Abstract
Many acyclovir-resistant herpes simplex virus isolates from patients contain insertions or deletions in homopolymeric sequences in the thymidine kinase (TK) gene (tk). Viruses that have one (G8) or two (G9) base insertions in a run of seven G's (G string) synthesize low levels of active TK (TK-low phenotype), evidently via ribosomal frameshifting. These levels of TK can suffice to permit reactivation from latently infected mouse ganglia, but in a majority of ganglia, especially with the G9 virus, reactivation of virus that has reverted to the TK-positive phenotype predominates. To help address the relative contributions of translational mechanisms and reversion in reactivation, we generated viruses with a base either inserted or deleted just downstream of the G string. Both of these viruses had a TK-low phenotype similar to that of the G8 and G9 viruses but with less reversion. Both of these viruses reactivated from latently infected trigeminal ganglia, albeit inefficiently, and most viruses that reactivated had a uniformly TK-low phenotype. We also generated viruses that have one insertion in a run of six C's or one deletion in a run of five C's. These viruses lack measurable TK activity. However, they reactivated from latently infected ganglia, albeit inefficiently, with the reactivating viruses having reverted to the wild-type TK phenotype. Therefore, for G-string mutants, levels of active TK as low as 0.25% generated by translational mechanisms can suffice for reactivation, but reversion can also contribute. For viruses that lack TK activity due to mutations on other homopolymeric sequences, reactivation can occur via reversion.
Although acyclovir (ACV) is an effective antiviral agent, herpes simplex virus (HSV) that is resistant to treatment can be a serious clinical problem in immunocompromised patients. It has been estimated that ACV-resistant (ACVr) HSV appears in as many as 5% of this population upon treatment (4, 10). Clinically significant drug resistance implies evasion of drug action but retention of pathogenesis. The vast majority of mutations associated with clinical drug resistance occur in the viral thymidine kinase (TK) gene (tk), which encodes the enzyme that selectively activates ACV. The phenotypes of clinical isolates with mutant TKs typically fall into one of three categories: altered substrate specificity, low TK activity (TKL), or lack of measurable TK activity (TK−) (11, 12, 26). That viruses lacking TK activity are pathogenic has been puzzling. Although TK is not required for growth in cell culture, it is essential for pathogenesis in mouse models of HSV infection, in particular, for reactivation of laboratory strains of HSV-1 from latently infected mouse trigeminal ganglia (3, 6, 9, 21). Many of the mutations associated with the mutant TK phenotypes are insertions or deletions of bases in homopolymeric sequences in the gene (11, 12, 26).
The most common mutation in ACVr viruses is a single G insertion into a run of seven G's known as the “G string” (11, 12, 19, 26). Double G insertions into the G string are also frequently observed (11-13, 17). Viruses that have either of these genotypes have a TKL phenotype (14, 15, 19). The active TK from viruses with a single G insertion occurs via an unusual ribosomal frameshift on the G string that is solely dependent on the G-rich nature of the G string and is possibly the result of a direct interaction between the G string and the rRNA (18; A. Griffiths and D. M. Coen, unpublished results). Preliminary in vitro evidence suggests that −1 ribosomal frameshifting occurs via a similar mechanism on a G string with a double G insertion (A. Griffiths and D. M. Coen, unpublished results). We have shown previously that viruses with the single G insertion reactivated from latency in mouse trigeminal ganglia, and occasionally, the virus that reactivated was uniformly TKL (14). These studies showed that the low levels of TK generated from viruses with the single G insertion were sufficient to support reactivation but that most of the viruses that reactivated contained some virus with the wild-type TK phenotype (TK+), suggesting that reversion may also play a role in the reactivation of viruses with the G8 genotype (14). Indeed, a virus that is TK+ should have a strong growth advantage in the ganglion. Reversion was especially evident with viruses that carried double G insertions into the G string, with about 3% of plaques apparently TK+ (13, 15), and this is consistent with the genetic instability of a homopolymeric sequence increasing with its length (22). All virus populations that reactivated from ganglia latently infected with a G9 virus contained TK+ virus. It was therefore not possible to ascertain whether the TKL phenotype of the G9 virus, presumably generated by a net −1 ribosomal frameshift, was sufficient to support reactivation. The mixed TK phenotypes of these viruses draw an interesting parallel to those of clinical isolates; many, if not all, ACVr clinical isolates are comprised of mixtures of viruses with multiple TK phenotypes (11, 24).
While viruses carrying one of the three mutations listed above are among the most frequently observed in isolates from patients, mutations on other homopolymeric sequences in tk are also associated with drug-resistant HSV disease (11, 12). However, it is not known whether these viruses generate active TK or reactivate from latency.
In this paper, we address the contributions of ribosomal frameshifting and reversion toward the reactivation of mutants with altered sequences around the G string. Additionally, we have generated recombinant viruses that carry ACVr mutations on other homopolymeric sequences and analyzed their TK activities and reactivation from latency.
MATERIALS AND METHODS
Plasmid construction.
