Abstract
The cellulose binding elicitor lectin (CBEL) from Phytophthora parasitica nicotianae contains two cellulose binding domains (CBDs) belonging to the Carbohydrate Binding Module1 family, which is found almost exclusively in fungi. The mechanism by which CBEL is perceived by the host plant remains unknown. The role of CBDs in eliciting activity was investigated using modified versions of the protein produced in Escherichia coli or synthesized in planta through the potato virus X expression system. Recombinant CBEL produced by E. coli elicited necrotic lesions and defense gene expression when injected into tobacco (Nicotiana tabacum) leaves. CBEL production in planta induced necrosis. Site-directed mutagenesis on aromatic amino acid residues located within the CBDs as well as leaf infiltration assays using mutated and truncated recombinant proteins confirmed the importance of intact CBDs to induce defense responses. Tobacco and Arabidopsis thaliana leaf infiltration assays using synthetic peptides showed that the CBDs of CBEL are essential and sufficient to stimulate defense responses. Moreover, CBEL elicits a transient variation of cytosolic calcium levels in tobacco cells but not in protoplasts. These results define CBDs as a novel class of molecular patterns in oomycetes that are targeted by the innate immune system of plants and might act through interaction with the cell wall.
INTRODUCTION
During their whole life, plants are exposed to pathogenic microorganisms in their environment. Similar to animals, they have developed various defense mechanisms to avoid disease and death. Besides being induced by contact with pathogenic microorganisms, active defense reactions can also be triggered by treatment with microbial compounds called elicitors, which may be characteristic of a whole group of organisms or limited to specific strains of a microbial species (Bonas and Lahaye, 2002; Montesano et al., 2003; Jones and Takemoto, 2004). Such compounds have been characterized from fungi, oomycetes, bacteria, and viruses. Among eukaryotic plant pathogens, the genus Phytophthora contains >60 species that are pathogenic on a wide array of plants, causing economically important diseases worldwide. Many elicitors of various structures have been isolated from Phytophthora species, among them a cell wall–derived heptaglucan (Sharp et al., 1984), extracellular (glyco)proteins such as a transglutaminase (Brunner et al., 2002), elicitins (Ricci et al., 1989; Kamoun et al., 1997), GP32 (Bailleuil et al., 1996), and cellulose binding elicitor lectin (CBEL) (Villalba-Mateos et al., 1997).
In recent years, remarkable similarities between defense mechanisms triggered by elicitors in plants and what is known as the innate immune response in animals have been found (Gomez-Gomez and Boller, 2000; Parker, 2003; Nürnberger et al., 2004; Zipfel and Felix, 2005). A model has emerged in which discrimination from self is achieved through receptors that recognize pathogen-associated molecular patterns (PAMPs) (Janeway and Medzhitov, 2002). Such patterns correspond to motifs or domains with conserved structural traits found in widely occurring compounds of microbes but not present in their hosts and essential for microbial fitness. High-affinity binding sites in plants have been described for several general elicitors of bacterial, fungal, and oomycete origin, such as flagellin, chitin fragments, a β-heptaglucoside, and cryptogein (Bourque et al., 1999; Gomez-Gomez and Boller, 2000; Bradley Day et al., 2001), and a few of them have been cloned. Four peptides were recently designated as PAMPs in phytopathogenic microorganisms, similar to those involved in the innate immune response in mammals and insects. These include a stretch of 22 amino acids represented by the peptide flg22 from a conserved domain in bacterial flagellin (Felix et al., 1999), the Pep-13 domain from the cell wall elicitor GP42 of Phytophthora sojae (Brunner et al., 2002), the RNA binding motif RNP-1 of bacterial cold-shock proteins (Felix and Boller, 2003), and the N terminus of bacterial elongation factor Tu (Kunze et al., 2004).
We reported previously on the characterization and cloning of CBEL, a cell wall glycoprotein from Phytophthora parasitica var nicotianae (Ppn), the causal agent of the black shank disease of tobacco (Nicotiana tabacum) (Villalba-Mateos et al., 1997). This glycoprotein is widespread in the genus Phytophthora (Khatib et al., 2004) and is present during the growth of Ppn in vitro and in planta (Séjalon-Delmas et al., 1997). CBEL is a potent elicitor in the Ppn host plant tobacco, in which it induces local hypersensitive response (HR)–like lesions, defense responses, and protection against subsequent infection with the oomycete. It is also active in various nonhost plants, among them Arabidopsis thaliana (Khatib et al., 2004). Using Arabidopsis mutants affected in the signaling pathways that involve salicylic acid, jasmonic acid, or ethylene, it was shown that the three pathways are triggered by the elicitor and that its necrosis-inducing activity depends on ethylene and jasmonic acid (Khatib et al., 2004). The protein moiety of CBEL is composed of two direct repeats of Cys-rich domains, connected by a linker. Each repeat contains a motif that closely resembles the fungal type I cellulose binding domain (CBD) consensus pattern found in cellulases from various fungi (Gilkes et al., 1991). CBEL is able to bind to crystalline cellulose and tobacco cell walls in vitro in a dose-dependent manner, but in contrast with cellulases, it does not possess any detectable enzyme activity on various polysaccharides (Villalba-Mateos et al., 1997). Phenotypic characterization of Ppn strains suppressed in CBEL expression revealed that this glycoprotein is involved in organized polysaccharide deposition in the cell wall and in adhesion of the mycelium to cellulosic substrates (Gaulin et al., 2002). Thus, the role of CBEL in the biology of Ppn and its occurrence among various species of Phytophthora, as well as the wide range of plants responding to the elicitor, support the view that CBEL might contain a PAMP of eukaryotic plant pathogens. According to this concept, it is assumed that conserved structural motifs of the molecule account for its activity.
In this study, we investigated the role of the CBDs of CBEL in elicitor activity. It was evaluated both by infiltrating tobacco leaves with recombinant forms of the protein produced in Escherichia coli and by producing the protein in planta through the potato virus X (PVX) expression system (Jones et al., 1999). Deletion and mutational analysis, along with the use of synthetic peptides, allowed us to assign the role of PAMP to the CBDs of CBEL.
RESULTS
Elicitor Activity of CBEL Produced in E. coli or in Planta
To study the relationship between the structure of CBEL and its elicitor activity, we set up its expression in two heterologous systems.
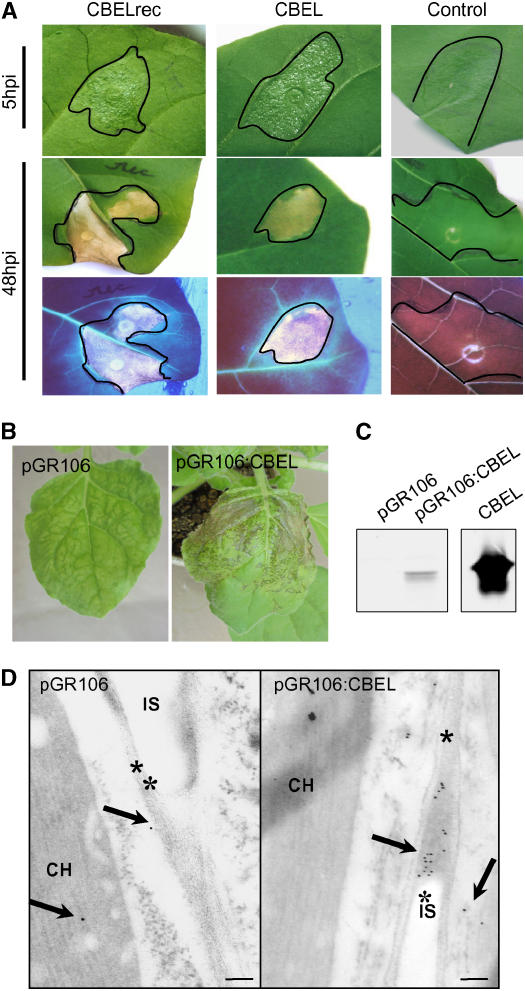
In the first approach, CBEL was produced in E. coli using the pFLAG-ATS vector (pATS:CBEL), which carries the OmpA secretion sequence, directing the recombinant protein to the periplasmic space. Upon isopropylthio-β-galactoside induction of BL21 cells containing pATS:CBEL, the corresponding protein was expressed and accumulated as insoluble inclusion bodies. After solubilization, efficient refolding of the denatured protein was achieved by adapting a protocol described by Berdichevsky et al. (1999). After dialysis against distilled water, the recombinant protein (CBELrec) appeared as a single band on SDS-PAGE after Coomassie blue staining (data not shown), with final amounts of 4 mg of protein per liter of cell culture. The elicitor activity of CBELrec was assessed by infiltrating a 200 nM protein solution into the mesophyll of tobacco leaves, with two leaves on three different plants for each assay. The infiltrated area turned slightly bright on the abaxial face at 4 to 5 h after infiltration, started to desiccate at 9 to 12 h after infiltration, and was fully desiccated by 24 h after infiltration. The necrosis turned brown thereafter and remained strictly limited to the infiltrated area (Figure 1A). UV light examination revealed that both the necrotic area and the surrounding tissues displayed a blue autofluorescence indicative of the accumulation of defense-related aromatic compounds. These symptoms and their time course are similar to those observed previously with native CBEL at the same protein concentration (Villalba-Mateos et al., 1997), indicating that the recombinant form of CBEL produced in E. coli is as active as the native protein and that the glycan part is not required for its elicitor activity.
