Abstract
Tripartite motif 5α (TRIM5α) restricts some retroviruses, including human immunodeficiency virus type 1 (HIV-1), from infecting the cells of particular species. TRIM5α is a member of the TRIM family of proteins, which contain RING, B-box, coiled-coil (CC), and, in some cases, B30.2(SPRY) domains. Here we investigated the abilities of domains from TRIM proteins (TRIM6, TRIM34, and TRIM21) that do not restrict HIV-1 infection to substitute for the domains of rhesus monkey TRIM5α (TRIM5αrh). The RING, B-box 2, and CC domains of the paralogous TRIM6 and TRIM34 proteins functionally replaced the corresponding TRIM5αrh domains, allowing HIV-1 restriction. By contrast, similar chimeras containing the components of TRIM21, a slightly more distant relative of TRIM5, did not restrict HIV-1 infection. The TRIM21 B-box 2 domain and its flanking linker regions contributed to the functional defectiveness of these chimeras. All of the chimeric proteins formed trimers. All of the chimeras that restricted HIV-1 infection bound the assembled HIV-1 capsid complexes. These results indicate that heterologous RING, B-box 2, and CC domains from related TRIM proteins can functionally substitute for TRIM5αrh domains.
Retroviruses encounter dominant postentry restrictions in cells of particular species. Human immunodeficiency virus type 1 (HIV-1) infection is blocked in the cells of Old World monkeys, whereas simian immunodeficiency virus infection is blocked in most New World monkey cells (11, 12, 29). The restriction is mediated by dominant host factors that are species specific and saturable (2, 3, 5, 7, 22, 33). The viral determinant for susceptibility to these restrictions is the capsid (CA) protein (5, 8, 18, 22, 23, 37).
A genetic screen of a rhesus monkey cDNA library identified a major restriction factor, tripartite motif 5α (TRIM5α) (34). Expression of rhesus monkey TRIM5α (TRIM5αrh) in permissive cells is sufficient to confer potent restriction of HIV-1 infection (34). Human TRIM5α (TRIM5αhu) is less potent in restricting HIV-1 but demonstrates strong antiviral activity against a gamma retrovirus, N-tropic murine leukemia virus (N-MLV); thus, TRIM5α accounts for the N-MLV-blocking activity found in human cells (9, 17, 25, 38). Variation in TRIM5α proteins in primate species accounts for the observed differences in the restriction of retroviruses (12, 28, 31, 32).
TRIM5α is a member of the large family of TRIM proteins (26). Members of this family all contain a RING, a B-box, and a coiled-coil (CC) domain and thus are also called RBCC proteins (26). The C-terminal portions of these proteins are more variable; many cytoplasmic TRIM proteins, like TRIM5α, contain a B30.2(SPRY) domain. Although some members of this protein family have been linked to diverse processes such as transcription regulation, apoptosis, inflammation, cell polarity determination, and antiviral activities, the molecular and cellular functions of most TRIM proteins remain unknown (16, 27).
Previous studies have provided some insight into the contribution of each of the TRIM5α domains to antiretroviral activity. Variation between the B30.2(SPRY) domains of TRIM5α from humans and Old World monkeys determines the observed species-specific potency of HIV-1 restriction (24, 36, 39). Recently, the B30.2 domain has been shown to be necessary for the specific association of monkey TRIM5α with assembled HIV-1 CA complexes (35). Deletion or disruption of the RING domain diminishes, but does not abolish, the antiviral activity of TRIM5αrh, whereas deletion or disruption of the B-box domain completely eliminates HIV-1-restricting ability (15, 24). The RBCC domains of human and Old World monkey TRIM5α proteins are functionally interchangeable (36, 39). Here, by generating a series of chimeras between TRIM5αrh and other human TRIM family members, we investigated the abilities of heterologous TRIM domains to substitute functionally for the RBCC domains of TRIM5αrh.
MATERIALS AND METHODS
Plasmid construction.
The coding sequences of the chimeric TRIM proteins were amplified by overlapping PCR extension (13, 14) with a sequence encoding a hemagglutinin (HA) tag sequence incorporated into the 3′ primers. The spliced PCR fragments were then digested and cloned into the EcoRI and ClaI sites of the pLPCX vector (Stratagene) as previously described (34). The TRIM21 B-box 2 point mutants were made by either QuikChange mutagenesis (Stratagene) or overlapping PCR extension with primers specifying the corresponding nucleic acid change.
Creation of cells stably expressing TRIM chimeras.
Recombinant viruses were produced in 293FT cells by cotransfecting the pLPCX plasmids expressing TRIM variants with the pVPack-GP and pVPack-VSV-G packaging plasmids (Stratagene). The pVPack-VSV-G plasmid encodes the vesicular stomatitis virus G envelope glycoprotein, which allows efficient entry into a wide range of vertebrate cells. The resulting virus particles were used to transduce 2 × 105 HeLa cells in six-well plates. The HeLa cells were then selected in 1 μg/ml puromycin (Sigma).
Immunoblotting.
HeLa cells stably expressing the TRIM proteins were lysed with phosphate-buffered saline (PBS) containing 1% NP-40 and a protease inhibitor cocktail (Roche). The lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotted with the horseradish peroxidase-conjugated 3F10 antibody (Roche) against the HA epitope or an antibody against β-actin (Sigma).
Infection with viruses expressing GFP.
Recombinant HIV-1 and N-MLV expressing green fluorescent protein (GFP) were made as previously described (25, 34). For infection, 3 × 104 cells were seeded in 24-well plates and incubated with the viruses for 60 h. Cells were then washed with PBS, fixed with 3.7% formaldehyde, and subjected to fluorescence-activated cell sorting (FACS) analysis with a FACScan (Becton Dickinson).
Cross-linking of TRIM chimeras.
