Abstract
UL25 is one of seven herpes simplex virus-encoded proteins involved specifically in DNA encapsidation. Its role appears to be to stabilize the capsid so that DNA is prevented from escaping once it has entered. To clarify the function of UL25, we have examined capsids with the goal of defining where it is located. Analysis of trypsin-treated capsids showed that UL25 is sensitive to cleavage like other proteins such as the major capsid and portal proteins that are exposed on the capsid surface. Internal proteins such as the scaffolding protein and protease were not affected under the same experimental conditions. Capsids were also examined by electron microscopy after staining with gold-labeled antibody specific for UL25. Images of stained capsids demonstrated that most labeled sites (71% in C capsids) were at capsid vertices, and most stained C capsids had label at more than one vertex. A quantitative immunoblotting method showed that the capsid contents of UL25 were 56, 20, and 75 copies per capsid in A, B, and C capsids, respectively. Finally, soluble UL25 protein was found to bind in vitro to purified capsids lacking it. The amount of bound UL25 corresponded to the amount present in B capsids, and bound UL25 was found by immunoelectron microscopy to be located predominantly at the capsid vertices. The results are interpreted to suggest that five UL25 molecules are found at or near each of the capsid vertices, where they are exposed on the capsid surface. Exposure on the surface is consistent with the view that UL25 is added to the capsid as DNA is packaged or during late stages of the packaging process.
Like replication of double-stranded DNA bacteriophage, herpesvirus replication involves a step in which progeny DNA molecules are introduced into a preformed capsid (4, 29). DNA encapsidation takes place in the infected-cell nucleus, where the virus DNA is synthesized and where progeny capsids are assembled. Once a capsid is filled with DNA, it transits to the cytoplasm, where it acquires tegument and membrane layers prior to exiting the host cell.
Studies of herpes simplex virus 1 (HSV-1) replication have led to the identification of seven genes involved specifically in DNA encapsidation (UL6, UL15, UL17, UL25, UL28, UL32, and UL33 genes) (2, 4, 5, 13, 15, 27, 28, 30, 38). When cells are infected with a mutant unable to produce any one of the seven gene products, capsids are assembled normally and the virus DNA is replicated but capsids are not filled. Concatemeric DNA and closed capsids accumulate in the infected-cell nucleus, indicating that the missing gene is involved specifically in DNA encapsidation and not, for instance, in capsid formation or DNA replication. Homologs for six of the seven HSV-1 DNA processing/packaging proteins are encoded by all herpesviruses (6).
Analysis of the DNA encapsidation process has defined the functions for three of the processing/packaging proteins, UL6, UL15, and UL28. UL6 is the structural subunit of the portal, a funnel-shaped 12-mer found at one of the 12 capsid vertices (22). DNA enters the capsid through the portal, which may play an active role in the DNA translocation process. UL15 and UL28 are the components of terminase, a multisubunit enzyme that acts to cleave the DNA concatemer into genome length units and to provide the energy required for translocating DNA into the capsid. Homologs of the HSV-1 UL6 and UL15 proteins are encoded in the genomes of double-stranded DNA bacteriophages such as lambda and T4 (12, 14, 16, 26, 33).
UL25 differs from the other processing/packaging proteins, as indicated by observation of the metabolism of virus DNA in cells infected with a null mutant virus. Whereas a mixture of concatemeric and genome length segments is found in cells infected with the UL25 null mutant KUL25NS, only concatemeric DNA is seen in cells infected with mutants deficient in any of the other six packaging genes (15). This result is interpreted to suggest that UL25 may function at a late stage of the DNA-packaging process after genome length DNA segments have been excised from the concatemer. In particular, it is suggested that UL25 may act after DNA has entered to stabilize the capsid against the pressure created by the packaged DNA (15, 36).
Biochemical studies have generally supported the mechanism described above for UL25 function. UL25 is found to be present as a minor structural component of the HSV-1 capsid. The copy number in B capsids, for instance, was reported to be 42 ± 17 compared to 955 for the major capsid protein (25). DNA-containing capsids (C capsids) were found to have a higher UL25 content than capsids lacking DNA (i.e., procapsids, A capsids and B capsids), as one would expect if it functions to stabilize the filled capsid against internal DNA pressure (11, 25, 40). Using immunoelectron microscopy, Ogasawara et al. (25) demonstrated localization of UL25 at one or more capsid vertices. The same authors reported specific binding of UL25 to HSV-1 DNA and an ability of UL25 to bind the major capsid protein (UL19) and one of the two triplex proteins (UL38) (25). Recently, a high-resolution structure has been determined for a substantial portion of UL25 (3).
We have examined the location of UL25 in the HSV-1 capsid with a view to determining whether it is exposed primarily outside or inside the capsid shell. Intact capsids were treated with the proteolytic enzyme trypsin and tested to determine whether UL25 is sensitive to digestion, as expected if it is located on the capsid surface, or resistant, as expected if it is inside. Immunoelectron microscopy and studies involving addition of UL25 to intact capsids were also undertaken to define the location of UL25 in the capsid.
MATERIALS AND METHODS
Cells, viruses, and capsid isolation.
Experiments involving wild-type virus and capsids were performed with the 17MP strain of HSV-1. Virus stocks were prepared by growth on Vero cell monolayers maintained in 150-cm2 flasks at 34°C on minimal essential medium containing 10% calf serum and antibiotics (22). For capsid preparation, the stock virus was used to infect BHK-21 cells (23), as capsid yields were found to be higher than in Vero cell infections. Capsids that lack UL25 were isolated after infection of Vero cells with the UL25-null mutant KUL25NS (multiplicity of infection = 5), which was maintained by passage in complementing cell line 8-1 (15).
Capsids were isolated, beginning with BHK-21 cells (10 150-cm2 flasks) that were infected while actively dividing and incubated for 20 h at 37°C. Infected cells were harvested by scraping, pelleted, resuspended in 30 ml distilled water, and frozen at −20°C. Cells were then thawed, adjusted to 1% Triton X-100-20 mM Tris-HCl, pH 7.5, containing protease inhibitors (0.1 volume of a solution prepared by dissolving one tablet of Roche Diagnostics Complete in 5 ml phosphate-buffered saline [PBS]), and incubated at 4°C for 1 h. The resulting nuclei were harvested by low-speed centrifugation, resuspended in 50 ml TNE (0.01 M Tris-HCl, 0.5 M NaCl, 1 mM EDTA, pH 7.5), and lysed by sonication (10 s; probe sonicator). The lysate was adjusted to 20 mM MgCl2 plus 500 μg/ml DNase I and incubated at 30°C until the viscosity dropped (5 to 10 min). The lysate was then cleared by brief centrifugation (16,000 × g for 5 min) and the virus pelleted into a 5-ml cushion of 35% sucrose (SW28 rotor; 23,000 rpm for 1 h). The pellets were suspended in 3 ml TNE and adjusted to 1 mM dithiothreitol. A, B, and C capsids were purified from the suspension by density gradient ultracentrifugation on 5-ml gradients of 25% to 50% sucrose prepared in TNE. Gradients were centrifuged for 40 min at 24,000 rpm in a Beckman SW55Ti rotor. Capsids from individual bands were removed from the gradient with a Pasteur pipette and pelleted by centrifugation (23,000 rpm for 1 h in a Beckman SW55Ti rotor). After resuspension in TNE, capsid species were rebanded separately by centrifugation on 600-μl gradients as previously described (20), pelleted, and resuspended in ∼0.3 ml TNE. The protein concentration was in the range of 0.25 to 0.50 mg/ml for both wild-type and UL25-negative capsids. Protease inhibitors were present in all capsid preparations except for B capsids used in trypsin digestion experiments.
