Abstract
A mutation's effect on fitness or phenotype may in part depend on the interaction of the mutation with the environment. The resulting phenotype or fitness is important, since it determines the adaptive potential of a species. To date, most studies have focused on alterations to protein-coding regions of the genome and their consequential fitness effects. Non-protein-coding regulatory regions have been largely neglected, although they make up a large and important part of an organism's genome. Here, we use human immunodeficiency virus type 1 as a model system to investigate fitness effects of random mutations in noncoding DNA-binding sites of the transcriptional promoter. We determined 242 fitness values for 35 viral promoter mutants with one, two, or three mutations across seven distinct cellular environments and identified that (i) all mutants have an effect in at least one cellular environment; (ii) fitness effects are highly dependent on the cellular environment; (iii) disadvantageous and advantageous mutations occur at high and similar frequencies; and (iv) epistatic effects of multiple mutations are rare. Our results underline the evolutionary potential of regulatory regions and indicate that DNA-binding sites evolve under strong selection, while at the same time, they are very plastic to environmental change.
Non-protein-coding regulatory regions comprise a significant part of an organism's genome and determine gene expression, affect disease susceptibility (44) and contain crucial information for an organism's complexity (6, 22). However, compared to our knowledge on protein-coding regions, our knowledge on the evolution of non-protein-coding regulatory regions is scarce.
A key function of regulatory regions is controlling gene expression, which is a tightly regulated process, as the levels of different gene products have to be tuned towards a well-balanced state. Regulation of gene expression depends on the interaction of transcription factors and cofactors with DNA-binding sites that are located upstream of the protein-coding regions of the genome. Due to the short length of DNA-binding sites (on average, 5 to 10 base pairs) and the relatively strict motifs that are necessary for binding a specific transcription factor, mutations in DNA-binding sites may have a large impact on a gene's expression profile and, therefore, its phenotype (31, 33, 63, 68).
Epistasis between DNA-binding sites may also significantly contribute to a gene's expression profile. Epistasis occurs when two or more loci interact and the collective contribution of different loci to phenotype or fitness deviates from their combined individual effects. Their combined effects may then be either stronger (synergistic) or weaker (antagonistic) than expected (13, 18, 66). Although epistasis was found to have a small role in Caenorhabditis elegans (47), it is widespread in protein-coding regions of many different organisms e.g., Drosophila (65), Escherichia coli (16), Aspergillus niger (12), bacteriophage ϕ6 (5), and vesicular stomatitis virus (54). In addition, epistasis has been found among genes that confer drug resistance in human immunodeficiency virus type 1 (HIV-1) (4). What the role of epistasis is for regulatory regions, however, is unclear.
Due to easy genetic manipulation and availability of different host-cell environments, HIV-1 offers a unique opportunity to investigate the fitness effect of mutations in DNA-binding sites across several environments. A retrovirus, such as HIV-1, exploits the host transcription machinery to initiate viral transcription by binding several host-cell transcription factors to specific DNA-binding sites encoded in its 5′ long terminal repeat (LTR). In this way, HIV-1 can initiate and regulate transcription and thus sustain virus propagation (21, 38, 61, 63). It is estimated that at least 2,000 transcription factors are encoded by the human genome (28, 62), and expression of these factors depends on, e.g., cell type, stage of development, and activation state. Therefore, optimal DNA-binding site composition of the HIV-1 LTR is likely to vary with transcription factor availability in the specific host-cell type that is infected.
In this study, we aimed to unravel the evolutionary potential of DNA-binding sites. To do so we determined the fitness effect (i.e., relative growth rate) of random single, double and triple nucleotide changes in DNA binding sites of the transcriptional promoter of HIV-1. To assess the influence of the host-cell environment, a mutation's fitness effect was measured across seven distinct host-cell environments. In addition, we directly test for the presence of epistasis between DNA-binding sites by constructing double mutants from combinations of the single-mutant set.
MATERIALS AND METHODS
Mutagenesis.
We constructed 15 viruses containing a single point mutation, 15 viruses with two point mutations, and 5 viruses with three point mutations within the transcriptional promoter of HIV-1. Random mutations were blindly chosen from all available possibilities. Eight of the mutants containing two mutations were constructed out of single mutants (also see section on epistasis). The other seven mutants contained two or three mutations that were randomly distributed across the transcriptional promoter. Mutagenesis was performed on the pBlue3′LTR intermediate plasmid by a PCR strategy with a mutagenic primer as described previously (23). Molecular clones containing one, two, or three nucleotide changes were obtained by insertion of the respective XhoI-HindIII fragment of pBlue3′LTR into the 3′LTR of the HIV-1 molecular clone pLAI. The 3′LTR from positions −147 to −1 is subsequently inherited in both LTRs of the viral progeny after the first round of virus replication. To prevent the occurrence of extra mutations in the genome which may influence the obtained fitness measurements we took three precautionary measures: (i) the region containing the mutation was separately cloned into the HIV-1 genome, thereby preventing the introduction of additional mutations during the mutagenesis PCR; (ii) all virus stocks to be utilized in competition experiments were produced in transfected C33A cells that support production of viral particles but not multiple rounds of HIV-1 infection, thereby excluding mistakes made by reverse transcription, the step at which the majority of mutations are introduced into the HIV-1 genome; and (iii) each competition was repeated between two and five times with virus stocks produced by at least two different plasmid preparations.
Cell lines and PBMCs.
