Abstract
Cell and cell wall growth are mutually dependent processes that must be tightly coordinated and controlled. LRR-extensin1 (LRX1) of Arabidopsis thaliana is a potential regulator of cell wall development, consisting of an N-terminal leucine-rich repeat domain and a C-terminal extensin-like domain typical for structural cell wall proteins. LRX1 is expressed in root hairs, and lrx1 mutant plants develop distorted root hairs that often swell, branch, or collapse. The aberrant cell wall structures found in lrx1 mutants point toward a function of LRX1 during the establishment of the extracellular matrix. To identify genes that are involved in an LRX1-dependent developmental pathway, a suppressor screen was performed on the lrx1 mutant, and two independent rol1 (for repressor of lrx1) alleles were isolated. ROL1 is allelic to Rhamnose Biosynthesis1, which codes for a protein involved in the biosynthesis of rhamnose, a major monosaccharide component of pectin. The rol1 mutations modify the pectic polysaccharide rhamnogalacturonan I and, for one allele, rhamnogalacturonan II. Furthermore, the rol1 mutations cause a change in the expression of a number of cell wall–related genes. Thus, the lrx1 mutant phenotype is likely to be suppressed by changes in pectic polysaccharides or other cell wall components.
INTRODUCTION
The plant cell wall is a rigid but pliable structure that confers protection, cell cohesion, and mechanical strength but is also important for the communication between individual cells. The main components of the primary cell wall of dicotyledonous plants are a cellulose-xyloglucan network considered to be the main load-bearing structure that is embedded in a matrix of pectic polysaccharides (Carpita and Gibeaut, 1993). The pectic matrix has three major components: homogalacturonan (HGA), rhamnogalacturonan I (RG I), and rhamnogalacturonan II (RG II) (Ridley et al., 2001). HGA is a homopolymer consisting of (1→4)-α-linked galacturonic acid, which is often partially methylesterified. Upon demethylesterification, HGA can be cross-linked by Ca2+, which promotes gel formation and cell wall rigidification. RG I is a rod-like heteropolymer with a backbone of repeating (1→2)-α-l-rhamnose-(1→4)-α-galacturonic acid disaccharide units containing numerous side chains attached to the rhamnose (Rha) residues, including galactans and arabinans. RG II is a complex but conserved heteropolysaccharide consisting of an HGA backbone decorated with four characteristic side chains composed of different monosaccharides. RG II can dimerize via the formation of borate diester links through apiose residues, contributing to the tensile strength of the wall (O'Neill et al., 2001). In addition to their effect on wall strength and cell adhesion, pectins also control wall porosity (Baron-Epel et al., 1988), which in turn regulates the mobility of cell wall modifying proteins and, thus, cell wall expansion. The porosity of the cell wall is thought to be influenced by RG I, a hypothesis that is corroborated by the association of RG I with cell wall growth (Ridley et al., 2001; Willats et al., 2001a; McCartney et al., 2003).
A sophisticated sugar biosynthetic machinery is required to synthesize the monosaccharides that form the cell wall carbohydrates (Reiter and Vanzin, 2001; Seifert, 2004). Pectins are synthesized in the Golgi apparatus by glycosyltransferases using nucleotide sugars as donor substrates (Scheible and Pauly, 2004). Many glycosyltransferases have been identified, but only a few are believed to be involved in pectin biosynthesis (Bouton et al., 2002; Iwai et al., 2002; Lao et al., 2003). Rha, a major component of RG I and RG II, has been hypothesized to be synthesized in Arabidopsis thaliana by a family of three highly similar Rhamnose Biosynthesis (RHM) proteins that convert UDP-d-Glc to UDP-l-Rha (Reiter and Vanzin, 2001). RHM2 was shown to be required for the biosynthesis of the pectinaceous seed coat mucilage mainly composed of RG I (Usadel et al., 2004; Western et al., 2004). Recently, B. Link and W.-D. Reiter (unpublished data) have demonstrated the biochemical activity of RHM1, with the in vitro conversion of UDP-d-Glc to UDP-l-Rha by recombinant RHM1.
During cell wall expansion, many polysaccharides need to be rearranged. This is conducted by a number of specific hydrolytic enzymes, such as the xyloglucan endotransglucosylases/hydrolases that act on xyloglucans and the polygalacturonases that act on pectins, and by other proteins, such as the nonhydrolyzing expansins (Cosgrove, 1999). Relatively little is known about the mechanisms that control cell wall expansion and assembly. Recently, proteins were identified that potentially function in this process. Cell wall–associated kinases (WAKs) were shown to be essential for cell wall expansion, since a reduction of WAK levels results in inhibition of cell elongation (Lally et al., 2001; Wagner and Kohorn, 2001). WAKs were found to bind the cell wall through pectin, establishing a physical link between the intracellular and the extracellular compartment that might serve as a signaling conduit (Wagner and Kohorn, 2001). Glycosylphosphatidylinositol-anchored proteins, such as COBRA, localize to the cell surface where they function in cell wall matrix remodeling. Mutations in COBRA strongly affect cellulose microfibril orientation (Roudier et al., 2005), and a general reduction in glycosylphosphatidylinositol-anchored proteins in the pnt1 mutant causes changes in the cellulose and pectin content and aberrant deposition of pectin, xyloglucans, and callose (Gilmor et al., 2005). Finally, arabinogalactan proteins (AGPs) are predicted to have adhesive and signaling properties since they can bind to pectins (Majewska-Sawka and Nothnagel, 2000; Willats et al., 2001a) and might also interact with WAKs. Cell wall properties, including extensibility, are also influenced by structural cell wall proteins that are oxidatively cross-linked in the extracellular matrix. Upon pathogen attack, wounding, mechanical stress, or after termination of cell growth, these proteins can be insolubilized to reinforce the cell wall and lock it in its final shape (Carpita and Gibeaut, 1993; Showalter, 1993; Ringli et al., 2001).
In Arabidopsis, we characterized LRR-extensin1 (LRX1), a gene involved in the regulation of cell wall formation. LRX1 is a member of a family of 11 genes coding for extracellular proteins consisting of an N-terminal leucine-rich repeat (LRR) domain and a structural extensin moiety at the C terminus (Baumberger et al., 2003a; Ringli, 2005). LRR domains are involved in protein–protein interactions and are found in many proteins playing a role in disease resistance, signaling pathways, or the regulation of extracellular enzymes (Forsthoefel et al., 2005, and references therein). The C-terminal moiety of LRX proteins is composed of [Ser-Hyp4]n repeats characteristic of Hyp-rich glycoproteins, which can modify the properties of the cell wall and might also play a role in connecting the cell wall with the plasma membrane by anchoring target proteins (Knox, 1995; Cassab, 1998). LRX1 is specifically expressed in root hairs and localizes to the cell wall where it is insolubilized (Baumberger et al., 2001). Root hairs are thin, long protrusions from specialized root epidermal cells (trichoblasts) that elongate by tip growth, a polarized form of cell growth (Foreman and Dolan, 2001). lrx1 mutant root hairs are short, form branches, swell, and frequently collapse (Baumberger et al., 2001). LRX1 synergistically interacts with its paralogous gene LRX2, and the lrx1 phenotype is enhanced in lrx1 lrx2 double mutants. Ultrastructural analysis revealed severe deficiencies in the cell wall architecture of lrx1 lrx2 double mutant plants, indicating a role of LRX1 and LRX2 during cell wall formation (Baumberger et al., 2003b).
To identify genes that are likely to be involved in the same developmental process as LRX1, we performed a suppressor screen on the lrx1 mutant. Here, we report the characterization of two allelic rol1 (for repressor of lrx1) mutants that compensate for the absence of LRX1. The ROL1 locus encodes RHM1, which is involved in the formation of UDP-l-Rha, and the rol1 mutations cause a modification of RG I and, in one rol1 allele, of RG II. Furthermore, the expression of several cell wall–related genes is altered in the rol1 mutants. Because l-Rha is an abundant component of pectic polysaccharides but absent from other cell wall components, our data suggest that LRX1 might be an extracellular regulator involved in the formation of the pectin matrix.