Two molar guanidine-HCl (GuHCl) was used to remove pentons selectively from capsids, beginning with B capsids isolated as described above from HSV-1-infected BHK-21 cells (23). Purified B capsids (100 μg) were suspended in 600 μl TNE containing 2 M GuHCl plus 10 mM dithiothreitol and incubated at 4°C for 45 min. The solution was then underlaid with 50 μl TNE containing 50% sucrose plus 2 M GuHCl, and the capsids were pelleted by centrifugation (600-μl tube) (20) at 23,000 rpm in an SW55.1 rotor. After centrifugation, the supernatant was carefully removed, the pellet was resuspended in 100 μl TNE, and the capsids were purified by sucrose density gradient centrifugation on a 600-μl gradient of 20% to 50% sucrose as described previously (20). Capsids prepared by treatment of B capsids with 2 M GuHCl as described above are called G capsids.
Trypsin treatment of B capsids.
B capsids (50 μl) were added to 5 μl TNE containing trypsin adjusted to yield a final concentration of 20 μg/ml, 2 μg/ml, or 0.2 μg/ml, and the mixtures were incubated for 45 min at 37°C. Capsids were then isolated by centrifugation on a 600-μl gradient of 20% to 50% sucrose prepared in TNE containing protease inhibitors. Centrifugation was for 40 min at 23,000 rpm in a Beckman SW55Ti rotor operated at 4°C. After centrifugation, gradients were photographed and then fractionated by bottom puncture of the tube; capsid-containing fractions were identified by dot blotting (2-μl specimen applied to wetted membrane and stained with 1% Ponceau S). Capsids were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblotting as previously described (17). Antibodies used for immunoblotting were as follows: UL25, monoclonal antibody (MAb) 2D9 (diluted 1:2,000) (15); UL19, MAb 3E8 (culture supernatant; diluted 1:500) (21); UL38, MAb 4A11 (culture supernatant; diluted 1:5,000; prepared as described in reference 24 using glutathione S-transferase-UL38 as the immunogen); UL26.5, MAb MCA406 (Serotec, Inc.; diluted 1:4,000); UL18, MAb 1D2 (diluted 1:10,000) (24); VP24, MAb 9-2 (culture supernatant; diluted 1:100,000) (31); UL6, MAb 1C9 (diluted 1:10,000) (22). When blots were stained with a second antibody, the original antibody was first removed by incubation of the blot for 30 min at 50°C in 100 ml 100 mM 2-mercaptoethanol, 2% sodium dodecyl sulfate, and 60 mM Tris-HCl, pH 7.2. Before reprobing, the blot was washed six times in PBS containing 0.5% Triton X-100 and tested for the presence of residual chemiluminescence.
Preparation of UL25 inclusion bodies.
Insoluble complexes composed of UL25 were isolated from Spodoptera frugiperda (Sf9) cells infected with BAC-UL25, a recombinant baculovirus encoding UL25 (15). Previously described procedures were used for growth of Sf9 cells in culture and for infecting them with BAC-UL25 (multiplicity of infection = 5; infection for 64 h) (39). Except as noted, all steps of inclusion body isolation were carried out at 4°C and all buffers contained protease inhibitors (see above). Infected cells were harvested by centrifugation (∼2.0 ml packed cells), resuspended in 2 volumes of PBS, and lysed by three cycles of freezing and thawing. The lysate was then centrifuged for 5 min at 16,000 × g and the supernatant discarded. The pellet, which contains UL25 inclusion bodies, was extracted sequentially with (i) 1 ml of TNE, 10 mM dithiothreitol, and 2% Triton X-100 (30 min on ice) to solubilize cellular membranes and (ii) 1 ml TNE, 20 mM MgSO4, 5 mg/ml DNase I (10 min at room temperature) to remove cellular DNA. After each extraction, inclusion bodies were harvested by centrifugation into a pellet (16,000 × g for 5 min). The pellet obtained after DNase I treatment was resuspended by sonication (bath sonicator) in 0.5 ml TNE and stored at −80°C in 20-μl aliquots. The protein concentration of individual preparations ranged from ∼5.0 to 7.5 mg/ml. Experiments were performed with a stock solution prepared by mixing one 20-μl aliquot with 80 μl dissociation buffer (1% SDS, 1% dithiothreitol, 40 mM EDTA, 20% sucrose) and boiling for 2 min.
Determination of UL25 copy number.
The UL25 copy numbers in A, B, and C capsids were determined by quantitative estimation of band intensities in Coomassie-stained SDS-PAGE gels and in stained immunoblots. Band intensities were determined by scanning gels in a flatbed scanner followed by quantitation using UN-SCAN-IT (version 5.1; Silk Scientific). Bands containing known amounts of bovine serum albumin (BSA) were used to estimate the protein concentration of a stock UL25 solution (see above) and the concentrations of stock preparations of A, B, and C capsids. The major capsid protein (UL19) band was used for quantitation. Dilutions of the stock UL25 solution were then stained in the same immunoblot used for capsids, and the integrated band intensities of UL25 standards were determined to estimate the amount of UL25 in capsid lanes. The number of capsids present was determined from the formula UL19 amount (grams) × 6.02 × 1,023/149,075 × 955, where 149,075 is the molecular weight (MW) of UL19 and 955 is the number of UL19 molecules per capsid. The number of UL25 molecules was determined from UL25 amount (grams) × 6.02 × 1023/62,666, where 62,666 is the MW of UL25.
Capsid labeling with UL25-specific antibody.
Immunoelectron microscopy was performed with capsids adsorbed to carbon-Formvar-coated copper grids. The procedure used for labeling was the same as that described previously (22) except that (i) the primary antibody was the UL25-specific rabbit polyclonal serum described by McNab et al. (15) and (ii) the secondary antibody was conjugated to 15-nm-diameter gold beads (EY Laboratories, San Mateo, CA). Labeled specimens were observed and photographed in a Philips 400T electron microscope operated at 80 keV. Bound gold beads were counted using electron microscope negatives that were digitized and displayed in Photoshop 6.0. Individual beads were assigned a vertex or nonvertex location by first identifying the most likely capsid vertex. Lines were then drawn along the two sides of the capsid image, and the lines intersected at the candidate vertex. A bead was assigned a vertex location if greater than one-half of its image was within the angle created by the two lines.
In vitro binding of UL25 to capsids.
Separate studies were performed to measure the binding of [35S]Met-labeled UL25 and UL25 present in insect cell lysates. In both studies, binding was performed with capsids lacking UL25 which were prepared from Vero cells infected as described above with the UL25 null mutant virus KUL25NS.
