Abstract
Despite replicating to very high titers, coxsackieviruses do not elicit strong CD8 T-cell responses, perhaps because antigen presentation is inhibited by virus-induced disruption of host protein trafficking. Herein, we evaluated the effects of three viral nonstructural proteins (2B, 2BC, and 3A) on intracellular trafficking. All three of these proteins inhibited secretion, to various degrees, and directly associated with the Golgi complex, causing trafficking proteins to accumulate in this compartment. The 3A protein almost completely ablated trafficking and secretion, by moving rapidly to the Golgi, and causing its disruption. Using an alanine-scanning 3A mutant, we show that Golgi targeting and disruption can be uncoupled. Thus, coxsackieviruses rely on the combined effects of several gene products that target a single cellular organelle to successfully block protein secretion during an infection. These findings have implications for viral pathogenesis.
Following their entry into a permissive cell, viruses quickly initiate a cascade of events that modify the intracellular space, making it an environment conducive to viral gene expression, genome replication, and, ultimately, the rapid spread of infectious particles to other susceptible cells within the host. To accomplish this, viral genomes often encode polypeptides that have very specific, often nonoverlapping functions ranging from the shutoff of host cell transcription and translation to inhibiting the antiviral immune response by curbing cytokine secretion and antigen presentation (9). While the often very large genomes of DNA and RNA viruses can encode a small army of gene products to accomplish these tasks (2), smaller viruses must rely on a more limited number of multifunctional polypeptides.
RNA viruses in the Picornaviridae family fall into this latter category, possessing very small genomes (∼7.5 kilobases) that encode no more than a dozen gene products. Despite having such a limited number of genes, these viruses are remarkably efficient at causing a wide range of diseases, including (but not limited to) the common cold (human rhinovirus), foot-and-mouth disease (foot-and-mouth-disease virus), poliomyelitis (poliovirus), hepatitis (hepatitis A virus), and myocarditis, pancreatitis, and meningitis (coxsackievirus) (38). How do these viruses, with their limited coding capacities, achieve the rapid and complete subversion of multiple cell types within the host? The answer lies in the expression of a large ∼250 kDa viral polyprotein (from a single long open reading frame) (Fig. 1A) that is processed by virally encoded proteinases (33) to yield several precursors and mature cleavage products, each with highly specific functions during the viral life cycle (17). Interestingly, many of the nonstructural proteins have membrane-associative properties (e.g., 2B, 2BC, 2C) and act, individually or in concert with each other, to form cytoplasmic vesicles (5, 27, 29, 32) that compartmentalize the host cell and provide an environment suitable for RNA replication. Concurrent with these changes, protein trafficking within the cell is severely limited. This inhibition of protein trafficking is not required for the production of infectious virus (4, 11), suggesting that these viruses may have retained this function in response to other evolutionary pressures, such as those imposed by the immune system. Consistent with this, these viruses inhibit the cell surface display of major histocompatibility complex class I-viral peptide complexes (10), the secretion of antiviral cytokines (11), and the surface expression of antiviral cytokine receptors (24). For several picornaviruses, most notably poliovirus type 1, it has been proposed that the 3A protein is responsible for this phenomenon, specifically blocking anterograde trafficking from the endoplasmic reticulum (ER) to the Golgi (12). Major histocompatibility complex class I transport inhibition has also been observed for foot-and-mouth-disease virus (28), and it has been proposed that the 2BC protein from this virus, rather than 3A, inhibits ER-to-Golgi trafficking (20). Interestingly, both the 2B and 3A proteins of coxsackievirus B3 (CVB3) have been shown to inhibit protein secretion (35, 37), but the exact mechanism behind this phenomenon is not known. Thus, the capacity to inhibit protein trafficking is conserved among many members of the Picornaviridae, but it is clear that the viral protein(s) responsible for this process may not be absolutely conserved. Indeed, a recent analysis of the 3A polypeptides from several different picornaviruses indicates that not all 3A proteins are capable of inhibiting protein secretion (8).
FIG. 1.
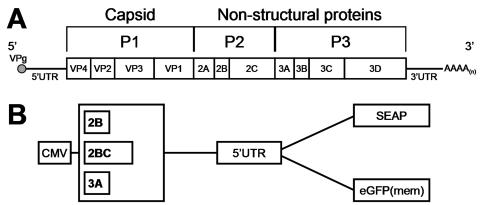
CVB3 genome organization and plasmid reporter constructs. (A) Schematic representation of the CVB3 genome. Shown is the positive-sense viral RNA containing a single open reading frame, from the product of which viral proteins are processed co- and posttranslationally. The 5′ UTR contains the internal ribosome entry site (IRES) used to direct translation of the marker proteins shown in panel B. (B) Six dicistronic plasmid constructs were prepared to examine the effects of viral proteins on protein trafficking. Two reporter proteins were employed, placental SEAP and eGFP(mem), which translocates from the cytoplasm to the plasma membrane; for each reporter, three viral proteins were cloned upstream. Two control constructs were also prepared (not shown); these expressed the reporter proteins but no viral gene.
Much of the published data suggesting that protein trafficking inhibition could function to evade the host's immune response came from in vitro analyses and cell culture studies. However, there is evidence suggesting that picornaviruses evade the immune response in vivo. Several studies have attempted to isolate picornavirus-specific CD8 T cells, but their detection, to date, has relied on extensive restimulation of the cells ex vivo (7, 19). This led us to propose that, compared to other well-characterized antiviral responses (e.g., that of lymphocytic choriomeningitis virus), the CD8 T-cell response to a picornavirus infection is greatly diminished. Indeed, using the mouse model of CVB3 infection, our laboratory has shown that this is the case (30).
The study whose results are presented here sought to understand the potential mechanism(s), at the cellular level, employed by CVB3 to inhibit host protein trafficking. By using two different experimental approaches, we show that three CVB3 proteins (2B, 2BC, and 3A) separately inhibit protein secretion but to various degrees. Confocal microscopy analyses of cells expressing 2B, 2BC, or 3A, along with an enhanced green fluorescent protein (eGFP) marker targeted to the plasma membrane, revealed that each of these three proteins possesses the ability to inhibit protein trafficking through the Golgi complex, by directly associating with this organelle. Interestingly, wild-type (wt) 3A possessed the additional function of destroying the Golgi complex itself, and studies with a mutant form of this polypeptide [3A (P19A)], previously shown to lack the ability to inhibit protein secretion (37), suggested that the proline at position 19 in 3A is important for its Golgi-disruptive function but not for its localization to this structure. Data from studies using infection time courses with wild-type CVB3 demonstrated that 3A targeting and Golgi disruption occur very early during a viral infection. Although no individual viral protein was capable of inhibiting ER with respect to Golgi secretion, data from studies using a recombinant DsRed(mem) CVB3 suggest that a synergy between viral gene products is required for this process. Our study provides important insights into the molecular and cell biology behind protein trafficking inhibition (and, probably, immune evasion) by CVB, highlighting viral genes that could be targeted, by reverse genetics, to generate one or more recombinant CVB3 vaccine strains capable of eliciting robust, long-lasting, protective CD8 T-cell responses.