RESULTS
Isolation of Suppressors of the lrx1 Mutation
The lrx1-1s allele (subsequently referred to as lrx1) used for ethyl methanesulfonate (EMS) mutagenesis was produced by the excision of the En-1 transposon from the original lrx1-1 allele (Baumberger et al., 2001). The excision resulted in a deletion of 6 bp or, on the protein level, the deletion of two amino acids and a change in a third residue (Diet et al., 2004). This has a strong impact on LRX1 function and results in a mutant phenotype that is very similar to the initially isolated lrx1-1 allele containing the En-1 insertion (Baumberger et al., 2001). Twenty-three individual plants were isolated from the EMS-mutagenized M2 population that displayed a wild type–like phenotype. These mutants were called rol (for repressor of lrx1). Mapping and allelism tests revealed the presence of two allelic, recessive mutants, rol1-1 and rol1-2, that were selected for detailed analysis. The subsequently cloned ROL1 gene (see below) was sequenced in the remaining 21 rol mutants, but none of them showed a nucleotide change. Accordingly, we assume that no additional rol1 alleles are present in the rol mutant collection.
rol1 Mutations Suppress the lrx1 Root Hair Formation Phenotype
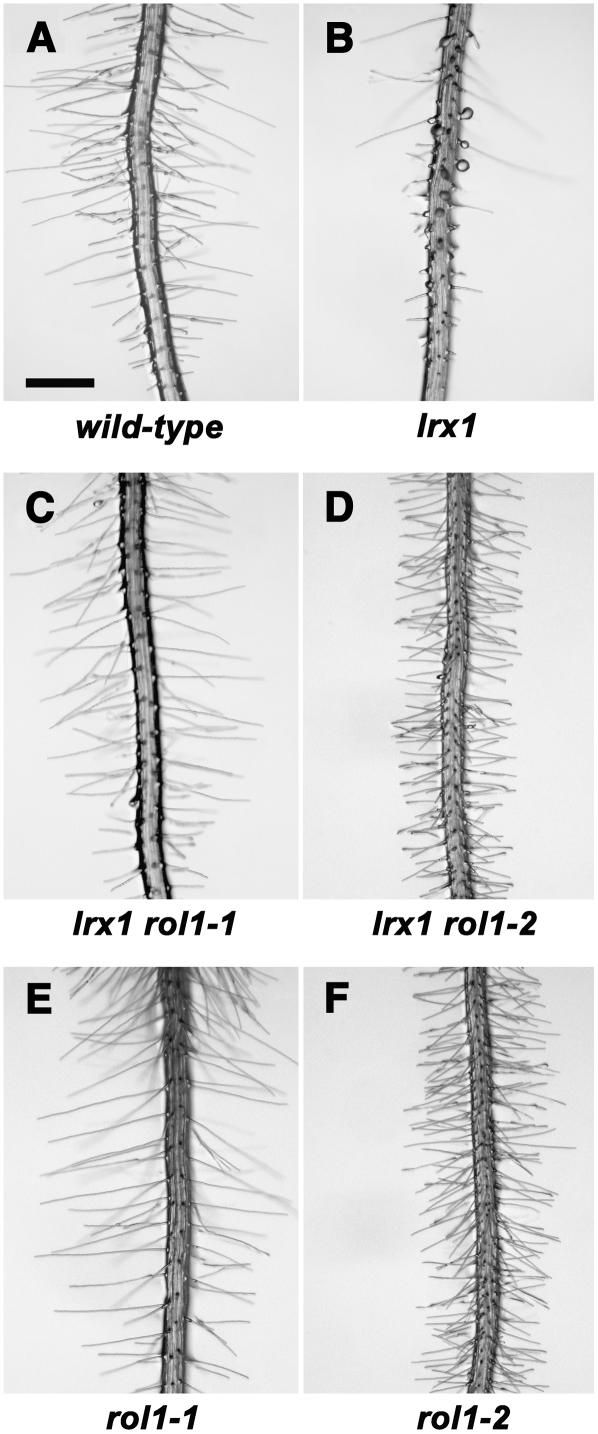
While wild-type seedlings display regular, thin, and long root hairs, lrx1 mutants are affected in root hair formation, with many short, collapsed, distorted, and sometimes branched root hairs (Baumberger et al., 2001; Figures 1A and 1B). By contrast, root hairs of lrx1 rol1-1 double mutants had a restored wild-type phenotype. Root hair length was comparable to the wild type, and branching or collapsing of root hairs was suppressed (Figure 1C). lrx1 rol1-2 seedlings were characterized by somewhat shorter root hairs compared with wild-type or lrx1 rol1-1 plants (Figure 1D). The length of root hairs was compared between the lrx1 rol1 mutants and wild-type plants. In lrx1 rol1-1 seedlings, root hairs were not significantly different from the wild type, but in lrx1 rol1-2, they were 32% shorter (Table 1). In addition, primary roots of lrx1 rol1-2 plants were shorter (Table 2), with an apparently higher root hair density compared with the wild type. To determine whether the latter phenotype is due to the formation of ectopic root hairs or to reduced cell expansion of trichoblasts of the root epidermis, we measured the length of wild-type and lrx1 rol1-2 trichoblasts. There was a 41% reduction in trichoblast length in lrx1 rol1-2 seedlings (0.115 ± 0.015 mm in lrx1 rol1-2 versus 0.195 ± 0.042 mm in the wild type), while the pattern of root hair formation appeared to be normal in the mutant. Thus, the higher density of root hairs in lrx1 rol1-2 plants can be explained by the reduced length of the trichoblasts.
Figure 1.
Phenotypes of the Different Mutant Lines.
Seedlings were grown in a vertical orientation for 4 d on half-strength Murashige and Skoog (MS) medium. Root hairs are shown of the wild type (A), the lrx1 mutant (B), which develops shorter and misshaped root hairs, the lrx1 rol1-1 double mutant (C), the lrx1 rol1-2 double mutant (D), with a suppressed lrx1 root hair phenotype, the rol1-1 single mutant (E), and the rol1-2 single mutant (F), with denser and slightly shorter root hairs compared with the wild type. Bar = 0.5 mm.
Table 1.
Length of Wild-Type and lrx1 rol1 Mutant Root Hairs
| Genotype | Root Hair Length (mm) |
|---|---|
| Wild type | 0.66 ± 0.10 |
| lrx1 rol1-1 | 0.59 ± 0.10 |
| lrx1 rol1-2 | 0.45 ± 0.06 |
Table 2.
Root Length Phenotype of the lrx1 rol1-2 Double Mutant
| Genotype | Root Length (mm) |
|---|---|
| Wild type | 19.20 ± 2.07 |
| lrx1 | 19.60 ± 2.54 |
| lrx1 rol1-2 | 15.10 ± 1.12 |
| lrx1 rol1-2, gen. RHM1a | 19.95 ± 2.01 |
lrx1 rol1-2 double mutant complemented with a wild-type genomic clone of RHM1.
To investigate the effect of the rol1 mutations on root hair development in the absence of the lrx1 mutation, the two lrx1 rol1 lines were backcrossed to wild-type Columbia (LRX1/LRX1) plants. For lrx1 rol1-1, only lrx1 and wild-type phenotypes were found in the F2 population derived from the backcross, whereas for lrx1 rol1-2, one-quarter of the F2 plants displayed the mutant phenotype of the lrx1 rol1-2 double mutant. This was confirmed in the F3 population of individual rol1-1 and rol1-2 single mutant lines. Hence, the rol1-1 mutation does not have an obvious effect on root hair development in the LRX1/LRX1 wild-type background, whereas the rol1-2 phenotype develops also in the absence of the lrx1 mutation and is thus epistatic to lrx1 (Figures 1E and 1F). In addition to the stunted root phenotype of rol1-2, both rol1 mutants revealed a mutant phenotype in cotyledons. Instead of a smooth cotyledon surface observed in wild-type seedlings, both rol1 mutants develop a rough surface. This phenotype is more pronounced in rol1-2 seedlings and is limited to cotyledons (see Supplemental Figure 1 online).
The rol1 Mutations Also Suppress the lrx1 lrx2 Double Mutant Root Hair Phenotype
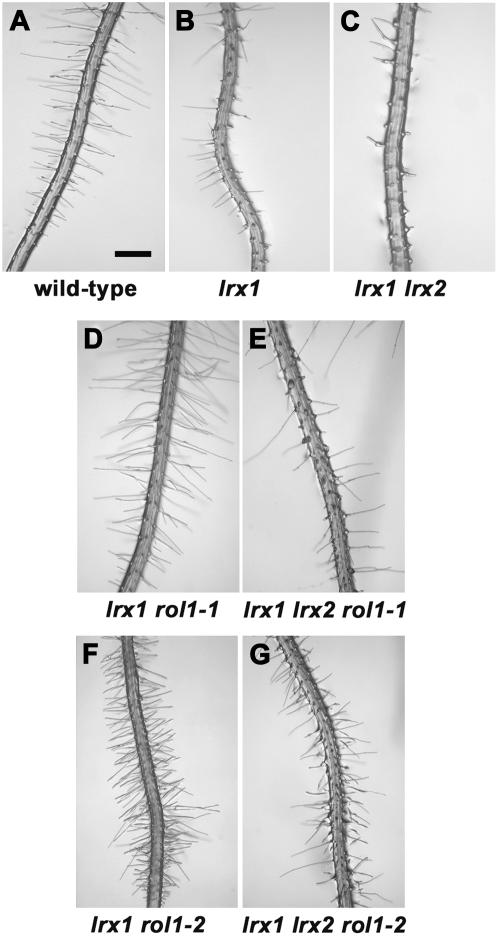
LRX2 is the paralog of and synergistically interacts with LRX1 during root hair cell wall formation. Its overexpression can suppress the lrx1 mutant phenotype (Baumberger et al., 2003b). This prompted us to investigate the role of LRX2 in the suppression of the lrx1 mutant phenotype by constructing lrx1 lrx2 rol1 triple mutants. The lrx1 rol1 mutants were crossed with an lrx1 lrx2 double mutant to create triple mutants. While lrx2 single mutants are indistinguishable from the wild type, lrx1 lrx2 double mutants develop an enhanced lrx1 phenotype with very few root hairs (Baumberger et al., 2003b; Figures 2A to 2C). The rol1 mutations suppressed the lrx1 lrx2 double mutant phenotype to different extents. lrx1 lrx2 rol1-1 triple mutants displayed a phenotype that was intermediate between the lrx1 mutant and the lrx1 rol1-1 double mutant (Figures 2B, 2D, and 2E). The lrx1 lrx2 rol1-2 triple mutants developed root hairs that are characteristic of the rol1-2 mutant, even though not all root hairs were formed properly (Figures 2F and 2G). Although the addition of the lrx2 mutation decreased the effectiveness of rol1 suppression, the effect of the rol1 mutations is not dependent on LRX2.