Cell lysates prepared in 1% NP-40-PBS-protease inhibitor cocktail were incubated with various concentrations (final concentrations of 0, 0.2, 0.4, 0.8, and 2.0 mM) of glutaraldehyde (Sigma) at room temperature for 5 min, after which excess glycine was added to quench the reaction. The cross-linked lysates were then subjected to SDS-PAGE and Western blotting. The blots were probed with a horseradish peroxidase-conjugated anti-HA antibody (Roche).
CA-NC binding assay.
HIV-1 CA-nucleocapsid (CA-NC) was expressed, purified, and assembled as previously described (19, 35). TRIM-expressing HeLa cells growing to confluence on 100-mm tissue culture dishes were washed once with PBS, followed by detachment with PBS buffer containing 10 mM EDTA. Cells were then collected by brief centrifugation and resuspended in hypotonic lysis buffer (10 mM Tris [pH 7.4], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol) (1). Lysates were made by Dounce homogenization and cleared of nuclei and cell debris by centrifugation at 13,200 rpm for 10 min at 4°C. The salt concentration of the supernatant was adjusted to 150 mM by adding 10× PBS buffer. A fraction of the supernatant (250 μl) was then mixed with 5 μl of 0.3 mM CA-NC proteins. Following a 1-h incubation at room temperature on a rocking platform, samples were taken to test the protein level in the total input. The remainder of the reaction mixture was layered onto a 3.5-ml 70% sucrose cushion (in PBS-5 mM dithiothreitol) and ultracentrifuged at 30,000 rpm for 1 h at 4°C in an SW55 rotor (Beckman). The sucrose cushion was then aspirated off, and the pellet was resuspended in SDS loading buffer and subsequently analyzed by Western blotting to detect the presence of TRIM proteins. The amount of CA-NC in the pellet was also examined by Western blotting with an anti-p24 antibody (1:5,000; ImmunoDiagnostics, Inc.).
RESULTS
Replacement of the TRIM5αrh domains with the RBCC domains of closely related TRIM proteins.
In humans, the TRIM5 gene is located in a cluster of paralogs (TRIM6, TRIM34, and TRIM22) on chromosome 11. The TRIM6 and TRIM34 proteins exhibit the closest relationships with TRIM5α, with 54.9% and 56.9% identity to TRIM5αrh, respectively. As is true of many cytoplasmic TRIM proteins, both TRIM6 and TRIM34 contain RING, B-box, CC, and carboxy-terminal B30.2(SPRY) domains (Fig. 1A).
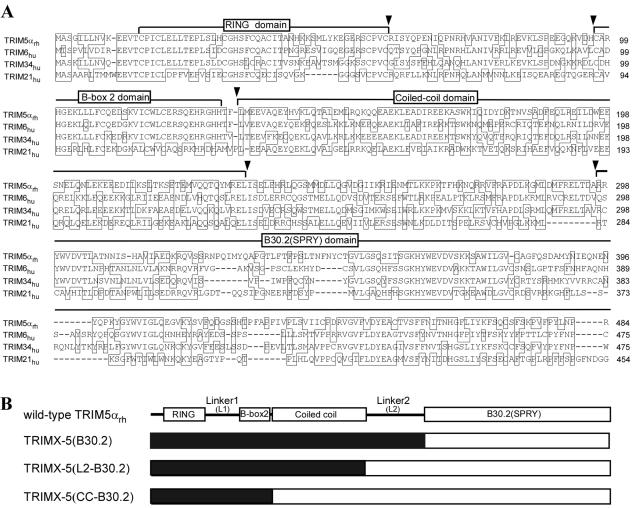
FIG. 1.
TRIM5-related proteins and chimeras. (A) Alignment of the amino acid sequences of TRIM5αrh and human TRIM6, TRIM34, and TRIM21 (DNAStar). Identical residues at each position are boxed. Gaps are indicated by dashes. The boundaries of conserved structural domains are indicated above the sequences. Arrowheads indicate the locations of the junctions of the chimeras used in this study. (B) Schematic illustrations of chimeras composed of other TRIM family members (TRIMX, where X = 6, 34, or 21) and TRIM5αrh. Black segments indicate sequences that are derived from the heterologous human TRIM proteins; white segments are derived from TRIM5αrh.
TRIM5αrh and TRIM5αhu block HIV-1 infection with high and modest potency, respectively (34). The RBCC domains of TRIM5αrh and TRIM5αhu are functionally interchangeable with respect to HIV-1 infection (36, 39). None of the closely related human TRIM family proteins exhibited anti-HIV-1 activities (Table 1 and data not shown). To investigate to what extent the RBCC domains of these closely related proteins can complement TRIM5αrh function, a series of chimeric proteins between TRIM5αrh and either human TRIM6 or human TRIM34 were made and their activities were characterized (Fig. 1B). The chimeric proteins and wild-type TRIM5αrh were expressed in HeLa cells by using an LPCX retroviral vector as previously described (34). All of the chimeras carry a C-terminal HA tag derived from influenza virus; the wild-type TRIM6 and TRIM34 proteins are tagged at the N terminus with a FLAG epitope. Addition of the FLAG tag at the N or C terminus did not affect the ability, or lack thereof, of these TRIM proteins to restrict HIV-1 infection (data not shown). Some variation in the steady-state level of expression of the chimeric proteins was observed in a Western blot assay (Fig. 2A).
TABLE 1.