The human lymphocytic SupT1, MT2, MT4, and C8166 T-cell lines were cultured in RPMI 1640 (Gibco BRL) supplemented with 10% fetal calf serum, penicillin (100 units/ml), and streptomycin (100 units/ml). The cervix carcinoma cell line C33A (ATCC HTB31) was cultured in Dulbecco's modified Eagle's medium (Gibco BRL) with the same supplements. Some SupT1 T-cell line cultures were supplemented every three days with tumor necrosis factor alpha (TNF-α; 50 ng/ml). Peripheral blood mononuclear cells (PBMCs) were isolated from fresh buffy coats (Sanquin Central Laboratory Blood Bank, Amsterdam, The Netherlands) by standard Ficoll-Hypaque density centrifugation. PBMCs isolated from a single donor were pooled and frozen in multiple vials and thawed when needed. After thawing, the PBMCs were activated with 3 μg/ml phytohemagglutinin and cultured in RPMI medium containing 10% fetal calf serum, penicillin (100 units/ml), and streptomycin (100 units/ml) with recombinant interleukin-2 (100 units/ml). All cell lines and PBMCs were kept at 37°C and 5% CO2.
Virus stocks.
C33A cells were calcium phosphate-transfected with 5 μg of the pLAI molecular clones to produce virus. The virus concentration was determined by the capsid (CA)-p24 enzyme-linked immunosorbent assay (23).
Competition experiments.
Competition experiments were performed to determine mutant virus fitness relative to the parental wild-type (wt) virus pLAI. A total of 106 cells was infected with virus stocks of the mutant and the wt (1 ng CA-p24). On average, cell cultures expanded from 106 to 107 cells over 3 to 4 days of culture. Infections were monitored for viral replication by measuring CA-p24 production in the culture supernatant and by monitoring syncytia formation by microscopic inspection, with the peak of infection typically reached between days 4 to 6 of culture. Competition experiments were repeated three to five times and each competition was continued for three passages by isolating virus at peak infection and subsequently infecting a fresh batch of host cells. Competition experiments were performed under seven different conditions (SupT1, MT2, MT4, and C8166, the SupT1 T-cell line with TNF-α, and PBMCs from two donors), which are referred to as cellular environments throughout the article. Total cell DNA was isolated before passage from approximately 0.25 × 106 cells as described previously (60). Proviral LTR sequences were PCR amplified and sequenced with the −21M13 Big Dye Primer cycle sequencing kit (ABI) to determine the ratio of both competitors (29) and calculate relative fitness (60).
It is known for vesicular stomatitis virus that when infection frequencies become too high (multiplicity of infection [MOI] > 1), competition between viruses may suffer from density-dependent selection, which can result in aberrant estimates of relative fitness (41). In our experiments, the initial MOI is small (0.001), so we do not expect density-dependent selection to play an important role. To directly check this we have tested infection with initial MOIs of 0.0001 and 0.01 for a number of mutants, and this did not affect our fitness estimates. In addition, the relatively small number of viral generations within the competition experiments (viral generation time is approximately 2 days) (46, 51), compared to the large initial number of infected cells, makes it so that de novo mutations during the competition experiment will not affect our fitness estimates. Even if such mutations are advantageous, they will not reach substantial copy numbers over the duration of the experiment. Direct evidence for insensitivity of our method for de novo mutations comes from the high reproducibility of fitness estimates between replicates and the fact that we never observed any new mutations in our sequencing. Also, recombination is unlikely to affect our results, because effective recombination for HIV depends on multiple infection of cells (3), and as indicated, the MOI in our experiments is low. Furthermore, recombination between the wild type and single mutants is neutral, as it does not create new genotypes, and we did not observe recombinant genotypes in any of our sequencing data.
Fitness calculation and statistics.
For each competition experiment, we compute the relative fitness (Wm) of the mutant, by comparing the n-fold expansion of the mutant and wild type (30) as follows: Wm = {ln[Nm(T) × d/Nm(0)]/ln[Nw(T) × d/Nw(0)]}, in which Nm(0) and Nw(0) are the initial densities of the mutant and wild-type genotype, respectively, and Nm(T)and Nw(T) are the corresponding densities at the end of the experiment and d is the dilution factor. The relative fitness Wm of a mutant is a dimensionless factor that scales the growth rate of the mutant compared to the wild type within a specific cellular environment. For simplicity, we assume that mutations affect the Malthusian (i.e., exponential) net replication rate, as we cannot distinguish between effects on replication rate and death rate (36). Since mutant-to-wild type ratios can be determined with an accuracy of within 5%, we added this measurement error to the variance of each fitness value.
The contributions of genotype, environment, and the interaction between environment and genotype to relative fitness were assessed in a linear model with genotype, environment, and genotype by environment as fixed factors.
The constructed double mutants were tested to correlate with expected additive fitness values of the single mutations (Wi + Wj − 1). Since we used Malthusian growth rates, an additive model is theoretically more appropriate than a multiplicative model; under the additive model, absence of epistasis corresponds to absence of linkage disequilibrium (S. P. Otto, personal communication). All statistics were determined with the programs Prism 3.0 and SPSS 12.0.1.
DNA-binding site predictions.
To screen for DNA-binding sites, we used the Web-based program Matinspector (7). Core and matrix probabilities were set at 0.75. The core sequence consists of the highest conserved nucleotide motif, typically encompassing 4 nucleotides, in the complete sequence (matrix) that defines the DNA-binding sites, which is typically around 5 to 10 nucleotides long.
RESULTS
Single nucleotide changes affect fitness in an environment dependent manner.
In order to study the effect of mutations in noncoding regions with regards to viral fitness, we introduced random mutations into the transcriptional promoter of HIV-1, which contains a variety of transcription factor binding sites (Fig. 1). Fifteen mutant viruses were constructed with a single nucleotide substitution, seven viruses were constructed with two substitutions, and five viruses were constructed with three substitutions. The relative fitness of each mutant virus was determined by performing in vitro competition experiments against the parental wt molecular clone. The wt strain is the high-fitness molecular clone pLAI (45). Competitions were performed by introducing the mutant and wt viruses in equal amounts into a host cell culture environment and were continued for three passages. At peak infection, cellular DNA was isolated and frequency of the competitors was determined by sequencing analysis. The change in frequency for the mutant was used to calculate the relative fitness of the mutant (29, 30, 60).
