Abstract
Homologs of the UL25 gene product of herpes simplex virus (HSV) have been identified in all three subfamilies of the Herpesviridae. However, their exact function during viral replication is not yet known. Whereas earlier studies indicated that the UL25 protein of HSV-1 is not required for cleavage of newly replicated viral DNA but is necessary for stable encapsidation (A. R. McNab, P. Desai, S. Person, L. Roof, D. R. Thompson, W. W. Newcomb, J. C. Brown, and F. L. Homa, J. Virol. 72:1060-1070, 1998), viral DNA packaging has recently been demonstrated to occur in the absence of UL25, although at significantly decreased levels compared to wild-type HSV-1 (N. Stow, J. Virol. 75:10755-10765 2001). To clarify the functional role of UL25 we analyzed the homologous protein of the alphaherpesvirus pseudorabies virus (PrV). PrV UL25 was found to be essential for viral replication, as a mutant virus lacking the UL25 protein required UL25-expressing cells for productive propagation. In the absence of the UL25 protein, newly replicated PrV DNA was cleaved and DNA-containing C-type capsids were detected in infected cell nuclei. However, although capsids were frequently found in close association with the inner nuclear membrane, nuclear egress was not observed. Consequently, no capsids were found in the cytoplasm, resulting in an inhibition of virion morphogenesis. In contrast, the formation of capsidless enveloped tegument structures (L particles) in the cytoplasm was readily observed. Thus, our data demonstrate that the PrV UL25 protein is not essential for cleavage and encapsidation of viral genomes, although both processes occur more efficiently in the presence of the protein. However, the presence of the PrV UL25 protein is a prerequisite for nuclear egress. By immunoelectron microscopy, we detected UL25-specific label on DNA-containing C capsids but not on other intranuclear immature or defective capsid forms. Thus, the PrV UL25 protein may represent the hitherto missing trigger that allows primary envelopment preferably of DNA-filled C capsids.
Herpesvirus assembly is a multistep process involving many protein-protein, as well as protein-DNA, interactions which occur in at least two distinct subcellular compartments. While capsid assembly and DNA packaging take place in the nucleus of infected cells, the addition of most of the tegument, as well as final envelopment, proceed in the cytoplasm (reviewed in references 42 and 43). In recent years, an increasing amount of data has indicated that the intranuclear stages of herpesvirus assembly, namely, DNA replication, capsid formation, and DNA cleavage/packaging follow pathways similar to those described for the better-studied large double-stranded DNA bacteriophages such as P22, T4, T7, and λ (26). The similarities even extend to capsid structure, which, together, point to a common ancestry of the eukaryotic Herpesviridae and the prokaryotic Caudovirales (8).
In a generalized model of cleavage and packaging of genomic DNA of double-stranded DNA viruses (reviewed in reference 9), the terminase enzyme binds to specialized sites in the concatemeric DNA and subsequently cleaves the duplex DNA to generate one mature genome end. The terminase with the attached concatemeric DNA then binds to the portal complex, a dodecameric ring structure located at one vertex in the capsid which forms a channel. The interaction of the portal with the terminase-genome complex generates the packaging motor, which pumps the DNA against growing mechanical forces into the capsid, fueled by ATP hydrolysis, resulting in removal of the internal scaffold structure. When the full-length genome is packaged, the terminase executes the second cleavage to complete the packaging process. Since the terminase remains bound to the DNA concatemer, it can subsequently bind to another procapsid to initiate a second round of packaging. To keep the tightly condensed DNA in the head, the portal channel has to be properly sealed, preventing DNA leakage. This can either be achieved by a conformational change of the portal protein, as shown for bacteriophage Φ29 (16), or by addition of head completion proteins, as described, e.g., for phage λ (53). Tail fibers are added to the DNA-filled and sealed bacteriophage head, while in herpesviruses mature capsids bud at the inner nuclear membrane, and are subsequently released into the cytoplasm.
For herpesviruses, four different capsid forms have been described. The procapsid, a spherical and porous particle with an internal scaffold structure, most likely constitutes the precursor for the icosahedral A, B, and C capsids (19, 46, 47, 55). C capsids contain the viral DNA genome and are characterized electron-microscopically by their electron-dense core. A capsids appear translucent, containing neither DNA nor scaffold proteins. They may be the product of an abortive packaging event. It is not completely clear whether B capsids, which are characterized by their ring-like internal scaffold structure, are dead-end products in which the scaffold is trapped or whether they eventually mature into C capsids (52, 55, 56).