Summary of HIV-1 infection in cells expressing TRIM proteins and chimeras
| TRIM variant | HIV-1 infectiona
|
||
|---|---|---|---|
| X = 6 | X = 34 | X = 21 | |
| TRIMX | +++ | ++++ | +++ |
| TRIMX-5(B30.2) | + | +++ | ++++ |
| TRIMX-5(L2-B30.2) | ++++ | + | ++++ |
| TRIMX-5(CC-B30.2) | + | + | ++++ |
| TRIMX-5(B-box-B30.2) | NDb | ND | ++ |
| TRIMX-5(L1-B30.2) | ND | ND | + |
| TRIM5-X(B-box 2) | ND | ND | +++ |
Following incubation with HIV-1-GFP, the level of infection in cells expressing the indicated TRIM proteins was assessed by measuring GFP-positive cells. A level of infection comparable to that seen in cells transduced with the empty LPCX vector is designated +++. Levels of infection that range from 30 to 60% of the level observed in the LPCX-transduced control cells are designated ++. Levels of infection that range from 1 to 29% of the level in LPCX-transduced cells are designated +. The designation ++++ indicates a level of infection greater than 120% of the level in LPCX-transduced cells.
ND, not determined.
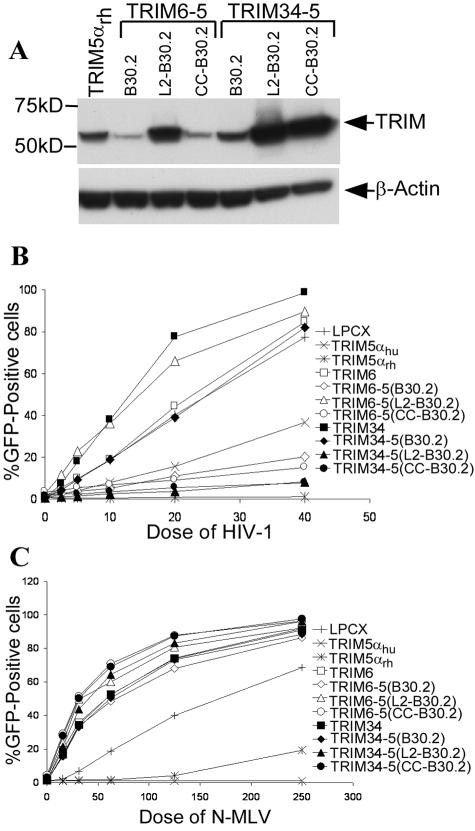
FIG. 2.
Expression and antiretroviral activity of TRIM chimeras. (A) Expression of chimeric TRIM proteins. The wild-type TRIM5αrh protein and all of the chimeras contain C-terminal HA tags. Lysates were made from HeLa cells and subjected to Western blotting with an anti-HA antibody (Roche). The blot was also probed for β-actin to control for the loading amount. (B and C) Effects of the TRIM proteins on retroviral infection. HeLa cells expressing the parental and chimeric TRIM proteins or control HeLa cells transduced with the empty LPCX vector were incubated with various amounts of HIV-1-GFP (B) or N-MLV-GFP (C). Infected GFP-positive cells were counted by FACS. Ten dose units of HIV-1 corresponds to ∼3,000 cpm reverse transcriptase units; 100 dose units of N-MLV corresponds to ∼400 cpm reverse transcriptase units. The results of a typical experiment are shown. The experiment was performed three times with similar results.
To assess antiretroviral activity, the HeLa cells expressing the TRIM variants were incubated with a recombinant HIV-1 vector expressing GFP. The infected, GFP-positive cells were counted by FACS. Compared with the control cells transduced with the LPCX vector, cells expressing TRIM5αrh were very resistant to HIV-1-GFP infection (Fig. 2B). TRIM5αhu inhibited HIV-1 infection but was not as potent as TRIM5αrh in this regard. Expression of human TRIM6 and TRIM34 did not result in any decreases in HIV-1 infection; in fact, HeLa cells expressing TRIM34 were more susceptible to HIV-1-GFP infection than control cells transduced with the empty LPCX vector. This enhancement of HIV-1 infection results from dominant-negative effects of TRIM34 on the endogenous TRIM5αhu in the HeLa cells (X. Li and J. Sodroski, unpublished data). A chimeric construct, TRIM6-5(B30.2), in which the human TRIM6 B30.2 domain is replaced with that of TRIM5αrh, inhibited HIV-1 infection (Fig. 2B and Table 1). A related construct, TRIM6-5(L2-B30.2), which in addition contains the TRIM5αrh L2 linker region, did not restrict HIV-1 infection. Conversely, the equivalent TRIM34-TRIM5 chimeras containing the TRIM5αrh B30.2 domain blocked HIV-1 infection only when the L2 linker of TRIM5αrh was included in the construct [compare TRIM34-5(B30.2) and TRIM34-5(L2-B30.2) in Fig. 2B and Table 1]. These results indicate that context can influence the efficiency with which the domains of these TRIM proteins can be interchanged. Chimeras that contained the CC, L2, and B30.2 regions of TRIM5αrh with the RING and B-box 2 domains of either TRIM6 or TRIM34 blocked HIV-1 infection [TRIM6-5(CC-B30.2) and TRIM34-5(CC-B30.2) in Fig. 2B]. The levels of expression of the chimeric proteins did not explain the observed differences in anti-HIV-1 activity (Fig. 2A). Thus, the RING, B-box 2, and CC domains of TRIM6 and TRIM34 can functionally replace the corresponding domains of TRIM5αrh with respect to inhibition of HIV-1 infection.
We also tested the abilities of the chimeras to restrict N-MLV infection. As expected, N-MLV infection was potently blocked by TRIM5αhu and partially blocked by TRIM5αrh (Fig. 2C). Because these experiments were performed with human cells, which express TRIM5αhu and thus already limit N-MLV infection, the TRIM5αrh-mediated N-MLV restriction is somewhat greater than that observed for TRIM5αrh expressed in nonhuman cells (9, 17, 25, 38). None of the chimeric TRIM proteins restricted N-MLV infection. N-MLV infection was more efficient in HeLa cells expressing TRIM6, TRIM34, and the chimeric proteins than in HeLa cells transduced with the empty LPCX vector. This increase in N-MLV infection results from dominant-negative effects of the TRIM6 and TRIM34 proteins on the endogenous TRIM5αhu protein in HeLa cells (Li and Sodroski, unpublished). These results indicate that the modest restricting ability of TRIM5αrh for N-MLV infection does not tolerate substitution of RBCC domains, even those derived from closely related TRIM proteins.