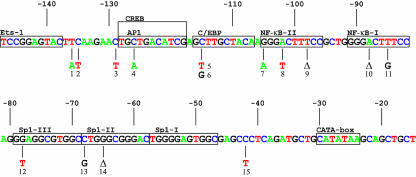
FIG. 1.
Mutations introduced into the transcriptional promoter of HIV-1. The introduced single-nucleotide changes are indicated below the sequence, a deletion is indicated by a delta. All mutations were randomly chosen except for mutant 9 (T-98 deleted), which was taken from a previous study (63). This LTR domain encodes a large variety of transcription factor binding sites and the generally accepted sites are boxed. Nucleotide positions are relative to the transcription start site.
To assess the influence of the cellular environment on fitness, competitions were performed in seven cell culture environmental settings, i.e., four distinct laboratory human lymphocytic T-cell lines (SupT1, MT-2, MT-4, and C8166), one of the T-cell lines (SupT1) with added inflammatory cytokine TNF-α, and two batches of freshly isolated PBMCs obtained from two healthy donors. SupT1 is a human leukemic T-cell line, and MT-2, MT-4, and C8166 are T-cell lines that have been immortalized through transformation by the human T-cell leukemia-lymphoma virus type I (57). The cells have different genetic backgrounds and are immortalized and activated in different ways and consequently contain different pools of transcription factors. For instance, the SupT1 T-cell line contains no basal levels of activated transcription factors that interact with the NF-κB DNA-binding sites (p50/p65). The addition of TNF-α to the SupT1 T-cell culture activates and releases these factors into the nucleus. Although exact differences between the T-cell lines have not been assessed, several differences are known for transcription factors ETS1, C/EBP, GATA, NF-κB, STAT5, and SP1 (1, 9, 37, 40, 49).
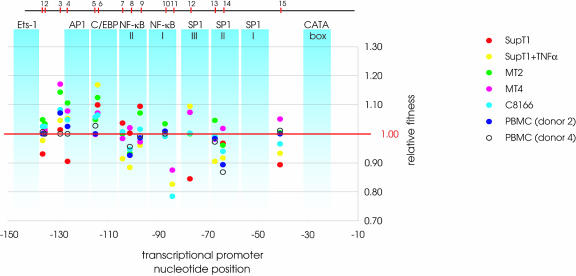
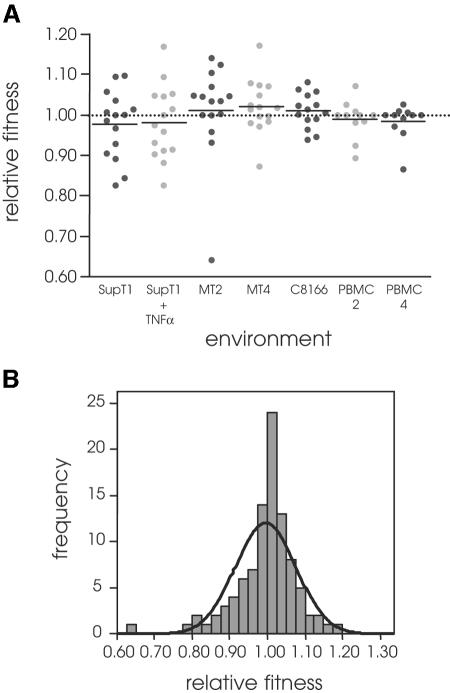
We first determined the contribution of each source of variation (genotype, environment, and genotype by environment) to relative fitness. All three factors significantly contribute to fitness (Table 1). The strong interaction between genotype and environment is illustrated by the changing relative fitness of mutant 4 (C-126→A), which had a disadvantageous effect in the SupT1 T-cell line, was neutral in PBMC donor 4, and was beneficial in the MT2, MT4, and C8166 T-cell lines, the SupT1 T-cell line with TNF-α, and PBMC donor 2 (see Fig. 3 and Table 2). The mean fitness effects of viruses with a single mutation were similar for each environment (analysis of variance P = 0.612) (Fig. 2a) and were close to neutrality when measured over all environments (0.996 ± 0.008 [mean ± standard error of the mean {SEM}]) (Fig. 2).
TABLE 1.
Contribution of each source of variation to relative fitness of +1 mutants
| Source of variation | SS type IIIa | df | Fb |
|---|---|---|---|
| Genotype | 1.322 | 14 | 51.278 |
| Environment | 0.159 | 5 | 14.414 |
| Genotype by environment | 1.535 | 75 | 11.117 |
| Error | 1.028 | 558 |
SS, sum of squares.
P < 0.0001.
FIG. 3.
Relative fitness of viruses with a single-nucleotide substitution measured in seven different cellular environments. Relative fitness values for all mutants are plotted on the nucleotide position within the transcriptional promoter. Different colors depict different cellular environments in which mutations were tested. The generally accepted DNA-binding sites are highlighted as light blue bars. For standard errors and significance see Table 1.
TABLE 2.