In herpes simplex virus type 1 (HSV-1), six conserved gene products have been shown to be essential for cleavage-encapsidation: UL6 encodes the portal protein (48, 51), whereas the products of the UL15 and UL28 genes probably form the subunits of the terminase (7, 54, 65). The products of the UL17 (57), UL32 (38), and UL33 (4) genes are also essential, but their precise roles during capsid maturation have not yet been determined. Mutants of all these genes show a common phenotype: newly replicated concatemeric DNA is not cleaved into unit-length genomes, and, accordingly, in infected cell nuclei no C capsids are detectable while scaffold-containing immature B capsids accumulate. In contrast, a mutant in a seventh gene, UL25, exhibits a different phenotype. Although concatemeric DNA was processed into unit-length genomes, no C capsids were found, indicating that the genome was not stably packaged (41). In agreement with this finding, in cells infected by a UL25-negative HSV-1 mutant, increased amounts of A capsids were present, suggesting that UL25 may function as a head completion protein or plug which seals the portal channel after DNA packaging (41). In line with this proposal, the HSV-1 UL25 protein has recently been reported to interact with the capsid shell and viral DNA, further suggesting a role in anchoring of the genome to the capsid (50).
However, this hypothesis for UL25 function has been challenged. Stow (60) reported that in cells infected with the HSV-1 UL25 null mutant KUL25NS, cleavage was less efficient but that the fraction of viral DNA that was processed properly was also stably packaged (60). Southern blot analyses of terminal fragments revealed that the L-terminal fragments were overrepresented while S termini were underrepresented, indicating that in the absence of UL25 the left genome end was produced normally while generation of the second (right) genome end was impaired (60). Although directionality of DNA packing has not been proven, these data indicate that the packaging reaction, which most likely generates first the L terminus, is initiated normally in KUL25NS-infected cells but is not completed in the majority of events. In line with this, the UL25 mutant was less impaired in packaging of HSV-1 amplicon DNA up to approximately 100 kb in size, indicating that UL25 is necessary for efficiently completing the final stages of packaging of the full-length genome but not for either cleavage or encapsidation of the smaller amplicon DNA (60). Thus, it was hypothesized that HSV-1 UL25 might fulfill a function similar to that of gene product D (gpD) in bacteriophage λ, where, upon packaging of 10 to 50% of the viral genome, the capsid undergoes significant changes in size and shape. At this stage, gpD is added as a trimer onto the surface and its presumed role is to stabilize the partially filled capsid to prevent the release of packaged genomic DNA (53; reviewed in reference 17). In the absence of gpD, only a fraction of capsids were filled (25), but the requirement for gpD could be overcome by partial deletion of the phage genome (53, 59). The two other head completion proteins, gpW and gpFII, are added only in the presence of gpD, which then triggers the addition of the preformed tail. Interestingly, although DNA-containing C capsids were found in cells infected with HSV-1 KUL25NS, these were not translocated to the cytoplasm, indicating that UL25 may be essential for a step beyond packaging (60).
To clarify the role of UL25 in the herpesvirus replication process, we analyzed the function of the homologous protein in PrV. The PrV UL25 protein has recently been identified as a 57-kDa minor capsid component (27). Based on reports that HSV-1 mutants with a temperature-sensitive phenotype in the UL25 gene region were found to be defective not only in capsid assembly but also in penetration (1, 3), Kaelin and coworkers (27) wanted to investigate whether UL25 might be involved in transport of capsids along microtubules toward the nuclear pore during virus entry. They showed that UL25, besides exhibiting a distinct nuclear localization, also colocalizes with microtubules. However, no mutant viruses were built to substantiate these findings on a functional basis.
Here, we show that UL25 is essential for PrV replication in cell culture. Although in mutant virus-infected cells cleavage of concatemeric DNA occurs and DNA is packaged into the nascent capsid, these nucleocapsids were not translocated to the cytoplasm. However, they were often found adjacent to the inner nuclear membrane, indicating that PrV UL25 is necessary for primary envelopment, which initiates nuclear egress.
MATERIALS AND METHODS
Cells and viruses.
All virus mutants used in this study were derived from PrV strain Kaplan (PrV-Ka) (28), whose genome has been cloned recently as bacterial artificial chromosome (BAC) clone pPrV-ΔgB (36) in Escherichia coli. Viruses were propagated in rabbit kidney (RK13) or pig kidney (PSEK) cells in minimum essential medium supplemented with 10% or 5% fetal calf serum, respectively. Generation of PrV-ΔUL17F and the corresponding RK13-UL17 cells has been described recently (30).
Preparation of UL25-specific antiserum.