Figure 1.
Elicitor Activity of CBEL Produced in E. coli or in Planta.
(A) Symptoms observed on tobacco leaves after infiltration of recombinant CBEL produced in E. coli. Leaves of 2-month-old tobacco plants were infiltrated with ∼100 μL of a 200 nM solution of purified CBELrec, native CBEL, or BSA as a control. Symptoms were observed at 5 and 48 h after inoculation (hpi) under white light (top) or UV light (bottom).
(B) Systemic symptoms observed on N. benthamiana plants after inoculation with A. tumefaciens strains carrying the pGR106 or pGR106:CBEL vector. Photographs were taken at 11 d after inoculation.
(C) Protein gel blot analysis of CBEL production in planta. Leaf samples were collected at 12 d after inoculation, and total proteins were extracted and subjected to protein gel blot analysis using an anti-CBEL polyclonal antiserum. The quantity of total protein corresponds to 3 cm2 of leaf area. Purified native CBEL (20 ng) was used as a control.
(D) Immunogold labeling of leaves at 11 d after inoculation with pGR106 (left) or pGR106:CBEL (right). Labeling was achieved with a purified polyclonal antiserum against CBEL and gold-conjugated goat antiserum to rabbit IgG. Sections were contrasted with uranyl acetate, and photographs were taken at ×25,000 magnification. Arrows show gold particles located in the cell wall; asterisks indicate the plant cell wall. CH, chloroplast; IS, intercellular space.
The second approach consisted of producing CBEL in planta by means of a viral expression vector derived from PVX. This virus infects Nicotiana benthamiana, and the elicitor activity of the heterologous proteins produced is visible either as a local HR-like necrosis or as systemically spreading necrotic lesions. The entire CBEL coding sequence, including its own secretion signal, was cloned into the binary PVX vector pGR106 (Lu et al., 2003), resulting in plasmid pGR106:CBEL.
This plasmid was introduced into Agrobacterium tumefaciens to allow the delivery of PVX into N. benthamiana plants via inoculation with the bacteria. Control plants inoculated with bacteria containing the empty vector pGR106 (PVX) developed the chlorotic mosaic symptoms and leaf curling characteristic of PVX infection in systemically infected leaves at 11 d after inoculation (Figure 1B). Inoculation with bacteria carrying pGR106:CBEL first induced systemic PVX mosaic symptoms in the upper noninoculated leaves, followed rapidly by localized necrotic lesions that became confluent soon afterward (Figure 1B). Protein gel blot analysis with polyclonal antibodies against CBEL showed that leaves developing necrosis contained substantial amounts of CBEL (Figure 1C).
Immunocytochemistry was used to localize CBEL in leaf tissues developing small necroses. Immunogold labeling using a purified CBEL antibody showed that the protein deposition occurred in the cell wall and junctions of pGR106:CBEL-infected parenchyma cells, whereas only a few gold particles were observed in the control samples (Figure 1D). A faint background labeling of chloroplasts and cytoplasm could be observed in both pGR106:CBEL-infected samples and the pGR106-infected controls. Together, these results show that CBEL retains its elicitor activity when expressed in plant cells and that its endogenous oomycete signal peptide is functional in directing the protein to the extracellular space. They also suggest that CBEL binds to the plant cell wall in situ, as no labeling was observed in the intercellular spaces.
Aromatic Amino Acids in the CBD of CBEL Are Important for Elicitor Activity
Studies Using CBEL Produced in Planta
Because the PVX-based expression system does not require purification of the proteins under study, it is very convenient for screening a large array of modified proteins. This approach was first used to search for structural motifs involved in the elicitor activity of CBEL.
The occurrence of two CBDs homologous with those found in fungal cellulases is one of the main characteristics of CBEL. Structure–function relationship studies on the CBD of cellobiohydrolase I, CBDCBHI (also named CBDCel7A), from the filamentous fungus Trichoderma reesei have demonstrated an essential role of the aromatic amino acids Y5, Y31, and Y32 for its binding to cellulose (Linder et al., 1995b). Based on sequence alignment of the two CBDs (CBD1 and CBD2) of CBEL with the T. reesei CBD (Figure 2A), the amino acids Y51 and Y52 in CBD1 and F187 and Y188 in CBD2 were predicted to be surface-exposed and involved in the cellulose binding of CBEL. Hence, these residues were replaced individually or in pairs by Ala through PCR-based site-directed mutagenesis. The mutant fragments were cloned into pGR106, resulting in constructs whose expression products were named Y51A, Y52A, Y51A_Y52A, Y188A, F187A_Y188A, and Y52A_Y188A (Figure 2B). The various plasmids were then introduced into A. tumefaciens for N. benthamiana inoculation. Three independent experiments were performed on 4-week-old plants; in each experiment, three plants were inoculated with each PVX construct, and symptoms were scored from 9 to 15 d after inoculation. Representative phenotypes of the five classes of the symptom scale are shown at the top of Figure 2C. Whereas control plants inoculated with pGR106 displayed systemic mosaic symptoms after 9 d, 90% of N. benthamiana leaves inoculated with pGR106:CBEL exhibited local or spreading necrosis at this time. The lesions spread rapidly, affecting 100% of the plants after 11 d and causing plant death after 15 d. Lesions were less extended and developed less rapidly with all mutant forms, notably those involving residue Y188; they appeared as localized spots between 11 and 13 d after inoculation and only rarely did they spread throughout the whole plant. No additive effect of these mutations was observed. Hence, this symptom scoring shows that the necrosis-inducing activity of CBEL is significantly reduced by point mutations of aromatic residues potentially involved in cellulose binding.
Figure 2.
Site-Directed Mutagenesis Analyses of CBEL Using the PVX Expression System.
(A) Alignment of CBD1 and CBD2 of CBEL and the cellobiohydrolase I CBD from T. reesei. The deduced amino acid sequences of CBDs from CBEL (CBD1, CBD2) and cellobiohydrolase I CBD from T. reesei (CBDCBHI) were aligned according to the ClustalW and BoxShade sequence analysis software programs. Single-letter codes are used for amino acid residues, black or gray boxes indicate identical or similar residues, and dashes indicate gaps introduced to allow optimal alignment of the sequence. Asterisks mark the positions of the aromatic amino acids of the CBD from T. reesei implicated in cellulose binding affinity.
(B) Scheme indicating the positions of mutations introduced into the CBEL sequence. Y51A, Y52A, and Y188A correspond to a single amino acid exchange of Tyr-51, Tyr-52, and Tyr-188 with Ala, respectively. Y51A_Y52A, F187A_Y188A, and Y52A-Y118A are double point-mutated versions of CBEL.
(C) Comparative analysis of necrosis-inducing activity of the mutated versions of CBEL. At top is the symptom scale used in the experiments, corresponding to the following phenotypes: no symptoms (I), typical mosaic disease symptoms (II), localized necrosis (III), confluent or spreading necrosis (IV), and death of the plant (V). Observations were conducted every 2 d from 9 to 15 d after inoculation (DPI). Control plants were inoculated with A. tumefaciens harboring the empty PVX expression vector (pGR106). Test plants were inoculated with agroinfected PVX strain containing a wild-type (pGR106:CBEL), point-mutated (pGR106:Y51A, pGR106:Y52A, pGR106:Y188A), and double mutated (pGR106:Y51A_Y52A, pGR106:F187A_Y188A, pGR106:Y52A_Y188A) DNA construct. Each box symbolizes one inoculated plant.
