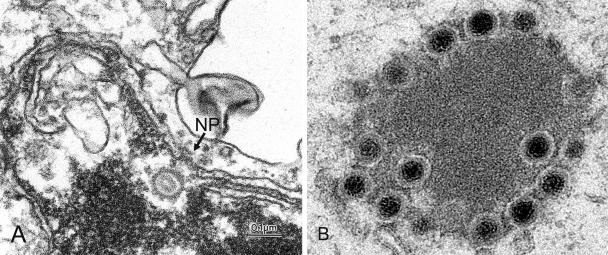
The presence of herpes simplex virus (HSV) capsids attached to invaginated cytoplasmic vesicles led Stackpole (15) to propose that capsids undergo envelopment at the inner nuclear membrane, de-envelopment at the outer nuclear membrane, and finally reenvelopment at cytoplasmic membranes. Over the years this model attracted numerous adherents principally on the basis of evidence that the extracellular virions lack proteins present in intracellular virions accumulating in the perinuclear space (11, 14). An alternative hypothesis was recently presented by Wild et al. (17) on the basis of the observation that nuclear pores become grossly enlarged in cells infected with wild-type virus. Two hypotheses have emerged. The first is that HSV virions undergo envelopment at the inner nuclear membrane, de-envelopment at the outer nuclear membrane or extensions thereof, and reenvelopment in cytoplasmic organelles (double envelopment model). The second hypothesis is that virions mature and egress the cell via two pathways. A minority, primarily early in infection, becomes enveloped at the inner nuclear membrane and is transported in vesicles to the extracellular space. The majority enters the cytoplasm late in infection through enlarged nuclear pores and becomes enveloped at cytoplasmic membranes, mainly Golgi and post-Golgi.
Challenges to existing theories are the fabric of science, and no other recent controversy has generated as much discussion as the challenge to the double envelopment hypothesis. The letter by Mettenleiter and Minson and the response by Wild (10) sustain the respective models but do not define the problems associated with each model or the data necessary to reaffirm or reject them.
The double envelopment model requires that virions accumulating in the perinuclear space fuse with the outer nuclear membrane or extensions thereof. HSV encodes a quartet of glycoproteins that execute the entry of virus into cells by fusion of the envelope with the plasma membrane and cell-to-cell fusion (4, 6, 9, 13, 16). Thus, glycoprotein D (gD) interacts with either the herpesvirus entry mediator or nectin 1 receptor, whereas gH, which contains structural elements shared with viral fusion proteins (7), and gB and gL execute the fusion (5). This quartet plays no role in the de-envelopment of virions, since mutants lacking any one of the four glycoproteins, while not infectious, nevertheless egress the cells in a manner similar to that of wild-type virus.
Other viral proteins, notably gK and UL20, are involved in viral egress, in that in their absence virus particles accumulate in the perinuclear space or cytoplasm. These proteins, however, appear to inhibit fusion performed by the quartet (2), and their presence in the viral envelope is debated. SNARE proteins, responsible for fusion of cellular vesicles, by their topology and necessary cytoplasmic cofactors cannot perform the fusion in the perinuclear space required by the double envelopment model. A common strategy of HSV is to sequester and redirect cellular proteins to perform at times novel functions (e.g., the binding of protein phosphatase 1α by γ34.5, the binding of cdc2 by UL42 [1, 8]). Putative cellular protein partners capable of fusing viral envelope with the outer nuclear membrane or vesicles derived from its extensions in the absence of one or more of the glycoprotein quartet have not been identified. In essence, the evidence required to support the double envelopment model is the definition of the mechanism by which enveloped virions accumulating in the perinuclear space could become de-enveloped, resulting in the release of capsids into the cytoplasm.
The egress of capsids though the nuclear pores proposed by Wild et al. (17) challenges the de-envelopment step of the Stackpole pathway. Capsids could egress through enlarged nuclear pores passively along with nuclear components and organelles or actively by engaging the nuclear export machinery of the cell. Unregulated egress of proteins or RNA from nuclei of infected cells has not been reported. On the other hand, while the evidence supports a specific association of capsids with the pores on the cytoplasmic side of the nuclear membrane, there is no evidence for an association with pores at the nuclear side. Thus, capsids encoded by two diverse viral mutants accumulate in large numbers at nuclear pores on the cytoplasmic side, but not juxtaposed at random to nuclear membranes (3, 11). In contrast, capsids frequently line the nuclear side of the inner nuclear membrane but not specifically at nuclear pores. At present, only mutants lacking the US3 protein kinase fail to transport capsids from the nucleus to the perinuclear space (12), but the effect of US3 may be on the integrity of the nuclear envelope and not specifically on the mechanism of capsid transport. The key evidence required to support the single envelopment model is identification of nuclear transport proteins that could interact with capsids and demonstration that the transport is abolished by mutagenesis of the capsid partner or genetic manipulation of the nuclear transport protein.
REFERENCES
- 1.Advani, S. J., R. R. Weichselbaum, and B. Roizman. 2001. cdc2 cyclin-dependent kinase binds and phosphorylates herpes simplex virus 1 UL42 DNA synthesis processivity factor. J. Virol. 75:10326-10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avitabile, E., G. Lombardi, T. Gianni, M. Capri, and G. Campadelli-Fiume. 2004. Coexpression of UL20p and gK inhibits cell-cell fusion mediated by herpes simplex virus glycoproteins gD, gH-gL, and wild-type gB or an endocytosis-defective gB mutant, and downmodulates their cell surface expression. J. Virol. 78:8015-8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batterson, W., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, W. Z., S. Person, C. DebRoy, and B. H. Gu. 1988. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1. An analysis of linker insertion mutants. J. Mol. Biol. 201:575-588. [DOI] [PubMed] [Google Scholar]
- 5.Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. [DOI] [PubMed] [Google Scholar]
- 6.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gianni, T., P. L. Martelli, R. Casadio, and G. Campadelli-Fiume. 2005. The ectodomain of herpes simplex virus glycoprotein H contains a membrane alpha-helix with attributes of an internal fusion peptide, positionally conserved in the Herpesviridae family. J. Virol. 79:2931-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He, B., M. Gross, and B. Roizman. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 62:1486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mettenleiter, T. C., and T. Minson. 2006. Egress of alphaherpesviruses. J. Virol. 80:1610-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roop, C., L. Hutchinson, and D. C. Johnson. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 67:2285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 75:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stackpole, C. W. 1969. Herpes-type virus of the frog renal adenocarcinoma. I. Virus development in tumor transplants maintained at low temperature. J. Virol. 4:75-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wild, P., M. Engels, C. Senn, K. Tobler, U. Ziegler, E. M. Schraner, E. Loepfe, M. Ackermann, M. Müller, and P. Walther. 2005. Impairment of nuclear pores in bovine herpesvirus 1-infected MDBK cells. J. Virol. 79:1071-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]