Relative fitness of viruses with a single nucleotide substitution
| Mutation no. | Substitution | Relative fitness (mean ± SE) in indicated cellular environment
|
||||||
|---|---|---|---|---|---|---|---|---|
| SupT1 | SupT1 + TNF-α | MT2 | MT4 | PBMC
|
||||
| C8166 | Donor 2 | Donor 4 | ||||||
| 1 | T-136→A | 0.929 ± 0.028b | 0.975 ± 0.008b | 1.046 ± 0.013b | 1.025 ± 0.033 | 1.023 ± 0.009 | 1.0 ± 0.006 | 1.006 ± 0.006 |
| 2 | C-135→T | 1.007 ± 0.006 | 1.015 ± 0.004b | 1.035 ± 0.011b,e | 1.014 ± 0.029 | 1.025 ± 0.007 | 1.0 ± 0.006 | 1.0 ± 0.006 |
| 3 | C-129→T | 1.014 ± 0.009 | 1.046 ± 0.035 | 1.141 ± 0.003d,e | 1.171 ± 0.016d,e | 1.082 ± 0.005b | 1.072 ± 0.004d,e | 1.0 ± 0.004 |
| 4 | C-126→A | 0.905 ± 0.015c,e | 1.052 ± 0.007c,e | 1.104 ± 0.019d,e | 1.079 ± 0.017b | 1.048 ± 0.011b | 1.025 ± 0.005d,e | 1.0 ± 0.005 |
| 5 | C-115→T | 1.097 ± 0.019d,e | 1.169 ± 0.029d,e | 1.124 ± 0.029c,e | 1.071 ± 0.014c,e | 1.063 ± 0.021b | NDf | ND |
| 6 | C-115→G | 1.057 ± 0.021b | 1.048 ± 0.010b | 1.048 ± 0.018b | 0.998 ± 0.030 | 1.058 ± 0.014c,e | 1.0 ± 0.000 | 1.026 ± 0.004b |
| 7 | G-105→A | 1.036 ± 0.019 | 0.912 ± 0.021d,e | 0.999 ± 0.004 | 0.984 ± 0.007b | 1.006 ± 0.008 | ND | ND |
| 8 | A-102→T | 1.000 ± 0.023 | 0.882 ± 0.024b,e | 0.932 ± 0.002c,e | 1.020 ± 0.015 | 0.946 ± 0.004b | 0.924 ± 0.009c,e | 0.955 ± 0.009b |
| 9 | T-98→Δg | 1.094 ± 0.011c,e | 0.959 ± 0.004d,e | 1.070 ± 0.007d,e | 0.971 ± 0.010b | 1.014 ± 0.003a | 0.984 ± 0.004c,e | 0.992 ± 0.005 |
| 10 | A-88→Δ | 1.0 ± 0.004 | 0.994 ± 0.018 | 1.035 ± 0.018a | 0.997 ± 0.018 | 0.991 ± 0.009 | 1.009 ± 0.002 | 1.0 ± 0.000 |
| 11 | T-85→G | 0.826 ± 0.001d,e | 0.826 ± 0.001d,e | 0.641 ± 0.108a | 0.873 ± 0.019b | 0.778 ± 0.014d,e | ND | ND |
| 12 | G-78→T | 0.844 ± 0.051b | 1.094 ± 0.007d,e | 1.0 ± 0.000 | 1.073 ± 0.020b | 1.001 ± 0.007 | ND | ND |
| 13 | C-68→G | 0.985 ± 0.010 | 0.903 ± 0.038b,e | 1.045 ± 0.018d | 0.980 ± 0.012b | 0.988 ± 0.007 | 0.982 ± 0.005b | 0.972 ± 0.004d,e |
| 14 | G-65→Δ | 0.967 ± 0.015b | 0.915 ± 0.013c,e | 0.959 ± 0.009b | 1.018 ± 0.003a | 0.939 ± 0.013a | 0.893 ± 0.010c,e | 0.866 ± 0.010c,e |
| 15 | C-42→T | 0.891 ± 0.008d,e | 0.931 ± 0.023c,e | 1.003 ± 0.015 | 1.049 ± 0.013a | 0.964 ± 0.003a | 1.0 ± 0.006 | 1.010 ± 0.003 |
P < 0.05.
P < 0.01.
P < 0.001.
P < 0.0001.
Relative fitness remained significantly different from neutrality after sequential Bonferroni corrections.
Not determined.
Δ, deletion.
FIG. 2.
Distribution of relative fitness values of random HIV-1 LTR mutants. (A) Fitness was measured relative to the wild type for 15 viruses with a single-nucleotide substitution in seven cellular environments. The dotted line indicates fitness 1. (B) Relative fitness of all mutants yields a distribution that fits a normal distribution (indicated by the bold line; Kolmogorov-Smirnov test, P = 0.077).
Our competition experiments give a good indication as to which mutants really have a fitness effect, i.e., have fitness different from neutrality. Within each competition experiment, the mutant and wild type are facing equal conditions, and copy numbers are large enough to cancel out stochastic effects. If we use as a heuristic criterion for nonneutrality either the mutant or the wild type consistently reaching a frequency above 85% after three rounds of competition, we find that 65% (63 out of 97) of fitness effects of mutants deviate from neutrality (31 positive and 32 negative). If we statistically test for nonneutrality, we find that 32 (out of 97) fitness effects are significantly different from neutrality (two-tailed t test with P < 0.05; also, applying sequential Bonferroni's correction for multiple testing) (Fig. 3; Table 2) (50). However, applying a Bonferroni's correction will greatly increase the number of type II statistical errors. A type II error occurs when a nonneutral fitness effect would, by lack of statistical power, not pass the significance test and falsely be categorized as neutral. Thus, by applying the correction method, we underestimate the number of nonneutral fitness effects. However, if Bonferroni's correction is not applied, type I errors will obscure the results. A type I error occurs when a neutral fitness effect is falsely attributed as nonneutral. Therefore, one overestimates the number of nonneutral fitness effects. It is possible to side-step Bonferroni's correction by estimating the possible number of type I errors that result from a t test. If we follow the approach used in reference 15 and apply a noncorrected two-tailed t test we can subsequently estimate the type I statistical error. This calculation predicts that approximately five (i.e., 97 × 0.05) neutral fitness effects are falsely attributed as nonneutral. Thus, we find that around 58 (out of 97) mutant fitness effects are nonneutral, which roughly corresponds to our heuristic estimate. In any case the number of nonneutral fitness effects is surprisingly high.
Single nucleotide changes can strongly affect binding affinity.