Plasmid pAG5 contains the entire BamHI P fragment from HSV-1 strain KOS in pBluescript SK(+) (Promega) (15). Plasmids engineered to carry the ACVr mutations were generated by site-directed mutagenesis using the QuikChange kit (Stratagene) according to the manufacturer's instructions. For each plasmid, two complementary oligonucleotides (Integrated DNA Technologies) were synthesized to facilitate mutagenesis; however, only the forward primers are listed here: pTKG7aC, GTTCTGGCTCCTCATATCGGGGGGGAGGCCTGGGAGCTCACATGC; pTKG7dG, GCCGTTCTGGCTCCTCATATCGGGGGGGAGCTGGGAGCTCAC; pTKC6+1C, GGGCAGCATGACCCCCCCAGGCCGTGCTGGCG; and pTK2C5−1C, GGCCCCCGAGTTGCTGGCCCCAACGGCGACCTGTATAACG. The presence of desired mutations and the absence of unwanted mutations were confirmed by sequencing of the tk gene.
Cells and viruses.
African green monkey (Vero) and TK− human osteosarcoma (143B) cells were obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium, supplemented with 10% bovine fetal serum, at 37°C and 5% CO2. Viruses that are not novel to this study are as follows: wild-type HSV-1 strain KOS, clinical isolate 615.9, and KOS-derived mutants KG111, LS-95/-85, LS-111/-101/-56/-46, 615.9, LS-29/-18, and tkLTRZ1 (1, 5-8, 20, 24).
Construction of recombinant viruses.
The method used to generate recombinant viruses has been described previously (15). Briefly, plasmid midi-prep DNA (Wizard Prep; Promega), virion “mini-prep” (7) tkLTRZ1 DNA, and transfection reagent (Effectene; QIAGEN) were added to 50% confluent Vero cells. The use of tkLTRZ1, a recombinant KOS strain that has an insertion in tk of the Moloney murine leukemia virus long terminal repeat (LTR) upstream of lacZ, permits blue/white screening for recombinant viruses. In addition, tkLTRZ1 does not reactivate from explanted mouse trigeminal ganglia (references 3, 14, 15, and 21 and this study). The entire BamHI “P” fragment of this virus has been sequenced and shown to be identical to that of KOS, except for the LTR-lacZ sequences (14). Following the appearance of sufficient cytopathic effect, the cells were harvested. Recombinants were cloned by limiting dilution using a blue/white screen in 96-well trays, requiring two rounds of screening until a single “white” plaque was observed in a well. Virus from this well was amplified, DNA was prepared, and the tk gene was sequenced to confirm the presence of the mutation. Two independently isolated recombinant viruses were generated from separate transfections with each mutant plasmid: TKG7aC.1 and TKG7aC.2, TKG7dG.1 and TKG7dG.2, TK6C+1C.1 and TK6C+1C.2, and TK2C5−1C.1 and TK2C5−1C.2.
Plaque autoradiography.
Quantitative plaque autoradiography was performed as described previously (15), using [3H]thymidine (Moravek) to radioactively label plaques. The assay was calibrated with viruses that generate known amounts of active TK polypeptide.
Assays of acute and latent infection in mice.
Male 8-week-old randomly bred CD-1 mice (Charles River Laboratories) were infected on scarified corneas with 2 × 106 PFU of strain KOS or 7 × 107 PFU of mutant, as described previously (5, 23). Acute viral replication was monitored by assaying virus in the tear film at 1 day postinfection (p.i.) and in ganglion homogenates at 3 days p.i. Reactivation from latently infected ganglia was measured by enzymatically dissociating ganglia harvested at 30 days p.i. and culturing on Vero cells (23). These cells were screened for 10 days for the appearance of cytopathic effect, replated, and screened for a further 7 days. Viruses that reactivated were amplified on Vero cells and subjected to plaque autoradiography. Additionally, viral DNA was prepared from these viruses and the tk genes were sequenced as previously described (14, 15).
RESULTS
Construction of viruses that have mutations immediately downstream of the G string.