Figure 2.
Suppression of the lrx1 lrx2 Double Mutant Phenotype by rol1-1 and rol1-2.
rol1-1 and rol1-2 can partially suppress the lrx1 lrx2 double mutant phenotype. Mutants were vertically grown for 4 d. Root hairs are shown of the wild type (A), lrx1 (B), which develops shorter and misshaped root hairs, lrx1 lrx2 (C), with an enhanced root hair phenotype, lrx1 rol1-1 (D), lrx1 lrx2 rol1-1 (E), and lrx1 rol1-2 (F) and lrx1 lrx2 rol1-2 (G), with suppressed lrx1 and lrx1 lrx2 root hair phenotypes, respectively. Bar = 0.5 mm.
Map-Based Cloning of the rol1 Gene
The rol1-1 and rol1-2 mutations were mapped to chromosome 1 south of nga111. The region containing the rol1 gene was delimited by the two markers uzu63 (BAC F3F9; position 38,200) and uzu58 (BAC F9K20; position 60,500) for rol1-1 and uzu67 and uzu68 (BAC T30F21; positions 29,700 and 58,300, respectively) for rol1-2 (Figure 3A). The genes encoded in this region were sequenced, and for both rol1-1 and rol1-2, a point mutation was identified in the RHM1 gene (At1g78570) (Figure 3B). To demonstrate that the mutations found in the rol1 alleles are responsible for the suppression of the lrx1 phenotype, both lrx1 rol1-1 and lrx1 rol1-2 plants were transformed with a 4.2-kb genomic fragment of the RHM1 gene containing the coding region, 1.5 kb of upstream promoter sequence, and 600 bp of 3′ noncoding sequence. Of each lrx1 rol1 mutant, 10 independent primary transformants (T1) were propagated to the T2 generation. The T2 populations segregated in a 3:1 ratio for the lrx1 to lrx1 rol1 phenotype. A DNA gel blot experiment demonstrated that the lrx1 phenotype cosegregated with the presence of the T-DNA (data not shown). The T2 seedlings developing the lrx1 root hair phenotype also displayed a reversion of the cotyledon and, in transgenic lines of the lrx1 rol1-2 genetic background, restoration of root length to that of lrx1 and wild-type seedlings (Table 2). This demonstrates that the presence of a wild-type copy of RHM1 in either lrx1 rol1-1 or lrx1 rol1-2 mutants restores the original lrx1 phenotype (abnormal root hair development, wild-type cotyledons, and wild-type root length) and, hence, that the suppression of the lrx1 phenotype in the lrx1 rol1 mutants is caused by the mutant RHM1 gene.
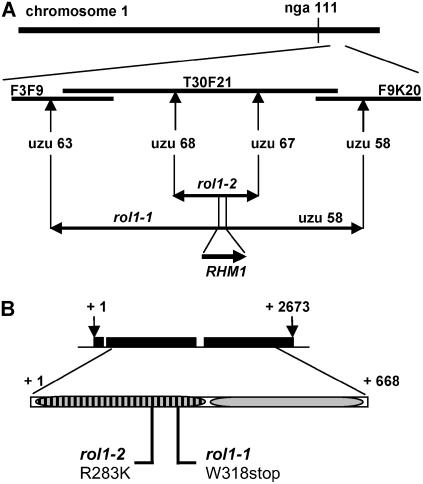
Figure 3.
Map-Based Cloning of the rol1 Locus.
(A) rol1-1 and rol1-2 were independently mapped on chromosome 1, south of nga111, to an interval of 100 and 38 kb, respectively, on BAC T30F21 using self-made uzu (cleaved-amplified polymorphic sequence [CAPS] and simple sequence length polymorphism [SSLP]) markers.
(B) The RHM1 gene (At1g78570) encodes a 2673-nucleotide transcript encoding a protein of 668 amino acids. Sequencing revealed point mutations at positions corresponding to the N-terminal dehydratase domain. rol1-1 is a nonsense mutation at position 318, and rol1-2 is a missense mutation changing an Arg at position 283 to a Lys. Numbers indicate nucleotide (above) or amino acid positions (below). Black boxes indicate the three exons of the RHM1 gene. Striped box, dehydratase domain of RHM1; shaded box, epimerase/reductase domain of RHM1.
RHM1 belongs to a subclass of the short chain dehydrogenase/reductase family of enzymes and, together with the highly similar RHM2 and RHM3, has been hypothesized to catalyze the conversion of UDP-d-Glc to UDP-l-Rha (Reiter and Vanzin, 2001). RHM1 is predicted to consist of an N-terminal dehydratase and a C-terminal epimerase/reductase domain, and its proposed activity has recently been demonstrated by assaying the recombinant enzyme in vitro (B. Link and W.-D. Reiter, unpublished data). rol1-1 contains a G-to-A mutation that results in the introduction of a stop codon near the end of the dehydratase domain at Trp-318. The rol1-2 mutant harbours a G-to-A mutation that leads to the replacement of Arg-283 by Lys (Figure 3B).
To investigate the effect of the rol1 mutations on RHM1 expression, quantitative real-time RT-PCR analysis was performed on wild-type, lrx1, lrx1 rol1-1, and lrx1 rol1-2 seedlings. Root RNA was used as starting material, and expression levels were normalized against ACTIN2 expression as an internal standard. Compared with the wild type and lrx1, which have comparable expression levels, the expression of the RHM1 gene was found to be barely detectable in lrx1 rol1-1 plants (RHM1 message was only detected in one of three experiments) but was present at wild-type levels in lrx1 rol1-2 (Figure 4A).
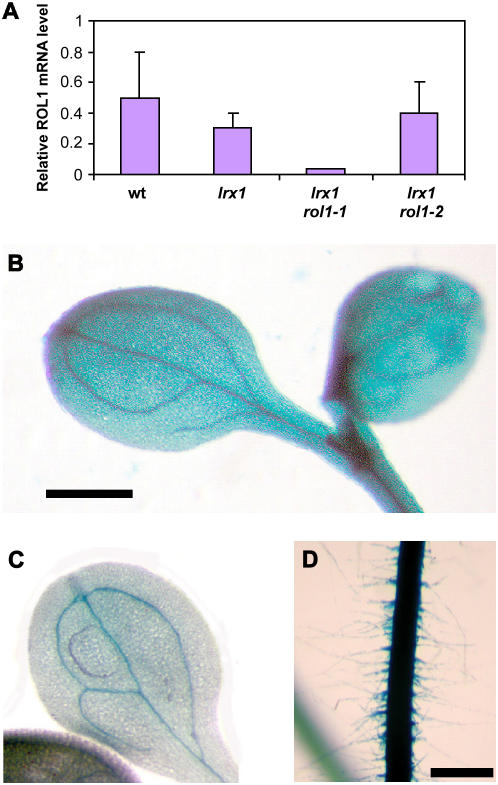
Figure 4.
Expression Analysis of RHM1.
(A) Quantitative RT-PCR was used to analyze the expression level of RHM1 in roots of 1-week-old seedlings grown in a vertical orientation. RHM1 expression was strongly reduced in lrx1 rol1-1 plants but was comparable to wild-type expression in lrx1 and lrx1 rol1-2 seedlings. Three independent analyses were performed; the error bars indicate the se.
(B) to (D) Expression analysis by an RHM1 promoter:GUS fusion construct in transgenic Arabidopsis seedlings. GUS staining was stopped after 3 h. All tissues of the seedling show GUS activity, confirming ubiquitous expression of RHM1. Cotyledons of seedlings of transgenic lines with high (B) and low (C) GUS activity are shown. (D) shows the root of a seedling with high GUS activity. Bars = 1 mm ([B] and [C]) and 0.5 mm (D).