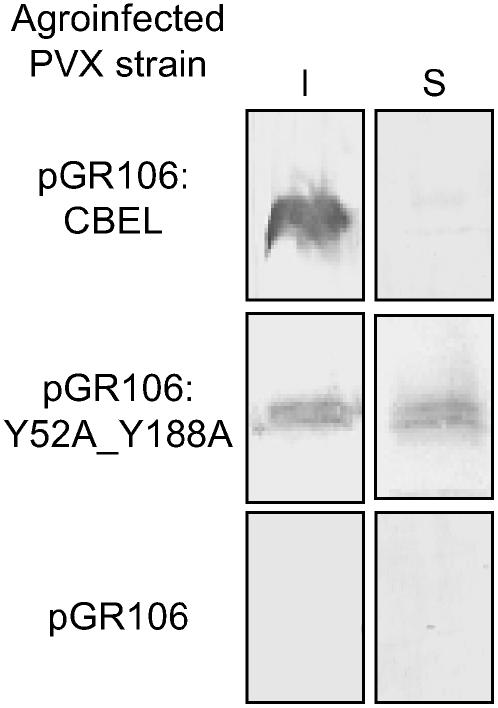
To determine whether the point mutations affected the ability of CBEL to bind to plant cell walls, proteins were extracted from systemically infected leaves at 14 d after inoculation, and the soluble and insoluble fractions were analyzed separately. As shown in Figure 3, the wild-type CBEL was recovered only in the insoluble fraction, indicating that it was associated with cell wall components, whereas the double mutant version Y52A_Y188A was recovered in both the soluble and insoluble fractions. Substitution of Y52 or of Y188 alone by Ala did not result in a significant reduction of this cell wall binding capacity (data not shown). Together, these results show that a single mutation on the aromatic amino acids of either CBD is sufficient to strongly decrease the elicitor activity of CBEL, whereas binding to the plant cell wall is affected only when both CBDs are mutated at functionally equivalent positions.
Figure 3.
Affinity of Wild-Type CBEL and Double Mutant Protein to the Plant Cell Wall.
Insoluble (I) and soluble (S) proteins were extracted from PVX systemically infected leaves at 14 d after inoculation and subjected to protein gel blot analysis using an anti-CBEL polyclonal antiserum. The specificity of labeling is shown with extracts from plants inoculated with bacteria carrying the empty vector pGR106.
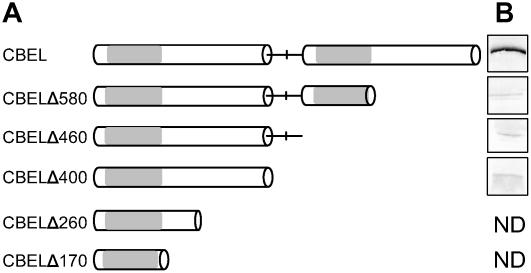
The fact that CBD double mutant versions of CBEL retained residual necrosis-inducing activity suggested that other motifs of CBEL might also be involved in elicitation. To determine the minimal length required for CBEL activity, constructs progressively shortened by C-terminal deletions were tested using the PVX system on N. benthamiana plants (Figure 4A). All plants inoculated with the various deleted constructs developed only systemic mosaic symptoms typical of PVX infection, comparable to control plants inoculated with the empty vector. Protein gel blot analyses were performed at 12 d after inoculation to check the presence of the truncated CBEL proteins (Figure 4B). Whereas a major protein band was revealed in leaves systemically infected with pGR106:CBEL, the protein band was very faint or undetectable in all cases of truncated CBEL versions. This finding indicates that the failure of CBEL deletion mutants to elicit necrosis was probably the result of insufficient accumulation of the proteins in the leaves.
Figure 4.
Expression of CBEL Truncated Proteins in Plant Cells via PVX.
(A) Schemes of the protein constructs. The two direct Cys-rich repeats are represented as white boxes and separated by a line corresponding to the Thr/Pro linker containing a central Met. Gray boxes symbolize the CBD subdomains.
(B) Protein gel blot analysis of CBEL accumulation in N. benthamiana systemically infected leaves. Protein extracts were obtained from inoculated plants at 14 d after inoculation and analyzed by use of an anti-CBEL polyclonal antiserum. ND, not determined.
In summary, PVX results demonstrate that necrosis on N. benthamiana plants is mediated by amino acids implicated in the interaction of CBEL with plant cell walls.
Studies Using Recombinant CBEL
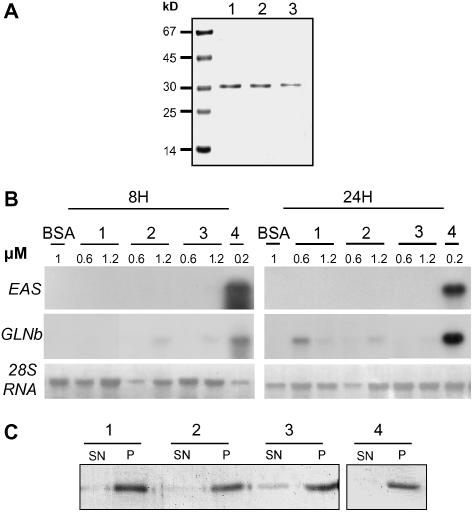
Three proteins with mutated CBDs (Y52Arec, Y188Arec, and Y52A_Y188Arec), previously tested using the PVX system, were also produced and purified to homogeneity from E. coli inclusion bodies (Figure 5A). They were infiltrated into the mesophyll of tobacco leaves, and symptoms were monitored from 24 h to 5 d after inoculation. At a concentration of 0.2 μM, equivalent to the concentration at which unmodified CBEL (CBELrec or CBEL) induced necrosis, none of them induced macroscopic symptoms; only at much higher concentrations (1.2 to 2.4 μM) was a faint fluorescence observed. The infiltrated areas (six areas per assay) were collected at 8 and 24 h after infiltration, and total RNA was extracted for RNA gel blot analyses (Figure 5B). CBELrec at 0.2 μM induced the accumulation of defense gene transcripts such as 5-epi-aristolochene synthase, also called sesquiterpene cyclase (EAS) and basic glucanase as efficiently as the native glycoprotein (Villalba-Mateos et al., 1997). By contrast, the Y52Arec, Y188Arec, and Y52A_Y188rec mutated forms of CBEL failed to induce defense gene expression at this concentration. Even at higher concentrations up to 1.2 μM, the mutated proteins were inactive except in rare cases in which a slight induction was observed. These results confirm the data obtained by PVX expression showing that mutations either in Y52 or Y188 impair CBEL recognition by the plant cell.
Figure 5.
Elicitor and Cellulose Binding Activity of CBEL Mutant Recombinant Proteins.
(A) Purified CBEL mutant proteins. Y52Arec (lane 1), Y188Arec (lane 2), and Y52A_Y118Arec (lane 3) were expressed in E. coli and purified from inclusion bodies as described in Methods. Proteins from purified extract (5 μg) were subjected to SDS-PAGE analysis and revealed by Coomassie blue staining.
(B) Defense gene induction by CBEL mutant proteins in tobacco leaves. Leaf infiltrations were performed with 0.6 or 1.2 μM solutions of each protein (Y52Arec [lane 1], Y188Arec [lane 2], Y52A_Y188Arec [lane 3], 0.2 μM CBELrec [lane 4], or 1 μM BSA as a control). Total RNA was isolated from tobacco leaves at 8 and 24 h after treatment and analyzed for the expression of defense genes encoding sesquiterpene cyclase (EAS) and basic glucanase (GLNb). Equal loading of the gel was checked on the membrane by visualization of rRNAs (28S RNA) under UV light (λ = 254 nm).
(C) Cellulose binding assays. Recombinant mutant proteins (lane 1, Y52Arec; lane 2, Y188Arec; lane 3, Y52A_Y188Arec [124 ng]) and CBELrec (lane 4 [140 ng]) were incubated with 400 ng of cellulose. Proteins in the supernatant (SN) or solubilized from the cellulose pellet (P) were analyzed by SDS-PAGE, and the proteins were visualized by silver staining.
To test whether such mutations would affect the CBEL cellulose binding affinity, the purified Y52Arec, Y188Arec, and Y52A_Y188Arec were assayed for their interaction with crystalline cellulose in vitro. Proteins were incubated for 1 h at room temperature in a 2% cellulose suspension according to Villalba-Mateos et al. (1997). Subsequent centrifugation resulted in the cellulose-bound (pellet) and unbound (supernatant) protein fractions. Their analysis by SDS-PAGE revealed that part of the double mutated protein (Y52A_Y188Arec) was in the supernatant, whereas unmodified CBELrec was detected only in the pellet. The two mutant proteins containing a single Ala exchange (Y52Arec and Y188Arec) were detected preferentially in the cellulose pellet (Figure 5C). Hence, the simultaneous mutation of the two CBDs slightly decreased the affinity of CBEL for cellulose, whereas mutation of only one CBD had no effect on binding.