To explain the strong, environmentally driven fitness effects of single point mutations, we examined whether the mutations affect the predicted affinity of the DNA-binding sites. First, we used the program Matinspector (48) to determine the core and matrix probability of the known DNA-binding sites shown in Fig. 1. We subsequently used this threshold value (core and matrix probability, 0.75) to screen for additional binding sites that could in principle be functional in macrophages and T lymphocytes. This analysis suggests that the HIV-1 transcriptional promoter might in fact be densely packed with overlapping DNA-binding sites, with a total of 86 potential sites in the 147 nucleotide sequence segment shown in Fig. 1. Each of our 15 random single mutations affects on average three to four sites, and creates two new sites (see Table S1 in the supplemental material). These statistics underline the potential sensitivity of DNA-binding sites to genetic change. They also provide a possible explanation for the strong dependence of the fitness effects on the cellular environment, since the functionality of the (potential) binding sites depends on the presence of specific host cellular transcription factors. Furthermore, the dense packing of binding sites may also explain why mutations in between the commonly accepted DNA-binding sites (i.e., mutations 1, 2, 3, and 15 in Fig. 1) significantly deviate from being neutral in several environments.
Additive fitness versus epistasis.
Next, we examined fitness effects of random double and triple mutations (Fig. 4). Similar to viruses with a single-nucleotide change the contribution of each source of variation to relative fitness was significant while genotype and genotype by environment contributed the most (see Tables S2 and S3 in the supplemental material). The average fitness effect over all environments was negative both for two mutations (0.924 ± 0.023 [mean ± SEM]) and three mutations (0.903 ± 0.023 [mean ± SEM]).
FIG. 4.
Relative fitness of viruses with one, two, and three point mutations. The average fitness effect within each mutation class is indicated by a horizontal black line. The average fitness effect of a single nucleotide substitution is 0.996 ± 0.008 [mean ± SEM] with a 95% CI of 1.012 to 0.979, that of two substitutions is 0.9241 ± 0.023 [mean ± SEM] with a 95% CI of 0.971 to 0.877, and that of three substitutions is 0.9032 ± 0.023 [mean ± SEM] with a 95% CI of 0.951 to 0.856.
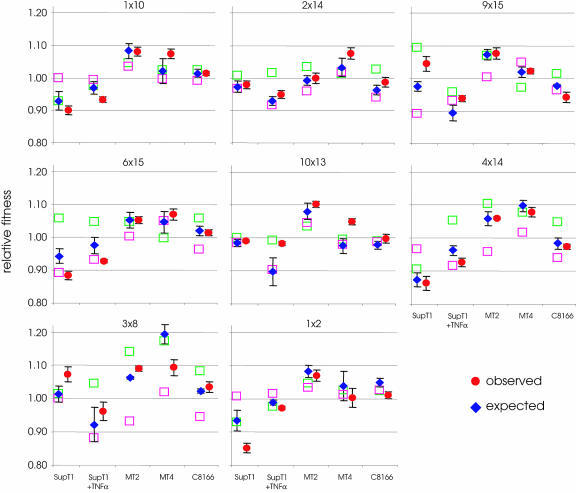
To directly test for epistasis, we constructed eight additional, nonrandom double mutants by combining mutations from the single-mutant set. These double mutants were assayed in five cellular environments, and the observed fitness was compared with the expected additive fitness based on the single mutants' fitness values (Fig. 5). The complete set of double mutants strongly correlates with a model of additive fitness, (n = 40, r2 = 0.71, P < 0.0001). In individual t tests, 2 out of 40 fitness values deviated significantly from the additive fitness model (mt1 multiplied by mt2 and mt9 multiplied by mt15 for SupT1 cells; P values of 0.02 and 0.03, respectively). However, after sequential Bonferroni's correction, none of the interactions were significantly different from additivity, and the two hits could well be type I statistical errors (i.e., we estimate the type I error to be 40 × 0.05, 2 false positives).
FIG. 5.
The relative fitness of a virus with two nucleotide substitutions corresponds well to an additive model. The fitness values of the single-mutant virus are indicated as green and purple squares, and their identity is shown above the graphs (see Fig. 1 and Table 1 for mutation numbers). Bars on circles represent standard errors. The five cellular environments in which relative fitness was measured are indicated below the graphs.
DISCUSSION
In this study, we demonstrate the sensitivity of non-protein-coding DNA-binding sites to random mutations and changes in the cellular environment. Single nucleotide substitutions in the transcriptional promoter of the HIV-1 molecular clone pLAI were sufficient to trigger a strong, host-cell dependent fitness effect. Surprisingly, positive and negative fitness effects are balanced, which is in sharp contrast to fitness distributions of random point mutations in protein-coding regions, where beneficial mutations are typically rare (5, 12, 26, 27, 39, 43, 55). Furthermore, even after applying the rather strict sequential Bonferroni's criterion, 13 out of 15 mutants had a significant fitness effect in at least one of the environments, while if we use our heuristic competitive criterion directly obtained from the competition experiments, 12 mutants had an advantageous fitness effect in at least a single environment. In any way, in contrast to protein-coding regions, silent mutations seem rare. Finally, in protein-coding regions, a substantial fraction of mutations will have strong deleterious effects, e.g., mutations may lead to a stop codon or a frameshift, causing premature termination or erroneous translation. Such strong deleterious mutations did not occur in our data set.
Another important finding in our study is the lack of epistasis. Epistasis is commonly observed in various organisms, such as Drosophila (65), yeast (56), bacteriophage ϕ6 (5), vesicular stomatitis virus (54), Escherichia coli (16), and Aspergillus niger (12). In addition, epistasis has been found in HIV-1 among genes that confer drug resistance (4). However, these examples are all based on interactions between protein-coding regions, and it is yet unclear whether epistasis is important for a gene's expression profile as well. We conclude that, at least in our data set, strong epistatic effects are rare. Whether this is a common feature of regulatory regions requires further analysis and may for instance depend on the complexity of the transcriptional network (14, 17, 24, 25, 53, 70). It should be stressed that our method assesses only whether epistatic effects are common in randomly generated mutants. Therefore, even if epistatic effects seem rare, they might still play an essential role and affect the adaptive rate.