Substitution of TRIM21 domains for those of TRIM5αrh.
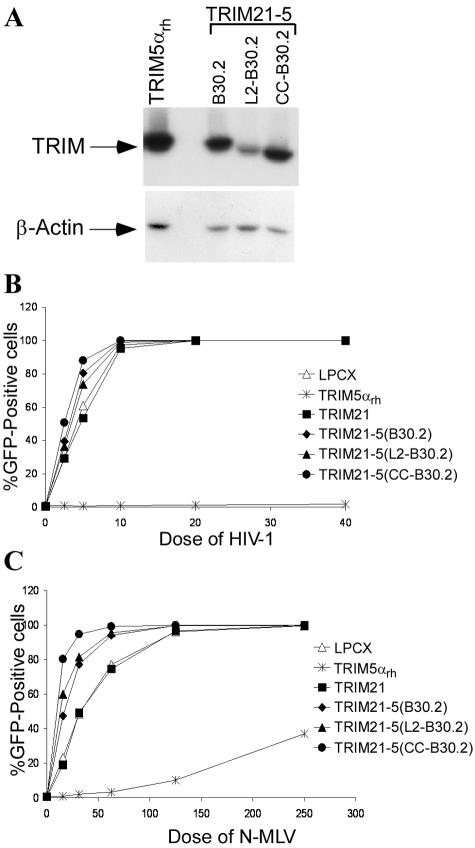
Beyond the TRIM5 paralogs (TRIM6, TRIM34, and TRIM22), TRIM21 is the closest human relative of TRIM5 (31). To investigate whether RBCC domains from the more distantly related TRIM21 protein could complement TRIM5α function, chimeric proteins analogous to those described above for TRIM6 and TRIM34 were constructed between human TRIM21 and TRIM5αrh. The stable expression of the TRIM21-TRIM5αrh chimeric proteins in HeLa cells was verified by Western blotting (Fig. 3A). Despite the efficient expression of the chimeric proteins, the cells were susceptible to infection by HIV-1 and N-MLV (Fig. 3B and C, respectively). As had been observed for some of the TRIM6 and TRIM34 chimeras, the HeLa cells expressing the TRIM21-TRIM5 chimeras were slightly more infectible by both HIV-1 and N-MLV; this effect may be due to slight dominant-negative activity of the TRIM21-TRIM5 chimeric proteins on the endogenous TRIM5αhu protein in the HeLa cells.
FIG. 3.
Expression and antiretroviral activities of TRIM21-TRIM5αrh chimeras. (A) Expression of TRIM21-TRIM5αrh chimeras was examined by Western blotting of HeLa lysates containing the indicated TRIM proteins with an anti-HA antibody (Roche). (B and C) Infection with HIV-1-GFP and N-MLV-GFP, respectively, of HeLa cell lines stably expressing the indicated parental and chimeric proteins.
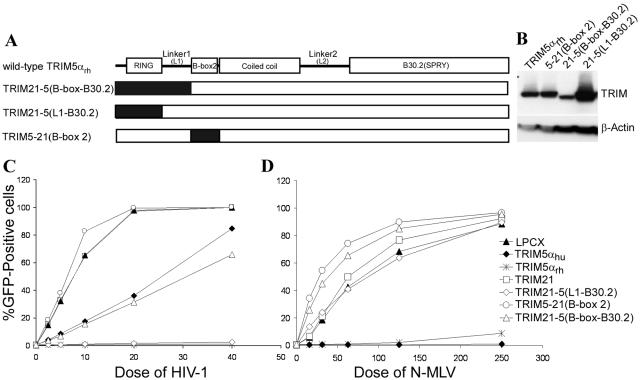
To identify the TRIM21 region responsible for the lack of antiviral function of the chimeras, additional chimeric proteins containing only the RING domain, only the B-box 2 domain, or the RING domain and L1 linker region of TRIM21 were constructed (Fig. 4A). The chimeric proteins were expressed efficiently in HeLa cells (Fig. 4B). The cells were incubated with the HIV-1-GFP and N-MLV-GFP recombinants to determine the antiviral activity of the TRIM21-TRIM5αrh chimeras. The TRIM21-5(L1-B30.2) protein, which contains only the RING domain of TRIM21, potently restricted HIV-1 infection (Fig. 4C and Table 1). An intermediate level of HIV-1 restriction was observed for the TRIM21-5(B-box-B30.2) protein, which contains the RING domain and the adjacent L1 linker region of TRIM21. The TRIM5-21(B-box 2) protein, which contains only the B-box 2 domain of TRIM21, did not inhibit HIV-1 infection. Thus, the RING domain of TRIM21 can functionally substitute for the TRIM5αrh RING domain with respect to inhibition of HIV-1 infection. By contrast, chimeras containing the TRIM21 B-box 2 domain do not exhibit HIV-1-restricting ability.
FIG. 4.
Effects of the TRIM21 RING and B-box 2 domains on the antiretroviral activity of TRIM5αrh. (A) Structures of the chimeric proteins involving the TRIM21 RING and B-box 2 domains. The RING domain, the B-box 2 domain, and the RING domain plus the L1 linker region of TRIM5αrh were individually replaced with the corresponding region from human TRIM21 (shaded black). (B) Expression of the TRIM21-TRIM5αrh chimeras. HeLa lysates were analyzed by Western blotting as described in the legend to Fig. 2. (C and D) Antiretroviral activities of TRIM21-TRIM5αrh chimeras. HeLa cells expressing the chimeric proteins were incubated with HIV-1-GFP (C) and N-MLV-GFP (D) and then analyzed for GFP expression by FACS.
The HeLa cells expressing the TRIM21-TRIM5αrh chimeras were incubated with N-MLV-GFP. Figure 4D shows that none of the TRIM21-TRIM5αrh chimeras inhibited N-MLV infection.