Together, the results observed with recombinant CBEL confirm those obtained with PVX expression (i.e., that amino acids putatively involved in cellulose binding are important for elicitor activity).
CBDs of CBEL Mediate Defense Responses in Plants
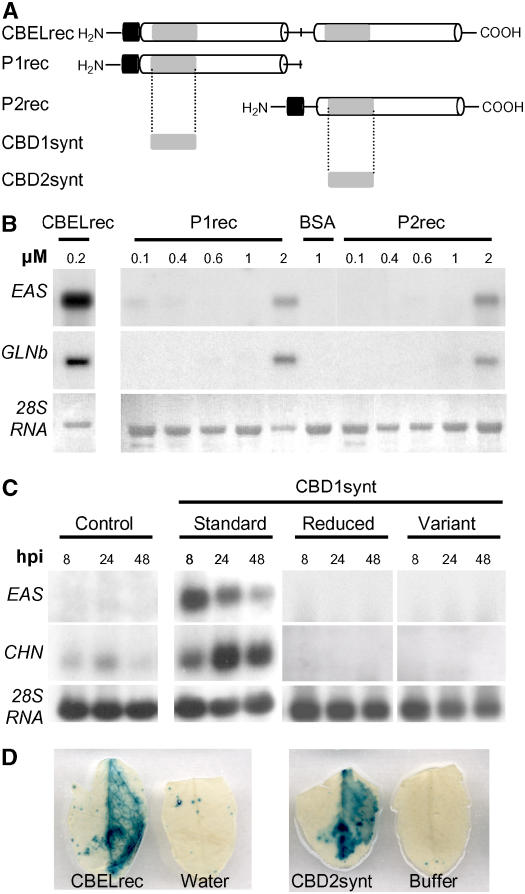
To more precisely define the CBEL domain involved in elicitor activity, the two moieties of CBEL containing their respective CBD (CBD1 or CBD2) were produced in E. coli (Figure 6A). Considering the protein as a repeated structure organized around the central Met residue, constructs for the production of either the N-terminal half (P1rec) or the C-terminal half (P2rec) of CBEL were cloned into the pFLAG:ATS vector and the proteins were purified from inclusion bodies as described above. After infiltration of P1rec and P2rec into tobacco mesophyll, no macroscopic symptom could be observed on leaves, in contrast with the areas infiltrated with CBELrec. However, under UV light examination, a faint blue autofluorescence, which is indicative of the accumulation of aromatic defense-like compounds, was detected after infiltration with either polypeptide. Dose–response experiments, in which the infiltrated P1rec and P2rec concentrations ranged from 0.1 to 2 μM, showed that EAS and GLN gene expression was induced at a concentration of 2 μM (Figure 6B). Thus, its appears that P1rec and P2rec each contains functional domains recognized by the plant cell to elicit defense responses.
Figure 6.
Elicitor Activity of CBDs.
(A) Schemes of CBEL moieties tested for elicitor activity. CBELrec, P1rec (amino acids 1 to 144), and P2rec (amino acids 144 to 268) were produced in E. coli. CBD1synt (amino acids 5 to 41) and CBD2synt (amino acids 142 to 177) were synthesized chemically. The two direct Cys-rich repeats are represented as white boxes and separated by a line corresponding to the Thr/Pro linker containing a central Met. Gray boxes symbolize the CBD subdomains, and the black boxes indicate the FLAG peptide localized at the N-terminal part of the recombinant proteins.
(B) Dose response of defense gene expression encoding sesquiterpene cyclase (EAS) and basic glucanase (GLNb) in tobacco leaves upon treatment with CBELrec, P1rec, and P2rec. Total RNA was extracted at 24 h after inoculation with P1rec (0.1 to 2 μM), P2rec (0.1 to 2 μM), CBELrec (0.2 μM), and BSA (1 μM). For the RNA gel blot analyses, equal loading of the gels was checked on the membranes by visualization of rRNAs (28S RNA) under UV light (λ = 254 nm).
(C) Tobacco leaf infiltrations were performed with 5 μM solutions of the synthetic peptide CBD1synt (standard), peptide CBD1synt preincubated in 50 mM DTT (reduced), peptide CBD1synt in which Cys residues were replaced by Ser residues (variant), or 250 μM sodium phosphate buffer as a control. Total RNA was isolated from tobacco leaves at 8, 24, and 48 h after inoculation (hpi) after treatment and analyzed for expression of defense genes encoding sesquiterpene cyclase (EAS) and chitinase (CHN). For the RNA gel blot analyses, equal loading of the gels was checked on the membranes by visualization of rRNAs (28S RNA) under UV light (λ = 254 nm).
(D) PR1–β-glucuronidase (GUS) A. thaliana leaves were infiltrated with 0.2 μM CBELrec or a 5 μM solution of the synthetic peptide CBD2synt as indicated. GUS activity was revealed at 48 h after inoculation.
To screen smaller sections of P1rec and P2rec, peptides corresponding to the CBD1 and CBD2 domains of CBEL (CBD1synt and CBD2synt, respectively) were chemically synthesized (Figure 6A). The elicitor activity of CBD1synt was analyzed by RNA gel blot upon infiltration of a 5 μM solution into tobacco leaves. As shown in Figure 6C, the peptide induced the expression of two defense genes coding EAS and chitinase, with maxima at 8 and 24 h after infiltration, respectively. The elicitor activity of CBD2synt was assessed by infiltration into leaves of transgenic Arabidopsis plants expressing the uidA gene under the control of a PR1 promoter. GUS activity staining was detected 48 h after infiltration with CBELrec (0.2 μM) or CBD2synt (5 μM) with a similar intensity, as shown in Figure 6D. These findings demonstrate that the CBDs are sufficient to activate plant defense reactions. The fact that CBD concentrations greater than CBEL concentrations are required for similar elicitor effects correlates with the fact that only a small proportion of the synthesized CBDs (<10%) had the ability to bind cellulose (see Supplemental Figure 1 online). This probably reflects incorrect folding of the synthetic peptide as a result of the conditions needed to solubilize these hydrophobic peptides containing several Cys residues. Indeed, a synthetic variant of CBD1 in which the Cys residues were replaced by Ser, as well as a reduced version of CBDsynth, proved inactive upon infiltration of tobacco leaves (Figure 6C).
CBEL Induces Calcium Fluxes in BY-2 Tobacco Cells but Not in Protoplasts
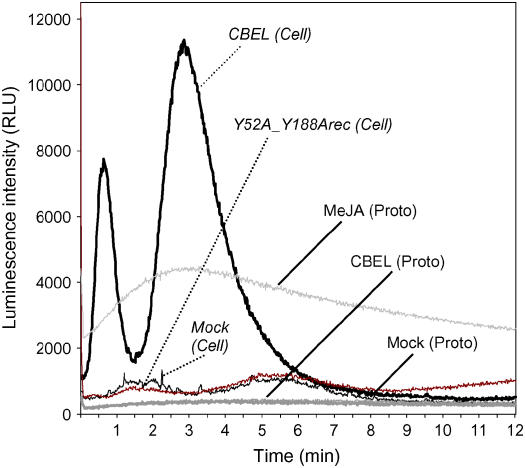
Because CBEL and its CBDs interact with cellulose, the question of whether the plant cell wall could be involved in elicitor perception was addressed. To test this hypothesis, an additional experiment was undertaken, based on the well-documented ability of elicitors to trigger a transient increase of calcium in plant cells before defense induction. A tobacco BY-2 cell suspension stably expressing the aequorin gene, and protoplasts derived from these cells, were used to analyze cytosolic Ca2+ concentration ([Ca2+]cyt) changes in response to CBEL. As shown in Figure 7, CBEL treatment (1.5 μM) induced a transient increase in [Ca2+]cyt in a bimodal manner. The first peak was observed within 40 s after CBEL addition and was followed by a second peak after 3 min of incubation. The[Ca2+]cyt returned to a basal level after 8 min. Such a [Ca2+]cyt increase was not observed when the double mutated CBEL (Y52A_Y188Arec) was used (1.5 μM) (Figure 7). Nor was it observed when BY-2 protoplasts were treated with 1.5 μM CBEL (Figure 7). This lack of response could not be attributed to a protoplast luminescence defect, because protoplasts had retained their ability to respond to external stimuli, such as methyl jasmonate (Figure 7).