For generation of a UL25-specific antiserum, the 861-bp SalI subfragment of BamHI fragment 9, which comprises the C-terminal part of the UL25 open reading frame (see Fig. 1C), was cloned into the SalI-cleaved prokaryotic expression vector pGEX-4T-3 (Amersham Biosciences, Freiburg, Germany). Correct orientation and in-frame cloning with the glutathione S-transferase (GST) open reading frame was verified by sequencing. The approximately 50-kDa GST-UL25 fusion protein was eluted from a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel and used for immunization of a rabbit, as described recently (32). Serum obtained after the fifth immunization was used throughout this study.
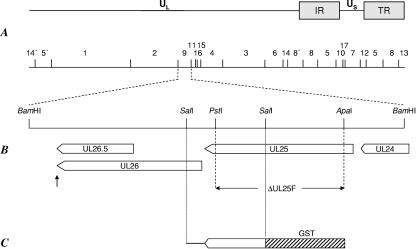
FIG. 1.
Construction of PrV-ΔUL25F. (A) Map of the PrV genome with the unique long (UL), unique short (US), and inverted repeat (IR, TR) sequences. BamHI restriction sites are indicated, and fragments are numbered according to size. (B) Enlargement of BamHI fragment 9. The UL24, UL25, UL26, and UL26.5 genes (shown as pointed rectangles) are transcribed into 3′-coterminal mRNAs with a common poly(A) addition signal (shown as an arrow pointing up). Relevant restriction sites for construction of the UL25 deletion mutant, PrV-ΔUL25F, are indicated, as is the extent of the introduced deletion. (C) A 0.9-kb SalI subfragment of BamHI fragment 9 was used for prokaryotic expression of GST-UL25.
Generation of a UL25-complementing cell line, UL25 deletion mutant PrV-ΔUL25F, and UL25 rescue mutant PrV-ΔUL25FR.
For generation of a UL25-complementing cell line, the complete UL25 open reading frame was amplified by PCR using Pfx polymerase (Invitrogen, Karlsruhe, Germany), primers UL25For (5′-CACAGAATTCGCGCGGCCCATGTCCCC-3′) (nucleotides [nt] 58936 to 58919; GenBank accession number BK001744 [31]; UL25 start codon shown in bold) and UL25Rev (5′-CACATCTAGAGCTCAGGCGGCGGCGAAC-3′) (nt 57305 to 57322; GenBank accession number BK001744; UL25 stop codon shown in bold), and cloned BamHI fragment 9 as template. The primer sequences contained restriction enzyme recognition sites for EcoRI (UL25For, shown in italics) and XbaI (UL25Rev, shown in italics), which were used for cloning of the 1.6-kb PCR product into appropriately cleaved eukaryotic expression vector pcDNA3 (Invitrogen). The resulting plasmid was used for transfection of RK13 cells using Superfect transfection reagent (QIAGEN, Hilden, Germany). Transfected cell clones were selected in medium containing 500 μg G418 (Invitrogen) per ml and tested for UL25 expression by indirect immunofluorescence with the anti-UL25 serum. One cell clone, designated RK13-UL25, was used for isolation of mutant PrV-ΔUL25F.
For construction of a PrV UL25 deletion mutant, the 1.6-kb EcoRI/XbaI insert of pcDNA-UL25 was cloned into appropriately cleaved pUC19 (New England Biolabs, Frankfurt, Germany) after inactivation of the resident PstI site, resulting in pUC19ΔP-UL25. This plasmid was then digested with PstI and ApaI, thereby removing 1,398 bp of UL25 coding sequences, which were replaced by a kanamycin resistance gene flanked by flp recombinase recognition target (FRT) sites derived from plasmid pKD13 (12). The complete insert of this plasmid was amplified by PCR using pUC-specific primers (New England Biolabs), and the resulting PCR product was used for mutagenesis of the BAC clone PrV-ΔgB, as described previously (36). After isolation of kanamycin-resistant clones, the kanamycin cassette was removed by flp recombinase provided by plasmid pCP20 (11). The gB gene was finally restored by cotransfection (20) of BAC pPRV-ΔUL25F DNA and plasmid pUC-B1BclI (36) into RK13-UL25 cells. The UL25 rescue mutant, PrV-ΔUL25FR, was isolated after cotransfection of RK13 cells with genomic DNA of PrV-ΔUL25F and cloned BamHI fragment 9 of PrV-Ka.
For the deletion and rescue mutants, several plaque isolates of the transfection progeny were screened by Southern blot analysis (data not shown), and one of each was randomly chosen for further characterization. Correct deletion of UL25-specific sequences was verified by sequencing of the PCR product generated with primers UL25For and UL25Rev on PrV-ΔUL25F DNA (data not shown).