We have shown previously that viruses with single G (G8) or double G (G9) insertions into the G string generated low levels of TK (15, 19) and that the low levels of TK expressed by the G8 virus can be sufficient to support reactivation from latency (14). However, viruses that had reverted to the wild-type TK phenotype were frequently observed following reactivation from latency with the G8 virus, and the frequency of reversion was so high with the G9 viruses studied by us and others that it made it impossible to determine whether the TKL phenotype contributed to reactivation (13-15). As it seemed likely that that the length of the G string affected its genetic stability, we hypothesized that reversion could be decreased while retaining a TKL phenotype by maintaining the G string at its wild-type length, seven G's. In vitro data suggested that the net +1 ribosomal frameshift functions on a wild-type G string (18). Two viruses were generated that each required a ribosomal frameshift to generate active TK while maintaining the wild-type length of the G string (independent isolates were generated for each mutation) (Fig. 1). Virus TKG7aC has a single C inserted immediately downstream of the G string, thereby requiring a net +1 ribosomal frameshift for the generation of active TK, as does the G8 virus. Virus TKG7dG has a single G deleted immediately downstream of the G string, thereby requiring a net −1 ribosomal frameshift for the generation of active TK. Although in vitro experiments demonstrate that the ribosomal frameshift occurs on the G string (18; A. Griffiths and D. M. Coen, unpublished results), as yet we do not know on which nucleotide the ribosomal frameshifts occur. Therefore, there are several possible amino acid sequences that could be synthesized following a frameshift, and these sequences are listed in Fig. 2. However, we considered that a small number of amino acid changes would be tolerated with minimal effect on enzyme activity as, based on the crystal structure of TK, the amino acids encoded by the G string are physically distant from the nucleoside and triphosphate binding sites (2, 27, 28). Also, it has been shown that viruses that have three nucleotides added to the G string (G10) and, therefore, an extra amino acid have wild-type TK activity (13, 15).
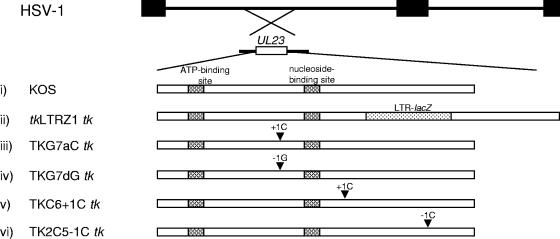
FIG. 1.
Structure of the tk genes of viruses used in this study. The top two lines represent the HSV genome and the location of the tk gene (UL23). Below are schematic diagrams of the tk genes of the viruses used in this study. Above the arrowheads are the mutations, and the arrowheads indicate the approximate position of the mutation in tk. (i) KOS (cross-hatched boxes, functional sites of TK); (ii) tkLTRZ1 (tk with LTR-lacZ inserted into the PstI site [dotted box]); (iii) TKG7aC (tk with a single C inserted immediately downstream of the G string of KOS tk); (iv) TKG7dG (tk with a single G deleted immediately downstream of the G string of KOS tk); (v) TKC6+1C (tk with a single C inserted into the C chord); (vi) TK2C5−1C (tk with a single C deleted in a run of five C's).
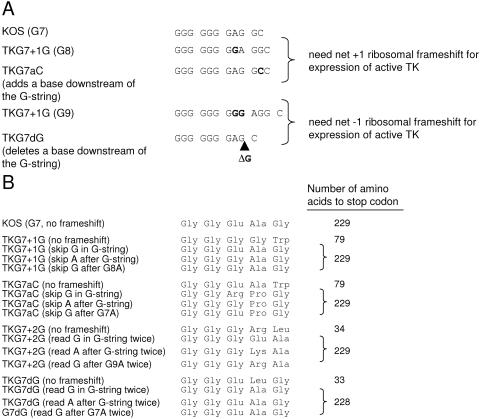
FIG. 2.
Sequences of viruses with mutations immediately downstream of the G string. (A) The first line shows the sequence of the G string in the wild-type virus. The nucleotides representing translated codons are indicated by spaces between the triplets. The second line shows the sequence in a virus that has a single G insertion in the G string, which requires a net +1 ribosomal frameshift for expression of active TK. The third line shows the sequence in a virus that has a single C added downstream of the G string, such that a net +1 ribosomal frameshift is required for expression of active TK. The fourth line shows the sequence in a virus that has a double G insertion in the G string, which requires a net −1 ribosomal frameshift for expression of active TK. The fifth line shows the sequence in a virus that has a single G deleted downstream of the G string, such that a net −1 ribosomal frameshift is required for expression of active TK. Inverted nucleotides are shown in boldface type. (B) Possible sequences of amino acids translated following ribosomal frameshifts on the G string. The left column lists the virus and a possible site of ribosomal frameshift. The middle column lists the amino acid sequence that would be translated. The right column shows the number of amino acids to the stop codon in the translated frame.
Viruses with mutations downstream of the G string generate low levels of active TK.
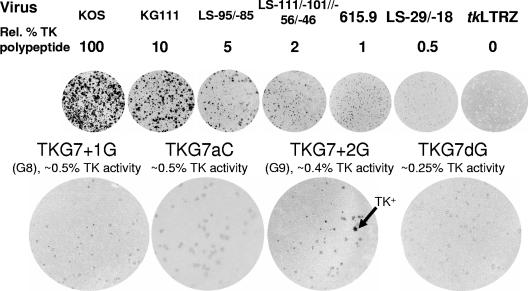
The levels of TK synthesized in cells infected with viruses TKG7aC and TKG7dG were measured using quantitative plaque autoradiography, calibrated using viruses that generate known levels of active TK (Fig. 3). Virus TKG7aC exhibited ∼0.5% of KOS TK activity (the previously reported G8 virus has ∼0.5% of activity [14]). No TK+ plaques were observed from >500 plaques examined. Virus TKG7dG exhibited ∼0.25% of KOS TK activity (most of the plaques from TK− cells infected with the previously reported G9 virus have ∼0.4% of activity [15]). No TK+ plaques were observed from >500 plaques examined. Thus, as anticipated, there was little reversion of these mutants containing a seven-nucleotide G string.