Figure 7.
Changes in Luminescence Intensity in Aequorin-Transformed Cells and Protoplasts upon CBEL Treatment.
Tobacco cells or protoplasts were treated with 1.5 μM CBEL [CBEL (Cell), CBEL (Proto)], 1.5 μM Y52A_Y188Arec [Y52A_Y188Arec (Cell)], 5 mM methyl jasmonate [MeJA (Proto)], or water [Mock (Cell), Mock (Proto)], and luminescence counts were recorded continuously at 1-s intervals for 12 min. Data correspond to one representative experiment out of three. RLU, relative light units.
Together with the various bioassays reported in this study, this experiment strongly supports the view that the cell wall and unmodified CBDs are involved in CBEL perception.
DISCUSSION
Of the various protein elicitors isolated from oomycetes, CBEL has the unique property to bind to cellulose and to plant cell walls, consistent with the modular structure of the protein comprising two CBDs (Villalba-Mateos et al., 1997). A structure–function relationship study was undertaken to evaluate the role of these domains in the induction of plant defense gene expression and the development of HR-like lesions. Two complementary approaches were developed, comprising the use of mutated versions of the entire CBEL molecule and synthetic peptides.
To investigate the importance of CBDs in the elicitor activity of CBEL, site-directed mutagenesis was performed. Because the CBDs of CBEL contain aromatic residues at positions that are highly conserved in all fungal CBDs and are known to be important in the cellulose binding affinity of these domains (Linder et al., 1995b), their contribution was evaluated by Ala replacement. Single mutations on either CBD1 or CBD2 (Y51A, Y52A, or Y188A) strongly reduced CBEL necrosis-inducing activity in planta. Double mutations (Y51A_Y52A, F187A_Y188A, or Y52A_Y188A) did not further decrease the activity of the elicitor. Infiltration assays using recombinant modified proteins fully confirmed that the integrity of each CBD must be preserved for full defense induction (i.e., necrosis and defense gene expression). Thus, mutations within the CBDs revealed the importance of conserved amino acids in these carbohydrate binding modules for elicitor activity. Interestingly, the data also show that the inductions of HR-like necrosis and defense gene expression are not correlated, because neither P1rec nor P2rec (corresponding to half of the native CBEL) induced necroses in infiltrated leaves, although they stimulated defense gene expression. This finding suggests that the two halves of CBEL have to be physically linked for the induction of necrosis. This is consistent with the previous finding that the inductions of defense gene expression and of necrosis by CBEL are regulated by different pathways in Arabidopsis (Khatib et al., 2004).
It was noticed that the simultaneous substitution of Y52 and Y188 by Ala only slightly reduced the binding affinity of CBEL to plant cell walls and crystalline cellulose. This might result from the fact that only one of the three surface-exposed aromatic residues was replaced in each CBD. It might also be attributable to a synergistic interplay between the two CBDs for binding to cellulose, which would result in stabilization of the CBEL–cellulose complex, as already described in the case of a genetically engineered double CBD of cellobiohydrolases from T. reesei (Linder et al., 1996). So, although they confirm that the aromatic amino acids indeed contribute to the cellulose binding and elicitor activity of CBEL, our results show that the two activities are not strictly correlated.
Synthetic peptides corresponding to the surface exposed CBDs of CBEL were used to check whether these domains are sufficient to act as elicitors in plants (i.e., tobacco and Arabidopsis). It was found that the synthetic CBD peptides did not induce necrosis but were still able to elicit defense. However, they generally had to be infiltrated at higher concentrations (5 versus 0.2 μM) to induce comparable defense gene expression (Figure 6C; see Supplemental Figure 2 online). Such differences could be attributable to several causes, including susceptibility to plant proteases or peptide stability. An example of peptide degradation is illustrated by flagellin-derived peptides that are not stable in suspension-cultured cells; thus, elicitor activity rapidly disappeared, except after heat treatment of the medium (Meindl et al., 2000). Correct folding of the synthetic peptides is also important. Indeed, CBD1synt and CBD2synt are long synthetic peptides (36 and 35 amino acids) containing four Cys residues involved in two disulfide bonds. Because the disulfide bonds of CBD are required for proper folding and cellulose binding activity (Gilkes et al., 1991), we investigated the possibility that the difference in specific activity between CBEL and the peptides might reflect misfolding of part of the synthetic CBDs. Upon incubation of the synthetic peptides with cellulose, it appeared that only a fraction was able to bind crystalline cellulose (see Supplemental Figure 1 online), suggesting that a substantial amount of the solution was not folded correctly, thereby explaining why a greater amount of CBDs was required to elicit defense responses. In addition, synthetic CBD conformation may differ from their native conformation in the context of the intact CBEL protein. The same situation was described for the synthetic pentapeptide TKLGE from the ethylene-inducing xylanase from Trichoderma viride, which was unable to elicit a hypersensitive response in tobacco but was shown to be essential for the elicitor activity of the protein (Rotblat et al., 2002). The possibilities that some regions outside of the two CBDs also play a role, and that the two CBDs should be physically linked to achieve optimal perception by the plant cell, also should be considered. This latter hypothesis is supported by the fact that P1rec and P2rec (corresponding to half of the native CBEL) displayed weaker elicitor activities than CBEL, even when they were infiltrated simultaneously into plant tissues. Despite these limitations, CBDs are sufficient for the activation of a biological response.
The family 1 carbohydrate binding modules (Pfam; PF00734) represented in CBEL by two CBDs are known to occur as structurally independent, well-defined, and very compact stable domains in most fungal glycanases (Linder et al., 1995a, 1995b; Bray et al., 1996). Importantly, CBM_1 is found in most fungi and has not been detected in higher plants (http://afmb.cnrs-mrs.fr/CAZY/CBM_1.html). In silico analysis of the occurrence of this module in sequenced annotated genomes of filamentous fungi showed that the genome of Neurospora crassa contains two to five times more CBDs than the genomes of the plant pathogens Magnaporthe grisea and Fusarium graminearum (see Supplemental Figure 3 online). The low occurrence of CBDs in fungal pathogens might reflect an adaptive mechanism that minimizes the perception and induction of basal plant defense responses.
The characteristic features of the CBDs from CBEL are in agreement with the definition of PAMPs, used when referring to microbial components that elicit innate immune responses. PAMPs are molecular patterns present in molecules that are unique to microbes and important for microbial fitness (Nürnberger et al., 2004). To date, CBEL is the only PAMP-containing molecule from phytopathogenic microorganisms for which a functional study in the corresponding microorganism was performed, by generating CBEL-silenced Phytophthora mutants (Gaulin et al., 2002). The phenotype of the mutants clearly showed that CBEL is involved in exogenous cellulose perception and Phytophthora cell wall organization. In this context, CBDs belonging to the CBM_1 family can be considered as PAMPs.
The mechanism of CBD perception in plants remains to be elucidated. It has been postulated that plants have evolved to recognize PAMPs, probably through the presence of pattern-recognition receptors (Ausubel, 2005), as demonstrated for flg22 and Pep-13 (Nürnberger et al., 1995; Bauer et al., 2001; Chinchilla et al., 2006). Because the aromatic amino acids involved in cellulose binding of CBDs are surface-exposed residues, they might easily interact with a putative receptor at the plasma membrane. However, one might also consider the possibility that the binding of CBEL to cellulose in plant cell walls is involved in its perception. Indeed, CBEL triggered a bimodal increase in [Ca2+]cyt when administered to aequorin-transformed BY-2 tobacco cells and not to tobacco protoplasts. Induction of biphasic increases of [Ca2+]cyt have been reported for tobacco cells treated with cryptogein and oligouronide elicitors (Lecourieux et al., 2002) and Pep-13–treated parsley (Petroselinum crispum) cells (Blume et al., 2000). In these studies, the induced calcium fluxes were shown to be tightly correlated to elicitor perception and were required for defense activation. Thus, the induction of calcium fluxes by an elicitor can be considered as a defense-related response, and in the case of CBEL, this response requires the presence of the cell wall. A close link between cell wall components and plant defense is indicated by several published data (reviewed in Vorwerk et al., 2004). Characterization of several Arabidopsis cell wall mutants carrying a mutation in the cellulose synthase gene CeSA3 (i.e., cev1 and eli1 mutants) further supports the notion that the cell wall can signal stress responses in plant–pathogen interactions (Ellis and Turner, 2001; Ellis et al., 2002, Cano-Delgado et al., 2003). Thus, it is likely that the interaction of a CBD with plant cellulose microfibrils in planta results in a local nonenzymatic disruption of plant cellulose microfibrils, as has been shown for the CBDs found in cellulases and expansin (Din et al., 1991; Shpigel et al., 1998; Levy et al., 2002). Such an alteration could be sufficient to trigger defense gene expression, whereas the simultaneous interaction of the two CBDs in CBEL with microfibrils could have an additional effect on cell wall integrity and lead to necrosis.