To determine the tissue specificity of RHM1 expression in Arabidopsis seedlings, 1.5 kb of the RHM1 promoter was fused to the β-glucuronidase reporter gene (uidA, subsequently referred to as GUS) and transformed into wild-type Columbia plants. Eight individual T1 transgenic plants were obtained and grown to the T2 generation. Whole seedlings were stained for GUS activity, which was found in all tissues (i.e., root including root hairs, hypocotyl, and cotyledons). In weakly expressing lines, mainly the vascular tissue showed activity, whereas in plants with higher GUS expression levels, whole cotyledons, the hypocotyl, and the root were stained (Figures 4B to 4D). The RHM1 expression pattern deduced from the promoter:GUS fusion experiment is consistent with the mutant phenotypes found in the roots and cotyledons of both rol1 alleles.
rol1 Mutations Have a Deleterious Effect on the Dehydratase Activity of RHM1
The effect of the point mutations in the rol1 mutants on the enzymatic activity of RHM1 was tested by an in vitro enzyme assay. Because expression of full-length wild-type RHM1 in Escherichia coli yielded very little soluble protein, the dehydratase domain of RHM1 (RHM1-D) was expressed separately and shown to catalyze the conversion of UDP-d-Glc to the reaction intermediate UDP-4-keto-6-deoxy-d-Glc, which was quantified by gas chromatography–mass spectrometry (GC-MS) (B. Link and W.-D. Reiter, unpublished data). Subsequently, the dehydratase domain of RHM1 encoded by rol1-1 and rol1-2, referred to as ROL1-1-D and ROL1-2-D, were expressed in E. coli and tested for their in vitro activity. Since rol1-1 contains a stop codon at position 318, ROL1-1-D was 43 amino acids shorter than RHM1-D. Although both ROL1-D proteins were successfully expressed, they failed to produce detectable amounts of UDP-4-keto-6-deoxy-d-Glc, whereas a wild-type control yielded the expected intermediate (data not shown). Thus, both rol1-1 and rol1-2 encode proteins with a dehydratase domain that is inactive in vitro, suggesting that both proteins fail to convert UDP-d-Glc to UDP-l-Rha in vivo.
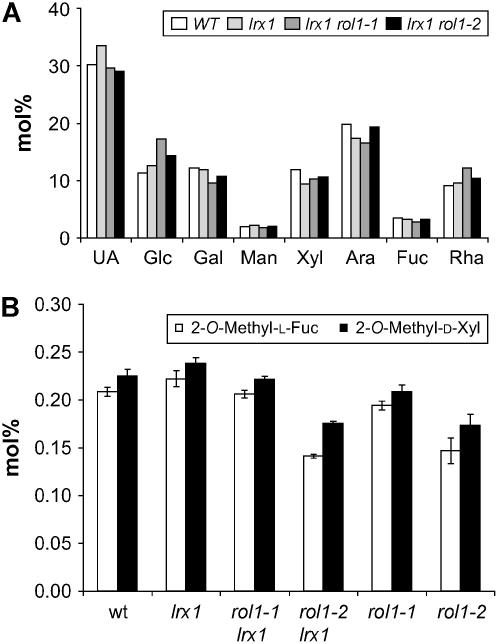
Monosaccharide Composition of the Extracellular Matrix
Because the rol1 mutations affect the biosynthesis of Rha, the abundance of Rha in cell wall material may be reduced. To test this possibility, the composition of the cell wall of root tissue of wild-type and mutant seedlings was determined. Although we did not find any significant differences (P < 0.1) in the amounts of the major sugars of cell wall material, including Rha (Figure 5A), rol1-2 and lrx1 rol1-2 showed an ∼30% reduction in the RG II–specific monosaccharides 2-O-methyl-d-xylose and 2-O-methyl-l-fucose (Figure 5B). These methylated sugars are diagnostic components of the two large Rha-containing side chains of RG II, and their reduced abundance may reflect a defect in the synthesis of RG II. The relative amount of both 2-O-methyl sugars was not significantly different between rol1-1 and wild-type lines.
Figure 5.
Monosaccharide Composition of Cell Wall Material.
Seedlings were grown for 1 week in a vertical orientation, and whole root material was collected for wall preparation.
(A) Amounts of major sugars in the cell wall. The average of two independent growth experiments is shown. UA, uronic acid.
(B) Amounts of 2-O-methyl-d-xylose and 2-O-methyl-l-fucose in the cell wall. The data of four independent experiments are shown; error bars indicate the se.
rol1-1 and rol1-2 Mutants Form Aberrant RG I
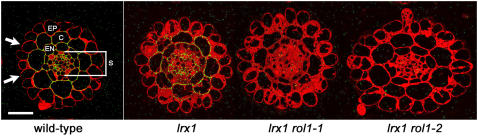
Although no significant alteration in the major sugars of cell wall material was detected in the rol1 mutants, it is possible that the structure of individual components of the cell wall is modified or that only particular tissues are affected. To test this, monoclonal antibodies against specific cell wall components were used to provide information about cell wall composition in planta. The absence of immunolabeling can indicate either the absence or masking of the epitope. Therefore, root transverse sections of wild-type, lrx1, lrx1 rol1, and rol1 single mutant seedlings were immunolabeled with antibodies that specifically detect different carbohydrate epitopes, mainly of pectin. The antibodies CCRC-M1 (Fuc-Gal side chains of xyloglucan; Puhlmann et al., 1994), JIM5 and JIM7 (highly and partially methylesterified epitope of HGA, respectively; Knox et al., 1990), LM2 (AGP side chains with GlcA; Yates et al., 1996), CCRC-M7 (AGPs; Steffan et al., 1995), and LM6 (arabinan side chains of RG I; Willats et al., 1998) showed no difference in labeling intensity (data not shown). A strong reduction in labeling was observed in lrx1 rol1-1 and lrx1 rol1-2 plants compared with the wild type or lrx1 with the LM5 antibody that detects the (1→4)-β-d-galactan side chains of RG I (Jones et al., 1997) (Figure 6). No difference in staining was observed between the wild type and lrx1. A comparison of wild-type and rol1 single mutants confirmed this observation (data not shown). In wild-type root tissue of the root hair–containing region, LM5 bound to walls of the cortex, endodermis, and stele but not to epidermal cells, including root hair forming cells (indicated by arrows in Figure 6). This is in line with previous findings that LM5 does not bind epidermal cells in root hair developing zones of the root (Willats et al., 2001b; McCartney et al., 2003).
Figure 6.
rol1 Mutants Are Affected in RG I.
Wild-type, lrx1, lrx1 rol1-1, and lrx1 rol1-2 plants were embedded and transverse sections of roots stained with the LM5 antibody, detecting galactan side chains of RG I. A strong reduction in staining (green fluorescence) in the lrx1 rol1-1 and lrx1 rol1-2 mutants compared with the wild type or lrx1 is observed. The sections are not of tissue of the identical developmental stage. Sections of lrx1 and lrx1 rol1-2 are of younger root tissue than those of the wild type and lrx1 rol1-1, as can be deduced from the degree of vacuolation of the trichoblasts. As labeling with LM5 is not found in epidermal cells, and labeling in the wild type and lrx1 is comparable; this difference is not relevant. Arrows indicate two root hair forming trichoblasts located over anticlinal walls of two cortex cells. EP, epidermis; C, cortex; EN, endodermis; S, stele. Bar = 100 μm.
rol1-1 and rol1-2 Have Different Effects on the Genome-Wide Gene Expression Profile
A microarray analysis of gene expression in roots of wild-type, lrx1, and the two lrx1 rol1 suppressor mutants was performed to assess the gene expression profiles (see Supplemental Table 1 online). ATH1 Arabidopsis whole-genome chips from Affymetrix containing ∼23,000 genes were used for hybridization with total root RNA. The microarray experiments were performed in biological triplicates, the data were analyzed as described in Methods, and induction/repression by a factor of 2 was chosen as the threshold value. First, the lrx1 mutant was compared with wild-type Columbia. No significant change in gene expression between these two lines could be found. Thus, the dramatic morphological difference between the wild type and lrx1 is not reflected by an extensive change in the gene expression profile.
In the next step, lrx1 and the two lrx1 rol1 mutants were compared. Considering the ubiquitous expression of RHM1 in roots, it was surprising not to find a single significant change in gene expression between lrx1 and lrx1 rol1-1. By contrast, the rol1-2 mutation produced a number of changes, with 38 induced and 19 repressed genes in lrx1 rol1-2 compared with lrx1 (Table 3).
Table 3.