Synthesis of [35S]methionine-labeled UL25 was carried out in a rabbit reticulocyte lysate using a coupled transcription and translation system (TNT T7Quick; Promega) according to the manufacturer's instructions using [35S]Met (1,000 mCi/mmol). A control plasmid expressing the luciferase gene was supplied with the kit. UL25 was expressed from the plasmid pGEM-UL25, which was constructed by digesting pAC-UL25 (15) with BamHI, purifying the UL25-containing fragment by gel electrophoresis, and ligating it into the BamHI site of the pGEM-4z vector (Promega). The [35S]methionine-labeled UL25 or luciferase (45 μl of the 50-μl in vitro translation reaction mixture) was mixed with 50 μl capsids (pooled A and B capsids) plus 400 μl PBS, and the reaction mixture was incubated for 90 min at room temperature with constant rotation. Capsids were isolated from the reaction mixture by centrifugation on a gradient of 20% to 50% sucrose in TNE (SW41 rotor, 24,000 rpm for 1 h at 4°C). After centrifugation, capsids were harvested from the gradient, precipitated by addition of an equal volume of 16% trichloroacetic acid, and analyzed by SDS-PAGE. Gels were stained with Coomassie blue and dried onto Whatman 3 M filter paper prior to autoradiography.
Binding of UL25 from insect cells was carried out beginning with Sf9 cells infected with BAC-UL25. Infected cells were suspended in 2 volumes of PBS and lysed by three cycles of freezing and thawing. Insoluble material was removed from the lysate by centrifugation at 30,000 rpm in a Beckman TL-100 ultracentrifuge (TLA rotor) for 30 min at 4°C. The supernatant (25 μl) was added to 100 μl KUL25NS A capsids plus 125 μl TNE and incubated for 1 h at 20°C. After incubation, capsids were harvested by pelleting through a 35% sucrose cushion and isolated by centrifugation on a 600-μl sucrose gradient as described above. Capsid bands were recovered by bottom puncture of the tube and analyzed by SDS-PAGE and immunoblotting. Primary antibodies were as follows: UL25, MAb 2D9 (diluted 1:2,000) (15); BSA, a mixture of MAbs 1N-11, 2-11, 3C-16, and 3N-5 (culture supernatants; diluted 1:500; gift from D. Benjamin, University of Virginia). Analysis of the Sf9 cell supernatant by sucrose density gradient centrifugation indicated that UL25 sedimented almost coincidently with BSA (MW, 69,293), suggesting that UL25 (MW, 62,666) is most probably a monomer in the Sf9 soluble fraction (W. W. Newcomb, unpublished observation).
RESULTS
Trypsin treatment of capsids.
The sensitivity of capsid-associated UL25 to digestion with trypsin was tested by treating HSV-1 B capsids in solution with three different concentrations of trypsin, 20 μg/ml, 2 μg/ml, and 0.2 μg/ml. After incubation, capsids were recovered by sucrose density gradient centrifugation and examined by Western blotting for evidence of UL25 digestion. The results showed extensive UL25 cleavage at all three trypsin concentrations tested (Fig. 1, UL25 panel). Intact UL25 was not detected in any of the three incubations, and digestion was particularly extensive at the two highest trypsin concentrations (lanes 2 to 4). Little evidence of digestion was observed if trypsin was omitted from reaction mixtures (lanes 1 and 6).
FIG. 1.
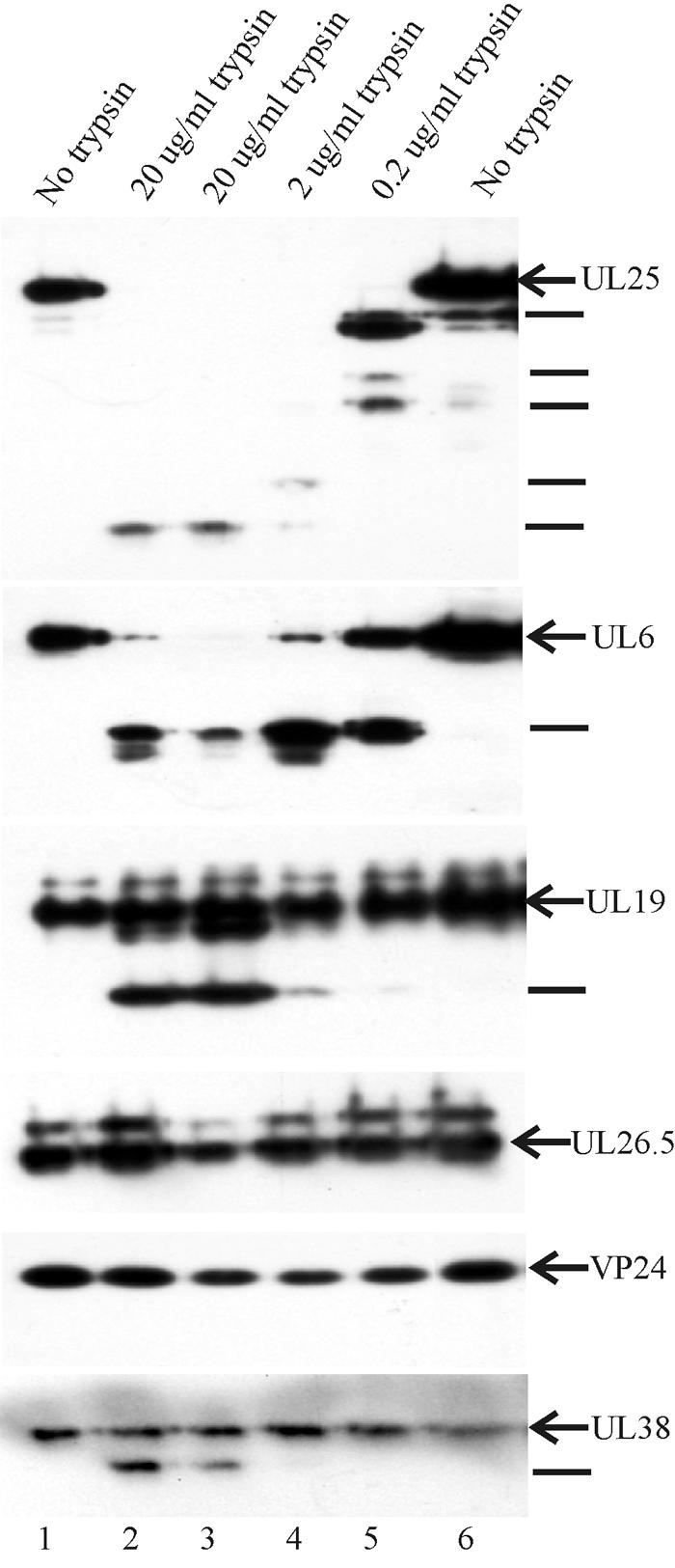
Immunoblot showing the extent to which UL25 and other HSV-1 capsid proteins are cleaved following treatment of B capsids with trypsin. Capsids were treated in vitro with the trypsin doses indicated at the top, purified, and subjected to SDS-PAGE followed by immunoblotting. The same blot was probed sequentially for all six proteins indicated at the right. Lines at the right indicate the positions of individual digestion products. Note evidence of digestion in the case of UL25, UL6, UL19, and UL38, but not UL26.5 or VP24.
Control experiments were performed with three proteins exposed on the capsid surface (UL6, UL19, and UL38) and two proteins (UL26.5 and VP24) expected to be protected inside the capsid shell (8, 35). The results showed significant cleavage of UL6, UL19, and UL38, with UL6 digestion the most extensive (Fig. 1, lanes 2 to 5). In contrast, cleavage was not detected with UL26.5 or VP24, as expected for these internal capsid proteins. Capsid-free UL26.5 and VP24 were both found to be sensitive to trypsin digestion in vitro (data not shown).
Immunoelectron microscopy.