Altogether, the data obtained in this work on CBEL, a general elicitor of original structure, showed that CBDs of the family 1 carbohydrate binding module are sufficient and necessary for the induction of plant defense gene expression. They add to already compelling evidence that the cell wall has a key position in the molecular dialogue between plants and microorganisms. Future work will aim to discover the precise mechanism by which CBDs are perceived by the plant cell.
METHODS
Plant Material
Tobacco (Nicotiana tabacum) plants were grown on vermiculite in a growth chamber at 75% hygrometry, with a photoperiod of 12 h of light at 25°C and 12 h of dark at 22°C. Plant leaves were infiltrated 7 weeks after seed germination. Nicotiana benthamiana plants were grown under the same conditions in pots containing a 1:2 sand:soil mix. Plants were challenged with recombinant PVX constructs at ∼4 to 6 weeks after seed germination.
BY-2 Cell Suspension Culture and Protoplast Isolation
Aequorin-transformed BY-2 tobacco cells were grown as described by Pauly et al. (2001). For protoplast isolation, aequorin-transformed BY-2 tobacco cells were resuspended into 25 mM Tris-MES buffer, pH 5.5, and 0.6 M mannitol supplemented with 1 mg/mL Pectolyase Y23 (MP Biomedicals), 10 mg/mL Cellulase RS (Onozuka), 2 mg/mL Driselase (Sigma-Aldrich), and 10 mg/mL BSA for protoplast isolation. One gram of BY-2 cells per 10 mL of enzyme cocktail was used for each experiment. The enzymatic digestion of the cell walls was performed for 2 h with mild agitation at 37°C. Protoplasts were then successively centrifuged (5 min, 4°C, 160g) and washed using 15 mL of 25 mM Tris-MES buffer, pH 5.5. The pellet was resuspended in 2 mL of 20% Ficoll PM-400 (Sigma-Aldrich) and then overlaid with 2 mL of 10% Ficoll and the same volume of 25 mM Tris-MES buffer, pH 5.5. The resulting step gradient was centrifuged at 160g for 30 min at 4°C. Intact protoplasts were collected from the 10/20% Ficoll interface, washed once with 25 mM Tris-MES buffer, pH 5.5, and diluted (5 × 106 protoplasts/mL).
Heterologous Expression of CBEL and Its Structural Derivatives in Escherichia coli
The CBEL coding region was amplified by PCR from cDNA and cloned into pATS (Sigma-Aldrich) between 5′ HindIII and 3′ EcoRI sites. pATS:CBEL was subjected to site-directed mutagenesis using the GenEditor in vitro site-directed mutagenesis system (Promega) according to the recommendations of the supplier. CBEL and mutants were expressed in E. coli BL21. Expression was induced by the addition of 500 μM isopropylthio-β-galactoside (4 h, 28°C). Recovery and solubilization of inclusion bodies were performed as described by Berdichevsky et al. (1999). The urea-solubilized inclusion bodies were dialyzed against 10 L of a sodium acetate buffer (100 mM, pH 5.2) at 4°C and subjected to CM-Sepharose chromatography using the same buffer. The column was eluted with a NaCl gradient from 0 to 1 M. The eluted proteins were combined and neutralized by dialysis against 100 volumes of water at 4°C for 16 h. Purified proteins were stored at 4°C, and concentration was estimated with the Bio-Rad protein assay kit using BSA as a standard.
Construction of Recombinant Agrobacterium tumefaciens Binary PVX Vectors
The construct pGR106:CBEL was based on the entire CBEL coding sequence, which was amplified by PCR using CBEL cDNA as a template. The ClaI-digested PCR fragment was ligated into pGR106 (Lu et al., 2003) (kindly provided by D.C. Baulcombe). pGR106:Y51A, pGR106:Y52A, pGR106:Y51A_Y52A, pGR106:Y188A, pGR106:F187A_Y188A, and pGR106:Y52A_Y118A vectors were based on pATS:Y51A, pATS:Y52A, pATS:Y51A_Y52A, pATS:Y188A, pATS:F187A_Y188A, and pATS:Y52A_Y188A vectors, respectively. In the first step, gene-specific primers complementary to the 5′ and 3′ ends of the CBEL native signal peptide, including restriction site overhangs for cloning into pUC19 and pGR106, were designed: PS-F, 5′-GAGCTCATCGATCCATTGCTATTCGCATTACCGTAGTCATCGCTGG-3′; and PS-R, 5′-TCTAGAGCGGCCGCGGTACCCTCGAGGCCGGCAGCATCAGAGCCACAGTTGC-3′.
After ligation into SacI and XbaI pUC19 restriction sites, vectors were digested by NaeI and KpnI. Digested fragments were ligated into pUC19:SP, resulting in a translational fusion of the CBEL signal peptide with the mutated CBEL open reading frame. In the third step, recombinant plasmids were digested by ClaI and the purified inserts were ligated into pGR106. Shortened C-terminal CBEL deletion vectors were obtained by PCR using pGR106:CBEL vector as a template. A ClaI site was introduced at the 5′ end of both forward and reverse primers: CBEL-F, 5′-GGGAAATCGATCCATGGCTATTCGCATTACCGTAGTCTTCGC-3′; PpΔ580-R, 5′-GGGAAATCGATTTATGGCGGCTGGATGCACTGGTAA-3′; PpΔ460-R, 5′-GGGAAATCGATTTAGGTTGGCGATGATGTCGTCGGCGTTGGC-3′; PpΔ400-R, 5′-GGGAAATCGATTTACGTCAAGATGCCCGAGACTGC-3′; PpΔ260-R, 5′-GGGAAATCGATTTAGCGCGTGCAGCATTCTCCGGGCTG-3′; PpΔ170-R, 5′-GGGAAATCGATTTAATCGAGGCACTGGTAGTAGTTGGC-3′.
The ClaI-digested PCR fragments were ligated into pGR106 that was linearized previously with the same enzyme. The binary expression constructs were introduced into A. tumefaciens strain GV3101 by electroporation, and transformed bacteria were selected on Luria-Bertani agar plates supplemented with tetracycline and kanamycin.
RNA Gel Blot Analysis
Total RNA was extracted from leaves by use of the Extract-all reagent (Eurobio). RNA gel blot analysis was performed with 15 μg of total RNA as described previously (Rickauer et al., 1997). The probes used in this study were pBS-TEAS for EAS (Facchini and Chappell, 1992), pNT517 for glucanase (Godiard et al., 1990), and pCHN50 for chitinase (Shinshi et al., 1987), all kindly provided by the authors cited. The corresponding inserts were labeled with [α-32P]dCTP by random priming using the RadPrime DNA labeling system (Life Technologies). After hybridization and washing, the membranes were exposed to Hyperfilm MP films (Amersham) at −80°C for 24 h. Each experiment, including sample preparation and RNA isolation, was performed three times independently.
Recombinant Protein Infiltration Assays, Binding Studies, and GUS Activity
Recombinant proteins were infiltrated into the mesophyll of fully expanded leaves of 2-month-old plants with a syringe without a needle. Two leaves per plant from three individual plants were infiltrated, and the corresponding area was delineated with a marker pen. The tissues corresponding to the different infiltrated areas (six per sample) were harvested, frozen in liquid nitrogen, and stored at −80°C until use.
In vitro cellulose binding studies using recombinant proteins and GUS enzyme assays were performed as described by Villalba-Mateos et al. (1997) and Jefferson et al. (1987), respectively.
Peptide Synthesis
Peptides (CBD1synt, 5′-SFGNCGSDAAGVSCCQSTQYCQPWNANYYQCLDLPA-3′; CBD2synt, 5′-PYGSCGSSNGATCCPSGYYCQPWNDSFYQCIQPPA-3′; and CBD1synt variant, 5′-SFGNSGSDAAGVSSSQSTQYSQPWNANYYQSLDLPA-3′) were synthesized by automated solid-phase synthesis using 9-fluorenylmethoxycarbonyl amino acids chemistry (Fmoc) and purified by reverse-phase HPLC (Bonnard et al., 2002). A major peak was collected and lyophilized, and the integrity of each purified peptide was checked by mass spectrometry.