Changes in Gene Expression in Irx1 rol1-2 Compared with Irx1
| Accession Number | Induction Factor | Gene Description |
|---|---|---|
| Genes Induced in Irx1 rol1-2 Compared with Irx1 | ||
| At2g32190 | 7.242 | Unknown protein |
| At2g47550 | 6.247 | Putative pectinesterase |
| At2g32210 | 5.679 | Unknown protein |
| At4g28850 | 4.446 | Xyloglucan endotransglycosylase AtXTH26, AtXTR18 |
| At5g57530 | 3.645 | Xyloglucan endotransglycosylase AtXTH12 |
| At4g13390 | 3.594 | Similar to class 1 extensins |
| At2g02990 | 3.531 | Ribonuclease AtRNS1 |
| At4g08040 | 3.126 | Strong similarity to 1-aminocyclopropane-1-carboxylic acid synthases |
| At1g54970 | 3.003 | Pro-rich protein AtPRP1 |
| At4g02330 | 2.874 | Expressed protein similar to pectinesterase |
| At5g45280 | 2.554 | Pectin acetylesterase |
| At5g44810 | 2.552 | Unknown protein |
| At2g35980 | 2.551 | Harpin-induced protein AtYLS9 |
| At4g22080 | 2.53 | Pectate lyase-like protein |
| At5g49080 | 2.513 | Extensin-like protein |
| At1g26250 | 2.498 | Extensin-like protein |
| At4g01700 | 2.479 | Putative chitinase |
| At4g25790 | 2.476 | Expressed pathogenesis-related protein |
| At3g22800 | 2.445 | LRR-extensin protein AtLRX6 |
| At5g44820 | 2.405 | Unknown protein |
| At4g19680 | 2.372 | Fe(II) transport protein AtlRT2 |
| At5g54490 | 2.37 | PINOID (PID) binding protein AtPBP1 |
| At3g50930 | 2.348 | AAA-type ATPase family protein |
| At5g22410 | 2.317 | Peroxidase ATP14a homolog |
| At2g29440 | 2.272 | Glutathione S-transferase AtGSTU6 |
| At3g01730 | 2.244 | Expressed protein |
| At2g46860 | 2.22 | Putative inorganic pyrophosphatase |
| At3g49960 | 2.218 | Peroxidase AtATP21a |
| At1g76470 | 2.214 | Putative cinnamoyl-CoA reductase |
| At2g39980 | 2.166 | Putative transferase protein |
| At1g59940 | 2.147 | Response regulator AtARR3 |
| At4g24340 | 2.105 | Phosphorylase family protein |
| At1g66160 | 2.1 | U-box containing protein |
| At4g34580 | 2.099 | Similar to SEC14 protein from Saccharomyces cerevisiae |
| At3g45970 | 2.099 | Expansin-like protein AtEXLA1 |
| At5g26340 | 2.087 | Hexose transporter-like protein |
| At5g67400 | 2.055 | Peroxidase PER73 |
| At1g65310 | 2.037 | Xyloglucan endotransglycosylase AtXTH17, AtXTR1 |
| Genes Repressed in Irx1 rol1-2 Compared with Irx1 | ||
| At2g25900 | 0.497 | Putative CCCH-type zinc finger protein AtCTH |
| At3g14660 | 0.489 | Putative cytochrome P450 CYP72A13 |
| At4g31730 | 0.487 | Gln dumper 1 AtGDU1 |
| At4g15390 | 0.486 | HSR201-like transferase protein |
| At5g52790 | 0.475 | CBS-domain containing protein |
| At3g49760 | 0.46 | bZIP transcription factor |
| At4g12550 | 0.451 | Putative cell wall–plasma membrane disconnecting CLCT protein |
| At5g55250 | 0.449 | S-adenosyl-l-methionine:carboxyl methyltransterase-like protein AtIAM I 1 |
| At2g02820 | 0.449 | MYB family transcription factor MYB88 |
| At3g06390 | 0.444 | Unknown integral membrane protein |
| At2g39310 | 0.44 | Putative myrosinase-binding protein |
| At5g63600 | 0.425 | Flavonol synthase-like protein |
| At2g28850 | 0.411 | Putative cytochrome P450 |
| At1g11080 | 0.406 | Ser carboxypeptidase |
| At3g52060 | 0.399 | Unknown protein |
| At2g28780 | 0.392 | Hypothetical protein |
| At1g21100 | 0.381 | O-methyltransferase |
| At3g01190 | 0.381 | Peroxidase AtPER27 |
| At1g33055 | 0.343 | Expressed protein |
Genes potentially involved in cell wall development are indicated in bold.
Interestingly, genes with known or predicted functions in cell wall development (indicated in bold in Table 3) were highly abundant in this collection (23 out of 57, or 40%). We further investigated this set of 23 cell wall–related genes in the lrx1 rol1-1 line using gene set enrichment analysis (GSEA; see Methods). This analysis determines whether the coordinate differential regulation of this set of genes was also found in lrx1 compared with lrx1 rol1-1 and whether the change in expression was significantly more consistent than the differences in expression seen in a randomly selected gene set of similar size. This subset was found significantly enriched in the set of regulated genes between lrx1 and lrx1 rol1-1 mutants (enrichment score = 0.89; P value < 0.005). This finding indicates that the cell wall–related genes modified in their expression by the rol1-2 mutation are also affected in the rol1-1 mutant, although in a subtler way. This shows that rol1-1 and rol1-2 have overlapping effects on gene expression.
DISCUSSION
Suppressor screens are valuable for identifying genes that are involved in the same process and can reveal relationships between genes that would not have been established by other methods (Huang and Sternberg, 1995; Prelich, 1999). They also allow for the identification of mutants such as rol1-1 that would not have been detected in normal forward genetic screens due to the absence of a visible phenotype. The lrx1 mutant, which is defective in root hair cell wall formation (Baumberger et al., 2001, 2003b), was EMS mutagenized, and rol mutants were identified that revert the lrx1 phenotype to the wild type and are thus likely to encode proteins that are related to LRX1 function.
Modification of Rha Biosynthesis Suppresses the lrx1 Phenotype
RHM1 codes for a 668–amino acid Rha biosynthetic protein that is part of the sugar interconversion pathway required for the synthesis of cell wall monosaccharides (Reiter and Vanzin, 2001; Seifert, 2004). RHM1 belongs to a subclass of the NAD(P)-dependent short chain dehydrogenase/reductase family of enzymes. Short chain dehydrogenase/reductase proteins share motifs such as the Rossmann-fold for cofactor binding and a highly conserved sequence motif (YxxxK), which is part of the catalytic triad (Kleiger and Eisenberg, 2002; Allard et al., 2004). In Arabidopsis, the three highly similar isoforms RHM1-RHM3 were identified as likely UDP-l-Rha synthases (Reiter and Vanzin, 2001), and RHM1 was shown to convert UDP-d-Glc to UDP-l-Rha (B. Link and W.-D. Reiter, unpublished data). Further evidence for a function of RHM proteins as UDP-l-Rha synthases is provided by rhm2 mutants that have a reduced amount of the pectinaceous seed coat mucilage mainly consisting of RG I (Usadel et al., 2004; Western et al., 2004). Since the cell wall composition of other tissues appears not to be affected in rhm2 mutants, the three RHM genes seem to be redundant in most parts of the plant. This conclusion is corroborated by the expression of all RHM genes in stems, roots, leaves, inflorescences, siliques, and seedlings as determined by RT-PCR (Usadel et al., 2004; Western et al., 2004). The RHM1 promoter:GUS analysis indicates that RHM1 is expressed in all tissues of seedlings. This expression pattern is supported by the observed rol1 mutant phenotypes in seedling roots, root hairs, and cotyledons.
The mutant version of RHM1 encoded by rol1-1 is truncated, and both rol1-1 and rol1-2 mutations lead to an inactive dehydratase domain in vitro. Furthermore, the expression level of rhm1 in rol1-1 mutants is very low, presumably due to nonsense-mediated mRNA decay. Thus, the lack of Rha biosynthesis through RHM1 causes the suppression of lrx1. Both rol1 mutants, however, show wild-type Rha levels in total cell wall material from roots, indicating that RHM2 and RHM3 are sufficient to compensate for the lack of functional RHM1. Nevertheless, RG I and RG II are affected in the rol1 mutants, suggesting a link between individual RHM isozymes and particular glycosidic linkages formed by Rha with other sugars, which could explain the modification of RG I and RG II in rol1 plants despite normal Rha levels.
rol1-1 and rol1-2 Display Different Phenotypes
Although both rol1 mutations suppress the lrx1 mutant phenotype, the two mutations have different effects on plant development. rol1-2 single mutants develop shorter root hairs and roots compared with the wild type, a phenotype that is not observed in rol1-1 plants. The microarray experiments revealed 57 genes with changes in the expression level by a factor of ≥2 in the lrx1 rol1-2 mutant compared with lrx1, while the effect of the rol1-1 mutation on gene expression is much more subtle. Thus, rol1-1 and rol1-2 are phenotypically distinct both on the morphological and the molecular level. The moderate influence of rol1-1 suggests that this mutation mainly affects the cell wall structure, whereas the effect of rol1-2 is more dramatic and might also include other processes besides the biosynthesis of Rha. In the rol1-1 mutant, only a truncated protein can be produced, which is enzymatically inactive at least in vitro. By contrast, the rol1-2 mutant encodes a full-length protein with an Arg-to-Lys substitution in the dehydratase domain. The mutated Arg residue is completely conserved between the RHM proteins of plants and dTDP-glucose 4,6-dehydratases from prokaryotes and has been shown to interact with the diphospho group of dTDP-glucose (Allard et al., 2004). Accordingly, this residue may be essential for substrate binding. The dehydratase domain encoded by rol1-2 is inactive in vitro, but the mutant protein is likely to be stable. It is possible that it binds to potential in vivo interaction partners of wild-type RHM1 and locks the protein complex in an inactive state, which has a stronger impact on plant development and thus explains the phenotypic differences between the two rol1 alleles.