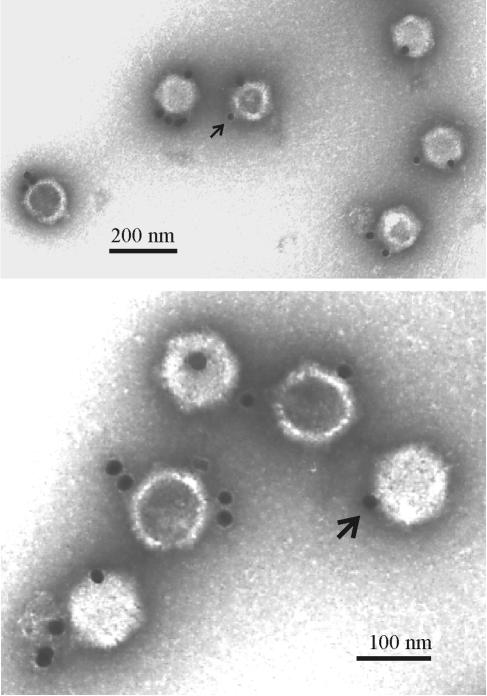
The location of UL25 was examined by immunoelectron microscopy of A, B, and C capsids. Capsids were incubated with polyclonal anti-UL25 serum followed by a gold bead-conjugated secondary antibody. Preparations were then examined by electron microscopy to identify the location of the gold bead.
The results showed that C capsids were well labeled (Fig. 2 and Table 1). At least one gold bead was observed on 74% of C capsids under conditions where 5% or fewer capsids were labeled in control experiments performed with nonspecific serum or with capsids lacking UL25 protein (Table 1). Compared to C capsids, a lower level of labeling was observed with A capsids (35%) and B capsids (23%). We suggest the proportions labeled may reflect the relative amount of UL25 present, which is highest in C capsids and lowest in B capsids (11, 25, 40). UL25 localization was determined after treatment of C capsids with DNase I to test whether antibody may be bound to UL25 associated with DNA protruding out of the capsid. The results showed little effect of DNase I treatment on the proportion of C capsids labeled (Table 1).
FIG. 2.
Electron micrographs showing HSV-1 C capsids after staining with antibody specific for UL25 followed by an antiantibody conjugated to gold beads. Note that most gold beads are found at capsid vertices (one is indicated by an arrow in each micrograph) and several capsids have label at more than one site.
TABLE 1.
Immunoelectron-microscopic localization of UL25 on the surface of the HSV-1 capsid
| Capsid typec | UL25-specific antiserumb
|
Control antiserum
|
||||
|---|---|---|---|---|---|---|
| Total capsids counted | No. of labeled capsidsa | % | Total capsids counted | No. of labeled capsidsa | % | |
| C | 226 | 167 | 74 | 190 | 10 | 5 |
| C + DNasee | 412 | 303 | 74 | 190 | 10 | 5 |
| B | 261 | 61 | 23 | 194 | 6 | 3 |
| A | 298 | 104 | 35 | 191 | 9 | 5 |
| A UL25 nulld | 362 | 5 | 2 | NDf | ND | ND |
Capsids containing one or more bound gold beads.
Polyclonal antiserum raised in rabbits (15).
Capsids were isolated from the nuclei of infected cells as described in Materials and Methods.
KUL25NS (15).
Capsids were treated with 20 μg/ml DNase I as described in Materials and Methods.
ND, not determined.
Capsids containing more than a single gold bead were observed with all three capsid types tested (Fig. 2 and Table 2). The proportion was highest in the case of C capsids, where 52% had more than one bound gold bead. The comparable proportions in A capsids and B capsids were 16% and 11%, respectively. Treatment of C capsids with DNase I had only a modest effect on the number of multiply labeled capsids (Table 2).
TABLE 2.
Immunoelectron-microscopic localization of UL25a
| Capsid type | Total capsids counted | No. of capsids (%) with indicated no. of bound gold beads
|
||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 or more | ||
| C | 226 | 59 (26) | 49 (22) | 59 (26) | 25 (11) | 20 (9) | 9 (4) | 6 (3) |
| C + DNaseb | 412 | 109 (26) | 130 (32) | 82 (20) | 53 (13) | 24 (6) | 8 (2) | 9 (2) |
| B | 261 | 200 (77) | 33 (13) | 15 (6) | 9 (3) | 4 (2) | 0 | 0 |
| A | 298 | 194 (65) | 60 (20) | 27 (9) | 14 (5) | 3 (1) | 3 (1) | 0 |
Capsids were isolated from 17MP-infected BHK-21 cells and stained with a UL25-specific polyclonal antiserum (15), followed by a gold-labeled secondary antibody as described in Materials and Methods.
Capsids were treated with 20 μg/ml DNase I.
Initial inspection of labeled capsids suggested that a high proportion of gold beads were located at or near a capsid vertex (Fig. 2). To put this observation on a quantitative basis, individual gold beads were classified as bound at a vertex or bound elsewhere on the capsid. Beads bound to labeled B and C capsids were classified, and two observers made independent counts. The results showed that 71% of beads were bound at a vertex and 29% elsewhere in C capsids (Table 3). For B capsids the percentages were 64% (vertex) and 36% (nonvertex).
TABLE 3.
Immunoelectron-microscopic localization of UL25: location of specifically bound gold beadsa
| Bead location | No of beads (%) with the indicated location in:
|
|
|---|---|---|
| C capsidsb | B capsids | |
| Vertex | 184 (71) | 53 (64) |
| Nonvertex | 76 (29) | 30 (36) |
| Total | 260 (100) | 83 (100) |
Capsids were labeled with anti-UL25 polyclonal antibody followed by gold-labeled secondary antibody as described in Materials and Methods. Beads were assigned a vertex or nonvertex location based on analysis of electron micrographs (such as Fig. 2) of stained capsids.
Capsids were prepared from the nuclei of 17MP-infected BHK-21 cells as described in Materials and Methods.
Effect of penton removal.
A biochemical study was carried out to test the idea that all or most of the UL25 is located at capsid vertices. The UL25 content in B capsids was compared with that in B capsids from which the pentons were removed by extraction with 2 M GuHCl. Treatment of B capsids in this way results in capsids, called G capsids, that are lacking all 12 pentons, the adjacent triplexes, and all of the scaffolding protein (23). The relative amount of UL25 present in B capsids compared to G capsids was measured by immunoblotting beginning with comparable amounts of the two capsid types. The results (Fig. 3) showed a substantial loss of UL25 as a result of GuHCl extraction. Quantitative analysis showed that the amount of UL25 remaining was between 3% and 4% in two experiments.
FIG. 3.