In vitro growth studies.
For analysis of one-step growth kinetics, RK13 or RK13-UL25 cells were infected with PrV-Ka, PrV-ΔUL25F, or PrV-ΔUL25FR at a multiplicity of infection (MOI) of 10 and incubated on ice for 1 h. Prewarmed medium was then added, and cells were further incubated for 1 h at 37°C. Thereafter, nonpenetrated virus was inactivated by low pH treatment, and cells were scraped into the medium immediately (0 h), as well as after 4, 8, 12, 24, and 36 h, and frozen at −70°C. After all samples were harvested, they were thawed at 37°C and centrifuged for 2 min at 13,000 rpm, and virus titers in the supernatant were determined on RK13-UL25 cells.
For plaque assays, RK13 or RK13-UL25 cells were infected with 100 PFU per well of PrV-Ka or PrV-ΔUL25FR, RK13-UL25 cells were infected with 100 PFU of PrV-ΔUL25F per well, and RK13 cells were infected with 1,000 PFU of PrV-ΔUL25F per well of a six-well culture dish. Two days after infection (p.i.), cells were fixed with ethanol and stained with a monoclonal antibody against gC (B16-c8 [49]). Fluorescent plaques were documented with a digital camera (C3040 Zoom; Olympus, Hamburg, Germany).
Preparation of virus DNA and Southern blotting.
RK13 and RK13-UL25 cells were infected with PrV-Ka, PrV-ΔUL25F, or PrV-ΔUL17F at an MOI of 5, and infected cell DNA was prepared approximately 16 h p.i. DNA was digested with BamHI, separated on 0.8% agarose gels, blotted onto nylon membranes, and probed with radioactively labeled genome-end-specific BamHI fragment 13 or 14′ (see Fig. 1 for location). Bound radioactivity was recorded with a phosphorimager (FLA-3000; Raytest, Straubenhardt, Germany).
Virus purification, virus fractionation, and immunoblotting.
Virus purification and preparation of infected cell lysates were done as previously described (34). Parallel blots were incubated with sera against UL25 (1:50,000 [this study]), UL19 (1:500,000 [32]), UL35 (1:100,000 [W. Fuchs, unpublished data]), UL6 (1:50,000 [B. G. Klupp, unpublished data]), UL17 (1:50,000 [30]), UL3 (1:50,000 [35]), US3 (1:100,000 [33]), UL31 (18), UL34 (32), and gH (29) and a monoclonal antibody against gB (b43-b6 [49]). Virus fractionation into capsid and envelope fractions was performed essentially as described previously (63). Briefly, purified PrV virions were incubated for 30 min on ice in 1% NP-40. The samples were subsequently layered onto a 30% sucrose cushion and centrifuged in a TLS55 rotor (Beckmann Instruments, Munich, Germany) at 45,000 rpm for 1 h at 4°C. The envelope/tegument fraction was collected from the top of the sucrose cushion, and the pellet containing capsids and part of the tegument was resuspended in phosphate-buffered saline. Proteins were separated on discontinuous SDS-polyacrylamide gels (37) and transferred onto nitrocellulose membranes. Parallel blots were incubated with sera against UL25 (1:50,000 [this study]), UL19 (1:500,000 [32]), UL6 (1:50,000 [unpublished data]), UL17 (1:50,000 [30]), UL37 (34), and gH (29).
Electron microscopy.
RK13 or RK13-UL25 cells were infected with PrV-ΔUL25F at an MOI of 1 and processed for electron microscopy as described previously (32). Counterstained ultrathin sections were analyzed with an electron microscope (Technai 12; Philips, The Netherlands). Immunolabeling was performed with the anti-UL25 serum and secondary gold-tagged anti-rabbit antibodies (GAR10, GAR 5; British Biocell International, Cambridge, United Kingdom) as previously described (21).
RESULTS
Isolation and characterization of PrV-ΔUL25F.
For functional characterization of the PrV UL25 protein, a deletion mutant was generated by mutagenesis of a BAC clone of the PrV genome in E. coli (Fig. 1). The recombinant PrV-ΔUL25F carried a deletion of codons 29 to 495 of the 534-amino-acid UL25 open reading frame and contained instead a 36-bp FRT site, a remnant of the mutagenesis reaction. This mutagenesis protocol was chosen to minimize negative effects on the up- and downstream genes UL24, UL26, and UL26.5, which are transcribed with UL25 into 3′-coterminal mRNAs (15). The FRT insert comprised stop codons that inhibit any possible expression of the remaining 39 C-terminal amino acids of UL25. Isolation of PrV-ΔUL25F was successful only with RK13-UL25 cells, indicating that PrV UL25 is essential for virus replication in cell culture.