MATERIALS AND METHODS
Construction of recombinant plasmids.
The 5′ untranslated region (UTR) of CVB3 was obtained using PCR from the plasmid pH3, an infectious CVB3 clone (GenBank no. U57056), generously provided by Kirk Knowlton (University of California, San Diego), and was cloned into the pCMVβ vector (BD Biosciences/Clontech; Mountain View, CA), replacing the β-galactosidase gene. The resulting vector, pCMV-UTR, contained restriction sites flanking the UTR. Two marker proteins were separately cloned downstream of the UTR: eGFP(mem) and secreted placental alkaline phosphatase (SEAP). eGFP was obtained by PCR from pEGFP-N1 (BD Biosciences/Clontech), and two complementary oligonucleotides were annealed and ligated to this fragment to construct the neuromodulin membrane targeting sequence (40) in frame with the eGFP coding sequence, generating eGFP(mem). SEAP was obtained from pSEAP2-control vector (BD Biosciences/Clontech). Plasmids encoding each of the marker proteins downstream of the UTR were used as recipients to generate dicistronic plasmids for the various picornaviral open reading frames (ORFs) described in the text. The viral ORFs (three CVB3 ORFs shown in Fig. 1B, as well as the 3A proteins from poliovirus type 1 and human rhinovirus 14; both kind gifts from Bert L. Semler, University of California, Irvine) were amplified by PCR, with 5′ start and 3′ stop codons being added. The PCR fragments were gel purified and cloned into the marker-bearing plasmids upstream of the UTR. Schematics of the CVB constructs are shown in Fig. 1B. The P19A mutant virus was generated using a QuikChange XL site-directed mutagenesis kit (Stratagene; La Jolla, CA) to incorporate 3A nucleotide changes in the context of pH3, the full-length infectious cDNA. To obtain bacterial expression of the 2BC protein, the 2BC ORF was cloned into pET15b (EMD Biosciences/Novagen, San Diego, CA). Sequences were verified by restriction digest and dideoxynucleotide sequencing.
Production of recombinant DsRed(mem) CVB3 virus.
The gene encoding a destabilized variant of the DsRed protein was obtained from pDsRed-Express-DR (BD Biosciences/Clontech), the destabilizing sequences were removed, and the neuromodulin membrane targeting sequence was added to the authentic DsRed sequence in a manner similar to that described above for eGFP(mem). A recombinant CVB3 expressing eGFP has been developed in this laboratory (14), and the eGFP in this recombinant was replaced by the DsRed(mem) ORF. The resulting plasmid was transfected into HeLa (RW) cells (a kind gift from Rainer Wessely, then at University of California, San Diego), from which infectious virus was derived. The virus titer was calculated by carrying out a plaque assay on HeLa (RW) monolayers as previously described (15).
DNA transfections and virus infection time courses.
HeLa (RW) cell monolayers were grown to approximately 75% confluence in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum supplemented with l-glutamine. For SEAP experiments, cells were grown in six-well culture dishes; for confocal microscopy studies, cells were grown on glass coverslips. DNA transfection was carried out using Lipofectamine with PLUS reagent (Invitrogen) in the presence of OptiMEM, according to the manufacturer's protocol. A total of 1 μg and 0.5 μg DNA was transfected for the SEAP and eGFP(mem) studies, respectively. For virus infection time courses, cells grown on coverslips were washed twice with saline solution, and virus was adsorbed at a multiplicity of infection (MOI) of 0.1 in OptiMEM for 30 min. Following adsorption, complete DMEM was added and the time course experiment began.
SEAP assays.
Triplicate transfections were carried out in six-well culture dishes of HeLa (RW) cells at ∼75% confluence. After transfection, the DNA-Lipofectamine PLUS reagent cocktail was replaced with 2 ml complete DMEM. At the indicated time points, 1 ml of medium was removed and stored at −80°C, the remaining 1 ml was removed, the cells were washed twice in saline solution, and 0.5 ml of 1× passive lysis buffer (Promega; Madison, WI) containing Complete Mini protease inhibitor cocktail (Roche Applied Science) was added. Following a 5 min lysis step, monolayers were harvested and stored at −80°C. Both medium and lysate fractions were thawed and centrifuged for 10 min at 16,000 × g, and equal portions of supernatant (0.25 μl lysate and 1.0 μl medium) were separately analyzed using a Great EscAPe SEAP chemiluminescence kit (BD Biosciences/Clontech). Chemiluminescence was measured on a LB96V MicroLumat Plus microplate luminometer (EG&G Berthold, Pforzheim, Germany) using a 10-s interval, and sample values were corrected for background by subtracting the alkaline phosphatase activity value obtained using mock-transfected cells. The corrected values were used to calculate the percentage of secreted SEAP in each sample (see legend to Fig. 2).
FIG. 2.
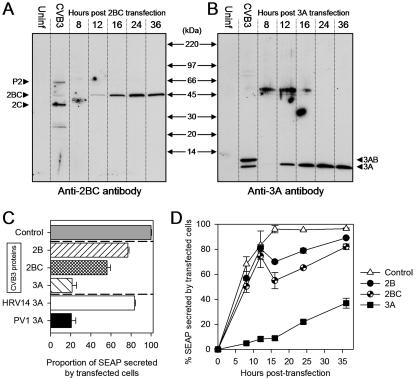
CVB3 proteins inhibit secretion of SEAP to different extents and with different kinetics. (A) 2BC protein expression profile. At the indicated times posttransfection, lysates from HeLa cells expressing the pCMV-2BC-UTR-SEAP construct (see Fig. 1) were harvested in sodium dodecyl sulfate lysis buffer and analyzed by Western blotting using an anti-2BC rabbit polyclonal antibody. A lysate harvested from cells infected with wt CVB3 (second lane) served as a marker; the anti-2BC serum detected the P2 precursor protein, as well as 2BC and the mature 2C polypeptide (arrowheads). Uninf, uninfected. (B) 3A protein expression profile. A similar analysis, using a polyclonal antibody directed against the amino terminus of 3A, was done using lysates from cells transfected with the pCMV-3A-UTR-SEAP construct (see Fig. 1). Again, a lysate from CVB3 infected cells was used as a marker (second lane). The anti-3A serum was capable of detecting the 3AB precursor protein, as well as the mature 3A polypeptide (arrowheads). (C) The three CVB3 protein-SEAP plasmids shown in Fig. 1 were transfected into HeLa (RW) cell monolayers, and the percentages of secreted SEAP activity were determined 16 h posttransfection (hatched bars). Also included were two constructs encoding the 3A protein from either human rhinovirus type 14 (open bar) or poliovirus type 1 (black bar) and a control that lacked any viral protein (gray bar). The experiment was done in triplicate, and all values for intracellular and extracellular SEAP were corrected against background alkaline phosphatase activity from mock transfections before the following formula was applied:  . The percentage of secreted SEAP in the control (gray bar) was ∼95% and was assigned the value 100; all other samples were then normalized to that value. (B) Secreted SEAP activity over a 36 h time course. The percentages of secreted SEAP were determined (as described for panel A) at 0, 8, 12, 16, 24, and 36 h posttransfection.