FIG. 3.
Quantitative plaque autoradiography of viruses. The top line shows the virus used. The line below shows the amount of active TK expressed by each mutant as a percentage of that expressed by wild-type strain KOS. Below, the images of the plates are presented. The next line shows the names of mutant viruses above the TK activities associated with these mutants (data for TKG7+1G have been published previously [14], and data for TKG7+2G have been published previously [15]). Below, the images of the plates are presented. Arrow, example of a plaque with the TK+ phenotype on the TKG7+2G plate. Rel., relative.
Replication of viruses TKG7aC and TKG7dG in mouse ganglia.
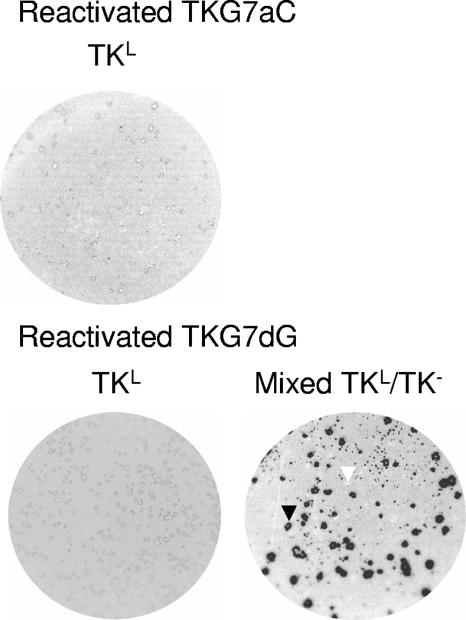
Separately, two independent isolates of each mutant virus were inoculated onto the scarified corneas of mice. Virus harvested in eye swabs was titrated at 1 day p.i., and few or no differences in replication were observed among the mutant viruses, KOS, and tkLTRZ1 (Table 1). At 3 days p.i., ganglia were harvested and titrated. Although robust replication of KOS was observed, virus was not detected in ganglia from animals infected with either TKG7aC or TKG7dG. Very little virus was detected in ganglia from animals infected with tkLTRZ1, which could be the result of low-level replication in the ganglia or contamination from the eye during the dissection of the ganglia (Table 1). To establish whether viruses TKG7aC and TKG7dG were able to reactivate from latency despite a frameshift mutation within tk, ganglia were harvested at 30 days postinoculation, enzymatically dissociated, and plated onto Vero cells that were monitored for the appearance of plaques. Two of 18 ganglia latently infected with TKG7aC.1 and 0 of 18 infected with TKG7aC.2 yielded virus (2/36 total), compared to all of those infected with KOS and none of those infected with tkLTRZ1 (Table 1). Both of the viruses that reactivated from ganglia infected with TKG7aC were amplified on Vero cells and analyzed by plaque autoradiography (Fig. 4). Both of these viruses appeared to be uniformly TKL. Sequencing confirmed that the G7aC mutation was maintained in the reactivated viruses.
TABLE 1.
Replication and reactivation of viruses with TKL phenotypes
| Virus | Inoculum (PFU) | Titer of virus in eye swabs at 1 day p.i. (mean PFU ± SE)b | Titer of virus in trigeminal ganglia at 3 days p.i. (mean PFU ± SE)b | No. of ganglia with reactivating virus/total no. of ganglia | TK phenotype(s) of reactivating virus |
|---|---|---|---|---|---|
| KOS | 2 × 106 | 4.9 × 105 ± 1.6 × 105 (4) | 1.1 × 104 (1) | 30/30 | TK+ |
| tkLTRZ1 | 7 × 107 | 1.8 × 106 ± 7.2 × 105 (4) | 2.7 ± 2.7 (3) | 0/34 | NDa |
| TKG7aC.1 | 7 × 107 | 2.0 × 105 ± 1.4 × 105 (2) | 0 (2) | 0/18 | ND |
| TKG7aC.2 | 7 × 107 | 1.3 × 106 ± 3.3 × 105 (2) | 0 (2) | 2/18 | TKL |
| TKG7dG.1 | 7 × 107 | 1.1 × 106 ± 5.2 × 105 (2) | 0 (1) | 2/14 | TKL and mixed TK+/TKL |
| TKG7dG.2 | 7 × 107 | ND | ND | 0/14 | ND |
ND, not done.
The number of samples titrated for each group is shown in parentheses.
FIG. 4.
Plaque autoradiography of viruses isolated following reactivation of TKL viruses. In the image of the mixed population, one example of a TK+ plaque and one of a TK− plaque are shown (black and white arrowheads, respectively).