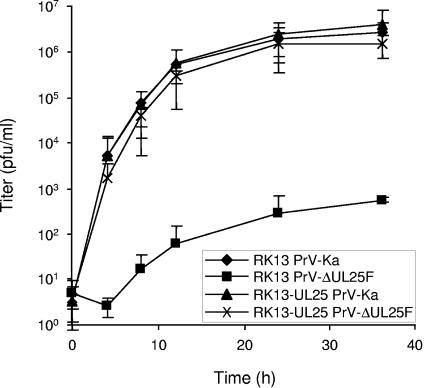
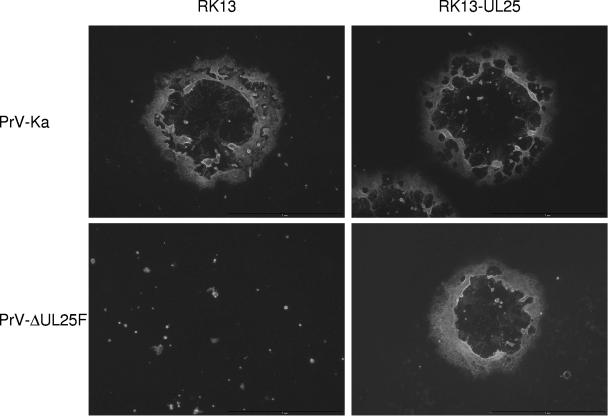
To analyze the growth defect of PrV-ΔUL25F in more detail, one-step growth kinetics and plaque assays were performed. Infection of noncomplementing RK13 cells with PrV-ΔUL25F resulted in only very few infectious particles, with maximum titers not exceeding 103 PFU per ml, while propagation of the mutant virus on RK13-UL25 cells resulted in infectious progeny comparable to wild-type PrV-Ka on both cell lines (Fig. 2). In plaque assays, only single infected cells or small syncytia of infected cells were observed on RK13 cells after infection by PrV-ΔUL25F, while plaque formation was as efficient as with PrV-Ka on the complementing cell line (Fig. 3). Since RK13-UL25 cells carry and express only the UL25 open reading frame and no other viral proteins, but are competent to rescue the defects of the mutant virus, the observed defects are due to the introduced UL25 deletion and not to other fortuitous alterations elsewhere in the genome. In the rescue mutant, the replication defects were also restored (data not shown).
FIG. 2.
One-step growth kinetics of PrV-ΔUL25F. RK13 or RK13-UL25 cells were infected with PrV-Ka or PrV-ΔUL25F at an MOI of 10. Cells and supernatant were harvested at the indicated time points, and infectious titers were determined on RK13-UL25 cells and plotted. Mean titers of three independent experiments and the corresponding standard deviations are shown.
FIG. 3.
Plaque formation of PrV-ΔUL25F. RK13 and RK13-UL25 cells were infected with PrV-Ka and PrV-ΔUL25F under plaque assay conditions and fixed 2 days p.i. Infected cells were visualized by indirect immunofluorescence with a glycoprotein C-specific monoclonal antibody.
Protein expression profiles in PrV-ΔUL25F-infected cells.