. The percentage of secreted SEAP in the control (gray bar) was ∼95% and was assigned the value 100; all other samples were then normalized to that value. (B) Secreted SEAP activity over a 36 h time course. The percentages of secreted SEAP were determined (as described for panel A) at 0, 8, 12, 16, 24, and 36 h posttransfection.
Immunofluorescence reagents and immunocytochemistry.
Hoechst 33342 was obtained from Molecular Probes. Rabbit anti-β-tubulin conjugated to Cy3 was obtained from Sigma-Aldrich (St. Louis, MO), mouse anti-GM130 (both unconjugated and fluorescein isothiocyanate conjugated) was obtained from BD Biosciences. Rabbit anti-calnexin, mouse anti-calnexin, and mouse anti-58K were obtained from Abcam (Cambridge, MA). Goat anti-mouse tetramethyl rhodamine isothiocyanate, goat anti-rabbit tetramethyl rhodamine isothiocyanate, chicken anti-mouse Alexa 647, and chicken anti-rabbit Alexa 647 were obtained from Molecular Probes. All primary antibodies were diluted 1:100; all secondary antibodies were diluted 1:250. Following transfection or infection, cells were washed in phosphate-buffered saline (PBS) and fixed either in ice-cold methanol:acetone (1:1 ratio) for 10 min at −20°C (tubulin, 2B, 2BC, and 3A immunostaining) or in 2% electron microscopy-grade formaldehyde (Polysciences Inc., Warrington, PA) in PBS for 2 to 3 min (all other immunostaining experiments). Cells were permeabilized with 0.5% Triton X-100 in PBS for 3 to 5 min and washed twice in PBS, and a 10% normal goat serum blocking solution (in PBS) was added for 1 h. Cells were washed again, incubated with primary antibody for 1 h, washed twice in PBS, and subjected to staining with secondary antibody for 30 min. Following a 5-min PBS wash, cells were counterstained with Hoechst 33342 (1:5,000 dilution in PBS) for 15 min. Coverglasses were mounted onto slides with ImmunoFluor anti-fade solution (MP Biomedicals, Irvine, CA) and sealed with Super Top Speed clear nail polish (Revlon, New York, NY).
Confocal microscopy.
Fixed and stained samples were prepared as described above and then viewed using a Rainbow Radiance 2100 laser scanning confocal system attached to a Nikon TE2000-U inverted microscope (Bio-Rad-Zeiss). Images were acquired using Laser Sharp 2000 software and then imported and further analyzed for quantitative colocalization using three independent software packages: Laser Sharp (Bio-Rad-Zeiss), LSM examiner (Zeiss), and Image J (NIH Imaging; http://rsb.info.nih.gov/ij). Colocalization between two fluorescently labeled reagents was quantified by obtaining the threshold range of real over background signal and then using the average real threshold range to calculate the correlation coefficients (M values) for at least 30 cells in four separate experiments.
Polyclonal antibodies against CVB3 3A and 2BC proteins.
Anti-3A polyclonal sera were produced by the antibody core facility of The Scripps Research Institute. A recombinant peptide consisting of a Cys-Gly dipeptide coupled to 10 amino acids (VYREIKISVA) from the amino-terminal region of CVB3 3A was generated and conjugated to keyhole limpet hemocyanin, and 200 μg (in a total of 1 ml) (RIBI adjuvant system; Corixa, Seattle, WA) of the keyhole limpet hemocyanin conjugate was injected subcutaneously into two New Zealand White rabbits. Multiple boosts were carried out. The 3A antiserum utilized for the studies presented here had been subjected to a sodium aluminosilicate (SAS) precipitation step. The pET15b-2BC plasmid described above was transformed into BL21(DE3) bacteria (Invitrogen) and plated on LB agar containing ampicillin. A single colony was inoculated into a 3 ml overnight culture in CircleGrow (MP Biomedicals) liquid medium-100 μg/ml ampicillin. The following day, bacteria were pelleted and resuspended in 3 ml Power Broth (US Biological; Swampscott, MA), 250 μl of which was inoculated into a 50 ml culture of Power Broth containing 2× ampicillin. At the logarithmic phase of bacterial growth (optical density at 600 nm, 0.4 to 0.7), the entire culture was inoculated into 1 liter of fresh Power Broth containing ampicillin. Upon arriving at logarithmic phase, recombinant protein expression was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) for a final concentration of 1 mM followed by incubation (with shaking) at 25°C for an additional 3 h. Cells were harvested by centrifugation and resuspended in equilibration buffer (50 mM sodium phosphate [pH 7.0], 500 mM NaCl, 10% glycerol). Following resuspension, a 1/10 final volume of 10× BugBuster reagent (EMD Biosciences/Novagen) was added to lyse the cells. Cells were rocked for 30 min at 25°C, and the insoluble protein fraction was pelleted by centrifugation (10,000 × g for 30 min). 2BC, expressed almost entirely in inclusion bodies, was solubilized in equilibration buffer containing 8 M urea. The resulting denatured protein pool consisted of at least 90% target protein, as analyzed by Coomassie blue staining. For antibody production, multiple subcutaneous injections of 200 μg recombinant protein (in a total of 1 ml RIBI adjuvant system) were carried out in two New Zealand White rabbits. The 2BC antiserum used here had been subjected to an SAS precipitation step, followed by protein G column purification.
RESULTS AND DISCUSSION
Construction of reporter plasmids to determine the effects of CVB3 polypeptides on protein trafficking and secretion.
The typical picornavirus genome encodes an array of polypeptides that have very specific functions in viral translation, RNA replication, and the modification of host cell processes, and the nonstructural proteins (regions P2 and P3 in Fig. 1A) have been implicated in the interruption of host protein trafficking. Therefore, we designed plasmids that allowed us to screen the CVB3 nonstructural proteins for their ability to (i) block protein secretion and (ii) modify trafficking of proteins destined for the plasma membrane. Blockade of protein secretion was evaluated using SEAP as a marker (3), and intracellular trafficking was visualized with eGFP(mem), an enhanced green fluorescent protein targeted to the plasma membrane (see Materials and Methods). As shown in Fig. 1B, six constructs were prepared in which the CVB3 proteins and marker proteins were expressed from dicistronic transcripts. The downstream cistron, whose translation is driven by the CVB3 5′ untranslated region (containing the internal ribosome entry site), expressed either SEAP or eGFP(mem), and the upstream cistron encoded one of the three indicated CVB3 proteins. In addition to these CVB3 constructs, we also generated two control constructs that do not express any viral gene products [pCMV-UTR-SEAP and pCMV-UTR-eGFP(mem)] and constructs that expressed the 3A protein from poliovirus type 1 or from human rhinovirus 14 (not shown).
Transient transfection of SEAP constructs reveals that CVB 2B, 2BC, and 3A affect protein secretion to various degrees.