Two of 14 ganglia latently infected with TKG7dG.1 and 0 of 14 infected with TKG7dG.2 yielded virus (2/28 total) (Table 1). Both of the viruses that reactivated from ganglia infected with TKG7dG were amplified on Vero cells and analyzed by plaque autoradiography (Fig. 4). One appeared to be uniformly TKL, and sequence analysis confirmed that this virus contained the G7dG genotype. The other appeared to be mixed TK+ and TKL, and this was consistent with sequencing data.
Generation of recombinant viruses carrying mutations at sites other than the G string in tk associated with clinical drug resistance.
The use of ACVr HSV clinical isolates with a known mutation in tk to investigate a phenotype is difficult without a pretherapy strain for comparison, e.g., different isolates with the same mutation have been reported to have different TK phenotypes (11). We therefore engineered into the laboratory HSV-1 strain KOS two other frameshift mutations that have been observed in isolates taken from patients receiving ACV therapy (Fig. 1). As we have done previously, we chose strain KOS, as it is known to be dependent upon TK for reactivation from latently infected mouse trigeminal ganglia (3, 6, 14, 15, 21). Two independent isolates were generated for each mutation. The generation of the recombinant viruses used a blue/white screening procedure, rather than ACV selection, to reduce the chance of selecting for an ACVr mutation at a second site. Importantly, the use of tkLTRZ1 as a starting point in the construction of the viruses eliminated a potential source of contaminating TK+ virus.
TK phenotypes of recombinant viruses.
To assess whether these viruses were able to synthesize active TK despite the introduction of mutations into homopolymeric sequences in the gene, as has been observed with other mutations, they were analyzed by plaque autoradiography. We have modified this technique to make it extremely sensitive—approximately 0.25% of wild-type TK activity can be detected. No TK activity could be detected from plaques infected with virus TKC6+1C or TK2C5−1C (Fig. 5). No TK+ plaques were observed from >300 plaques that were examined from each virus.
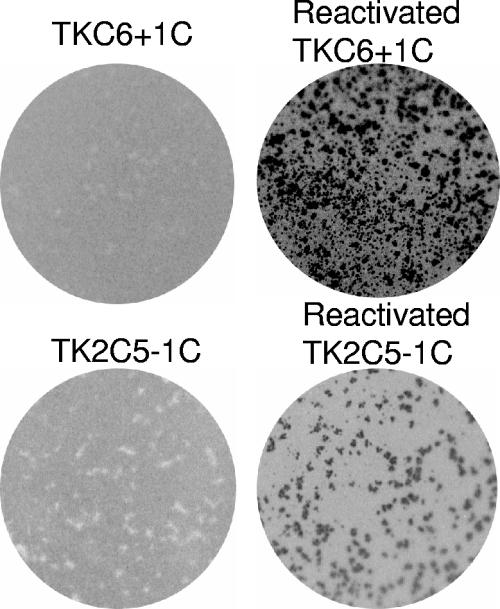
FIG. 5.
Plaque autoradiography of viruses TKC6+1C and TK2C5−1C and the viruses isolated following reactivation. Each row shows an image of a plate infected with the original virus on the left and a plate with the corresponding virus that reactivated on the right.
Reactivation of TKC6+1C and TK2C5−1C from latency.
To establish whether viruses TKC6+1C and TK2C5−1C were able to reactivate from latency despite a frameshift mutation within tk, ganglia were harvested from latently infected mice, enzymatically dissociated, and plated onto Vero cells that were monitored for the appearance of plaques. One of 13 ganglia latently infected with TKC6+1C.1 and 0 of 12 infected with TKC6+1C.2 yielded virus (1/25 total), compared to all of those infected with KOS and none of those infected with tkLTRZ1 (Table 2). One of 16 ganglia latently infected with TK2C5−1C.1 and 0 of 16 infected with TK2C5−1C.2 yielded virus (1/26 total) (Table 2). The viruses that reactivated were amplified on Vero cells, analyzed by plaque autoradiography, and shown to be TK+ (Fig. 5). Sequencing of the tk genes from these viruses showed that they had each reverted to the wild-type tk genotype.
TABLE 2.
Reactivation of viruses lacking measurable TK activity from latently infected mouse trigeminal ganglia
| Virus | Inoculum (PFU) | No. of ganglia with reactivating virus/total no. of ganglia | TK phenotype of reactivating virus |
|---|---|---|---|
| KOS | 2 × 106 | 30/30 | TK+ |
| tkLTRZ1 | 7 × 107 | 0/34 | NDa |
| TKC6+1C.1 | 7 × 107 | 1/13 | TK+ |
| TKC6+1C.2 | 7 × 107 | 0/12 | ND |
| TK2C5−1C.1 | 7 × 107 | 1/16 | TK+ |
| TK2C5−1C.2 | 7 × 107 | 0/16 | ND |
ND, not done.