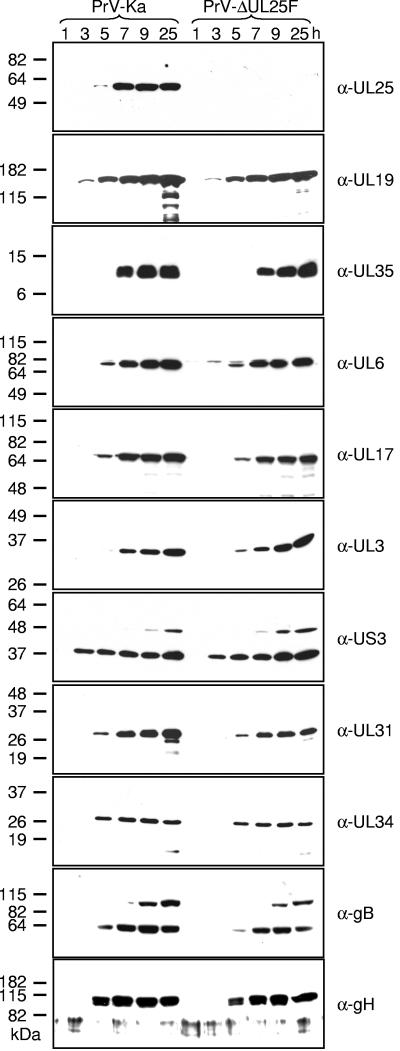
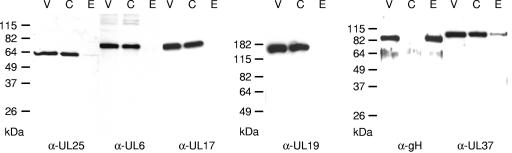
The PrV UL25 protein has been described recently as a 57-kDa minor capsid protein (27). To verify that PrV-ΔUL25F lacks the corresponding protein, we raised a UL25-specific antiserum against the C-terminal 220 amino acids by immunization of a rabbit with a GST-UL25 fusion protein (Fig. 1C). Serum obtained after the fifth immunization was used in this study. In Western blot analyses of PrV-Ka-infected RK13 cells, the serum reacted as expected with a single protein of 57 kDa, which was first detected at approximately 5 h p.i. (Fig. 4). No signal was found in cell lysates of PrV-ΔUL25F-infected RK13 cells, verifying specificity of the generated antiserum and correct deletion in the mutant virus. To investigate whether deletion of UL25 affected expression of other viral proteins, parallel blots were incubated with sera against other capsid or capsid-associated proteins (UL6, UL17, UL19, UL35), viral proteins targeted to the nucleus or nuclear membrane (UL3, UL31, UL34, US3), or membrane glycoproteins (gB, gH). All tested antisera reacted similarly with PrV-Ka- or PrV-ΔUL25F-infected cell lysates, indicating that synthesis of other proteins is not impaired in the absence of UL25.
FIG. 4.
Expression kinetics of mutant PrV-ΔUL25F. Western blot analysis of rabbit kidney cells infected with PrV-Ka or PrV-ΔUL25F at an MOI of 10 is shown. Cells were harvested at the indicated time points after infection, and proteins were separated on SDS-10% polyacrylamide gels. After electrotransfer onto nylon membranes, parallel blots were incubated with the indicated antisera. Molecular masses of marker proteins are indicated on the left.
C capsids are formed in the absence of PrV UL25.
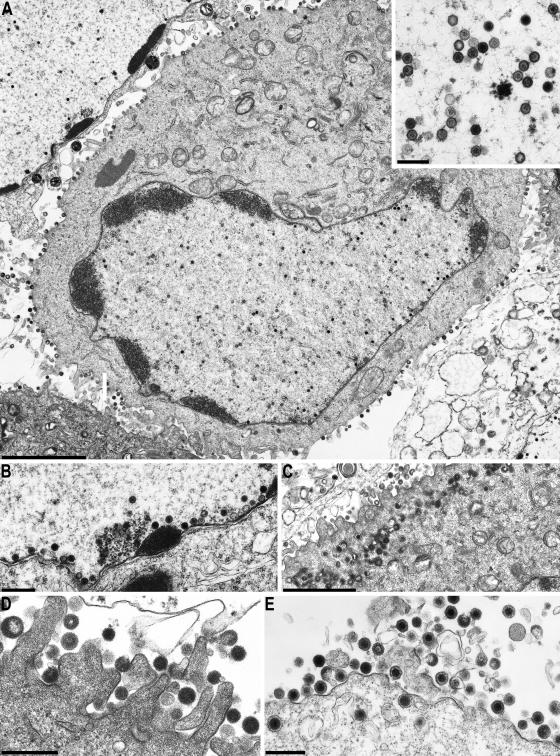
One-step growth kinetics already indicated that virtually no infectious particles were produced in the absence of UL25. In electron microscopic analyses of PrV-ΔUL25F-infected RK13 cells, no extracellular virions or nucleocapsids in the cytoplasm were detectable (Fig. 5A, C, and D). However, in infected cell nuclei, DNA-containing C-type capsids were clearly visible (Fig. 5A, A inset, and B), indicating that DNA encapsidation takes place in the absence of UL25. In addition, A and B capsids (Fig. 5A, inset) were present. Interestingly, although all capsid forms, including DNA-containing C capsids, which are characterized by their electron-dense core, were detected frequently in close proximity to the inner nuclear membrane (Fig. 5B), neither primary envelopment nor primary enveloped virions in the perinuclear space or virions or nucleocapsids in the cytoplasm were observed. This indicates that the UL25 protein is necessary for nuclear egress of capsids but not for encapsidation of viral DNA. In the cytoplasm and on the surface of PrV-ΔUL25F-infected RK13 cells, numerous capsidless tegument-containing particles (L, or light, particles [2, 40]) were detectable (Fig. 5C and D), illustrating that secondary envelopment of tegument proteins in the cytoplasm apparently proceeds undisturbed in the absence of UL25. In contrast, on complementing cells or in RK13 cells infected with the UL25 rescue mutant PrV-ΔUL25FR, all stages of capsid morphogenesis and virus assembly were easily observed and numerous mature virions lined the cell surface (Fig. 5E and data not shown).