PVX Expression Assay
Agrobacteria containing the various PVX constructs were allowed to grow for 2 d at 28°C on Luria-Bertani agar plates supplemented with tetracycline and kanamycin. The colonies were toothpick-inoculated by a perforation at each side of the central vein of one lower leaf of 4-week-old N. benthamiana plants. Culture of the inoculated plants continued under the conditions described above, and symptoms were scored daily on a scale ranging from mosaic chlorosis to death of the plant.
Aequorin Luminescence Measurements
At the end of the exponential phase (12 d), tobacco cells expressing cytosolic apoaequorin were washed with an incubation buffer (175 mM mannitol, 0.5 mM CaCl2, 0.5 mM K2SO4, and 2 mM MES adjusted to pH 5.8). The cells were then diluted with this buffer to obtain a packed cell volume of 20% (v/v). Washed cells were supplemented with 2.5 μM native coelenterazine (Molecular Probes) to perform in vivo reconstitution of aequorin and incubated overnight in the dark (150 rpm, 24°C). Purified protoplasts were incubated overnight at 4°C with 2.5 μM native coelenterazine. Bioluminescence measurements were made using a digital luminometer (Lumat LB9507; Berthold) as described by Pauly et al. (2001). Briefly, 100 μL of culture cells/protoplasts were transferred carefully to a luminometer glass tube, and the luminescence counts were recorded continuously at 1-s intervals. Results are expressed as relative light units per second.
Protein Extraction and Analysis
Fourteen days after inoculation, samples were taken from systemically infected N. benthamiana leaves (60 mg), immediately frozen in liquid nitrogen, and ground in a mortar with a pestle. Extraction was achieved in 50 mM Tris-HCl buffer, pH 6.8. After centrifugation of the suspension (5 min, 13,000 rpm, room temperature), the supernatant was recovered and the pellet was washed two times with fresh extraction buffer. Protein gel blot analyses were performed as described by Séjalon-Delmas et al. (1997). Molecular mass standards were purchased from Pharmacia, and silver nitrate staining was performed as described by Oakley et al. (1980).
Electron Microscopy and Immunogold Labeling
Small pieces were cut from systemically infected N. benthamiana leaves 11 d after inoculation and embedded in LR White resin as described previously (Gaulin et al., 2002). In the case of pGR106:CBEL-inoculated plants, only leaves with beginning point necrosis were selected to study undamaged tissue. Ultrathin sections (90 nm thickness) were cut with a diamond knife on an UltraCut E ultramicrotome (Reichert-Leica) and collected on gold grids. Immunogold labeling experiments were performed as described by Boudart et al. (2003) using polyclonal antibodies directed against the protein part of CBEL (dilution, 1:50) and goat anti-rabbit antibodies coupled to colloidal gold (Sigma-Aldrich) at a dilution of 1:400. After contrasting with 5% uranyl acetate, the sections were observed on an H600 electron microscope (Hitachi) at 75 kV.
Accession Number
Sequence data for CBEL can be found in the GenBank/EMBL data libraries under accession number X97205.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. HPLC Elution Profile of a C4 Column of CBD1synth before and after Incubation with Crystalline Cellulose, and Corresponding Histograms Showing the Proportions of the Two Peaks.
Supplemental Figure 2. Dose–Response Experiments Using CBEL and Its Recombinant and Synthetic Derivatives.
Supplemental Figure 3. CBM_1 Modules Repartition among Sequenced Plants, Filamentous Fungi, and Oomycetes.
Supplementary Material
Acknowledgments
We thank D.C. Baulcombe (Sainsbury Laboratory) for kindly providing pGR106, Y. Marco (Institut National de la Recherche Agronomique) for pNT517, J. Chappell (University of Lexington) for pBS-TEAS, F. Meins, Jr. (Friedrich Miescher Institute) for pCHN50, and A. Shapiro and B. Staskawicz (University of California) for the PR1-GUS line. We thank G. Borderies and M. Rossignol (Unité Mixte de Recherche 5546) for proteins and synthetic peptide analyses. We are grateful to A. Jauneau (Institut Fédératif de Recherche 40) and R. O'Connell (Max Planck Institute für Züchtungsforschung) for helpful discussions and to P. Rech (Université Paris VI) for constructive suggestions. E.G. and F.V.-M. received a grant from the French Ministry of Education and Research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Elodie Gaulin (gaulin@scsv.ups-tlse.fr).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.038687.
References
- Ausubel, F.M. (2005). Are innate immune signalling pathways in plants and animals conserved? Nat. Immunol. 6 973–979. [DOI] [PubMed] [Google Scholar]
- Bailleuil, F., Fritig, B., and Kauffman, S. (1996). Occurrence among Phytophthora species of a glycoprotein eliciting a hypersensitive response in tobacco and its relationships with elicitins. Mol. Plant Microbe Interact. 9 214–216. [Google Scholar]
- Bauer, Z., Gomez-Gomez, L., Boller, T., and Felix, G. (2001). Sensitivity of different ecotypes and mutants of Arabidopsis thaliana toward the bacterial elicitor flagellin correlates with the presence of receptor-binding sites. J. Biol. Chem. 49 45669–45676. [DOI] [PubMed] [Google Scholar]
- Berdichevsky, Y., Lamed, R., Frenkel, D., Gophna, U., Bayer, E.A., Yaron, S., Shoham, Y., and Benhar, I. (1999). Matrix-assisted refolding of single-chain Fv- cellulose binding domain fusion proteins. Protein Expr. Purif. 17 249–259. [DOI] [PubMed] [Google Scholar]
- Blume, B., Nürnberger, T., Nass, N., and Scheel, D. (2000). Receptor-mediated increase in cytoplasmic free calcium required for activation of pathogen defense in parsley. Plant Cell 8 1425–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonas, U., and Lahaye, T. (2002). Plant disease resistance triggered by pathogen-derived molecules: Refined models of specific recognition. Curr. Opin. Microbiol. 5 44–50. [DOI] [PubMed] [Google Scholar]
- Bonnard, E., Mazarguil, H., and Zajac, J.M. (2002). Peptide nucleic acids targeted to the mouse proNPFFA reveal an endogenous opioid tonus. Peptides 23 1107–1113. [DOI] [PubMed] [Google Scholar]
- Boudart, G., Charpentier, M., Lafitte, C., Martinez, Y., Jauneau, A., Gaulin, E., Esquerré-Tugayé, M.T., and Dumas, B. (2003). Elicitor activity of a fungal endopolygalacturonase in Nicotiana tabacum requires a functional catalytic site and cell wall localization. Plant Physiol. 131 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque, S., Binet, M.N., Ponchet, M., Pugin, A., and Lebrun-Garcia, A. (1999). Characterization of the cryptogein binding sites on plant plasma membranes. J. Biol. Chem. 274 34699–34705. [DOI] [PubMed] [Google Scholar]
- Bradley Day, R., Okada, M., Ito, Y., Tsukada, K., Zaghouani, H., Shibuya, M., and Stacey, G. (2001). Binding site for chitin oligosaccharides in the soybean plasma membrane. Plant Physiol. 126 1162–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, M.R., Johnson, P.E., Gilkes, N.R., McIntosh, L.P., Kilburn, D.G., and Warren, R.A. (1996). Probing the role of tryptophan residues in a cellulose-binding domain by chemical modification. Protein Sci. 5 2311–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, F., Rosahl, S., Lee, J., Rudd, J.J., Geiler, C., Kauppinen, S., Rasmussen, G., Scheel, D., and Nürnberger, T. (2002). Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO J. 21 6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Delgado, A., Penfield, S., Smith, C., Catley, M., and Bevan, M. (2003). Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34 351–362. [DOI] [PubMed] [Google Scholar]
- Chinchilla, D., Bauer, Z., Regenass, M., Boller, T., and Felix, G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din, N., Gilkes, N.R., Tekant, B., Miller, R.C., Jr., Warren, A.J., and Kilburn, D.G. (1991). Non hydrolytic disruption of cellulose fibres by the binding domain of bacterial cellulase. Biotechnology 9 1096–1099. [Google Scholar]