DISCUSSION
We are interested in mechanisms that permit HSV to evade antiviral chemotherapy yet retain pathogenicity. We have reported previously that ribosomal frameshifting and an internal ribosome entry site can support the expression of low levels of TK (15, 16, 19), an important phenotype that appears to permit pathogenesis in immunocompromised individuals while not activating effective amounts of drug. It has been noted that certain of these mutations that confer a TKL phenotype to the virus, particularly the G9 mutation, render the virus disposed to reversion to the TK+ phenotype (13, 15, 25). Indeed, although we have previously shown that virus reactivating from some G8 mutant-infected ganglia were completely TKL, it has been difficult to study these two mechanisms separately. Herein, we have addressed the contribution of the TKL phenotype to pathogenesis, in the absence of reversion, by asking whether a wild-type G string supports the expression of TK originally reported for mutant sequences. This study shows that the TKL phenotype is sufficient to support reactivation from latently infected mouse trigeminal ganglia, even when the level of TK is as low as ∼0.25% of that of the wild-type virus. Reactivation was also observed under conditions that minimized the frequency of reversion. Conversely, we also wanted to address the contribution of reversion to pathogenesis in the absence of TK. This study shows that for two viruses that lack measurable TK activity, TKC6+1C and TK2C5−1C, reversion to the TK+ phenotype is sufficient to permit reactivation from latently infected mouse trigeminal ganglia. We discuss below the importance of these observations with regard to the pathogenesis of drug-resistant HSV and the biology of the wild-type virus.
The wild-type-length G string supports ribosomal frameshifting.
In vitro data using dual-reporter constructs translated in rabbit reticulocyte lysate showed that the G7 sequence of wild-type tk was able to support net +1 ribosomal frameshifting (18). We have now shown that a wild-type-length G string behaves similarly in the context of viral infection. Additionally, the expression of active TK following an insertion immediately downstream of the G string suggests that the wild-type G string may support net −1 ribosomal frameshifting. These observations could mean that translation of the wild-type tk gene results in the generation of several polypeptides via ribosomal frameshifting into both the −1 and + 1 reading frames. We are currently investigating this possibility. Also, we have previously suggested that because of the relatively simple sequence requirements of the G8 G-string net +1 ribosomal frameshift, there may be other hitherto-unrecognized polypeptides generated in mammalian genomes (18). We now propose an increase in the number of potential polypeptides that may be generated following frameshifting on G strings, given that seven G's can suffice for ribosomal frameshifting and the frameshifts appear to be into either the −1 or + 1 reading frames.
Active TK generated despite single base insertions or deletions downstream of the G string can support reactivation from latently infected trigeminal ganglia.
We have previously shown that low levels of TK generated via ribosomal frameshifting, from a virus with a single G insertion (G8), was sufficient to support reactivation from latently infected mouse trigeminal ganglia (11 of 35 ganglia reactivated) (14). We addressed the possibility that reactivation of a TKL virus could be the result of a second-site mutation by introducing a mutation into the reactivated virus such that it did not synthesize active TK; this virus did not reactivate from latently infected mouse ganglia (14). A limitation of studying viruses with insertions into the G string was that virus populations that reactivated from some ganglia contained TK+ virus, presumably a result of reversion. In this study, we maintained the wild-type-length G string, added a base immediately downstream, and observed reactivation in only 2 of 36 ganglia infected with TKG7aC viruses. The virus that reactivated from both of these ganglia was uniformly TKL. Although these two experiments were not performed at the same time, the data suggest that the difference in frequency of reactivation between the two genotypes may be due largely to the increased instability of the G8 G string versus that of the G7 G string.
Viruses with a double G insertion in the G string (G9) were remarkably unstable, with ∼3% of plaques appearing TK+ (15). The remaining ∼97% of plaques were TKL. In that study, virus populations that reactivated from all ganglia contained TK+ virus, leaving us uncertain as to whether active TK generated via a net −1 ribosomal frameshift was sufficient to support reactivation from latently infected trigeminal ganglia. Grey and colleagues (13) also reported that a virus with a double G insertion into the G string is prone to reversion to the TK+ phenotype. Those authors also detected low levels of TK activity in lysates of cells infected with a clinical isolate carrying the double G mutation but concluded that the TK activity was likely a result of viruses that had reverted to the TK+ phenotype. Given that enzyme assays, unlike plaque autoradiography, are unable to discriminate between viruses with different TK phenotypes within a population, it remains at least possible that the clinical isolate studied by Grey and colleagues has a TKL phenotype, in addition to being prone to reversion. Previously, we observed that of 35 ganglia latently infected with G9 virus, 20 reactivated, and the two populations that reactivated that were analyzed by plaque autoradiography contained TK+ virus (15). In this study, virus was observed to reactivate from only 2 of 28 ganglia latently infected with virus TKG7dG (Table 1). As noted above, although the experiments were not performed at the same time, the data strongly suggest that the difference in frequency of reactivation between the two genotypes may be due largely to the increased instability of the elongated G string.
We interpret the low frequency of reactivation of these viruses as being a reflection of the very low levels of TK expressed. Although it is possible that viruses appearing to reactivate as uniformly TKL populations could contain low levels of TK+ virus, we consider this highly unlikely, given that we detected <0.2% of TK+ virus in these populations and given the strong selective advantage that TK+ viruses would have in the ganglion.