B-box 2 domain contributions to the function of TRIM5α chimeras.
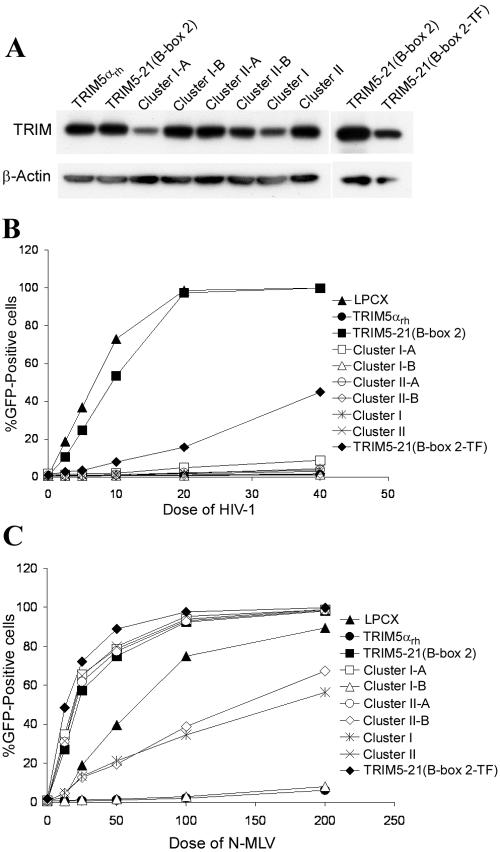
A total of 17 amino acids differ between the sequences of the nonfunctional TRIM5-21(B-box 2) chimera and wild-type TRIM5αrh (Fig. 1). To investigate which of these are responsible for the defectiveness of TRIM5-21(B-box 2), additional constructs were created and tested. As a guide, we used the nuclear magnetic resonance structure of a B-box domain from Xenopus laevis nuclear factor 7 (4); the amino acids that differ between TRIM5αrh and TRIM5-21(B-box 2) were mapped on this structure. These amino acids mostly clustered into two surface-exposed regions; cluster I is composed of Q109E110V114I115L118G127H129, and cluster II includes K103E120R121Q123E124. The amino acids in each region of TRIM5αrh were changed either individually or in combination to those found in TRIM21, and the expression and activities of these mutants were examined (Fig. 5A and B). All of the TRIM5αrh B-box 2 mutants retained the ability to restrict HIV-1 infection potently.
FIG. 5.
Expression and antiretroviral activities of TRIM5αrh B-box 2 variants. The following changes were introduced into the TRIM5αrh protein: cluster I-A, Q109E, E110K, V114A, I115L, and L118V; cluster I-B, G127D, and H129A; cluster II-A, E120A, R121Q, Q123R, and E124K; cluster II-B, K103R. Cluster I is a combination of the changes in cluster I-A and cluster I-B; cluster II is a combination of the changes in cluster II-A and cluster II-B. Numbers indicate the residue positions of substitutions based on the numbering of TRIM5αrh. The letter before the number is the amino acid residue present in the wild-type TRIM5αrh sequence, and the letter after the number corresponds to human TRIM21. The TRIM5-21(B-box 2-TF) mutant is identical to TRIM5-21(B-box 2) except that the MVP sequence immediately N terminal to the B-box 2-CC junction in the latter chimera has been changed to TF, the sequence in wild-type TRIM5αrh. (A) To examine the expression of the TRIM5αrh B-box 2 variants, HeLa lysates were analyzed by Western blotting as described in the legend to Fig. 2. (B and C) To assess the antiretroviral activity of TRIM5αrh B-box 2 variants, HeLa cells expressing the mutant proteins were incubated with HIV-1-GFP (B) and N-MLV-GFP (C) and then analyzed for GFP expression by FACS.
TRIM5-21(B-box 2) also differs from TRIM5αrh in the junction between the B-box 2 and CC domains. When the three amino acids (MVP) in this junctional region of TRIM5-21(B-box 2) were changed to those residues (TF) found in TRIM5αrh, the mutant protein [TRIM5-21(B-box 2-TF)] gained partial activity in restricting HIV-1 infection (Fig. 5B). Thus, at least part of the loss of anti-HIV-1 activity of TRIM5-21(B-box 2), relative to that of TRIM5αrh, is an effect of the suboptimal junction between the B-box 2 domain and the CC domain.
In contrast to HIV-1 restriction, most of the changes in the B-box 2 domain of TRIM5αrh were detrimental to the ability to restrict N-MLV infection (Fig. 5C). A conservative change of lysine 103 to arginine (cluster II-B) caused a more-than-10-fold decrease in the restrictive activity of TRIM5αrh. Mutant cluster I-B (G127D/H129A) was exceptional in retaining anti-N-MLV activity. Moreover, the cluster I-B changes also partially rescued the activity of mutant cluster I-A (cluster I in Fig. 5C). Thus, the partial restricting ability of TRIM5αrh for N-MLV infection is more sensitive to changes in the B-box 2 domain than is HIV-1 restriction.
Oligomerization of chimeric TRIM proteins.
TRIM proteins typically form homo-oligomers (26). The role of oligomerization in the function of most TRIM proteins is unknown. Some TRIM5αrh mutants are able to associate with wild-type TRIM5αrh and exert dominant-negative effects (15, 24), suggesting that the functional TRIM5α moiety may be a multimer. Recently, wild-type TRIM5α proteins from different primate species have been shown to form trimers (20). To determine whether the chimeric TRIM proteins in this study form oligomers, lysates from cells stably expressing these chimeras were incubated with the cross-linker glutaraldehyde. The multimerization of the chimeric TRIM proteins was examined by Western blotting. All of the chimeric proteins were cross-linked into gel-stable complexes with molecular masses of approximately 150 to 170 kDa, consistent with the formation of trimers (Fig. 6).