- Ellis, C., Karafyllidis, I., Wasternack, C., and Turner, J.G. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, C., and Turner, J.G. (2001). The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini, P.J., and Chappell, J. (1992). Gene family for an elicitor-induced sesquiterpene cyclase in tobacco. Proc. Natl. Acad. Sci. USA 89 11088–11092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, G., and Boller, T. (2003). Molecular sensing of bacteria in plants. The highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J. Biol. Chem. 278 6201–6208. [DOI] [PubMed] [Google Scholar]
- Felix, G., Duran, J.D., Volko, S., and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18 265–276. [DOI] [PubMed] [Google Scholar]
- Gaulin, E., Jauneau, A., Villalba, F., Rickauer, M., Esquerré-Tugayé, M.T., and Bottin, A. (2002). The CBEL glycoprotein of Phytophthora parasitica var nicotianae is involved in cell wall deposition and adhesion to cellulosic substrates. J. Cell Sci. 115 4565–4575. [DOI] [PubMed] [Google Scholar]
- Gilkes, N.R., Henrissat, B., Kilburn, D.G., Miller, R.C., Jr., and Warren, R.A. (1991). Domains in microbial beta-1,4-glycanases: Sequence conservation, function, and enzyme families. Microbiol. Rev. 55 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godiard, L., Ragueh, F., Froissard, D., Leguay, J., Grosset, J., Chartier, Y., Meyer, Y., and Marco, Y. (1990). Analysis of synthesis of several pathogenesis related proteins in tobacco leaves infiltrated with water and with compatible and incompatible isolates of Pseudomonas solanacearum. Mol. Plant Microbe Interact. 3 207–213. [Google Scholar]
- Gomez-Gomez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5 1003–1011. [DOI] [PubMed] [Google Scholar]
- Janeway, C.A., Jr., and Medzhitov, R. (2002). Innate immune recognition. Annu. Rev. Immunol. 20 197–216. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A., and Takemoto, D. (2004). Plant innate immunity—Direct and indirect recognition of general and specific pathogen-associated molecules. Curr. Opin. Immunol. 16 48–62. [DOI] [PubMed] [Google Scholar]
- Jones, L., Hamilton, A.J., Voinnet, O., Thomas, C.L., Maule, A.J., and Baulcombe, D.C. (1999). RNA–DNA interactions and DNA methylation in posttranscriptional gene silencing. Plant Cell 11 2291–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S., Lindqvist, H., and Govers, F. (1997). A novel class of elicitin-like genes from Phytophthora infestans. Mol. Plant Microbe Interact. 10 1028–1030. [DOI] [PubMed] [Google Scholar]
- Khatib, M., Lafitte, C., Esquerré-Tugayé, M.-T., Bottin, A., and Rickauer, M. (2004). The CBEL elicitor of Phytophthora parasitica var. nicotianae activates defence in Arabidopsis thaliana via three different signalling pathways. New Phytol. 162 501–510. [Google Scholar]
- Kunze, G., Zipfel, C., Robatzek, S., Niehaus, K., Boller, T., and Felix G. (2004). The N terminus of bacterial elongation factor TU elicits innate immunity in Arabidopsis plants. Plant Cell 16 3496–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourieux, D., Mazars, C., Pauly, N., Ranjeva, R., and Pugin, A. (2002). Analysis and effects of cytosolic free calcium increases in response to elicitors in Nicotiana plumbaginifolia cells. Plant Cell 10 2627–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, I., Shani, Z., and Shoseyov, O. (2002). Modification of polysaccharides and plant cell wall by endo-1,4-beta-glucanase and cellulose-binding domains. Biomol. Eng. 19 17–30. [DOI] [PubMed] [Google Scholar]
- Linder, M., Lindeberg, G., Reinikainen, T., Teeri, T.T., and Pettersson, G. (1995. a). The difference in affinity between two fungal cellulose-binding domains is dominated by a single amino acid substitution. FEBS Lett. 372 96–99. [DOI] [PubMed] [Google Scholar]
- Linder, M., Mattinen, M.L., Kontteli, M., Lindeberg, G., Stahlberg, J., Drakenberg, T., Reinikainen, T., Pettersson, G., and Annila, A. (1995. b). Identification of functionally important amino acids in the cellulose-binding domain of Trichoderma reesei cellobiohydrolase I. Protein Sci. 4 1056–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder, M., Salovuori, I., Ruohonen, L., and Teeri, T.T. (1996). Characterization of a double cellulose-binding domain. Synergistic high affinity binding to crystalline cellulose. J. Biol. Chem. 271 21268–21272. [DOI] [PubMed] [Google Scholar]
- Lu, R., Malcuit, I., Moffett, P., Ruiz, M., Peart, J.R., Wu, A.J., Rathjen, J.P., Bendahmane, A., Day, L., and Baulcombe, D.C. (2003). High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22 5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl, T., Boller, T., and Felix, G. (2000). The bacterial elicitor flagellin activates its receptor in tomato cells according to the address-message concept. Plant Cell 12 1783–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesano, M., Brader, G., and Palva, E.T. (2003). Pathogen derived elicitors: Searching for receptors in plants. Mol. Plant Pathol. 4 73–79. [DOI] [PubMed] [Google Scholar]
- Nürnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 198 249–266. [DOI] [PubMed] [Google Scholar]
- Nürnberger, T., Nennstiel, D., Hahlbrock, K., and Scheel, D. (1995). Covalent cross-linking of the Phytophthora megasperma oligopeptide elicitor to its receptor in parsley membranes. Proc. Natl. Acad. Sci. USA 6 2338–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, B.R., Kirsch, D.R., and Morris, N.R. (1980). A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal. Biochem. 105 361–363. [DOI] [PubMed] [Google Scholar]
- Parker, J.E. (2003). Plant recognition of microbial patterns. Trends Plant Sci. 8 245–247. [DOI] [PubMed] [Google Scholar]
- Pauly, N., Knight, M.R., Thuleau, P., Graziana, A., Muto, S., Ranjeva, R., and Mazars, C. (2001). The nucleus together with the cytosol generates patterns of specific cellular calcium signatures in tobacco suspension culture cells. Cell Calcium 30 413–421. [DOI] [PubMed] [Google Scholar]
- Ricci, P., Bonnet, P., Huet, J.C., Sallantin, M., Beauvais-Cante, F., Bruneteau, M., Billard, V., Michel, G., and Pernollet, J.C. (1989). Structure and activity of proteins from pathogenic fungi Phytophthora eliciting necrosis and acquired resistance in tobacco. Gene 183 555–563. [DOI] [PubMed] [Google Scholar]
- Rickauer, M., Brodschelm, W., Bottin, A., Véronési, C., Grimal, H., and Esquerré-Tugayé, M.T. (1997). The jasmonate pathway is involved differentially in regulation of different defence responses in tobacco cells. Planta 202 155–162. [Google Scholar]
- Rotblat, B., Enshell-Seijffers, D., Gershoni, J.M., Schuster, S., and Avni, A. (2002). Identification of an essential component of the elicitation active site of the EIX protein elicitor. Plant J. 32 1049–1055. [DOI] [PubMed] [Google Scholar]
- Séjalon-Delmas, N., Villalba Mateos, F., Bottin, A., Rickauer, M., Dargent, R., and Esquerré-Tugayé, M.T. (1997). Purification, elicitor activity, and cell wall localization of a glycoprotein from Phytophthora parasitica var. nicotianae, a fungal pathogen of tobacco. Phytopathology 87 899–909. [DOI] [PubMed] [Google Scholar]
- Sharp, J.K., Valent, B., and Albersheim, P. (1984). Purification and partial characterization of a beta-glucan fragment that elicits phytoalexin accumulation in soybean. J. Biol. Chem. 259 11312–11320. [PubMed] [Google Scholar]
- Shinshi, H., Mohnen, D., and Meins, F., Jr. (1987). Regulation of a plant pathogenesis-related enzyme: Inhibition of chitinase and chitinase mRNA accumulation in cultured tobacco tissues by auxin and cytokinin. Proc. Natl. Acad. Sci. USA 66 773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpigel, E., Roiz, L., Goren, R., and Shoseyov, O. (1998). Bacterial cellulose-binding domain modulates in vitro elongation of different plant cells. Plant Physiol. 117 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba-Mateos, F., Rickauer, M., and Esquerré-Tugayé, M.T. (1997). Cloning and characterization of a cDNA encoding an elicitor of Phytophthora parasitica var. nicotianae that shows cellulose-binding and lectin-like activities. Mol. Plant Microbe Interact. 10 1045–1053. [DOI] [PubMed] [Google Scholar]
- Vorwerk, S., Somerville, S., and Somerville, C. (2004). The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci. 9 203–209. [DOI] [PubMed] [Google Scholar]
- Zipfel, C., and Felix, G. (2005). Plants and animals: A different taste for microbes? Curr. Opin. Plant Biol. 8 353–360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.