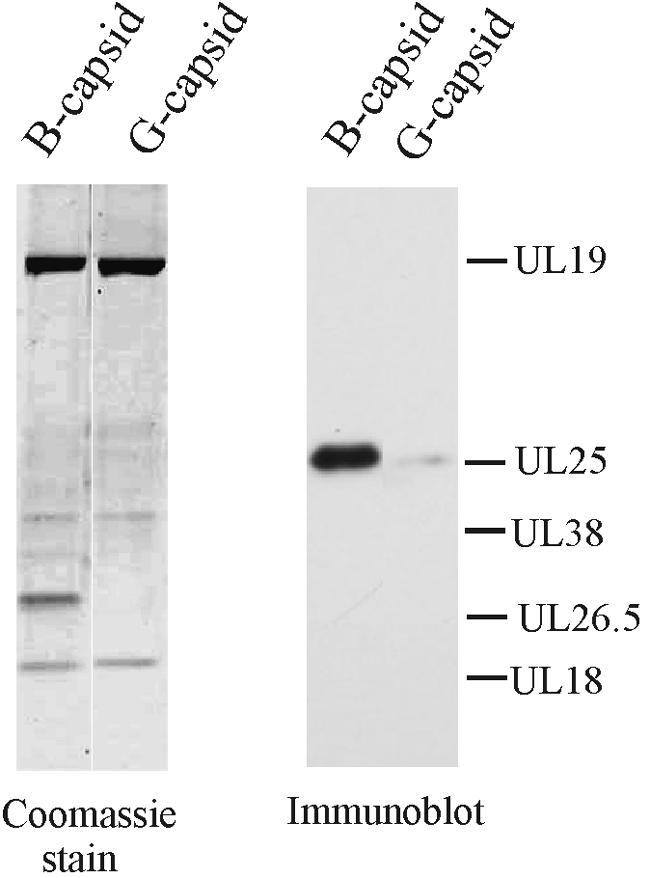
Immunoblot analysis of UL25 in B capsids and in B capsids after treatment with 2 M guanidine-HCl (G capsids). Sister SDS-PAGE gels were stained with Coomassie blue (left) and with antibody specific for UL25 (right). Note that nearly all UL25 is removed by exposure to guanidine-HCl, a procedure that causes removal of capsid pentons (23).
Capsid UL25 copy number.
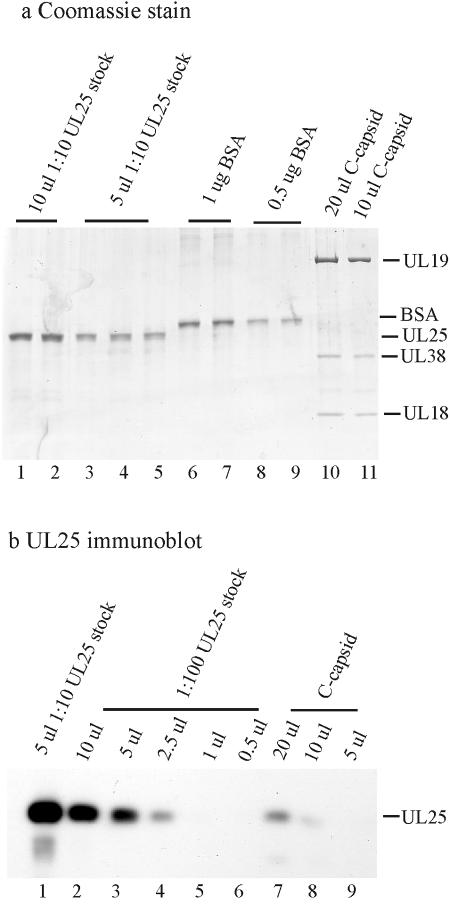
A quantitative immunoblotting method was employed to determine the UL25 copy number in A, B, and C capsids. The method depends on the observation that some UL25 protein is found in insoluble inclusion bodies when it is synthesized in Sf9 cells infected with a recombinant baculovirus containing the UL25 gene (15). By purifying inclusion bodies, we found that a highly enriched preparation of UL25 could be isolated (Fig. 4a, lanes 1 to 5). The protein concentration of such a preparation was estimated with reference to a BSA standard (Fig. 4a, lanes 6 to 9), and the UL25 stock was diluted for immunoblotting.
FIG. 4.
SDS-polyacrylamide gel electrophoresis (a) and immunoblot analyses (b) used to estimate the UL25 copy number in HSV-1 C capsids. Similar experiments were done with A and B capsids. Known amounts of BSA (a, lanes 6 to 9) were used as standards to estimate the protein concentrations of a UL25 stock solution (a, lanes 1 to 5) and HSV-1 C capsids (a, lanes 10 and 11). The immunoblot signals from known UL25 concentrations (b, lanes 1 to 6) were then used to measure the amount of UL25 present in C capsids (b, lanes 7 to 9).
The concentrations of capsid preparations were also determined with respect to the BSA standard (Fig. 4a, lanes 10 and 11), and capsid preparations were analyzed on the same immunoblot used for UL25 standards. The signal from UL25 dilutions was used to create a standard curve (Fig. 4b, lanes 1 to 6) from which the capsid UL25 signal could be interpreted (Fig. 4b, lanes 7 to 9; C capsids). Two independent determinations were made. Average values were found to be 56, 20, and 75 copies per capsid for A, B, and C capsids, respectively (Table 4). The results are in reasonably good agreement with a previous determination of 42 ± 17 for the UL25 copy number in B capsids (25) and with previous analyses indicating that the UL25 content is higher in C capsids than in A capsids and higher in A capsids than in B capsids (11, 25, 40, 41).
TABLE 4.
Quantitative immunoblot analysis of UL25 copy number per capsid
| Capsid typea | UL25 copies per capsid in:
|
||
|---|---|---|---|
| Expt 1 | Expt 2 | Avg | |
| A | 46 | 66 | 56 |
| B | 9 | 31 | 20 |
| C | 85 | 65 | 75 |
Capsids were prepared from BHK-21 cells infected with the 17MP strain of HSV-1 as described in Materials and Methods. Previously described methods were used for SDS-PAGE, blotting, and quantitation of blots (20).
Binding of UL25 to capsids in vitro.
Observations indicating that UL25 is exposed on the capsid surface (e.g., Fig. 1) suggested the possibility that externally added UL25 might bind specifically to capsids that lack it. This idea was tested by examining the ability of soluble UL25 to bind in vitro to UL25-negative capsids. The experiment was done in two ways. In one, the source of UL25 was [35S]methionine-labeled UL25 synthesized in a coupled in vitro transcription-translation system. In the other, UL25 was derived from the soluble fraction of Sf9 cells expressing UL25 after infection with a recombinant baculovirus encoding UL25.
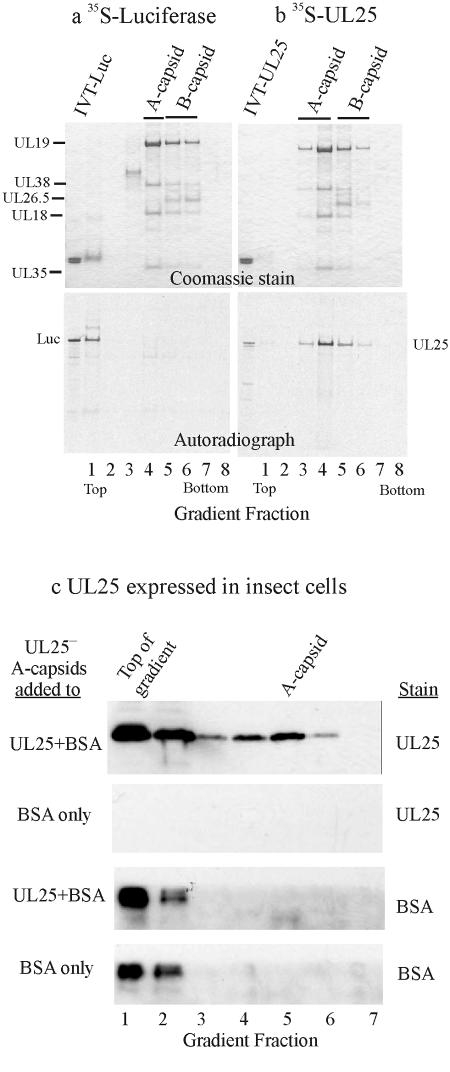
[35S]Met-labeled UL25 was synthesized in a commercial transcription-translation system derived from rabbit reticulocytes and programmed with a plasmid encoding UL25 (pGEM-UL25; see Materials and Methods). A control experiment demonstrated that [35S]Met-labeled UL25 was synthesized in the in vitro system (Fig. 5b, IVT-UL25 lane). An aliquot of the reaction mixture was added to pooled A and B capsids derived from the nuclei of cells infected with the UL25 null mutant, KUL25NS. After incubation to allow UL25 to attach, the capsids were separated from other reaction components by centrifugation on a sucrose gradient, and gradient fractions were analyzed by SDS-PAGE followed by autoradiography. The results showed that UL25 was present in both A and B capsid fractions but absent from other fractions, suggesting it was bound to capsids (Fig. 5b). In contrast, no binding was observed with a control protein, [35S]Met-labeled luciferase, as shown in Fig. 5a.