To evaluate the expression of the CVB3 proteins, it was necessary to employ reagents that permitted their detection in infected and transfected cells. Antibodies specific for individual CVB3 proteins are not generally available, and attaching an immunologically detectable “tag” can abrogate the biological function of picornaviral 3A proteins (8), so we developed polyclonal antisera against the CVB3 3A and 2BC proteins by peptide immunization or immunization with bacterially produced recombinant protein (see Materials and Methods). As shown in Fig. 2A (2nd lane), polyclonal antiserum induced by the 2BC protein recognized the P2, 2BC, and 2C gene products from infected cells; the 2B protein was not identifiable by Western blot analysis, although it was detectable in native form in transfected cells (see Fig. 4). In cells transfected with the 2BC plasmid, the 2BC protein was barely detected at 12 h posttransfection, but its expression level increased thereafter. The antiserum induced by 3A peptide immunization detected the 3AB and 3B proteins in infected cell extracts, as well as the 3A protein in cells transfected with the 3A-encoding plasmid (Fig. 2B); this viral protein was readily detected at 12 h posttransfection.
FIG. 4.
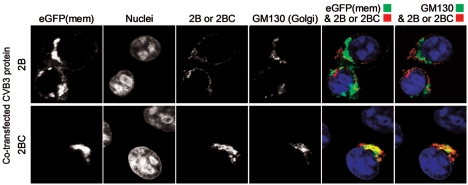
In transfected cells, the 2B and 2BC proteins accumulate in the Golgi and in a perinuclear compartment. HeLa (RW) cells were transfected with either the 2B or the 2BC plasmid (as indicated), and 16 h later, polyclonal anti-2BC sera were used to localize the 2B and 2BC proteins (red fluorescence) and to determine their colocalization with the Golgi marker GM130 (far-red fluorescence). Hoechst 33342 nuclear staining was employed to reveal nuclei. Merged images of eGFP(mem) and 2B or 2BC and of 2B-2BC and GM130 (pseudocolored green) are shown.
The effects of CVB3 2B, 2BC, and 3A on protein secretion are shown in Fig. 2C. Reporter plasmids were transfected into HeLa cells and, 16 h later, the extracellular and intracellular levels of SEAP were determined by chemiluminescence and were used to calculate the percentage of SEAP secreted in the presence or absence of each viral gene product. In the absence of any CVB3 protein expression, ∼95% of SEAP activity was detectable in the extracellular fraction; in Fig. 2C, this control has been assigned the value 100, and the proportions of secreted SEAP in the other samples have been normalized to this. CVB3 2B reduced secretion by ∼20%, which is similar to a result in a previous report (35). An inhibitory function of the 2BC precursor has not been previously reported, and we show here that it is even more effective than 2B, inhibiting SEAP secretion by ∼40 to 50%. The most dramatic inhibition was achieved with the CVB3 3A protein, which inhibited SEAP secretion by ∼80%, a result very similar to that obtained with a construct that expressed the poliovirus type 1 3A polypeptide (Fig. 2C), which has previously been shown to have a strong antisecretory effect (12). The 3A protein from another picornavirus, human rhinovirus 14, was much less effective, suggesting that not all picornaviral 3A proteins possess this activity and confirming a report published while this paper was in preparation (8). Next, a time course was carried out to determine how quickly these CVB3 proteins interrupted SEAP secretion. The onset of the antisecretory activity of 2BC occurred at 12 to 16 h posttransfection, coincident with the expression of the 2BC protein in these cells, as judged by Western blot analysis; the antisecretory effect of the 2B plasmid shows very similar kinetics, but we were unable to detect this protein by use of our antibody, even in infected cell extracts, so we cannot definitively correlate 2B expression with its antisecretory function. In contrast, the 3A protein acted very rapidly, inhibiting secretion by ∼80% (compared to control results) at 12 h posttransfection (Fig. 2D). At this time point, the 3A protein generated a much more intense signal on the Western blot than the 2BC construct (Fig. 2A and B); this apparent earlier and more abundant expression of 3A may explain, in part, its more rapid and profound impact on SEAP secretion. The reason(s) for the increased SEAP secretion observed at later time points in this assay are not known; we speculate that they may be related to threshold effects of protein concentration, combined with the dilutional effects of cell division, and possible cytopathic effects that release SEAP into the tissue culture medium.
CVB3 2B and 2BC inhibit protein trafficking through the Golgi, while 3A leads to disruption of this organelle.
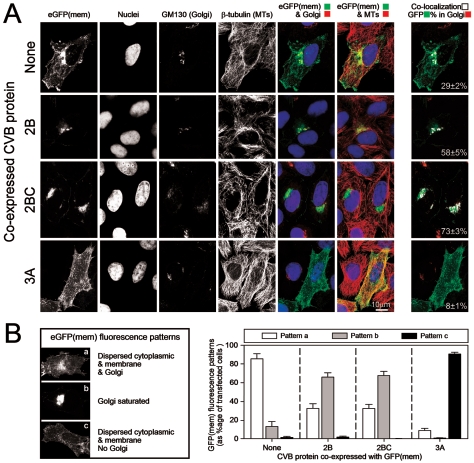
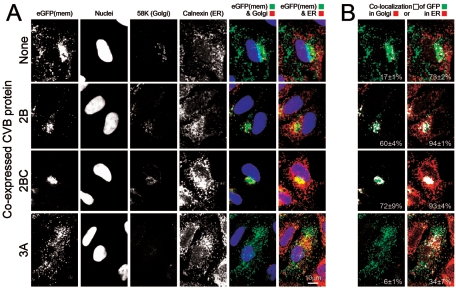
The data in Fig. 2 indicate that multiple CVB3 viral gene products (2B, 2BC, and 3A) mediate inhibition of protein secretion. We next sought to determine which step(s) in the intracellular secretion pathway are inhibited by each of these three proteins by transfecting cell monolayers with the eGFP(mem) dicistronic constructs shown in Fig. 1B. At 16 h posttransfection, laser scanning confocal microscopy was used to quantitate the proportion of eGFP in two main subcellular compartments: the Golgi complex and ER. Representative images are shown in Fig. 3A. In the absence of CVB3 protein expression (top row), eGFP(mem) is found in the plasma membrane (first image), in the Golgi (fifth image), and along microtubules (sixth image). We determined the percentage of eGFP(mem) protein colocalized to the Golgi complex by analyzing in detail, in four separate experiments, an average of 30 cells and computing the colocalization coefficient for each. Those values were averaged, and the resulting value, along with the standard error of the mean, is shown in each far right-hand panel, in which the colocalized points are displayed in white. In the absence of CVB3 protein expression, ∼30% of the eGFP(mem) protein is found within the Golgi complex. Transfection of the 2B-expressing eGFP(mem) dicistronic constructs (Fig. 3A, second row) resulted in an approximately twofold-higher level (∼60%) of eGFP(mem) protein in the Golgi apparatus, demonstrating that 2B causes an accumulation of trafficking protein in this subcellular compartment. Interestingly, 2BC expression (third row) resulted in a still higher colocalization of eGFP(mem) in the Golgi (∼75%), providing one possible mechanism to explain why 2BC is a more efficient inhibitor of SEAP protein secretion (Fig. 2). Additionally, in the presence of the 2B or 2BC polypeptide, virtually no eGFP(mem) protein is observed in vesicles tracking along microtubule structures between the Golgi complex and the plasma membrane (Fig. 3A, rows 2 and 3, sixth column), suggesting that much of the eGFP is prevented from transiting to the cell surface; consistent with this, there is reduced expression of eGFP(mem) at the plasma membrane. In addition, the accumulation of eGFP(mem) in the Golgi of cells transfected with 2B or 2BC implies that trafficking within the Golgi stacks, and retrograde trafficking from Golgi to ER, has been arrested. We also examined the effects of 3A polypeptide expression on trafficking of the eGFP(mem) marker protein (Fig. 3A, bottom row) and found that, in virtually all of the cells inspected, eGFP(mem) was highly dispersed. To more directly evaluate the effect of the 3A protein on the Golgi complex, we studied the distribution of the GM130 protein. GM130 is one of the earliest structural proteins necessary for Golgi biogenesis and, therefore, its dynamic nucleation (16); furthermore, this protein plays an important role in maintaining the integrity of the cis- and medial faces of the Golgi matrix (23). GM130 staining was very faint and diffuse, indicating that the Golgi complex was almost completely disrupted (Fig. 3A, bottom row, third column; compare transfected [eGFP-positive] cell with adjacent nontransfected cell). This result is surprising, as the 3A proteins of poliovirus (12, 32) and CVB (37) have been reported to localize in the ER and to prevent anterograde ER-to-Golgi trafficking. Our data indicate that the 3A protein, expressed alone, leads to disruption of the Golgi organelle, a major trafficking point in the secretory pathway; this would explain the profound reduction in SEAP secretion that we observed. Interestingly, microtubule staining in the presence of 3A expression shows that despite disrupting the Golgi, where the microtubule organizing center (MTOC) resides (39), the microtubule network remains intact (Fig. 3A, fourth row, fourth column). Immunostaining of two key components of the MTOC (γ tubulin and the centriole) confirmed that the integrity of the MTOC remains uncompromised in 3A-transfected cells (data not shown).