Among the 38 genes induced in lrx1 rol1-2 compared with lrx1, 20 genes (>50%) are presumed to be involved in cell wall–related processes. This is >5 times more than the estimated 2000 genes corresponding to <10% of the Arabidopsis genome that are predicted to have such a function (Yong et al., 2005). Among the 19 genes repressed in lrx1 rol1-2, three genes (16%) are presumed to be cell wall related. The cell wall alterations induced by the rol1-2 mutation seem to preferentially affect the expression of cell wall–related genes, which presumably reflects the ability of the plants to respond to alterations in the extracellular matrix. Although the changes in gene expression are more subtle in the lrx1 rol1-1 mutant, they affect the same cell wall–related genes that are differentially regulated in the lrx1 rol1-2 line.
Of the 20 cell wall–related genes induced in lrx1 rol1-2 compared with lrx1, seven are involved in the modification of xyloglucan or pectic polysaccharides. Four genes encode structural cell wall proteins (one Pro-rich protein and three extensins), which may strengthen the cell wall (Carpita and Gibeaut, 1993; Cassab, 1998). It remains to be shown whether the changes in gene expression are the cause of the visible phenotypes of rol1-2 plants. Since these modifications in gene expression are also induced by the rol1-1 mutation, even though to a lesser extent, they may contribute to the suppression of the lrx1 root hair phenotype. Higher expression of extensin genes might help to stabilize the aberrant and weakened root hair cell wall of the lrx1 mutant and thus prevent bulging and collapsing of the root hair structure. LRX6, a member of the LRX gene family, is also upregulated in the lrx1 rol1-2 line compared with lrx1. LRX6 is specifically expressed during lateral root development in wild-type plants (Baumberger et al., 2003a) and is not known to influence root hair formation. By contrast, LRX2, the paralog of LRX1, is not induced in the rol1 mutants and does not appear to play a role in the suppression of the lrx1 phenotype by the rol1 mutations. We were surprised to find no changes in gene expression between wild-type and lrx1 plants; however, trichoblasts account for only a small fraction of the total number of cells in the root (Dolan et al., 1994). For this reason, changes in gene expression in only this cell type might not be detectable. Alternatively, small changes in expression levels might be masked by the biological variance in gene expression (Hudson et al., 2003).
rol1 Suppressors Indicate a Possible Role of LRX1 during Pectin Matrix Formation
The involvement of the RHM proteins in pectin formation (Usadel et al., 2004; Western et al., 2004), the modifications of pectin in rol1 mutants, and the altered expression of a number of cell wall–related genes in the rol1 mutants suggest that structural changes in the cell wall cause suppression of the lrx1 mutant phenotype. Our data indicate a reduction in the two large Rha-containing side chains of RG II in the rol1-2 allele. This modification might account for the stunted root phenotype of the rol1-2 mutant, as changes in the RG II structure can affect plant growth (O'Neill et al., 2001). Our immunolocalization data indicate that both rol1 alleles contain modified RG I. Even though the effects of the rol1 mutations on root hair formation suggest an alteration of pectin in this cell type, the molecular basis of this modification is unknown. The LM5 antibody does not bind to root hair cells, possibly due to masking of the epitope. Alternatively, an unrecognized structure of pectin could be affected. Suppression of the lrx1 phenotype through modifications of pectin points toward a possible function of LRX1 in a pectin-related process. In future experiments, it will be useful to investigate whether the rol1 and lrx1 mutants change the pectin structure of root hairs and whether, for example, pore sizes might be affected in these lines. A method to measure porosity of pectin has been established for Chenopodium album (Fleischer et al., 1998) and can most likely be adapted for Arabidopsis to investigate this point. In a complementary approach, Fourier transform infrared spectroscopy (McCann and Carpita, 2005) might reveal changes in the molecular structure of cell walls of the different mutants. Another possibility is that the changes in gene expression caused by the rol1 mutations are the basis of suppression of the lrx1 phenotype. For example, the increased expression of structural cell wall proteins may lead to a stabilization of the weakened root hair cell wall of the lrx1 mutant. This hypothesis can be tested by analyzing plant lines that are mutated in these genes or that overexpress the respective proteins.
METHODS
Plant Material, EMS Mutagenesis, and Mapping
The lrx1-1s allele and the EMS mutagenesis procedure used for this mutant line are described by Diet et al. (2004). The lrx1 single and lrx1 lrx2 double mutants are in the Columbia genetic background. For vertical growth on plates, seeds were surface sterilized with a solution of 1% sodium hypochlorite and 0.03% Triton X-100, stratified 3 to 4 d at 4°C, and grown in a vertical orientation on the surface of half-strength MS medium containing 0.6% phytagel and 2% sucrose (Sigma-Aldrich) with a 16-h-light/8-h-dark cycle at 22°C. For crosses and propagation of the plants, seedlings were transferred to soil and grown in growth chambers with a 16-h-light/8-h-dark cycle at 22°C. Light microscopy observations were done on 4-d-old vertically grown seedlings with a Leica LZ M125 stereomicroscope. For measurements of the lengths of trichoblasts and root hairs, 50 cells of the mature root of more than five different seedlings per plant line were used. For the root length measurement, >30 roots per plant line were measured.
Molecular markers for all mutations were established to confirm the genotype of the different lines. The marker for lrx1 is described by Diet et al. (2004). The lrx2 mutation used in this study is a footprint allele (lrx2-2) caused by the excision of the En-1 transposon initially inserted at position 1478 of the coding region. The excision of En-1 resulted in the insertion of 4 bp (GTAC) and a frame shift in the beginning of the extensin coding region. On the DNA level, an RsaI restriction site polymorphism was introduced, which allows for the detection of the lrx2-2 mutation. For rol1-1 and rol1-2, CAPS markers were established using the primers 5′-ACTCCGGGTTCTGTGGGTAC-3′/5′-GATGTTGCCAAAGACATCTGC-3′ for rol1-1 and 5′-GTACCTCTGATCGTTAAAACGT-3′/5′-TTGTTCTTCACAAGGGAGAAG-3′ for rol1-2. The mismatches in the primers (underlined positions) introduce a KpnI site in wild-type DNA but not rol1-1 and an AclI site in rol1-2 but not wild-type DNA.
For mapping, the lrx1 rol1 mutants were crossed with Landsberg erecta (Ler) and propagated to the F2 generation. The F2 population containing the rol1-2 allele was selected for seedlings displaying the rol1-2 phenotype. Nine hundred mutant F2 seedlings were subsequently used for mapping the rol1-2 locus to <40 kb. For rol1-1, 500 F2 seedlings displaying a wild-type root hair phenotype were selected and screened by PCR for homozygous lrx1 mutant plants. These plants were assumed to be homozygous mutant for rol1-1 and were thus used for initial mapping. Once the approximate map position of rol1-1 was identified, F2 plants displaying an lrx1 mutant phenotype (i.e., being homozygous mutant lrx1) were selected, and those heterozygous Columbia/Ler in the region containing the rol1-1 locus were propagated to the F3 generation. As expected, seedlings of the F3 population segregated 3:1 for lrx1 versus wild-type root hairs. Five hundred wild type–like F3 seedlings were selected for detailed mapping. Mapping was performed using standard SSLP and CAPS markers developed based on the Columbia/Ler polymorphism databank (Jander et al., 2002).
Constructs and Plant Transformation
For the RHM1 promoter:GUS fusion construct, 1.5 kb of the promoter region was amplified by PCR using the primers RHM1GUSF 5′-GGATGTCGACGTATGAGTCTGTTG-3′ and RHM1GUSR 5′-TCGAAGTCGACGTGGAGTGAGTCT-3′, and the resulting fragment was digested with SalI for cloning into the pGPTV-Kan plant transformation vector cut with the same enzyme (Becker et al., 1992). For the RHM1 genomic clone used for the complementation test, 4.2 kb containing 1.5 kb of the promoter region, the coding region, and 600 bp of 3′ region were amplified by long-range PCR using the oligos RHM1genoF 5′-TCATGCGGCCGCGACCGAAGACCACCT-3′ and RHM1genofR 5′-ACGAGCGGCCGCAACGAGGAACGAA-3′. The PCR product was cloned into the TOPO 10 blunt end cloning vector (Invitrogen) for control sequencing and then cloned into the pART27 plant transformation vector (Gleave, 1992) by digesting with NotI. The binary vectors were transformed by electroporation into Agrobacterium tumefaciens GV3101. Plants were transformed by the floral dip method described by Clough and Bent (1998), and transgenic T1 plants were selected on 50 μg/mL kanamycin. The presence of the transgene was confirmed using primers specific for the kanamycin resistance gene NPTII. GUS staining was performed in 50 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1% Triton X-100, and 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid for 3 h at 37°C.