In vivo, within a single patient, HIV-1 will encounter different pools of transcription factors in variant cell types and in differentially activated cells, e.g., in different body compartments or due to viral or bacterial coinfections (10, 19, 35). Also within a single cell, the promoter may experience variable conditions that affect its activity (64). Thus, the local environment of the transcriptional promoter of HIV-1 can be envisaged as heterogeneous and multidimensional, as the pool of transcription factors varies both temporally and spatially. To overcome the hurdle of a highly unpredictable environment, HIV-1 may have evolved a generalist promoter that provides transcriptional activity across many different environments. Indeed, our data show that the wild-type virus displays good fitness across all tested environments, whereas in any specific environment, HIV-1 could, in principle, evolve to higher fitness.
It has been speculated that DNA-binding sites can evolve faster than protein-coding regions due to their small size. This is based on the finding that promoter sequences have diverged extensively among relatively closely related species, including gains and losses of binding sites and changes in the position of regulatory sequences relative to the transcription start site (32, 52, 59, 67, 69). Moreover, a comparison of well-characterized regulatory regions in mammals revealed that approximately one-third of the binding sites in humans are probably not functional in rodents (11). Classic neutral theory of evolution has been proposed for the evolution of DNA-binding sites (42, 58), but this process seems too slow to explain their rapid turnover (2, 6, 33, 34, 68). Their fast rate of evolution may instead be a consequence of the tight interplay between the genotype of a DNA-binding site and the transcription factors in the environment, where minor changes in either have a direct and strong effect on fitness as indicated by our experiments.
In this paper, we underline the evolutionary potential of regulatory regions as we show how sensitive DNA-binding sites are to mutations in their sequence and to changes in the cellular environment. From HIV-1 and other animal retroviruses, it is known that the transcriptional promoter is a major determinant for virulence and minor changes or rearrangements can have a significant impact on cell tropism and pathogenicity (8, 20, 38). Our data indicate that DNA-binding site evolution is very plastic to environmental change, and it may therefore, in general, play a pivotal role in the rapid adaptation to new environments.
Supplementary Material
Acknowledgments
We acknowledge Marloes Naarding and Nienke Westerink for expert technical assistance and Sara Magalhaes, Bill Paxton, Pleuni Pennings, Maus Sabelis, and Nadine Vastenhouw for discussions and critical reading of the manuscript. Furthermore, we thank Sally Otto for advice on how to handle the epistasis data and Emiel van Loon for advice on statistics.
This research was supported by NWO-ALW project number 811.35.001, as well as by ZonMw (vici-grant) and NWO-CW (TOP-grant) awarded to B.B.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Beck, Z., A. Bacsi, X. Liu, P. Ebbesen, I. Andirko, E. Csoma, J. Konya, E. Nagy, and F. D. Toth. 2003. Differential patterns of human cytomegalovirus gene expression in various T-cell lines carrying human T-cell leukemia-lymphoma virus type I: role of Tax-activated cellular transcription factors. J. Med. Virol. 71:94-104. [DOI] [PubMed] [Google Scholar]
- 2.Berg, J., S. Willmann, and M. Lassig. 2004. Adaptive evolution of transcription factor binding sites. BMC Evol. Biol. 4:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boerlijst, M. C., S. Bonhoeffer, and M. A. Nowak. 1996. Viral quasi-species and recombination. Proc. R. Soc. Lond. B 263:1577-1584. [Google Scholar]
- 4.Bonhoeffer, S., C. Chappey, N. T. Parkin, J. M. Whitcomb, and C. J. Petropoulos. 2004. Evidence for positive epistasis in HIV-1. Science 306:1547-1550. [DOI] [PubMed] [Google Scholar]
- 5.Burch, C. L., and L. Chao. 2004. Epistasis and its relationship to canalization in the RNA virus phi 6. Genetics 167:559-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll, S. B., J. K. Grenier, and S. D. Weatherbee. 2001. From DNA to diversity. Blackwell Scientific, Oxford, United Kingdom.
- 7.Cartharius, K., K. Frech, K. Grote, M. Haltmeier, A. Klingenhoff, M. Frisch, M. Bayerlein, and T. Werner. 2005. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21:2933-2942. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho, M., and D. Derse. 1991. Mutational analysis of the equine infectious anemia virus Tat-responsive element. J. Virol. 65:3468-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, B. K., M. B. Feinberg, and D. Baltimore. 1997. The κB sites in the human immunodeficiency virus type 1 long terminal repeat enhance virus replication yet are not absolutely required for viral growth. J. Virol. 71:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, K. R., M. E. Quinones-Mateu, Z. Toossi, and E. J. Arts. 2002. Impact of tuberculosis on HIV-1 replication, diversity, and disease progression. AIDS Rev. 4:165-176. [PubMed] [Google Scholar]
- 11.Dermitzakis, E. T., and A. G. Clark. 2002. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: conservation and turnover. Mol. Biol. Evol. 19:1114-1121. [DOI] [PubMed] [Google Scholar]