FIG. 5.
Analysis of UL25-negative capsids after exposure to UL25 in vitro. Pooled KUL25NS A and B capsids were allowed to bind [35S]Met-labeled UL25 and then purified by sucrose gradient centrifugation. Gradient fractions were then analyzed by SDS-PAGE followed by Coomassie staining (a and b, top panels) and autoradiography (bottom panels). Note that both A and B capsids bound UL25 (b) whereas neither bound the control protein luciferase (Luc) (a). IVT, in vitro transcription-translation. (c) Results obtained when KUL25NS A capsids were allowed to bind UL25 present in insect cell extracts. After incubation to permit binding to occur, capsids were purified by sucrose gradient centrifugation. Gradient fractions were then analyzed by SDS-PAGE followed by immunoblotting for UL25 (top panels) or BSA (bottom panels). All three analytical procedures were performed with the same blot. Note that capsids bound UL25 but not the control protein BSA.
UL25 protein synthesized in insect cells was presented to capsids in the form of a supernatant fraction prepared by cell lysis followed by high-speed centrifugation. Such supernatants, to which BSA was added as a control protein, were incubated with KUL25NS A capsids. After incubation, the reaction mixture was centrifuged on a sucrose gradient and the fractions analyzed by SDS-PAGE followed by immunoblotting. The results indicated that UL25 bound to A capsids since the UL25 peak coincided with the capsid peak (Fig. 5c, lanes 4 and 5). A control protein, BSA, did not bind to capsids when added either with the UL25-containing extract or separately (Fig. 5c).
Two further experiments were performed to examine the resemblance between UL25 bound to capsids in vitro and that found in capsids isolated from infected cells. In the first, we compared the amount of UL25 bound to A capsids in vitro with the amount found in capsids derived from infected cells. In the second, we used immunoelectron microscopy to determine the location of UL25 added to capsids in vitro. Both studies were done with UL25 derived from the soluble fraction of Sf9 cells expressing UL25 as described above.
UL25 binding was accomplished by mixing KUL25NS A capsids with the UL25-containing Sf9 cell soluble fraction and with dilutions of the soluble fraction. The amount of UL25 bound was determined after isolating capsids by sucrose gradient centrifugation and SDS-PAGE followed by immunoblotting as described above. The results showed that the amount of UL25 bound was dependent on the amount of extract added over a concentration range of 100-fold or greater (Fig. 6). There was evidence for saturation of binding sites at or near the concentration of the undiluted soluble fraction. The maximum binding level corresponded to a level approximately fourfold lower than that observed in A capsids derived from cells infected with wild-type HSV-1 (bar in Fig. 6).
FIG. 6.
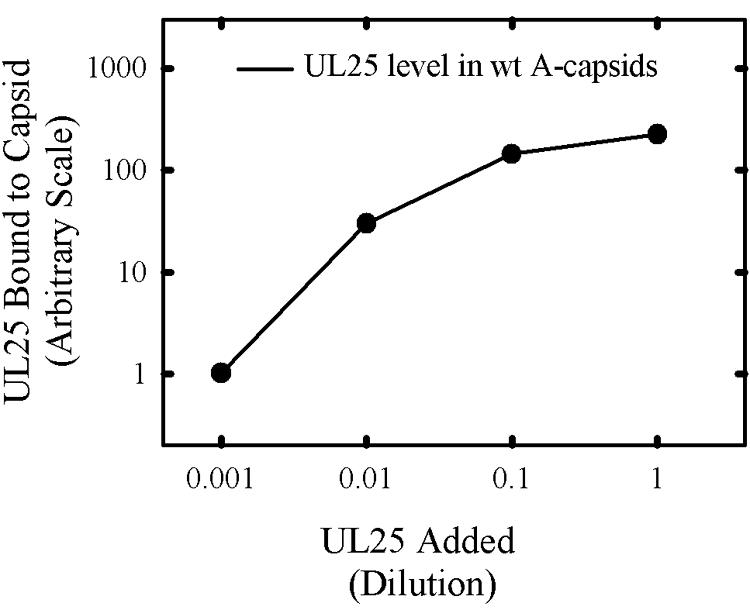
Effect of UL25 concentration on the amount bound to UL25-negative capsids. UL25 synthesized in insect cells was added to KUL25NS capsids and incubated as described in Materials and Methods. Capsids were then isolated by sucrose gradient centrifugation, and the amount of UL25 present was determined by immunoblotting. The amount of UL25 present in wild-type (wt) 17MP A capsids was estimated from capsids isolated in a companion sucrose gradient and analyzed on the same blot.
Immunoelectron microscopy of capsids containing UL25 bound in vitro was carried out using the same methods described above for capsids derived from infections with wild-type virus. Counts of stained capsids showed that 29% had one or more bound gold beads (Table 5). Control experiments performed with capsids incubated without UL25 showed 4% labeling. The location of UL25 on the capsid was determined as described above by analyzing images of labeled capsids. Of 186 gold beads classified in this way, 140 (75%) were bound at a capsid vertex while the remainder were found at other locations. This result is in reasonable agreement with the proportion of vertex-associated beads found in C capsids (71%) and B capsids (64%; Table 3), and it supports the view that most UL25 is bound at or near the capsid vertices.
TABLE 5.
Immunoelectron microscopy of UL25− A capsids after addition of UL25 protein in vitroa
| UL25 added | No. of capsids with UL25 label present (%) | Total capsids counted |
|---|---|---|
| Yes | 249 (29) | 863 |
| No | 17 (4) | 417 |
Experiments were performed with A capsids isolated from the nuclei of Vero cells infected with the KUL25 NS strain (UL25−) of HSV-1. The Materials and Methods section describes the procedures by which capsids were incubated with UL25-containing Sf9 cell extracts in vitro, stained with UL25-specific antibody followed by a gold bead-labeled secondary antibody, and visualized by electron microscopy. Data entries show the number of capsids containing one or more bound gold beads.