RNA Isolation and RT-PCR Analysis
Two hundred seedlings per plant line were grown in a vertical orientation on half-strength MS plates, and root total RNA was extracted using the TRIzol method (Gibco BRL). Ten micrograms of each RNA sample were reverse transcribed using oligo(dT) and a Superscript II RNase H reverse transcriptase kit (Invitrogen) following the manufacturer's recommendations. Quantitative real-time PCR was performed with an ABI PRISM 7700 sequence detection system (Applied Biosystems) using a SYBR Green PCR kit from Applied Biosystems and primers specific for the RHM1 gene. Relative mRNA abundance was calculated using the comparative Δ-Ct method and normalized to the corresponding ACTIN2 transcript levels.
Expression of Recombinant Protein and Activity Assay
The RHM1-D constructs were amplified by the primers 5′ Nhe I, 5′-TAAGTGCTAGCATGGCTTCGTACACTCCCAAGAACATTC-3′, and 3′ Bam HI, 5′-GATAAGGATCCTCAGTGGTGGTGGTGGTGGTGAGATAAAGTAGCTGCCAGCGAATTGTCCTC-3′, and cloned into the vector pET11a (Novagen) digested with NheI and BamHI. A His tag was added by the 3′ Bam HI primer, allowing the protein to be partially purified using standard nickel-nitrilotriacetic acid agarose columns. For protein expression, the constructs were transformed into Rosetta DE3 cells (Novagen). Protein expression and enzymatic analysis were performed as described by B. Link and W.-D. Reiter (unpublished data). In summary, bacterial cultures were grown to OD600 ≥ 0.6, and protein expression was induced with 0.3 mM isopropylthio-β-galactoside. The protein extract was purified over a Ni-NTA column (Qiagen), and protein activity was assayed in 20 mM glycine buffer, pH 8.5, 1 mM NAD+, and 1.6 mM UDP-glucose using 0.5 to 1 μg protein. After stopping the reactions, nucleotide sugars were hydrolyzed by adding an equal volume (100 μL) of 4 M trifluoroacetic acid and incubating them at 90°C for 30 min. The hydrolysate was mixed with 0.25 mL of 2 M ammonia and 4% NaBH4 and incubated at 40°C for 1 to 2 h. Next, 0.25 mL of acetic acid, 1 mL DMSO, 0.25 mL 1-methylimidazole, and 4 mL of acetic anhydride were added to the reactions to convert the reduced sugars to alditol acetates. The alditol acetates were extracted from the aqueous solutions using 1 mL dichloromethane, and the latter was used for injection into a 30-m SP-2330 column with 0.20-μm film thickness (Supelco) for analysis by GC-MS in selected ion monitoring mode. RHM1-D protein produced robust signals (>3 μM of product) in 20 min. Since the solubility of the truncated RHM1-D protein encoded by rol1-1 was very low, the protein was also expressed without a His tag, and bacterial extracts were used for the analysis. The result was the same for both approaches.
Preparation of Cell Wall Material and Sugar Composition Analysis
Cell wall material was prepared from roots of 1-week-old seedlings. Plant tissue was frozen in liquid nitrogen and macerated in 70% ethanol (aqueous) using a Retschmill (Retsch). The ground tissue was pelleted by centrifugation, and the resulting pellet washed with a 1:1 (v/v) mixture of chloroform and methanol. The pellet was then washed twice with acetone and dried. The cell wall material was hydrolyzed using 2 M trifluoroacetic acid, and the solubilized monosaccharides were converted into their corresponding alditol acetates followed by GC-MS analysis, and the uronic acid content was determined using the m-hydroxy-biphenyl assay as described by Usadel et al. (2004). The 2-O-methyl-sugars were quantified by selective ion monitoring at a mass-to-charge ratio of 117 with hydrolyzed 2-O-methyl-β-d-galactopyranosyl-(1→4)- d-glucose (Sigma-Aldrich) as an internal standard.
Immunolabeling
Reflection microscopy of silver-enhanced, immunogold-labeled, resin-embedded tissue sections was performed as described previously (Bush and McCann, 1999). All treatments of the wild type and mutants, including image acquisition and processing, were performed in parallel.
Microarray Analysis
To obtain total RNA, Columbia, lrx1, and the rol mutants (in the lrx1/lrx1 mutant background) were grown in a vertical orientation on half-strength MS medium, and root tissue was collected. RNA was extracted by the TRIzol method (Gibco BRL), further purified using the RNeasy kit (Qiagen), and tested for degradation by a lab on a chip analysis (Agilent Technologies). Double-stranded cDNA was synthesized with the SuperScript kit (Invitrogen) using 20 μg total RNA as starting material. Purification was done with the Affymetrix GeneChip sample cleanup module (Affymetrix). The probe was labeled with the ENZO-BioArray labeling kit (Loxo) and finally purified again with the Affymetrix GeneChip sample cleanup module. Hydrolysis of the labeled RNA and chip hybridization were performed as recommended by Affymetrix. All the experiments were performed in biological triplicates. After hybridization and scanning, probe cell intensities were calculated with Affymetrix MAS5 software (Affymetrix). Summarization and normalization for the respective probe sets were performed using dCHIP software (Li and Wong, 2003). Genes were filtered out from the resulting lists of normalized expression values in Genespring 7.2 when not showing present calls in all replicate measurements of at least one condition. Subsequently, the significance of changes in expression was tested in Genespring by an equal-variance t test, applying a Benjamini-Hochberg correction (Benjamini and Hochberg, 1995) with a false discovery rate of 0.05.
GSEA was performed as described by Subramanian et al. (2005). GSEA provides an enrichment score that measures the degree of enrichment of the gene set at the top (upregulated in lrx1 versus lrx1 rol1-1) or bottom (downregulated in lrx1 versus lrx1 rol1-1) of a rank-ordered gene list derived from the data set. The nominal P value is used to assess the significance of the enrichment score.
Accession Numbers
Sequence data from this article can be found in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MEXP-722. The locus identifiers are At1g12040 (LRX1) and At1g78570 (RHM1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Cotyledon Phenotype of rol1 Mutants.
Supplemental Table 1. Effect of lrx1 and lrx1 rol1 Mutations on Gene Expression.
Supplementary Material
Acknowledgments
We thank Andreas Patrigniani for technical assistance with the microarray analysis, Julia Schönfeld for the monosaccharide quantification, Kim Findlay and Susan Bunnewell for technical help with tissue sectioning, and Beat Keller for critical reading of the manuscript. This work was supported by Swiss National Science Foundation Grants 31-61419.00 and 3100A0-103891, by a Nachwuchsforscher-Stipendium of the University of Zürich (560027), and by the Functional Genomics Center (A.D. and C.R.). It was also supported by Biotechnology and Biological Science Research Council Grant 208/D10332 and European Union Grant QLK5-CT-2001-00443 (G.J.S.), by U.S. Department of Energy Grant DE-FG02-95ER20203 (W.-D.R.), and by the German Ministry for Science and Education GABI Grant 0312277D (M.P.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Christoph Ringli (chringli@botinst.unizh.ch).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.038653.