- 12.de Visser, J. A., R. F. Hoekstra, and H. van den Ende. 1997. An experimental test for synergistic epistasis and its application in Chlamydomonas. Genetics 145:815-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Visser, J. A., J. Hermisson, G. P. Wagner, M. L. Ancel, H. Bagheri-Chaichian, J. L. Blanchard, L. Chao, J. M. Cheverud, S. F. Elena, W. Fontana, G. Gibson, T. F. Hansen, D. Krakauer, R. C. Lewontin, C. Ofria, S. H. Rice, G. von Dassow, A. Wagner, and M. C. Whitlock. 2003. Perspective: evolution and detection of genetic robustness. Evol. Int. J. Org. Evol. 57:1959-1972. [DOI] [PubMed] [Google Scholar]
- 14.DiLeone, R. J., L. B. Russell, and D. M. Kingsley. 1998. An extensive 3′ regulatory region controls expression of Bmp5 in specific anatomical structures of the mouse embryo. Genetics 148:401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elena, S. F., L. Ekunwe, N. Hajela, S. A. Oden, and R. E. Lenski. 1998. Distribution of fitness effects caused by random insertion mutations in Escherichia coli. Genetica 102/103:349-358. [PubMed] [Google Scholar]
- 16.Elena, S. F., and R. E. Lenski. 1997. Test of synergistic interactions among deleterious mutations in bacteria. Nature 390:395-398. [DOI] [PubMed] [Google Scholar]
- 17.Gibson, G. 1996. Epistasis and pleiotropy as natural properties of transcriptional regulation. Theor. Popul. Biol. 49:58-89. [DOI] [PubMed] [Google Scholar]
- 18.Gibson, G., and G. Wagner. 2000. Canalization in evolutionary genetics: a stabilizing theory? Bioessays 22:372-380. [DOI] [PubMed] [Google Scholar]
- 19.Gomez-Gonzalo, M., M. Carretero, J. Rullas, E. Lara-Pezzi, J. Aramburu, B. Berkhout, J. Alcami, and M. Lopez-Cabrera. 2001. The hepatitis B virus X protein induces HIV-1 replication and transcription in synergy with T-cell activation signals: functional roles of NF-kappaB/NF-AT and SP1-binding sites in the HIV-1 long terminal repeat promoter. J. Biol. Chem. 276:35435-35443. [DOI] [PubMed] [Google Scholar]
- 20.Grez, M., M. Zörnig, J. Nowock, and M. Ziegler. 1991. A single point mutation activates the Moloney murine leukemia virus long terminal repeat in embryonal stem cells. J. Virol. 65:4691-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hines, R., B. R. Sorensen, M. A. Shea, and W. Maury. 2004. PU. 1 binding to ets motifs within the equine infectious anemia virus long terminal repeat (LTR) enhancer: regulation of LTR activity and virus replication in macrophages. J. Virol. 78:3407-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jasny, B. R., and L. Roberts. 2004. Genes in action. Science 306:629-650. [Google Scholar]
- 23.Jeeninga, R. E., M. Hoogenkamp, M. Armand-Ugon, M. de Baar, K. Verhoef, and B. Berkhout. 2000. Functional differences between the LTR transcriptional promoters of HIV-1 subtypes A through G. J. Virol. 74:3740-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, N. A., and A. H. Porter. 2000. Rapid speciation via parallel, directional selection on regulatory genetic pathways. J. Theor. Biol. 205:527-542. [DOI] [PubMed] [Google Scholar]
- 25.Kammandel, B., K. Chowdhury, A. Stoykova, S. Aparicio, S. Brenner, and P. Gruss. 1999. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Dev. Biol. 205:79-97. [DOI] [PubMed] [Google Scholar]
- 26.Keightley, P. D., and M. Lynch. 2003. Toward a realistic model of mutations affecting fitness. Evol. Int. J. Org. Evol. 57:683-685. [DOI] [PubMed] [Google Scholar]
- 27.Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, United Kingdom.
- 28.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 29.Larder, B. A., A. Kohli, P. Kellam, S. D. Kemp, M. Kronick, and R. D. Henfrey. 1993. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature 365:671-673. [DOI] [PubMed] [Google Scholar]
- 30.Lenski, R. E., M. R. S. Rose, and S. C. S. C. Tadler. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138:1315-1341. [Google Scholar]
- 31.Leung, T. H., A. Hoffmann, and D. Baltimore. 2004. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell 118:453-464. [DOI] [PubMed] [Google Scholar]
- 32.Liu, T., J. Wu, and F. He. 2000. Evolution of cis-acting elements in 5′ flanking regions of vertebrate actin genes. J. Mol. Evol. 50:22-30. [DOI] [PubMed] [Google Scholar]
- 33.Ludwig, M. Z., C. Bergman, N. H. Patel, and M. Kreitman. 2000. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403:564-567. [DOI] [PubMed] [Google Scholar]
- 34.MacArthur, S., and J. F. Brookfield. 2004. Expected rates and modes of evolution of enhancer sequences. Mol. Biol. Evol. 21:1064-1073. [DOI] [PubMed] [Google Scholar]
- 35.Mallon, R., J. Borkowski, R. Albin, S. Pepitoni, J. Schwartz, and E. Kieff. 1990. The Epstein-Barr virus BZLF1 gene product activates the human immunodeficiency virus type 1 5′ long terminal repeat. J. Virol. 64:6282-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maree, A. F., W. Keulen, C. A. Boucher, and R. J. de Boer. 2000. Estimating relative fitness in viral competition experiments. J. Virol. 74:11067-11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuo, Y., H. G. Drexler, A. Harashima, A. Okochi, N. Shimizu, and K. Orita. 2004. Transcription factor expression in cell lines derived from natural killer-cell and natural killer-like T-cell leukemia-lymphoma. Hum. Cell 17:85-92. [DOI] [PubMed] [Google Scholar]
- 38.Maury, W. 1998. Regulation of equine infectious anemia virus expression. J. Biomed. Sci. 5:11-23. [DOI] [PubMed] [Google Scholar]
- 39.Miralles, R., P. J. Gerrish, A. Moya, and S. F. Elena. 1999. Clonal interference and the evolution of RNA viruses. Science 285:1745-1747. [DOI] [PubMed] [Google Scholar]
- 40.Mohapatra, S., B. Chu, S. Wei, J. Djeu, P. K. Epling-Burnette, T. Loughran, R. Jove, and W. J. Pledger. 2003. Roscovitine inhibits STAT5 activity and induces apoptosis in the human leukemia virus type 1-transformed cell line MT-2. Cancer Res. 63:8523-8530. [PubMed] [Google Scholar]