FIG. 3.
Viral proteins 2B and 2BC inhibit trafficking out of the Golgi complex, while 3A disrupts this organelle completely. (A) The localization of eGFP(mem) protein following transfection of the four indicated eGFP(mem) dicistronic constructs is shown, along with staining of nuclei (Hoechst 33342), the Golgi marker GM130 (Alexa 647; far-red fluorescence), and β-tubulin (CY3 conjugate; red fluorescence). Colored images were merged (red pseudocoloring was used for the far-red GM130 signal), and colocalization analysis was done to determine the percentage of eGFP(mem) found in the Golgi complex (last column; white pixels). Colocalization percentages (with standard errors of the means) are given and were calculated using values from a minimum of 30 different transfected cells. (B) Three main eGFP(mem) patterns, termed a, b, and c, were observed. Between 50 and 100 cells were assessed for their phenotypic eGFP(mem) distribution, and the results were graphed for each of the four constructs examined.
Cells transfected with the eGFP(mem) dicistronic constructs displayed one of three main fluorescence patterns, characterized by eGFP(mem) distribution and Golgi labeling; representative examples are shown in Fig. 3B. Pattern a consists of eGFP(mem) protein dispersed throughout the cytoplasm, within the Golgi, and at the plasma membrane; pattern b consists of a majority of eGFP(mem) protein found within the Golgi complex; and pattern c consists of eGFP(mem) dispersed throughout the cytoplasm, in the absence of an intact Golgi apparatus. To quantitate the observed effects, and to better compare the effects of the three CVB3 dicistronic constructs, patterns for 50 to 100 different cells were determined, in four independent experiments, and the percentages of total cells showing each pattern were calculated (Fig. 3B, graph). The great majority of cells transfected with the control eGFP(mem) construct showed the pattern a distribution, indicating the normal trafficking pattern of the marker protein. In stark contrast, ∼60% of the 2B- and 2BC-transfected cells conformed to pattern b [Golgi saturated with eGFP(mem)], and ∼90% of cells transfected with the 3A construct showed a completely disrupted Golgi and diffuse cytoplasmic eGFP(mem) distribution (pattern c). Taken together, the data in Fig. 2 and Fig. 3 suggest that 2B and 2BC act to inhibit vesicular trafficking out of the Golgi complex (reducing protein secretion by ∼30 to 50%) and that the 3A polypeptide has Golgi-disruptive properties. Thus, at least three different CVB3 proteins functionally target the Golgi to disrupt protein secretion, at the level of blockade (2B and 2BC) and physical disruption (3A).
CVB3 2B and 2BC proteins accumulate in the Golgi complex, and in a perinuclear compartment, in transfected cells.
The availability of the 2BC-induced polyclonal antiserum allowed us to track the location of this protein in transfected cells. Although the 2B protein was not detected on Western blots using this antiserum, we also analyzed cells transfected with the 2B-encoding plasmid and, as shown in Fig. 4, the native viral protein was detected. Both the 2B and 2BC proteins caused the accumulation of eGFP(mem) in the Golgi apparatus, as previously observed, and this antibody revealed that the viral proteins were also present in this organelle; this was especially true for the 2BC precursor, much of which colocalized with the GM130 marker. The mature viral 2B protein was also distributed in a perinuclear compartment that did not stain for GM130 and probably is distinct from the Golgi complex. The ability of this antiserum to identify both 2B and 2BC proteins is advantageous for transfected cell studies but also means that these two proteins cannot be distinguished in virus-infected cells; therefore, equivalent analyses of virus-infected cells are not shown.
3A disrupts the entire Golgi network and does not prevent protein exit from the ER.
