjuk, L., Weber, H., Panitz, R., Manteuffel, R., and Wobus, U. (1995). Embryogenesis of Vicia faba L.: Histodifferentiation in relation to starch and storage protein synthesis. J. Plant Physiol. 147 203–218. [Google Scholar]
- Brunori, A.L., Forino, M.C., Frediani, M., and Ruberti, F. (1993). Cell number and polyploidy in the starchy endosperm of Triticum. J. Genet. Breed. 47 217–220. [Google Scholar]
- Caddick, M.X., Greenland, A.J., Jepson, I., Krause, K.P., Qu, N., Riddell, K.V., Salter, M.G., Schuch, W., Sonnewald, U., and Tomsett, A.B. (1998). An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nat. Biotechnol. 16 177–180. [DOI] [PubMed] [Google Scholar]
- Carson, F.L. (1990). Histotechnology: A Self-Instructional Text. (Chicago: ASCP Press).
- Chaudhury, A.M. (1993). Nuclear genes controlling male fertility. Plant Cell 5 1277–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheikh, N., and Jones, R.J. (1994). Disruption of maize kernel growth and development by heat stress (role of cytokinin/abscisic acid balance). Plant Physiol. 106 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F., and Foolad, M.R. (1997). Molecular organization of a gene in barley which encodes a protein similar to aspartic protease and its specific expression in nucellar cells during degeneration. Plant Mol. Biol. 35 821–831. [DOI] [PubMed] [Google Scholar]
- Coca, M.A., Almoguera, C., and Jordano, J. (1994). Expression of sunflower low-molecular-weight heat-shock proteins during embryogenesis and persistence after germination: Localization and possible functional implications. Plant Mol. Biol. 25 479–492. [DOI] [PubMed] [Google Scholar]
- DeRocher, A.E., and Vierling, E. (1994). Developmental control and small heat shock protein expression during pea seed maturation. Plant J. 5 93–102. [Google Scholar]
- Doan, D.N., Linnestad, C., and Olsen, O.A. (1996). Isolation of molecular markers from the barley endosperm coenocyte and the surrounding nucellus cell layers. Plant Mol. Biol. 31 877–886. [DOI] [PubMed] [Google Scholar]
- Dominguez, F., Moreno, J., and Cejudo, F.J. (2001). The nucellus degenerates by a process of programmed cell death during the early stages of wheat grain development. Planta 213 352–360. [DOI] [PubMed] [Google Scholar]
- Drea, S., Leader, D.J., Arnold, B.C., Shaw, P., Dolan, L., and Doonan, J.H. (2005). Systematic spatial analysis of gene expression during wheat caryopsis development. Plant Cell 17 2172–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffus, C.M., and Cochrane, M.P. (1993). Formation of the barley grain: Morphology, physiology, and biochemistry. In Barley: Chemistry and Technology, A.W. MacGregor and R.S. Bhatty, eds (St. Paul, MN: American Association of Cereal Chemists), pp. 31–72.
- Engelen-Eigles, G., Jones, R.J., and Phillips, R.L. (2001). DNA endoreduplication in maize endosperm cells is reduced by high temperature during the mitotic phase. Crop Sci. 41 1114–1121. [Google Scholar]
- Gerlach, D. (1977). Botanische Mikrotechnik. (Stuttgart, Germany: Thieme Verlag).