FIG. 6.
Cross-linking of TRIM protein chimeras. Lysates from HeLa cells stably expressing the chimeric TRIM proteins were cross-linked with increasing concentrations of glutaraldehyde (GA; 0, 0.2, 0.4, 0.8, and 2.0 mM). The reaction was allowed to proceed for 5 min at room temperature and then stopped with glycine. Following denaturation at 37°C for 30 min in SDS buffer, the samples were analyzed by SDS-PAGE and Western blotting with an anti-HA antibody.
Binding of TRIM chimeras to HIV-1 CA-NC.
The CA specificity of retroviral restriction mediated by TRIM5α (9, 25, 34, 39) suggests that TRIM5α might recognize the incoming CA of entering viruses. Recently, a good correlation has been observed between the abilities of TRIM5α proteins from different primate species to bind HIV-1 CA complexes assembled in vitro and their activities in restricting HIV-1 (35). To explore the mechanism underlying the viral restriction activities of the TRIM chimeras, a previously established binding assay (35) was used to examine the association between the TRIM chimeras and the in vitro-assembled HIV-1 CA-NC protein complex. In this assay, TRIM proteins that otherwise would not sediment through a 70% sucrose cushion can associate and cosediment with the assembled HIV-1 CA-NC complexes after ultracentrifugation. We confirmed that none of the TRIM chimeras pelleted through 70% sucrose cushions in the absence of added HIV-1 CA-NC complexes (data not shown). For most of the chimeras between TRIM5αrh and TRIM6, TRIM21, and TRIM34, a striking correlation between anti-HIV-1 activity and binding to the HIV-1 CA-NC complexes was observed (Fig. 7A). Any chimera that exhibited function in blocking HIV-1 associated to some degree with the CA-NC complexes. In general, the chimeras that did not restrict HIV-1 infection did not bind the HIV-1 CA-NC complexes; examples are TRIM6-5(L2-B30.2), TRIM21-5(B30.2), TRIM21-5(L2-B30.2), and TRIM34-5(B30.2), some of which were present at very high levels in the input lysates (Fig. 7A). Although it did not restrict HIV-1 infection, TRIM21-5(CC-B30.2) displayed a very weak association with the CA-NC complexes.
FIG. 7.
Binding of TRIM chimeras to HIV-1 CA-NC complexes. Hypotonic lysates of HeLa cells expressing different TRIM chimeric proteins were incubated with the in vitro-assembled HIV-1 CA-NC complexes. The total input (Input) from the lysate and the pellet (Pellet) from the sucrose gradient were tested for the presence of TRIM proteins which are C terminally tagged with HA. The bottom panel shows the amount of HIV-1 CA-NC protein pelleted through the 70% sucrose cushion.
Among the TRIM21-TRIM5 chimeras involving the RING and B-box 2 domains, TRIM21-5(L1-B30.2) bound the CA efficiently (Fig. 7B), consistent with its potent ability to restrict HIV-1 infection. TRIM5-21(B-box 2-TF), with intermediate HIV-1-restricting activity, also associated efficiently with the HIV-1 CA-NC complexes. TRIM21-5(B-box-B30.2), another chimera with modest HIV-1-restricting activity, bound detectably but weakly to the HIV-1 CA-NC complexes in this assay. The functionally inactive TRIM5-21(B-box 2) chimera interacted efficiently with the HIV-1 CA-NC complexes. The results are consistent with the expectation that the ability of a TRIM protein to associate with the HIV-1 CA complex is necessary but not sufficient for virus-restricting activity.
DISCUSSION
The cluster of human TRIM paralogs on chromosome 11 is composed of TRIM6, TRIM34, TRIM5, and TRIM22. The protein products of these genes exhibit between 51% and 60% identity in amino acid sequence (31). The next closest human relative to TRIM5 is TRIM21, which is encoded by a gene that resides on chromosome 11, outside the above paralogous cluster. All of the proteins possess RING, B-box 2, CC, and B30.2 domains, in addition to the linker regions (L1 and L2). Despite the similarities of these TRIM proteins, only TRIM5α exhibited inhibitory activity with respect to HIV-1 or N-MLV infection. We analyzed the function of chimeric proteins composed of the domains of TRIM5αrh and those of the other TRIM proteins to gain insight into the basis for the lack of antiretroviral activity of TRIM6, TRIM34, and TRIM21. The TRIM5αrh protein was chosen for these studies because of the potent ability to restrict HIV-1 infection; in HeLa cells, TRIM5αrh apparently can cooperate with the endogenous TRIM5αhu protein to block N-MLV infection significantly as well. Therefore, the use of TRIM5αrh allowed us to assess the impact of chimerism on the ability to block infection by two distantly related retroviruses.
Previous studies demonstrated that the B30.2 domain of TRIM5α specifies the differences in anti-HIV-1 potency observed between the TRIM5αhu and TRIM5αrh proteins (24, 36, 39). Similarly, replacement of the TRIM6 or TRIM34 B30.2 domain with that of TRIM5αrh, either without or with the L2 linker, respectively, allowed anti-HIV-1 activity of the chimeric protein. The mechanistic basis for this restoration of antiviral function may be the acquisition of specific binding to the HIV-1 CA. The RBCC domains of TRIM6 and TRIM34 are capable of mediating all of the additional functions required for antiviral activity. For example, the chimeric proteins all form homotrimers, a process that may contribute to the avidity of TRIM-CA interactions.