Previous reports have suggested that the 3A protein from several picornaviruses, including CVB3, acts to inhibit ER-to-Golgi trafficking (12, 37), with retention of marker protein in the ER. However, the results discussed above do not corroborate those findings and suggest that eGFP(mem) protein may be free to leave the ER, even in the presence of the 3A protein. Therefore, our next experiment was designed to accomplish two goals: (i) to determine whether CVB 3A-mediated Golgi disruption affects the entire Golgi network and (ii) to directly assess the colocalization of eGFP(mem) protein with the ER. In addition to exploiting the GM130 protein as a Golgi marker, we used a well-established global anchoring protein that also is found within this organelle. This marker, the 58K protein, acts as an anchor site for microtubules on the cytoplasmic face of the Golgi, coordinating interactions between microtubule-associated proteins and Golgi-associated proteins and thereby acting as a dynamic linker that directs the Golgi to reside near the MTOC (6). As shown in Fig. 5A, the 58K Golgi marker appears normal in cells expressing the 2B and 2BC proteins (rows 2 and 3, third column), but it is completely dispersed in cells expressing 3A (row 4, third column), consistent with a disassembly of the entire Golgi complex. The colocalization values for eGFP(mem) and the 58K protein (Fig. 5B) were similar to those obtained using the GM130 marker (compare column 7 in Fig. 3A with column 1 in Fig. 5B). The dramatic effect of the CVB3 3A protein on the 58K and GM130 markers suggests that the entire Golgi complex, a dynamic entity which can replenish itself from various directions, is absent. To determine whether 3A caused retention of trafficking proteins within the ER, immunostaining for calnexin (36) was carried out. High (∼90%) ER colocalization values were noted for eGFP(mem) in 2B- and 2BC-expressing cells. This was surprising because of the strong evidence (Fig. 3) that these proteins caused accumulation of eGFP(mem) in the Golgi. How can this paradox be resolved? We propose that these data reflect a well-established limit of resolution in use of the light microscopy approach; the Golgi complex is closely enveloped by ER, and so any protein present in the Golgi complex will appear also to colocalize with the ER. The association between these two organelles is clearly interdigitated (13, 21, 22, 26, 31), and because of continuous membrane insertion, it is very difficult to distinguish between them at certain faces of the Golgi. We confirmed this phenomenon by measuring the colocalization of the cis-Golgi protein GM130 with the ER in uninfected cells; 98 to 99% of this Golgi-specific protein was found to colocalize with the ER (not shown). Thus, in the presence of an intact Golgi apparatus, apparent ER localization should be interpreted with caution. This artifact does not affect cells expressing 3A, because they lack a discernible Golgi complex; and in these cells, only ∼35% of eGFP(mem) protein colocalized with the ER (Fig. 5B, bottom row), suggesting that the 3A protein does not cause marked retention of trafficking protein within the ER. As previously mentioned, our findings contrast with previously published studies of CVB3 3A in which the investigators reported a much higher colocalization of the marker protein (temperature-sensitive vesicular stomatitis virus-GP) with the ER (37). We propose that 3A, when expressed in isolation from its normal viral consorts, does not function to hold trafficking host proteins in the ER, as previously proposed; rather, the largely cytoplasmic, vesicular distribution of eGFP(mem) in these cells suggests that either (i) the entry of newly translated protein into the ER is blocked or, more likely, (ii) the eGFP(mem) protein is transported out of the ER but, in the absence of an intact destination (the Golgi), is randomly distributed throughout the cytoplasm. Our laboratory is currently examining these two possibilities.
FIG. 5.
Immunostaining of the 58K marker confirms 3A-mediated Golgi disruption, and calnexin staining results suggest that 3A does not inhibit protein exit from the ER. (A) Cells were transfected as described for Fig. 3. Anti-58K (red fluorescence) and anticalnexin ER (far-red fluorescence) antibodies were utilized along with Hoechst 33342 to stain nuclei. eGFP(mem) signal was merged either with the anti-58K Golgi or calnexin ER staining (pseudocolored red), as indicated. (B) Colocalized points (white pixels) were assessed and quantitated as described for Fig. 3, indicating the percentage of eGFP(mem) colocalizing with either the Golgi or ER subcompartment.
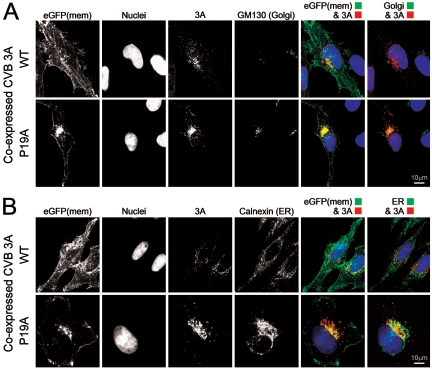
3A associates directly with the Golgi to mediate its disruption.
Studies with poliovirus (32) and CVB (37) have suggested that their 3A proteins may inhibit ER-to-Golgi trafficking by localizing to the ER subcompartment itself. However, recent data have suggested that not all picornavirus 3A proteins directly target this organelle (20), and our observation of Golgi disruption suggested—but did not prove—that the CVB3 3A protein might traffic beyond the ER. Thus, we used our anti-3A polyclonal antiserum to determine the localization of 3A in transfected cells. We also evaluated a 3A polypeptide harboring a single amino acid mutation (P19A), recently reported to drastically reduce the ability of CVB3 to inhibit protein secretion (37), in order to determine the effects of this mutation on 3A localization, Golgi integrity, and interruption of protein trafficking [using eGFP(mem)]. Two dicistronic constructs encoding wt or mutant 3A were separately transfected, and 16 h later the cells were immunostained with antibodies against 3A and GM130 (Fig. 6A, top row). wt 3A protein largely disrupted the Golgi complex but was detected in the region where the Golgi apparatus once resided (top row; compare the third column to the location of remaining GM130 signal in the fourth column), suggesting that wt 3A may have directly associated with components of the Golgi organelle prior to its disruption (see red 3A signal and pseudocolored green GM130 signal in merged image, top row, last column). Immunofluorescence studies of the P19A mutant 3A protein (Fig. 6A, bottom row) revealed that this protein localizes almost exclusively to the Golgi (see 3A/GFP merged image, fifth column) and in structures trafficking along microtubules (data not shown) in the absence of any disruption of the Golgi organelle itself. These results suggest that (i) when expressed in isolation, the CVB3 3A protein transits to the Golgi; (ii) the P19 residue is not required for this trafficking; and (iii) the P19 residue is required for Golgi disruption. Because our findings differ from published data, we considered it important also to directly evaluate CVB3 3A localization in the ER. Immunofluorescence studies with antibodies against 3A and calnexin (Fig. 6B) showed that wt 3A did not noticeably overlap with the ER, while the P19A mutant protein appeared to overlap, at least partially, with the ER. However, given that the P19A signal localized at or very near the GM130 Golgi marker, we propose that this apparent ER colocalization is, once again, a byproduct of the very close juxtaposition of the Golgi and ER subcompartments within the cell; it is not observed with the wt 3A protein, because the Golgi is disrupted. Taken together, the data in Fig. 6 suggest that when expressed in isolation, the 3A protein from CVB3 3A (Woodruff) is not retained in the ER and instead moves to and disrupts the Golgi apparatus. Thus, all three CVB3 proteins (2B, 2BC, and 3A) target the Golgi complex to stop protein transit through this organelle. The mechanism(s) by which the 2B and 2BC proteins inhibit secretion remains unknown. Interestingly, the P19A mutant causes most of the trafficking eGFP(mem) protein to be retained in this cellular subcompartment (Fig. 6A, bottom row, column 5), in a pattern very similar to that observed with 2B and 2BC expression (Fig. 3 and Fig. 5). Thus, after entering the Golgi apparatus, the 3A protein can interrupt trafficking to some extent, even in the absence of Golgi disruption.
FIG. 6.
Polyclonal antiserum against CVB 3A shows that this protein moves to the Golgi apparatus and mediates its disruption. (A) HeLa (RW) cells were transfected with either the wild-type (WT) 3A construct (top row) or P19A, a mutant 3A construct previously shown to be deficient in protein secretion inhibition (bottom row). At 16 h later, polyclonal anti-3A sera were used to localize both versions of 3A (red fluorescence) and to determine their colocalization with the Golgi marker GM130 (far-red fluorescence). Hoechst 33342 nuclear staining was employed to reveal nuclei. Merged images of eGFP(mem) and 3A and of 3A and GM130 (pseudocolored green) are shown. (B) The experiment was repeated, this time to evaluate ER localization using calnexin staining (far-red fluorescence, shown as green pseudocolor).