FIG. 5.
Electron microscopy of PrV-ΔUL25F-infected RK13 and RK13-UL25 cells. RK13 cells (A to D) or RK13-UL25 cells (E) were infected with PrV-ΔUL25F at an MOI of 1 and processed for electron microscopy at 14 h p.i. Bars represent 3 μm in panel A (300 nm in inset); 0.5 μm in panels B, D, and E; and 2 μm in panel C.
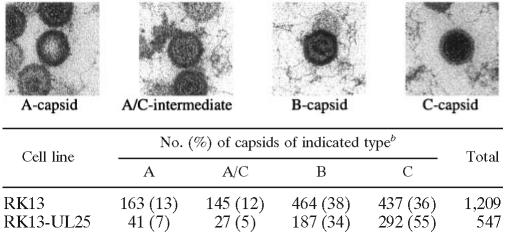
To estimate relative frequencies, the abundance of the different capsid forms was determined in PrV-ΔUL25F-infected RK13 and RK13-UL25 nuclei (Table 1). Capsids were differentiated by absence of either DNA or internal scaffold (A capsids), capsids apparently undergoing DNA packaging (A/C intermediate), capsids with clearly discernible internal scaffold (B capsids), and capsids with an electron-dense core (C capsids). The results indicated that fewer C capsids (36% versus 55%), similar numbers of B capsids (34% and 38%), and more A capsids (13% versus 7%) were present in noncomplementing cells, indicating that although DNA is packaged in absence of UL25, packaging appears to be less efficient and results in more abortive events. This is in line with the observation that intermediate capsid forms (designated A/C intermediates in Table 1), which are supposed to be in the process of DNA packaging, were more abundant in PrV-ΔUL25F-infected RK13 nuclei (12%) than in UL25-expressing cells (5%) (Table 1).
TABLE 1.
Quantitation of intranuclear capsids in PrV-ΔUL25F-infected cellsa
Representative electron micrographs for the different capsid forms are given. A-type capsids are identified by the lack of an internal structure. B capsids contain ring- or crescent-shaped scaffolding. C capsids are identified by an electron-dense core. A/C intermediates are capsids that are not assigned to any of the “classical” types; they are probably in the process of genome packaging.
Capsids were counted after infection by PrV-ΔUL25F of noncomplementing RK13 cells or complementing RK13-UL25 cells.
PrV UL25 is not required for cleavage of concatemeric DNA.
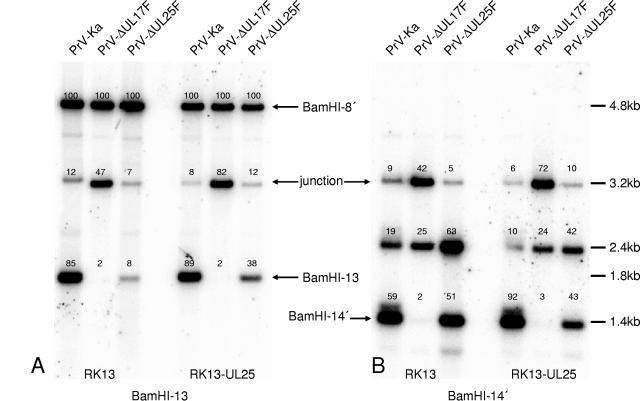
The electron microscopic images already indicated that DNA packaging is not inhibited in the absence of PrV UL25 but appears to be less efficient. To investigate whether cleavage is impaired in PrV-ΔUL25F-infected cells, Southern blot analyses with genome-end-specific fragments were performed. To this end, RK13 or RK13-UL25 cells were infected with PrV-Ka, PrV-ΔUL25F, or PrV-ΔUL17F, a mutant which had been identified previously as deficient in cleavage and encapsidation of viral DNA (30). Whole-cell DNA was isolated, digested with BamHI, transferred onto nylon membranes, and probed with genome-end-specific BamHI-fragment 13 (Fig. 6A) or 14′ (Fig. 6B), located at the US and UL ends, respectively (Fig. 1A). As evident in Fig. 6A, in DNA from PrV-Ka-infected cells, the 1.8-kb terminal fragment 13 is clearly detectable (85% of count compared to BamHI fragment 8′) while the 3.2-kb junction fragment, originating from head-to-tail fused genomes, yields a weaker hybridization signal (12%). Since BamHI fragment 13 is located in the inverted repeat regions, the probe also reacted with BamHI fragment 8′, which was used as an internal standard for quantification, and corresponding counts were set as 100%. In contrast, no terminal fragment 13 was detectable in DNA of RK13 cells infected with the cleavage-deficient mutant PrV-ΔUL17F, but the signal representing the junction fragment was significantly increased (47%). In DNA from PrV-ΔUL25F-infected RK13 cells, terminal BamHI fragment 13 was detectable but its signal was much less intense than after wild-type PrV-Ka infection (8% compared to 85% in PrV-Ka). However, the signal strength for the junction fragment did not increase in parallel, as in PrV-ΔUL17F DNA, indicating that cleavage of concatemeric DNA must have occurred efficiently.