A virus with an insertion into the C chord lacks TK activity.
We have recently shown that a virus that has a deletion in a run of six C's in tk, known as the “C chord,” has a TKL phenotype and the TK activity is dependent on an unusual internal ribosome entry site (IRES) in tk (16). Interestingly, the C chord itself was not necessary for the synthesis of the polypeptide generated via the IRES. It was therefore surprising that virus TKC6+1C did not generate active TK. As this virus carried an insertion into the C chord, rather than a deletion, the amino acids synthesized downstream of the mutations, which are not in the wild-type TK reading frame, are different between the two viruses. Therefore, it appears that these out-of-frame amino acids are important for the TK activity mediated via the IRES. Consistent with this idea, a virus that carries a stop codon in the mutant reading frame that would be synthesized downstream of the C6−1C mutation lacks TK activity (A. Griffiths and D. M. Coen, unpublished results).
Reversion to the TK+ phenotype in the context of an otherwise TK− virus can support reactivation from latency.
Other groups have suggested that viruses carrying frameshift mutations on the G string in tk, but lacking measurable TK activity, can retain some level of pathogenicity due to reversion to the TK+ phenotype (13, 25). However, more-sensitive assays have shown such viruses to be TKL, rather than TK− (references 15 and 19 and this study). In this report, we have shown that recombinant viruses that lack measurable TK activity due to mutations in other homopolymeric sequences are indeed able to reactivate from latently infected mouse trigeminal ganglia. The reactivating virus in each case was TK+. Although we cannot rule out the possibility that these viruses generate levels of TK below our detection levels, the data suggest that reversion due to the inability of the DNA polymerase to faithfully replicate homopolymeric sequences plays an important role in the pathogenesis of many ACVr viruses.
It seems clear that both the TKL phenotype and reversion to the TK+ phenotype can suffice to support reactivation from latently infected mouse ganglia. However, further experiments are needed to quantify the relative contributions of reversion and low TK activity toward reactivation and to determine at what stage (establishment or reactivation) these phenotypes make their contributions.
Acknowledgments
We dedicate this paper to the memory of Steve Sacks, who was a pioneer in studies of reversion in the pathogenesis of acyclovir-resistant mutants and who was a warm and wonderful colleague.
We thank Jean Pesola for assistance with the animal experiments.
This work was supported by grants PO1 NS35138, RO1 AI26126, and T32 AI07245 from the National Institutes of Health.
REFERENCES
- 1.Böni, J., and D. M. Coen. 1989. Examination of the roles of transcription factor Sp1-binding sites and an octamer motif in trans induction of the herpes simplex virus thymidine kinase gene. J. Virol. 63:4088-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, D. G., R. Visse, G. Sandhu, A. Davies, P. J. Rizkallah, C. Melitz, W. C. Summers, and M. R. Sanderson. 1995. Crystal structures of the thymidine kinase from herpes simplex virus type-1 in complex with deoxythymidine and ganciclovir. Nat. Struct. Biol. 2:876-881. [DOI] [PubMed] [Google Scholar]
- 3.Chen, S. H., W. J. Cook, K. L. Grove, and D. M. Coen. 1998. Human thymidine kinase can functionally replace herpes simplex virus type 1 thymidine kinase for viral replication in mouse sensory ganglia and reactivation from latency upon explant. J. Virol. 72:6710-6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christophers, J., J. Clayton, J. Craske, R. Ward, P. Collins, M. Trowbridge, and G. Darby. 1998. Survey of resistance of herpes simplex virus to acyclovir in northwest England. Antimicrob. Agents Chemother. 42:868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coen, D. M., A. F. Irmiere, J. G. Jacobson, and K. M. Kerns. 1989. Low levels of herpes simplex virus thymidine-thymidylate kinase are not limiting for sensitivity to certain antiviral drugs or for latency in a mouse model. Virology 168:221-231. [DOI] [PubMed] [Google Scholar]
- 6.Coen, D. M., M. Kosz Vnenchak, J. G. Jacobson, D. A. Leib, C. L. Bogard, P. A. Schaffer, K. L. Tyler, and D. M. Knipe. 1989. Thymidine kinase-negative herpes simplex virus mutants establish latency in mouse trigeminal ganglia but do not reactivate. Proc. Natl. Acad. Sci. USA 86:4736-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coen, D. M., S. P. Weinheimer, and S. L. McKnight. 1986. A genetic approach to promoter recognition during trans induction of viral gene expression. Science 234:53-59. [DOI] [PubMed] [Google Scholar]