The TRIM5αrh RING and B-box 2 domains are not essential for homotrimerization and CA binding (20, 35), but their role in retroviral restriction is still unknown. The TRIM6 and TRIM34 RING and B-box 2 domains effectively substituted for those of TRIM5αrh with regard to HIV-1 restriction, whereas the identical substitutions involving TRIM21 resulted in nonfunctional proteins. In the chimeras containing multiple TRIM21 domains and linker regions, the divergence of TRIM21 and TRIM5 may be too great to allow all of the intra- or intermolecular interactions required for retroviral restriction. Depending on the junctions of the chimeras, the individual TRIM21 RING and B-box 2 domains can at least partially complement antiretroviral function. The TRIM21 RING domain efficiently complemented the anti-HIV-1 function of TRIM5αrh, whereas only partial activity was achieved for one chimera [TRIM5-21(B-box 2-TF)] containing the TRIM21 B-box 2 domain. Previous deletion analysis has suggested that the B-box 2 domain is more critical than the RING domain for TRIM5αrh function (15, 24). The mechanistic role of the B-box 2 domain in the function of TRIM proteins is unknown. This domain contains seven conserved cysteine and histidine residues, but the nuclear magnetic resonance structure of the X. laevis Xnf7 B-box domain (4) indicates that only four of these residues are involved in coordinating a zinc atom and thus contributing to the integrity of the domain. The TRIM21 B-box 2 domain retains all seven of the conserved cysteines and histidines, so other elements of the TRIM21 B-box 2 domain determine its inability to complement TRIM5αrh antiviral function. Our studies indicate that the junction between the B-box 2 and CC domains can contribute to the functionality, or lack thereof, of the TRIM21-TRIM5 chimeras.
The identities of the L1 and L2 linker regions can also influence the function of the TRIM chimeras. As an example of the influence of the L2 region, a TRIM6 chimera with only the TRIM5αrh B30.2 domain restricted HIV-1 infection, whereas the analogous TRIM34 chimera did not. Addition of the TRIM5αrh L2 linker region diminished the anti-HIV-1 function of the TRIM6 chimera but potentiated the function of the TRIM34 chimera. The L2 linker region may be involved in correctly positioning the two adjacent functional elements, the CC and B30.2 domains; thus, the contribution of the TRIM5αrh L2 linker to function varies, depending on the flanking domains. Likewise, the inclusion of the TRIM21 L1 linker region in the TRIM21-5(B-box-B30.2) chimera decreased the potency of HIV-1 restriction compared with the TRIM21-5(L1-B30.2) chimera, which contains the TRIM5αrh L1 sequence. The L1 linker of TRIM21 has been suggested to influence the interaction of the RING and B-box 2 domains, on the basis of antibody recognition studies (10, 21). Thus, at a minimum, interdomain linkers in TRIM proteins may govern the spatial positions and orientation of critical functional elements. Moreover, the linker regions, although not conforming to known structural domains, may contribute to TRIM protein function directly.
The N-MLV-restricting function of TRIM5αrh was much more sensitive to substitution of heterologous RBCC domains than the HIV-1-restricting function of this protein. This could be due to quantitative or qualitative differences in the restriction of these retroviruses by TRIM5αrh. The less potent restriction of N-MLV infection by TRIM5αrh may exhibit greater sensitivity to other disruptive changes in the protein. Even for HIV-1 restriction, the chimeric TRIM6-TRIM5 and TRIM34-TRIM5 proteins are not as potent as TRIM5αrh, consistent with a quantitative explanation. It is also possible that there are qualitative differences in the intermolecular associations required for N-MLV and HIV-1 restriction. Combined quantitative and qualitative explanations are also possible. For example, as TRIM5α has been suggested to accelerate the disassembly of the restricted viral CA (35), N-MLV CAs may be more stable and require a greater number of TRIM5 intermolecular associations to achieve the requisite level of disassembly. Further studies will address these possibilities.
The anti-HIV-1 activities of the TRIM chimeras generally correlated with their abilities to bind the HIV-1 CA-NC complexes. This is consistent with a model in which TRIM5α binding to the viral CA is a prerequisite for TRIM5α-mediated retroviral restriction. Whether TRIM5α-CA binding involves a direct interaction or is mediated through other cellular factors requires further investigation.
The CC and B30.2 domains, and the intervening linker 2 region, have been shown to be important for TRIM5αrh binding to HIV-1 CA-NC complexes (35). Consistent with a model in which these C-terminal domains constitute a CA-binding unit, all of the chimeras that contained these sequences from TRIM5αrh bound HIV-1 CA-NC complexes, although to different degrees. TRIM21-5(CC-B30.2) associated with the HIV-1 CA-NC complexes only weakly, for example. Our results suggest that sequences N terminal to the CC can influence the efficiency of CA binding, either directly or indirectly. Further studies should shed light on the functional interplay of TRIM5α domains.
Although binding to the CA is a prerequisite for the antiviral activity of TRIM5α, the function of other domains not solely involved in CA binding also likely contributes. In this study and in other work (6, 35), a few TRIM variants that appear to bind HIV-1 CA-NC complexes efficiently exhibited minimal anti-HIV-1 activity. For example, the TRIM5-21(B-box 2) mutant trimerized and associated with HIV-1 CA-NC complexes but did not block HIV-1 infection. These results imply the existence of an “effector” function that is required, in addition to CA binding, for retroviral restriction.
These studies underscore the importance of specific RBCC domains in achieving potent levels of retrovirus restriction. The results obtained with the TRIM21 chimeras indicate that generic RBCC functions are not sufficient to complement the CA-binding function of the TRIM5α B30.2 domain. These results provide a natural explanation for the repeated evolution of antiretroviral restriction factors from the TRIM5/6/34/22 group. For example, in owl monkeys, an unusual restriction factor, TRIMCyp, is a fusion of a CA-binding moiety, cyclophilin A, and the RBCC domains of TRIM5 (20a, 28a). The requirement for rather specific TRIM5-related RBCC moieties for viral restriction explains the selection of TRIM5, and not another TRIM protein, as the fusion partner in the creation of TRIMCyp. Recently, another TRIM protein with antiretroviral activity has been shown to have evolved from the TRIM5/6/34/22 subfamily in cows (30). Again, certain properties of this family are apparently conducive to the convergent evolution of retroviral restriction factors.