DISCUSSION
Packaging DNA into the HSV-1 capsid is thought to involve the procapsid, an enzyme called terminase, and the concatemeric viral DNA produced by the DNA replication machinery (4). Encapsidation begins when terminase cleaves the DNA concatemer at a pac site and docks a DNA end onto the procapsid by way of the portal complex. DNA is then translocated through the portal channel and into the capsid in a process that requires energy derived from ATP hydrolysis. The terminase is thought to play a key role in DNA translocation, and the portal may also be actively involved. As DNA enters, the scaffolding protein exits the capsid cavity and the shell is transformed from the spherical morphology characteristic of the procapsid to the icosahedral structure of the mature capsid. Once a complete DNA genome has entered, the concatemer is cleaved at a second pac site and the portal channel is sealed to prevent DNA from exiting the capsid.
As described above, UL25 is thought to function late in the packaging process to retain DNA inside the capsid. It may, for instance, be involved in sealing the portal channel or in stabilizing the capsid against the internal pressure created by the packaged DNA. Functioning of UL25 late in the DNA injection process suggests that (i) it is likely to be located on the outside of the capsid and not inside with the mass of packaged DNA and (ii) it might be able to attach in vitro to capsids that lack it. Studies described here were carried out to test both expectations.
Location of UL25 in the capsid.
The location of UL25 was examined by determining its sensitivity to digestion when capsids were treated with trypsin, by immunoelectron microscopy, and by biochemical analysis of capsids from which the pentons have been removed. Experiments involving trypsin treatment of B capsids supported the view that UL25 is exposed on the capsid surface. It was cleaved under conditions where the internal scaffolding and protease proteins were unaffected (Fig. 1). In fact, of the capsid proteins tested UL25 was found to be the most sensitive to trypsin digestion. For instance, in contrast to the other proteins examined, intact UL25 was not detected after treatment of capsids with any of three trypsin concentrations tested. The absence of an undigested population supports the view that all UL25 molecules are exposed on the capsid surface.
Immunoelectron-microscopic studies were carried out to define the location of UL25 more precisely. The most revealing observations were as follows. (i) UL25 label was found at multiple distinct sites in C, B, and A capsids (Table 2). Individual capsids were observed with up to 10 gold beads, while labeling at multiple sites was not observed with control capsids lacking UL25. (ii) Most UL25 was found at the vertices in the two capsid types tested, B and C capsids (Table 3), in agreement with immunoelectron-microscopic findings of Ogasawara and colleagues, who also observed UL25 selectively located at the capsid vertices (25).
The immunoelectron-microscopic results reported here and those of earlier investigators (25, 41) support the view that UL25 is located at multiple sites on the capsid surface. The large number of multiply labeled capsids observed (52%, 11%, and 16% in C, B, and A capsids, respectively; Table 2) would not be expected if all UL25 molecules were located at a single site. Since 11 of the 12 capsid vertices are compositionally and geometrically identical, we suggest these may be the primary sites of UL25 protein. The compositionally distinct portal vertex may also contain UL25. As each vertex has fivefold symmetry, it is likely that the number of UL25 molecules present at each is five or a multiple of five. The experimental value for the UL25 copy number in C capsids (average = 75; Table 4) supports the view that five UL25 molecules are found at each vertex. Such a structure would imply a UL25 copy number of 55 (or 60 if UL25 is also present at the portal vertex). In contrast, a copy number of 110 would be expected if there were 10 UL25 molecules at each of the 11 compositionally identical vertices.
Label observed at nonvertex sites in immunoelectron-microscopic experiments suggests there may be a nonvertex population of UL25 molecules in the HSV-1 capsid. However, apparent nonvertex labeling may also result from the experimental variables involved in distinguishing vertex from nonvertex gold beads in capsid images such as those shown in Fig. 2. A gold bead attached at a vertex may be separated by the length of two antibody molecules and appear to be bound at a capsid face or edge. Tethering may also be to a vertex obscured by the capsid. We favor the view that all UL25 is associated with capsid vertices and that apparent nonvertex label results from factors such as those described above that cause beads to appear to be bound at nonvertex sites.
A vertex location for UL25 is also supported by the results of studies with capsids from which the pentons have been removed by extraction with 2 M GuHCl (i.e., G capsids). Removal of the pentons was accompanied by removal of the greatest part of UL25 (Fig. 3). This experiment admits the possibility that both vertex and nonvertex UL25 may be removed by GuHCl treatment, but it is also the result expected if all UL25 is found at capsid vertices. We interpret the small amount of unextractable UL25 (3% to 4%) to be an experimental background rather than evidence for a distinct population of UL25 molecules.
Location of UL25 at the capsid vertices is consistent with its proposed function to stabilize the capsid against pressure produced by the packaged DNA (15, 36). Previous studies have identified the vertices as the capsid sites most vulnerable to disruption by proteolytic digestion or by chaotropic agents such as guanidine-HCl (18, 23, 34). It is reasonable, therefore, that the vertices may be the sites most in need of stabilization against internal pressure. We suggest that UL25 be called a vertex reinforcement protein to emphasize its role in capsid stabilization.
The proposed addition of UL25 to the capsid at or near the end of the packaging process suggests it may function similarly to bacteriophage proteins that are added late. The T4 phage soc and hoc proteins are examples of such phage-encoded proteins. These bind to the capsid surface after DNA has entered and are thought to reinforce the capsid structure (1, 10). The location of UL25 at multiple distinct sites in the capsid suggests a similarity to soc, hoc, and other similar proteins, such as the phage lambda D protein (9) and L phage Dec protein (7), that are also located at multiple sites. P22 phage tail accessory proteins and lambda head completion proteins would appear to be distinct from UL25, as they are located specifically at the portal vertex (17, 37).
Measurements of the UL25 copy numbers in A, B, and C capsids as shown in Table 4 are in agreement with previous studies showing that the UL25 content is greatest in C capsids (11, 25, 32, 40, 41). The results add absolute copy numbers for A and C capsids to the previous determination for B capsids (25). The higher level of UL25 in C capsids is consistent with the view that UL25 is added incrementally as DNA is packaged or perhaps as packaging is completed, as suggested by Stow (36).
Binding of UL25 to capsids in vitro.
Experiments shown in Fig. 5 demonstrate that UL25 protein can be added in vitro to capsids that lack it. Soluble UL25 synthesized in a cell extract or in insect cells was found to bind KUL25NS capsids under conditions where control proteins did not. Immunoelectron-microscopic studies performed with the resulting capsids showed that most of the bound UL25 is located at capsid vertices as it is in capsids isolated from infected cells (see Results). It was of interest that the amount of UL25 bound to capsids in vitro was small, comparable to the level present in B capsids. We interpret this result to be consistent with the idea that UL25 is added incrementally as DNA enters the capsid. Since no DNA is present in the capsids used to measure UL25 attachment in vitro, it is expected that the amount of UL25 binding would be minimal.
It should be noted that studies by Stow demonstrated that the UL25 null mutant was less impaired in stably packaging HSV-1 amplicon DNA than in packaging its own genome (36). Interestingly, the amplicon genomes that were packaged were all significantly smaller (<100 kbp) than the full-length HSV-1 genome and were found to be retained in the nucleus. As suggested by Stow, one of the functions of UL25 may be in the translocation of the capsids out of the nucleus by the interaction of UL25 with the transport machinery. This function may require the presence of a critical number of UL25 molecules bound to the capsid shell, a level that would be reached only when a complete genome is packaged.