- 8.Davar, G., M. F. Kramer, D. Garber, A. L. Roca, J. K. Andersen, W. Bebrin, D. M. Coen, M. Kosz Vnenchak, D. M. Knipe, X. O. Breakefield, and O. Isacson. 1994. Comparative efficacy of expression of genes delivered to mouse sensory neurons with herpes virus vectors. J. Comp. Neurol. 339:3-11. [DOI] [PubMed] [Google Scholar]
- 9.Efstathiou, S., S. Kemp, G. Darby, and A. C. Minson. 1989. The role of herpes simplex virus type 1 thymidine kinase in pathogenesis. J. Gen. Virol. 70:869-879. [DOI] [PubMed] [Google Scholar]
- 10.Englund, J. A., M. E. Zimmerman, E. M. Swierkosz, J. L. Goodman, D. R. Scholl, and H. H. Balfour, Jr. 1990. Herpes simplex virus resistant to acyclovir. A study in a tertiary care center. Ann. Intern. Med. 112:416-422. [DOI] [PubMed] [Google Scholar]
- 11.Gaudreau, A., E. Hill, H. H. Balfour, Jr., A. Erice, and G. Boivin. 1998. Phenotypic and genotypic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J. Infect. Dis. 178:297-303. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert, C., J. Bestman-Smith, and G. Boivin. 2002. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist. Updates 5:88-114. [DOI] [PubMed] [Google Scholar]
- 13.Grey, F., M. Sowa, P. Collins, R. J. Fenton, W. Harris, W. Snowden, S. Efstathiou, and G. Darby. 2003. Characterization of a neurovirulent aciclovir-resistant variant of herpes simplex virus. J. Gen. Virol. 84:1403-1410. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths, A., S. H. Chen, B. C. Horsburgh, and D. M. Coen. 2003. Translational compensation of a frameshift mutation affecting herpes simplex virus thymidine kinase is sufficient to permit reactivation from latency. J. Virol. 77:4703-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths, A., and D. M. Coen. 2003. High-frequency phenotypic reversion and pathogenicity of an acyclovir-resistant herpes simplex virus mutant. J. Virol. 77:2282-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths, A., and D. M. Coen. 2005. An unusual internal ribosome entry site in the herpes simplex virus thymidine kinase gene. Proc. Natl. Acad. Sci. USA 102:9667-9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris, W., P. Collins, R. J. Fenton, W. Snowden, M. Sowa, and G. Darby. 2003. Phenotypic and genotypic characterization of clinical isolates of herpes simplex virus resistant to aciclovir. J. Gen. Virol. 84:1393-1401. [DOI] [PubMed] [Google Scholar]
- 18.Horsburgh, B. C., H. Kollmus, H. Hauser, and D. M. Coen. 1996. Translational recoding induced by G-rich mRNA sequences that form unusual structures. Cell 86:949-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang, C. B., B. C. Horsburgh, E. Pelosi, S. Roberts, P. Digard, and D. M. Coen. 1994. A net +1 frameshift permits synthesis of thymidine kinase from a drug-resistant herpes simplex virus mutant. Proc. Natl. Acad. Sci. USA 91:5461-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irmiere, A. F., M. M. Manos, J. G. Jacobson, J. S. Gibbs, and D. M. Coen. 1989. Effect of an amber mutation in the herpes simplex virus thymidine kinase gene on polypeptide synthesis and stability. Virology 168:210-220. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson, J. G., S. H. Chen, W. J. Cook, M. F. Kramer, and D. M. Coen. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161-169. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel, T. A., and K. Bebenek. 2000. DNA replication fidelity. Annu. Rev. Biochem. 69:497-529. [DOI] [PubMed] [Google Scholar]
- 23.Leib, D. A., D. M. Coen, C. L. Bogard, K. A. Hicks, D. R. Yager, D. M. Knipe, K. L. Tyler, and P. A. Schaffer. 1989. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J. Virol. 63:759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sacks, S. L., R. J. Wanklin, D. E. Reece, K. A. Hicks, K. L. Tyler, and D. M. Coen. 1989. Progressive esophagitis from acyclovir-resistant herpes simplex. Clinical roles for DNA polymerase mutants and viral heterogeneity? Ann. Intern. Med. 111:893-899. [DOI] [PubMed] [Google Scholar]
- 25.Sasadeusz, J. J., and S. L. Sacks. 1996. Spontaneous reactivation of thymidine kinase-deficient, acyclovir-resistant type-2 herpes simplex virus: masked heterogeneity or reversion? J. Infect. Dis. 174:476-482. [DOI] [PubMed] [Google Scholar]
- 26.Sasadeusz, J. J., F. Tufaro, S. Safrin, K. Schubert, M. M. Hubinette, P. K. Cheung, and S. L. Sacks. 1997. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J. Virol. 71:3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wild, K., T. Bohner, A. Aubry, G. Folkers, and G. E. Schulz. 1995. The three-dimensional structure of thymidine kinase from herpes simplex virus type 1. FEBS Lett. 368:289-292. [DOI] [PubMed] [Google Scholar]
- 28.Wild, K., T. Bohner, G. Folkers, and G. E. Schulz. 1997. The structures of thymidine kinase from herpes simplex virus type 1 in complex with substrates and a substrate analogue. Protein Sci. 6:2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]