- Gilroy, S., and Jones, D.L. (2000). Through form to function: Root hair development and nutrient uptake. Trends Plant Sci. 5 56–60. [DOI] [PubMed] [Google Scholar]
- Goetz, G., Fkyerat, A., Métais, N., Kunz, M., Tabacchi, R., Pezet, R., and Pont, V. (1999). Resistance factors to grey mould in grape berries: Identification of some phenolic inhibitors of Botrytis cinerea stilbene oxidase. Phytochemistry 52 759–767. [Google Scholar]
- Greenwood, J.S., Helm, M., and Gietl, C. (2005). Ricinosomes and endosperm transfer cell structure in programmed cell death of the nucellus during Ricinus seed development. Proc. Natl. Acad. Sci. USA 102 2238–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Jarvi, A.J., and Eslick, R.F. (1975). Shrunken endosperm mutants in barley. Crop Sci. 15 363–366. [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboj, J.S., Blake, P.S., and Baker, D.A. (1998). Cytokinins in the vascular saps of Ricinus communis. Plant Growth Regul. 25 123–126. [Google Scholar]
- Koch, K.E. (2004). Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 7 235–246. [DOI] [PubMed] [Google Scholar]
- Kumlehn, J., Serazetdinova, L., Hensel, G., Becker, D., and Loerz, H. (2006). Genetic transformation of barley (Hordeum vulgare L.) via infection of androgenetic pollen cultures with Agrobacterium tumefaciens. Plant Biotechnol. J. 4 251–261. [DOI] [PubMed] [Google Scholar]
- Kunne, C., Lange, M., Funke, T., Miene, H., Thiel, T., Grosse, I., and Scholz, U. (2005). CR-EST: A resource for crop ESTs. Nucleic Acids Res. 33 619–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E. (2004). Controlled cell death, plant survival and development. Nat. Rev. Mol. Cell Biol. 5 305–315. [DOI] [PubMed] [Google Scholar]
- Linnestad, C., Doan, D.N., Brown, R.C., Lemmon, B.E., Meyer, D.J., Jung, R., and Olsen, O.A. (1998). Nucellain, a barley homolog of the dicot vacuolar-processing protease, is localized in nucellar cell walls. Plant Physiol. 118 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, M.A., and Larkins, B.A. (1993). Endosperm origin, development, and function. Plant Cell 5 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and Walsh, B. (1998). Genetics and Analysis of Quantitative Traits. (Sunderland, MA: Sinauer Associates).
- Mariani, C., de Beuckeleer, M., Truttner, J., Leemans, J., and Goldberg, R.B. (1990). Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature 347 737–741. [Google Scholar]
- Mendez, E., Moreno, A., Colilla, F., Pelaez, F., Limas, G.G., Mendez, R., Soriano, F., Salinas, M., and de Haro, C. (1990). Primary structure and inhibition of protein synthesis in eukaryotic cell-free system of a novel thionin, gamma-hordothionin, from barley endosperm. Eur. J. Biochem. 194 533–539. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Norstog, K. (1974). Nucellus during early embryogeny in barley: Fine structure. Bot. Gaz. 135 97–103. [Google Scholar]
- Ober, E.S., Setter, T.L., Madison, J.T., Thompson, J.F., and Shapiro, P.S. (1991). Influence of water deficit in maize endosperm development. Enzyme activities and RNA transcripts of starch and zein synthesis, abscisic acid, and cell division. Plant Physiol. 97 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick, J.W., and Offler, C.E. (2001). Compartmentation of transport and transfer events in developing seeds. J. Exp. Bot. 52 551–564. [PubMed] [Google Scholar]
- Radchuk, V.V., Sreenivasulu, N., Radchuk, R.I., Wobus, U., and Weschke, W. (2005). The methylation cycle and its possible function in barley endosperm development. Plant Mol. Biol. 59 289–307. [DOI] [PubMed] [Google Scholar]
- Rolletschek, H., Koch, K., Wobus, U., and Borisjuk, L. (2005). Positional cues for the starch/lipid balance in maize kernels and resource partitioning to the embryo. Plant J. 42 69–83. [DOI] [PubMed] [Google Scholar]
- Reiser, L., and Fischer, R.L. (1993). The ovule and the embryo sac. Plant Cell 5 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Beers, K., Pruitt, R.E., and Gasser, C.S. (1992). Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 4 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez de la Vega, R.C., and Possani, L.D. (2005). Overview of scorpion toxins specific for Na+ channels and related peptides: Biodiversity, structure–function relationships and evolution. Toxicon 46 831–844. [DOI] [PubMed] [Google Scholar]
- Rolland, F., Moore, B., and Sheen, J. (2002). Sugar sensing and signaling in plants. Plant Cell 14 (suppl.), S185–S205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schussler, J.R., and Westgate, M.E. (1995). Assimilate flux determines kernel set at low water potential in maize. Crop Sci. 35 1074–1080. [Google Scholar]