Future studies may elucidate the precise contribution of each of the RBCC domains to TRIM protein function, including antiretroviral activities.
Acknowledgments
We thank Yvette McLaughlin and Sheri Farnum for manuscript preparation.
This work was supported by grants from the National Institutes of Health (AI063987, HL54785, and Center for AIDS Research award AI28691), the International AIDS Vaccine Initiative, the Bristol-Myers Squibb Foundation, and the late William F. McCarty-Cooper.
REFERENCES
- 1.Auewarakul, P., P. Wacharapornin, S. Srichatrapimuk, S. Chutipongtanate, and P. Puthavathana. 2005. Uncoating of HIV-1 requires cellular activation. Virology 337:93-101. [DOI] [PubMed] [Google Scholar]
- 2.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D. 2003. Restriction factors: a defense against retroviral infection. Trends Microbiol. 11:286-291. [DOI] [PubMed] [Google Scholar]
- 4.Borden, K. L., J. M. Lally, S. R. Martin, N. J. O'Reilly, L. D. Etkin, and P. S. Freemont. 1995. Novel topology of a zinc-binding domain from a protein involved in regulating early Xenopus development. EMBO J. 14:5947-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz-Griffero, F., X. Li, H. Javanbakht, B. Song, S. Welikala, M. Stremlau, and J. Sodroski. 10February2006. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology [Epub ahead of print.] doi: 10.1016/j.virol.2005.12.040. [DOI] [PubMed]
- 7.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatziioannou, T., S. Cowan, U. K. Von Schwedler, W. I. Sundquist, and P. D. Bieniasz. 2004. Species-specific tropism determinants in the human immunodeficiency virus type 1 capsid. J. Virol. 78:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hennig, J., L. Ottosson, C. Andresen, L. Horvath, V. K. Kuchroo, K. Broo, M. Wahren-Herlenius, and M. Sunnerhagen. 2005. Structural organization and Zn2+-dependent subdomain interactions involving autoantigenic epitopes in the Ring-B-box-coiled-coil (RBCC) region of Ro52. J. Biol. Chem. 280:33250-33261. [DOI] [PubMed] [Google Scholar]
- 11.Himathongkham, S., and P. A. Luciw. 1996. Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology 219:485-488. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73:10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 14.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 15.Javanbakht, H., F. Diaz-Griffero, M. Stremlau, Z. Si, and J. Sodroski. 2005. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5α. J. Biol. Chem. 280:26933-26940. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, K., C. Shiels, and P. S. Freemont. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223-7233. [DOI] [PubMed] [Google Scholar]
- 17.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kootstra, N. A., C. Munk, N. Tonnu, N. R. Landau, and I. M. Verma. 2003. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc. Natl. Acad. Sci. USA 100:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 20.Mische, C. C., H. Javanbakht, B. Song, F. Diaz-Griffero, M. Stremlau, B. Strack, Z. Si, and J. Sodroski. 2005. Retroviral restriction factor TRIM5α is a trimer. J. Virol. 79:14446-14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Nisole, S., C. Lynch, J. P. Stoye, and M. W. Yap. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 101:13324-13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ottosson, L., J. Hennig, A. Espinosa, S. Brauner, M. Wahren-Herlenius, and M. Sunnerhagen. 2006. Structural, functional and immunologic characterization of folded subdomains in the Ro52 protein targeted in Sjögren's syndrome. Mol. Immunol. 43:588-598. [DOI] [PubMed] [Google Scholar]
- 22.Owens, C. M., B. Song, M. J. Perron, P. C. Yang, M. Stremlau, and J. Sodroski. 2004. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 78:5423-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owens, C. M., P. C. Yang, H. Gottlinger, and J. Sodroski. 2003. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J. Virol. 77:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Caballero, D., T. Hatziioannou, A. Yang, S. Cowan, and P. D. Bieniasz. 2005. Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J. Virol. 79:8969-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saurin, A. J., K. L. Borden, M. N. Boddy, and P. S. Freemont. 1996. Does this have a familiar RING? Trends Biochem. Sci. 21:208-214. [PubMed] [Google Scholar]
- 28.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 102:2832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573. [DOI] [PubMed] [Google Scholar]
- 29.Shibata, R., H. Sakai, M. Kawamura, K. Tokunaga, and A. Adachi. 1995. Early replication block of human immunodeficiency virus type 1 in monkey cells. J. Gen. Virol. 76(Pt. 11):2723-2730. [DOI] [PubMed] [Google Scholar]
- 30.Si, Z., N. Vandegraaff, C. O'Huigin, B. Song, W. Yuan, C. Xu, M. Perron, X. Li, W. Marasco, A. Engelman, M. Dean, and J. Sodroski. 2006. Evolution of a cytoplasmic tripartite motif (TRIM) protein in cows that restricts retroviral infection. Proc. Natl. Acad. Sci. USA 103:7454-7459. (First published 28 April 2006; doi: 10.1073/pnas.0600771103.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song, B., B. Gold, C. O'Huigin, H. Javanbakht, X. Li, M. Stremlau, C. Winkler, M. Dean, and J. Sodroski. 2005. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5α exhibits lineage-specific length and sequence variation in primates. J. Virol. 79:6111-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song, B., H. Javanbakht, M. Perron, D. H. Park, M. Stremlau, and J. Sodroski. 2005. Retrovirus restriction by TRIM5α variants from Old World and New World primates. J. Virol. 79:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoye, J. P. 2002. An intracellular block to primate lentivirus replication. Proc. Natl. Acad. Sci. USA 99:11549-11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 35.Stremlau, M., M. Perron, M. Lee, L. Yuan, B. Song, H. Javanbakht, F. Diaz-Griffero, D. Anderson, W. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. USA 103:5514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stremlau, M., M. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction. Curr. Biol. 15:73-78. [DOI] [PubMed] [Google Scholar]