- 41.Novella, I. S., D. D. Reissig, and C. O. Wilke. 2004. Density-dependent selection in vesicular stomatitis virus. J. Virol. 78:5799-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohta, T. 2002. Near-neutrality in evolution of genes and gene regulation. Proc. Natl. Acad. Sci. USA 99:16134-16137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orr, H. A. 2003. The distribution of fitness effects among beneficial mutations. Genetics 163:1519-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pastinen, T., and T. J. Hudson. 2004. Cis-acting regulatory variation in the human genome. Science 306:647-650. [DOI] [PubMed] [Google Scholar]
- 45.Peden, K., M. Emerman, and L. Montagnier. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661-672. [DOI] [PubMed] [Google Scholar]
- 46.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 47.Peters, A. D., and P. D. Keightley. 2000. A test for epistasis among induced mutations in Caenorhabditis elegans. Genetics 156:1635-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quivy, V., E. Adam, Y. Collette, D. Demonte, A. Chariot, C. Vanhulle, B. Berkhout, R. Castellano, Y. de Launoit, A. Burny, J. Piette, V. Bours, and C. Van Lint. 2002. Synergistic activation of human immunodeficiency virus type 1 promoter activity by NF-kappaB and inhibitors of deacetylases: potential perspectives for the development of therapeutic strategies. J. Virol. 76:11091-11103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rice, W. R. 1989. Analyzing tables of statistical tests. Evol. Int. J. Org. Evol. 43:223-225. [DOI] [PubMed] [Google Scholar]
- 51.Rodrigo, A. G., E. G. Shpaer, E. L. Delwart, A. K. Iversen, M. V. Gallo, J. Brojatsch, M. S. Hirsch, B. D. Walker, and J. I. Mullins. 1999. Coalescent estimates of HIV-1 generation time in vivo. Proc. Natl. Acad. Sci. USA 96:2187-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romano, L. A., and G. A. Wray. 2003. Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation. Development 130:4187-4199. [DOI] [PubMed] [Google Scholar]
- 53.Sackerson, C., M. Fujioka, and T. Goto. 1999. The even-skipped locus is contained in a 16-kb chromatin domain. Dev. Biol. 211:39-52. [DOI] [PubMed] [Google Scholar]
- 54.Sanjuan, R., A. Moya, and S. F. Elena. 2004. The contribution of epistasis to the architecture of fitness in an RNA virus. Proc. Natl. Acad. Sci. USA 101:15376-15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanjuan, R., A. Moya, and S. F. Elena. 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc. Natl. Acad. Sci. USA 101:8396-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segre, D., A. Deluna, G. M. Church, and R. Kishony. 2005. Modular epistasis in yeast metabolism. Nat. Genet. 37:77-83. [DOI] [PubMed] [Google Scholar]
- 57.Smith, S. D., M. Shatsky, P. S. Cohen, R. Warnke, M. P. Link, and B. E. Glader. 1984. Monoclonal antibody and enzymatic profiles of human malignant T-lymphoid cells and derived cell lines. Cancer Res. 44:5657-5660. [PubMed] [Google Scholar]
- 58.Stone, J. R., and G. A. Wray. 2001. Rapid evolution of cis-regulatory sequences via local point mutations. Mol. Biol. Evol. 18:1764-1770. [DOI] [PubMed] [Google Scholar]
- 59.Takahashi, H., Y. Mitani, G. Satoh, and N. Satoh. 1999. Evolutionary alterations of the minimal promoter for notochord-specific Brachyury expression in ascidian embryos. Development 126:3725-3734. [DOI] [PubMed] [Google Scholar]
- 60.van Opijnen, T., R. E. Jeeninga, M. C. Boerlijst, G. P. Pollakis, V. Zetterberg, M. Salminen, and B. Berkhout. 2004. Human immunodeficiency virus type 1 subtypes have a distinct long terminal repeat that determines the replication rate in a host-cell-specific manner. J. Virol. 78:3675-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Opijnen, T., J. Kamoschinski, R. E. Jeeninga, and B. Berkhout. 2004. The human immunodeficiency virus type 1 promoter contains a CATA box instead of a TATA box for optimal transcription and replication. J. Virol. 78:6883-6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans, R. A. Holt, J. D. Gocayne, et al. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 63.Verhoef, K., R. W. Sanders, V. Fontaine, S. Kitajima, and B. Berkhout. 1999. Evolution of the HIV-1 LTR promoter by conversion of an NF-κB enhancer element into a GABP binding site. J. Virol. 73:1331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weinberger, L. S., J. C. Burnett, J. E. Toettcher, A. P. Arkin, and D. V. Schaffer. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122:169-182. [DOI] [PubMed] [Google Scholar]
- 65.Whitlock, M. C., and D. Bourguet. 2000. Factors affecting the genetic load in Drosophila: synergistic epistasis and correlations among fitness components. Evol. Int. J. Org. Evol. 54:1654-1660. [DOI] [PubMed] [Google Scholar]
- 66.Wilke, C. O., and C. Adami. 2003. Evolution of mutational robustness. Mutat. Res. 522:3-11. [DOI] [PubMed] [Google Scholar]
- 67.Wolff, C., M. Pepling, P. Gergen, and M. Klingler. 1999. Structure and evolution of a pair-rule interaction element: runt regulatory sequences in D. melanogaster and D. virilis. Mech. Dev. 80:87-99. [DOI] [PubMed] [Google Scholar]
- 68.Wray, G. A., M. W. Hahn, E. Abouheif, J. P. Balhoff, M. Pizer, M. V. Rockman, and L. A. Romano. 2003. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20:1377-1419. [DOI] [PubMed] [Google Scholar]
- 69.Wu, C. Y., and M. D. Brennan. 1993. Similar tissue-specific expression of the Adh genes from different Drosophila species is mediated by distinct arrangements of cis-acting sequences. Mol. Gen. Genet. 240:58-64. [DOI] [PubMed] [Google Scholar]
- 70.Yuh, C. H., H. Bolouri, and E. H. Davidson. 1998. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science 279:1896-1902. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.