- Simmonds, D.H., and O'Brien, T.P. (1981). Morphological and biochemical development of the wheat endosperm. Adv. Cereal Sci. Technol. 4 5–70. [Google Scholar]
- Smart, C.M. (1994). Gene expression during leaf senescence. New Phytol. 126 419–448. [DOI] [PubMed] [Google Scholar]
- Smidansky, E.D., Clancy, M., Meyer, F.D., Lanning, S.P., Blake, N.K., Talbert, L.E., and Giroux, M.J. (2002). Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc. Natl. Acad. Sci. USA 99 1724–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu, N., Altschmied, L., Panitz, R., Hähnel, U., Michalek, W., Weschke, W., and Wobus, U. (2002). Identification of genes specifically expressed in maternal and filial tissues of the barley caryopsis: A cDNA array analysis. Mol. Genet. Genomics 266 758–767. [DOI] [PubMed] [Google Scholar]
- Sturaro, M., Linnestad, C., Kleihofs, A., Olsen, O.A., and Doan, D.N.P. (1998). Characterization of a cDNA encoding a putative extensin from developing barley grains (Hordeum vulgare L.). J. Exp. Bot. 49 1935–1944. [Google Scholar]
- Sun, K., Hunt, K., Bernard, A., and Hauser, B.A. (2004). Ovule abortion in Arabidopsis triggered by stress. Plant Physiol. 135 2358–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P., Cammue, B.P., and Thevissen, K. (2002). Plant defensins. Planta 216 193–202. [DOI] [PubMed] [Google Scholar]
- Tomlinson, K., and Denyer, K. (2003). Starch synthesis in cereal grains. Adv. Bot. Res. 40 1–61. [Google Scholar]
- Thorne, J.H. (1985). Phloem unloading of C and N assimilates in developing seeds. Annu. Rev. Plant Physiol. 36 317–343. [Google Scholar]
- Vazquez, A., Becerril, B., Martin, B.M., Zamudio, F., Bolivar, F., and Possani, L.D. (1993). Primary structure determination and cloning of the cDNA encoding toxin 4 of the scorpion Centruroides noxius Hoffmann. FEBS Lett. 320 43–46. [DOI] [PubMed] [Google Scholar]
- Wang, A., Xia, Q., Xie, W., Datla, R., and Selvaraj, G. (2003). The classical Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development. Proc. Natl. Acad. Sci. USA 100 14487–14492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H.L., Offler, C.E., Patrick, J.W., and Ugalde, T.D. (1994). Cellular pathway of photosynthate transfer in the developing wheat grain. I. Delineation of the potential transfer pathway using fluorescent dyes. Plant Cell Environ. 17 257–266. [Google Scholar]
- Wang, M.B., Abbott, D.C., Upadhyaya, N.M., Jacobsen, J.V., and Waterhouse, P.M. (2001). Agrobacterium tumefaciens-mediated transformation of an elite Australian barley cultivar with virus resistance and reporter genes. Aust. J. Plant Physiol. 28 149–156. [Google Scholar]
- Waters, E.R., Lee, G.J., and Vierling, E. (1996). Evolution, structure and function of small heat shock proteins in plant. J. Exp. Bot. 47 325–338. [Google Scholar]
- Wehmeyer, N.N., Hernandez, L.D., Finkelstein, R.R., and Vierling, E. (1996). Synthesis of small heat-shock proteins is part of the developmental program of late seed maturation. Plant Physiol. 112 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weschke, W., Panitz, R., Sauer, N., Wang, Q., Neubohn, B., Weber, H., and Wobus, U. (2000). Sucrose transport into barley seeds: Molecular characterization of two transporters and implications for seed development and starch accumulation. Plant J. 21 455–467. [DOI] [PubMed] [Google Scholar]
- Westgate, M.E., and Boyer, J.S. (1986). Reproduction at low sink and pollen water potentials in maize. Crop Sci. 26 951–956. [Google Scholar]
- Williams, L.E., Lemoine, R., and Sauer, N. (2000). Sugar transporters in higher plants - A diversity of roles and complex regulation. Trends Plant Sci. 5 283–290. [DOI] [PubMed] [Google Scholar]
- Wobus, U., Sreenivasulu, N., Borisjuk, L., Rolletschek, H., Panitz, R., Gubatz, S., and Weschke, W. (2004). Molecular physiology and genomics of developing barley grains. Recent Res. Devel. Plant Mol. Biol. 2 1–29. [Google Scholar]
- Wobus, U., and Weber, H. (1999). Sugars as signal molecules in plant seed development. Biol. Chem. 380 937–944. [DOI] [PubMed] [Google Scholar]
- Wu, H., and Cheung, A.Y. (2000). Programmed cell death in plant reproduction. Plant Mol. Biol. 44 267–281. [DOI] [PubMed] [Google Scholar]
- Yang, J., Zhang, J., Huang, Z., Wang, Z., Zhu, Q., and Liu, L. (2002). Correlation of cytokinin levels in the endosperms and roots with cell number and cell division activity during endosperm development in rice. Ann. Bot. (Lond.) 90 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinselmeier, C., Westgate, M.E., Schussler, J.R., and Jones, R.J. (1995). Low water potential disrupts carbohydrate metabolism in maize (Zea mays L.) ovaries. Plant Physiol. 107 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.