3A associates with the Golgi early in CVB3 infection.
Figure 6 demonstrates that the CVB3 3A protein, expressed in isolation, targets the Golgi complex and interrupts protein trafficking. To test whether this occurs also during virus infection, HeLa (RW) cell monolayers were infected with wt CVB3, and the polyclonal antibody was used to examine the localization of the 3A protein at several time points postinfection (Fig. 7A). wt 3A protein first became detectable by immunofluorescence at approximately 3 h postinfection (Fig. 7A, first row, third column) and was localized to a region that corresponded to the Golgi complex (top row; compare first and third columns and merged image in fourth), which had not yet been completely disrupted. By 5 h postinfection (Fig. 7A, second row), infected cells lacked a discernible Golgi organelle (third column), and 3A localized in a reticular pattern reminiscent of the ER (first image and merged panel, last column). Thus, during CVB3 infection, 3A first localizes to the Golgi complex, disrupts this organelle, and thereafter takes on a vesicular distribution that is a hallmark feature of the rearranged vesicles upon which RNA replication takes place.
FIG. 7.
3A localizes to the Golgi early in CVB3 infection, and trafficking proteins ultimately accumulate in the ER. (A) Cells were infected with wild-type (WT) CVB3 at an MOI of 0.1, and at early (3 h; top row) or late (5 h; bottom row) time points postinfection, the cells were fixed and stained with anti-3A serum (red fluorescence), Hoechst 33342, and GM130 antibody (far-red fluorescence). GM130 was pseudocolored green in the merged images (last column). (B) A DsRed(mem)-expressing recombinant CVB3 was used to infect HeLa (RW) cells at an MOI of 0.1. At an early time point (3 h; top row) and a later time point (5 h; bottom row), cells were fixed and stained with Hoechst 33342, anti-GM130 (conjugated to fluorescein isothiocyanate, green fluorescence), and anticalnexin (far-red fluorescence) to examine the localization of transiting DsRed(mem) protein. Calnexin was pseudocolored green in the merged images. DsRed(mem) colocalization with Golgi or ER is shown in white pixels.
During a viral infection, protein trafficking is inhibited at a step before the Golgi.
2B, 2BC, and 3A can act independently to inhibit protein trafficking through the Golgi, but the data in Fig. 7A indicate that, during a viral infection, their effects are modulated. Therefore, we sought to determine the cumulative effects of CVB3 infection on protein trafficking. To accomplish this, we generated a recombinant CVB3 expressing the DsRed(mem) protein (see Materials and Methods). Following infection of HeLa (RW) monolayers with CVB3 DsRed(mem) at an MOI of 0.1, we observed cells at early (∼3 h) and late (∼5 h) times postinfection and assessed the localization of transiting DsRed(mem) protein by immunostaining both the Golgi marker GM130 and the ER marker calnexin (Fig. 7B). At an early time point postinfection (top row), weak labeling of the GM130 marker was observed, suggesting an incomplete disruption of the Golgi complex (top row, third column) similar to what was observed for wt CVB3, as shown in Fig. 7A. Interestingly, the DsRed(mem) protein localized to a perinuclear region that resembled the remainder of the Golgi (first column), indicating a stoppage of transiting protein at, or very near, this organelle early during an infection. Colocalization analysis of DsRed(mem) with the Golgi, as well as with calnexin (ER) (top row, last two columns), revealed that the DsRed(mem) protein resided in an ER subcompartment closely associated with remnants of the Golgi. At a later time postinfection (5 h; bottom panel), the Golgi was completely absent, while an increased quantity of DsRed(mem) marker protein in the ER was observed (see colocalization results, bottom row, last column). Thus, during infection, trafficking protein eventually accumulates at a step before the Golgi complex.
Conclusions.
In conclusion, our studies confirm and extend previous work and show that several picornaviral polypeptides have antisecretory effects. Why should a virus with such a small genome encode such apparently redundant functions? RNA viruses are highly susceptible to the deleterious effects of random, polymerase-generated mutations, and having multiple gene products with overlapping antisecretory functions afford picornaviruses some evolutionary flexibility while maintaining their ability to evade the host cellular immune response. The requirements for successful immune evasion are stringent, because antiviral T cells can exert their in vivo antiviral effects with extraordinary rapidity (18); thus, if viruses are to survive, they must express their countermeasures even more rapidly. It is, therefore, intriguing that the viral 2BC precursor protein—not previously known to have antisecretory activity—is likely to be expressed earlier than its product, 2B, to which such functions have been ascribed. Herein, we report for the first time that 2BC has antisecretory effects and that these effects are greater than those of 2B. We propose, therefore, that 2BC plays a major role in early immune evasion and that the part played by 2B is less critical; this protein subserves other vital functions later in the viral life cycle, including acting as a “viroporin” to increase intracellular membrane permeability and facilitating the release of progeny virions (1, 25, 34). However, the antisecretory effect of 2BC is incomplete (Fig. 2) and is dramatically enhanced by the effects of the 3A protein, which targets, and disrupts, the Golgi apparatus. As the Golgi begins to disperse in an infected cell, transiting protein becomes trapped in the ER (Fig. 7). Why is this ER localization of transiting protein observed in infected cells but not in 3A-transfected cells (Fig. 5)? We speculate that other viral proteins, perhaps 2B, 2BC, or a precursor of the mature 3A molecule, may play a role in ER localization. Finally, our data underline the fact that the effects of one viral protein, expressed in isolation, may not accurately represent its activity in a biological context.
Acknowledgments
We thank Annette Lord for excellent secretarial support, Bert L. Semler (University of California, Irvine) for the gift of plasmids pT7PV1 and pT7HRV14, and Peter Sims (Dept. of Molecular and Experimental Medicine) for use of a luminometer.
This work was supported by National Institutes of Health awards R01 AI-42314 (J.L.W.) and F32 AI-065095 (C.T.C.).
Footnotes
This is manuscript number 17878-MIND from the Scripps Research Institute.
REFERENCES
- 1.Agirre, A., A. Barco, L. Carrasco, and J. L. Nieva. 2002. Viroporin-mediated membrane permeabilization. Pore formation by nonstructural poliovirus 2B protein. J. Biol. Chem. 277:40434-40441. [DOI] [PubMed] [Google Scholar]
- 2.Alcami, A. 2003. Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 3:36-50. [DOI] [PubMed] [Google Scholar]
- 3.Berger, J., J. Hauber, R. Hauber, R. Geiger, and B. R. Cullen. 1988. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene 66:1-10. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, H. D., P. Sarnow, and D. Baltimore. 1986. Genetic complementation among poliovirus mutants derived from an infectious cDNA clone. J. Virol. 60:1040-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 6.Bloom, G. S., and T. A. Brashear. 1989. A novel 58-kDa protein associates with the Golgi apparatus and microtubules. J. Biol. Chem. 264:16083-16092. [PubMed] [Google Scholar]
- 7.Cho, S. P., B. Lee, and M. K. Min. 2000. Recombinant polioviruses expressing hepatitis B virus-specific cytotoxic T-lymphocyte epitopes. Vaccine 18:2878-2885. [DOI] [PubMed] [Google Scholar]
- 8.Choe, S. S., D. A. Dodd, and K. Kirkegaard. 2005. Inhibition of cellular protein secretion by picornaviral 3A proteins. Virology 337:18-29. [DOI] [PubMed] [Google Scholar]
- 9.Cornell, C. T., and B. L. Semler. 2004. Gene expression and replication of picornaviruses, p. 93-117. In R. A. Meyers (ed.), Encyclopedia of molecular cell biology and molecular medicine, 2nd ed. Wiley-VCH, Weinheim, Germany.