The proposed role of UL25 to stabilize DNA inside the capsid makes it attractive to speculate that UL25 may also be involved in uncoating the genome at the beginning of a new infection. Loss or degradation of UL25, for instance, may cause the capsid vertices to become destabilized and DNA to be extruded through one or more of them. In support of this view, we note that DNA is lost specifically through the vertices when C capsids are treated in vitro with 0.5 M guanidine-HCl (19). In this context, it is of interest that capsid-associated UL25 is found to be highly susceptible to proteolytic digestion (Fig. 1). Cleavage of UL25 therefore has the potential to serve as an initiating event if loss of UL25 is functionally involved in DNA uncoating.
Acknowledgments
We gratefully acknowledge David Radoff, Oneida Mason, Evonne Johnson, and Jamie Huffman for help with aspects of this study.
The work was supported by NIH awards AI041644-09 (J.C.B.) and AI060836 (F.L.H.).
REFERENCES
- 1.Aebi, A., R. van Driel, R. K. L. Bijlenga, B. ten Heggeler, R. van der Broek, A. C. Steven, and P. R. Smith. 1977. Capsid fine structure of T-even bacteriophages. Binding and localization of two dispensable capsid proteins into the P23 surface lattice. J. Mol. Biol. 110:687-698. [DOI] [PubMed] [Google Scholar]
- 2.Al-Kobaisi, M. F., F. J. Rixon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology 180:380-388. [DOI] [PubMed] [Google Scholar]
- 3.Bowman, B. R., R. L. Welschhaus, H. Jayaram, N. D. Stow, V. G. Preston, and F. A. Quiocho. 2006. Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1. J. Virol. 80:2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, J. C., M. A. McVoy, and F. L. Homa. 2002. Packaging DNA into herpesvirus capsids, p. 111-153. In A. Holzenburg and E. Bogner (ed.), Structure-function relationships of human pathogenic viruses. Kluwer Academic/Plenum Publishers, London, United Kingdom.
- 5.Chang, Y. E., A. P. Poon, and B. Roizman. 1996. Properties of the protein encoded by the UL32 open reading frame of herpes simplex virus type 1. J. Virol. 70:3938-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davison, A. J. 2002. Evolution of the herpesviruses. Vet. Microbiol. 86:69-88. [DOI] [PubMed] [Google Scholar]
- 7.Gilcrease, E. B., D. A. Winn-Stapley, F. C. Hewitt, L. Joss, and S. R. Casjens. 2005. Nucleotide sequence of the head assembly gene cluster of bacteriophage L and decoration protein characterization. J. Bacteriol. 187:2050-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 9.Imber, R., A. Tsugita, M. Wurtz, and T. Hohn. 1980. The outer surface protein of bacteriophage lambda. J. Mol. Biol. 139:277-295. [DOI] [PubMed] [Google Scholar]
- 10.Ishii, T., Y. Yamaguchi, and M. Yanagida. 1978. Binding of the structural protein soc to the head shell of bacteriophage T4. J. Mol. Biol. 120:533-544. [DOI] [PubMed] [Google Scholar]
- 11.Kaelin, K., S. Dezelee, M. J. Masse, F. Bras, and A. Flamand. 2000. The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and to microtubules. J. Virol. 74:474-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanamaru, S., K. Kondabagil, M. G. Rossmann, and V. B. Rao. 2004. The functional domains of bacteriophage t4 terminase. J. Biol. Chem. 279:40795-40801. [DOI] [PubMed] [Google Scholar]
- 13.Lamberti, C., and S. K. Weller. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226:403-407. [DOI] [PubMed] [Google Scholar]
- 14.Maluf, N. K., Q. Yang, and C. E. Catalano. 2005. Self-association properties of the bacteriophage lambda terminase holoenzyme: implications for the DNA packaging motor. J. Mol. Biol. 347:523-542. [DOI] [PubMed] [Google Scholar]
- 15.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell, M. S., and V. B. Rao. 2004. Novel and deviant Walker A ATP-binding motifs in bacteriophage large terminase-DNA packaging proteins. Virology 321:217-221. [DOI] [PubMed] [Google Scholar]
- 17.Murialdo, H., X. Xing, D. Tzamtzis, A. Haddad, and M. Gold. 2003. The product of the bacteriophage lambda W gene: purification and properties. Biochem. Cell Biol. 81:307-315. [DOI] [PubMed] [Google Scholar]
- 18.Newcomb, W. W., and J. C. Brown. 1991. Structure of the herpes simplex virus capsid: effects of extraction with guanidine-HCl and partial reconstitution of extracted capsids. J. Virol. 65:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newcomb, W. W., and J. C. Brown. 1994. Induced extrusion of DNA from the capsid of herpes simplex virus type 1. J. Virol. 68:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newcomb, W. W., F. L. Homa, and J. C. Brown. 2005. Involvement of the portal at an early step in herpes simplex virus capsid assembly. J. Virol. 79:10540-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid assembly. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 22.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 24.Newcomb, W. W., B. L. Trus, N. Cheng, A. C. Steven, A. K. Sheaffer, D. J. Tenney, S. K. Weller, and J. C. Brown. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 74:1663-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 75:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orlova, E. V., B. Gowen, A. Droge, A. Stiege, F. Weise, R. Lurz, M. van Heel, and P. Tavares. 2003. Structure of a viral DNA gatekeeper at 10 Å resolution by cryo-electron microscopy. EMBO J. 22:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel, A. H., F. J. Rixon, C. Cunningham, and A. J. Davison. 1996. Isolation and characterization of a herpes simplex virus type-1 mutant defective in the UL6 gene. Virology 217:111-123. [DOI] [PubMed] [Google Scholar]
- 28.Poon, A. P. W., and B. Roizman. 1993. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J. Virol. 67:4497-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rixon, F. J. 1993. Structure and assembly of herpesviruses. Semin. Virol. 4:135-144. [Google Scholar]
- 30.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheaffer, A. K., W. W. Newcomb, J. C. Brown, M. Gao, S. K. Weller, and D. J. Tenney. 2000. Evidence for controlled incorporation of herpes simplex virus type 1 UL26 protease into capsids. J. Virol. 74:6838-6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson, A. A., Y. Tao, P. G. Leiman, M. O. Badasso, Y. He, P. J. Jardine, N. H. Olson, M. C. Morais, S. Grimes, D. L. Anderson, T. S. Baker, and M. G. Rossmann. 2000. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steven, A. C., C. R. Roberts, J. Hay, M. E. Bisher, T. Pun, and B. L. Trus. 1986. Hexavalent capsomers of herpes simplex virus type 2: symmetry, shape, dimensions and oligomeric status. J. Virol. 57:578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steven, A. C., and P. G. Spear. 1996. Herpesvirus capsid assembly and envelopment, p. 312-351. In R. Burnett, W. Chiu, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 36.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang, L., W. R. Marion, G. Cingolani, P. E. Prevelige, and J. E. Johnson. 2005. Three-dimensional structure of the bacteriophage P22 tail machine. EMBO J. 24:2087-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tengelsen, L. A., N. E. Pederson, P. R. Shaver, M. W. Wathen, and F. L. Homa. 1993. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J. Virol. 67:3470-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomsen, D. R., L. L. Roof, and F. L. Homa. 1994. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J. Virol. 68:2442-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thurlow, J. K., F. J. Rixon, M. Murphy, P. Targett-Adams, M. Hughes, and V. G. Preston. 2005. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J. Virol. 79:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thurlow, J. K., M. Murphy, N. D. Stow, and V. G. Preston. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 80:2118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]