References
- Allard, S.T.M., Cleland, W.W., and Holden, H.M. (2004). High resolution X-ray structure of dTDP-glucose 4,6-dehydratase from Streptomyces venezuelae. J. Biol. Chem. 279 2211–2220. [DOI] [PubMed] [Google Scholar]
- Baron-Epel, O., Gharyal, P.K., and Schindler, M. (1988). Pectins as mediators of wall porosity in soybean cells. Planta 175 389–395. [DOI] [PubMed] [Google Scholar]
- Baumberger, N., Doesseger, B., Guyot, R., Diet, A., Parsons, R.L., Clark, M.A., Simmons, M.P., Bedinger, P., Goff, S.A., Ringli, C., and Keller, B. (2003. a). Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice: A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol. 131 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger, N., Ringli, C., and Keller, B. (2001). The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev. 15 1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger, N., Steiner, M., Ryser, U., Keller, B., and Ringli, C. (2003. b). Synergistic interaction of the two paralogous Arabidopsis genes LRX1 and LRX2 in cell wall formation during root hair development. Plant J. 35 71–81. [DOI] [PubMed] [Google Scholar]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20 1195–1197. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57 289–300. [Google Scholar]
- Bouton, S., Leboeuf, E., Mouille, G., Leydecker, M.T., Talbotec, J., Granier, F., Lahaye, M., Höfte, H., and Truong, H.N. (2002). QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14 2577–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, M.S., and McCann, M.C. (1999). Pectic epitopes are differentially distributed in the cell walls of potato (Solanum tuberosum) tubers. Physiol. Plant. 107 201–213. [Google Scholar]
- Carpita, N.C., and Gibeaut, D.M. (1993). Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3 1–30. [DOI] [PubMed] [Google Scholar]
- Cassab, G.I. (1998). Plant cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 281–309. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1999). Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 391–417. [DOI] [PubMed] [Google Scholar]
- Diet, A., Brunner, S., and Ringli, C. (2004). The enl mutants enhance the lrx1 root hair mutant phenotype of Arabidopsis thaliana. Plant Cell Physiol. 45 734–741. [DOI] [PubMed] [Google Scholar]
- Dolan, L., Duckett, C.M., Grierson, C., Linstead, P., Schneider, K., Lawson, E., Dean, C., Poethig, S., and Roberts, K. (1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120 2465–2474. [Google Scholar]
- Fleischer, A., Titel, C., and Ehwald, R. (1998). The boron requirement and cell wall properties of growing and stationary suspension-cultured Chenopodium album L. cells. Plant Physiol. 117 1401–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman, J., and Dolan, L. (2001). Root hairs as a model system for studying plant cell growth. Ann. Bot. (Lond.) 88 1–7. [Google Scholar]
- Forsthoefel, N.R., Cutler, K., Port, M.D., Yamamoto, T., and Vernon, D.M. (2005). PIRLs: A novel class of plant intracellular leucine-rich repeat proteins. Plant Cell Physiol. 46 913–922. [DOI] [PubMed] [Google Scholar]
- Gilmor, C.S., Lukowitz, W., Brininstool, G., Sedbrook, J.C., Hamann, T., Poindexter, P., and Somerville, C. (2005). Glycosylphosphatidyl inositol-anchored proteins are required for cell wall synthesis and morphogenesis in Arabidopsis. Plant Cell 17 1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20 1203–1207. [DOI] [PubMed] [Google Scholar]
- Huang, L.S., and Sternberg, P.W. (1995). Genetic dissection of developmental pathways. In Methods in Cell Biology, H.F. Epstein and C.C. Shaker, eds (San Diego, CA: Academic Press), pp. 97–122. [DOI] [PubMed]
- Hudson, M.E., Lisch, D.R., and Quail, P.H. (2003). The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A signaling pathway. Plant J. 34 453–471. [DOI] [PubMed] [Google Scholar]
- Iwai, H., Masaoka, N., Ishii, T., and Satoh, S. (2002). A pectin glucuronyltransferase gene is essential for intercellular attachment in the plant meristem. Proc. Natl. Acad. Sci. USA 99 16319–16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., and Last, R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L., Seymour, G.B., and Knox, J.P. (1997). Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1→4)-β-D-galactan. Plant Physiol. 113 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger, G., and Eisenberg, D. (2002). GXXXG and GXXXA motifs stabilize FAD and NAD(P)-binding Rossmann folds through Cα-H….O hydrogen bonds and van der waals interactions. J. Mol. Biol. 323 69–76. [DOI] [PubMed] [Google Scholar]
- Knox, J.P. (1995). The extracellular matrix in higher plants. 1. Developmentally regulated proteoglycans and glycoproteins of the plant cell surface. FASEB J. 9 1004–1012. [DOI] [PubMed] [Google Scholar]
- Knox, J.P., Linstead, P.J., King, J., Cooper, C., and Roberts, K. (1990). Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 181 512–521. [DOI] [PubMed] [Google Scholar]
- Lally, D., Ingmire, P., Tong, H.Y., and He, Z.H. (2001). Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell 13 1317–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao, N.T., Long, D., Kiang, S., Coupland, G., Shoue, D.A., Carpita, N.C., and Kavanagh, T.A. (2003). Mutation of a family 8 glycosyltransferase gene alters cell wall carbohydrate composition and causes a humidity-sensitive semi-sterile dwarf phenotype in Arabidopsis. Plant Mol. Biol. 53 687–701. [DOI] [PubMed] [Google Scholar]
- Li, C., and Wong, W.H. (2003). DNA-chip analyzer (dCHIP). The analysis of gene expression data. In Methods and Software, G. Parmigiani, E.E. Garrett, I. Irizarry, and S.L. Zeger, eds (New York: Springer), pp. 120–141.
- Majewska-Sawka, A., and Nothnagel, E.A. (2000). The multiple roles of arabinogalactan proteins in plant development. Plant Physiol. 122 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann, M.C., and Carpita, N.C. (2005). Looking for invisible phenotypes in cell wall mutants of Arabidopsis thaliana. Plant Biosyst. 139 80–83. [Google Scholar]
- McCartney, L., Steele-King, C.G., Jordan, E., and Knox, J.P. (2003). Cell wall pectic (1→4)-β-D-galactan marks the acceleration of cell elongation in the Arabidopsis seedling root meristem. Plant J. 33 447–454. [DOI] [PubMed] [Google Scholar]
- O'Neill, M.A., Eberhard, S., Albersheim, P., and Darvill, A.G. (2001). Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294 846–849. [DOI] [PubMed] [Google Scholar]
- Prelich, G. (1999). Suppression mechanisms: Themes from variations. Trends Genet. 15 261–266. [DOI] [PubMed] [Google Scholar]
- Puhlmann, J., Bucheli, E., Swain, M.J., Dunning, N., Albersheim, P., Darvill, A.G., and Hahn, M.G. (1994). Generation of monoclonal antibodies against plant cell-wall polysaccharides. 1. Characterization of a monoclonal antibody to a terminal α-(1→2)-linked fucosyl-containing epitope. Plant Physiol. 104 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter, W.D., and Vanzin, G.F. (2001). Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Mol. Biol. 47 95–113. [PubMed] [Google Scholar]
- Ridley, B.L., O'Neill, M.A., and Mohnen, D.A. (2001). Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57 929–967. [DOI] [PubMed] [Google Scholar]
- Ringli, C. (2005). The role of extracellular LRR-extensin (LRX) proteins in cell wall formation. Plant Biosyst. 139 32–35. [Google Scholar]
- Ringli, C., Keller, B., and Ryser, U. (2001). Glycine-rich proteins as structural components of plant cell walls. Cell. Mol. Life Sci. 58 1430–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier, F., Fernandez, A.G., Fujita, M., Himmelspach, R., Borner, G.H.H., Schindelman, G., Song, S., Baskin, T.I., Dupree, P., Wasteneys, G.O., and Benfey, P.N. (2005). COBRA, an Arabidopsis extracellular glycosylphosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell 17 1749–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible, W.R., and Pauly, M. (2004). Glycosyltransferases and cell wall biosynthesis: Novel players and insights. Curr. Opin. Plant Biol. 7 285–295. [DOI] [PubMed] [Google Scholar]
- Seifert, G.J. (2004). Nucleotide sugar interconversions and cell wall biosynthesis: How to bring the inside to the outside. Curr. Opin. Plant Biol. 7 277–284. [DOI] [PubMed] [Google Scholar]
- Showalter, A.M. (1993). Structure and function of plant cell wall proteins. Plant Cell 5 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan, W., Kovac, P., Albersheim, P., Darvill, A.G., and Hahn, M.G. (1995). Characterization of a monoclonal antibody that recognizes an arabinosylated (1→6)-β-D-Galactan epitope in plant complex carbohydrates. Carbohydr. Res. 275 295–307. [DOI] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo, P., Mootha, V.K., Mukherjee, S., Ebert, B.L., Gillette, M.A., Paulovich, A., Pomeroy, S.L., Golub, T.R., Lander, E.S., and Mesirov, J.P. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel, B., Kuschinsky, A.M., Rosso, M.G., Eckermann, N., and Pauly, M. (2004). RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiol. 134 286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, T.A., and Kohorn, B.D. (2001). Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 13 303–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western, T.L., Young, D.S., Dean, G.H., Tan, W.L., Samuels, A.L., and Haughn, G.W. (2004). MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol. 134 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats, W.G.T., Marcus, S.E., and Knox, J.P. (1998). Generation of a monoclonal antibody specific to (1→5)-α-L-arabinan. Carbohydr. Res. 308 149–152. [DOI] [PubMed] [Google Scholar]
- Willats, W.G.T., McCartney, L., and Knox, J.P. (2001. b). In situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta 213 37–44. [DOI] [PubMed] [Google Scholar]
- Willats, W.G.T., McCartney, L., Mackie, W., and Knox, J.P. (2001. a). Pectin: Cell biology and prospects for functional analysis. Plant Mol. Biol. 47 9–27. [PubMed] [Google Scholar]
- Yates, E.A., Valdor, J.F., Haslam, S.M., Morris, H.R., Dell, A., Mackie, W., and Knox, J.P. (1996). Characterization of carbohydrate structural features recognized by anti-arabinogalactan-protein monoclonal antibodies. Glycobiology 6 131–139. [DOI] [PubMed] [Google Scholar]
- Yong, W.D., et al. (2005). Genomics of plant cell wall biogenesis. Planta 221 747–751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.