- 10.Deitz, S. B., D. A. Dodd, S. Cooper, P. Parham, and K. Kirkegaard. 2000. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc. Natl. Acad. Sci. USA 97:13790-13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dodd, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2001. Poliovirus 3A protein limits interleukin-6 (IL-6), IL-8, and beta interferon secretion during viral infection. J. Virol. 75:8158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farquhar, M. G., and G. E. Palade. 1998. The Golgi apparatus: 100 years of progress and controversy. Trends Cell Biol. 8:2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feuer, R., I. Mena, R. R. Pagarigan, M. K. Slifka, and J. L. Whitton. 2002. Cell cycle status affects coxsackievirus replication, persistence, and reactivation in vitro. J. Virol. 76:4430-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harkins, S., C. T. Cornell, and J. L. Whitton. 2005. Analysis of translational initiation in coxsackievirus B3 suggests an alternative explanation for the high frequency of R+4 in the eukaryotic consensus motif. J. Virol. 79:987-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasap, M., S. Thomas, E. Danaher, V. Holton, S. Jiang, and B. Storrie. 2004. Dynamic nucleation of Golgi apparatus assembly from the endoplasmic reticulum in interphase HeLa cells. Traffic 5:595-605. [DOI] [PubMed] [Google Scholar]
- 17.Leong, L. E.-C., C. T. Cornell, and B. L. Semler. 2002. Processing determinants and functions of cleavage products of picornavirus polyproteins, p. 187-197. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 18.Liu, F., and J. L. Whitton. 2005. Cutting edge: re-evaluating the in vivo cytokine responses of CD8+ T cells during primary and secondary viral infections. J. Immunol. 174:5936-5940. [DOI] [PubMed] [Google Scholar]
- 19.Mandl, S., L. J. Sigal, K. L. Rock, and R. Andino. 1998. Poliovirus vaccine vectors elicit antigen-specific cytotoxic T cells and protect mice against lethal challenge with malignant melanoma cells expressing a model antigen. Proc. Natl. Acad. Sci. USA 95:8216-8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moffat, K., G. Howell, C. Knox, G. J. Belsham, P. Monaghan, M. D. Ryan, and T. Wileman. 2005. Effects of foot-and-mouth disease virus nonstructural proteins on the structure and function of the early secretory pathway: 2BC but not 3A blocks endoplasmic reticulum-to-Golgi transport. J. Virol. 79:4382-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mogelsvang, S., B. J. Marsh, M. S. Ladinsky, and K. E. Howell. 2004. Predicting function from structure: 3D structure studies of the mammalian Golgi complex. Traffic 5:338-345. [DOI] [PubMed] [Google Scholar]
- 22.Munro, S. 2005. The Golgi apparatus: defining the identity of Golgi membranes. Curr. Opin. Cell Biol. 17:395-401. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura, N., C. Rabouille, R. Watson, T. Nilsson, N. Hui, P. Slusarewicz, T. E. Kreis, and G. Warren. 1995. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131:1715-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neznanov, N., A. Kondratova, K. M. Chumakov, B. Angres, B. Zhumabayeva, V. I. Agol, and A. V. Gudkov. 2001. Poliovirus protein 3A inhibits tumor necrosis factor (TNF)-induced apoptosis by eliminating the TNF receptor from the cell surface. J. Virol. 75:10409-10420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nieva, J. L., A. Agirre, S. Nir, and L. Carrasco. 2003. Mechanisms of membrane permeabilization by picornavirus 2B viroporin. FEBS Lett. 552:68-73. [DOI] [PubMed] [Google Scholar]
- 26.Rothman, J. E., and L. Orci. 1992. Molecular dissection of the secretory pathway. Nature 355:409-415. [DOI] [PubMed] [Google Scholar]
- 27.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. Hong, H. P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 75:9808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanz-Parra, A., F. Sobrino, and V. Ley. 1998. Infection with foot-and-mouth disease virus results in a rapid reduction of MHC class I surface expression. J. Gen. Virol. 79:433-436. [DOI] [PubMed] [Google Scholar]
- 29.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slifka, M. K., R. R. Pagarigan, I. Mena, R. Feuer, and J. L. Whitton. 2001. Using recombinant coxsackievirus B3 to evaluate the induction and protective efficacy of CD8+ T cells in controlling picornaviral infection. J. Virol. 75:2377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storrie, B. 2005. Maintenance of Golgi apparatus structure in the face of continuous protein recycling to the endoplasmic reticulum: making ends meet. Int. Rev. Cytol. 244:69-94. [DOI] [PubMed] [Google Scholar]
- 32.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Summers, D. F., and J. V. Maizel, Jr. 1968. Evidence for large precursor proteins in poliovirus synthesis. Proc. Natl. Acad. Sci. USA 59:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Kuppeveld, F. J., J. G. Hoenderop, R. L. Smeets, P. H. Willems, H. B. Dijkman, J. M. Galama, and W. J. Melchers. 1997. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 16:3519-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Kuppeveld, F. J., W. J. Melchers, K. Kirkegaard, and J. R. Doedens. 1997. Structure-function analysis of coxsackie B3 virus protein 2B. Virology 227:111-118. [DOI] [PubMed] [Google Scholar]
- 36.Wada, I., D. Rindress, P. H. Cameron, W. J. Ou, J. J. Doherty, D. Louvard, A. W. Bell, D. Dignard, D. Y. Thomas, and J. J. Bergeron. 1991. SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J. Biol. Chem. 266:19599-19610. [PubMed] [Google Scholar]
- 37.Wessels, E., D. Duijsings, R. A. Notebaart, W. J. Melchers, and F. J. van Kuppeveld. 2005. A proline-rich region in the coxsackievirus 3A protein is required for the protein to inhibit endoplasmic reticulum-to-Golgi transport. J. Virol. 79:5163-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitton, J. L., C. T. Cornell, and R. Feuer. 2005. Host and virus determinants of picornavirus pathogenesis and tropism. Nat. Rev. Microbiol. 3:765-776. [DOI] [PubMed] [Google Scholar]
- 39.Zeligs, J. D. 1979. Association of centrioles with clusters of apical vesicles in mitotic thyroid epithelial cells. Are centrioles involved in directing secretion? Cell Tissue Res. 201:11-21. [DOI] [PubMed] [Google Scholar]
- 40.Zuber, M. X., S. M. Strittmatter, and M. C. Fishman. 1989. A membrane-targeting signal in the amino terminus of the neuronal protein GAP-43. Nature 341:345-348. [DOI] [PubMed] [Google Scholar]