FIG. 6.
Impairment of cleavage of concatemeric DNA in the absence of PrV UL25. RK13 or RK13-UL25 cells were infected with PrV-Ka, PrV-ΔUL17, or PrV-ΔUL25F at an MOI of 5 and harvested at 16 h p.i. Whole-cell DNA was isolated, digested with BamHI, separated on a 0.8% agarose gel, and blotted onto a nylon membrane. Parallel blots were probed with labeled genome-end-specific BamHI fragments 13 (A) and 14′ (B). Bound radioactivity was recorded by a phosphorimager and quantified. Values were calculated with respect to bound radioactivity present on BamHI fragment 8′, set as 100% for each lane. Identification of fragments and their sizes are indicated. “Junction” refers to the junction fragment derived from head-to-tail-fused termini.
Hybridization of a parallel blot with BamHI fragment 14′ gave a different result (Fig. 6B). Here, in DNA isolated from PrV-ΔUL25F- or PrV-Ka-infected cells, terminal fragment 14′ was present in similar amounts (51% to 59%), whereas in PrV-ΔUL17F-infected cells it was not produced. In the latter case, the junction fragment was overrepresented compared to DNA of cells infected by the other two viruses (42% compared to 9 and 5%, respectively). The cleavage defect of PrV-ΔUL25F concerning the production of terminal fragment 13′ could be at least partially rescued after propagation on RK13-UL25 cells. Cleavage of concatemeric DNA in PrV-ΔUL17F could be rescued after propagation on RK13-UL17 cells (30). Thus, these data show that the cleavage reactions that produce the left (UL) end of the genome occur with equal efficiency in PrV-Ka- and PrV-ΔUL25F-infected cells, whereas there is a defect in creating the right (US) end of the genome, represented by BamHI fragment 13 in PrV-ΔUL25F infection. The hybridization probe containing BamHI fragment 14′ also hybridized with a third fragment, of 2.4 kb, in all DNAs analyzed. Cloning and sequencing of the respective DNA fragment revealed that it contains two terminal BamHI fragments 14′ fused in a tail-to-tail orientation. Its provenance is unclear. However, it was reproducibly found in greater amounts in PrV-ΔUL25F-infected cells than in PrV-ΔUL17- or PrV-Ka-infected RK13 or RK13-UL25 cells.
Subviral localization of PrV UL25.
Kaelin et al. identified PrV UL25 as a minor capsid component (27), although recent studies indicate that it may represent a rather abundant protein in purified virion preparations (45). To study the localization of UL25 in the virus particle, we chose two experimental procedures. First, purified virions were separated into a capsid/tegument and a tegument/envelope fraction (63). As shown in Fig. 7, antiserum against glycoprotein gH reacted only with complete virion and envelope fractions, whereas antiserum against the major capsid protein UL19 recognized its target in whole virion and capsid fractions. A similar reactivity was observed using antisera against capsid-associated protein UL6 (the portal protein) or UL17 (30). The UL37 protein, which is part of the capsid-proximal inner tegument (34, 42, 43), was present primarily in the capsid fraction, and only a faint signal was detected in the “envelope” fraction. The majority of the UL25 protein was detectable in the whole-virus and capsid fractions, with only a very faint signal in the envelope/tegument preparation, indicating that UL25 is tightly bound to the capsid.
FIG. 7.
Virus fractionation. Purified virion preparations (V) were separated into capsid (C) and envelope (E) fractions and analyzed by immunoblotting with sera against the major capsid protein UL19, the portal protein UL6, the capsid component UL17, envelope glycoprotein H (gH), or the UL37 tegument protein, as well as with the UL25-specific serum. Molecular mass markers are indicated on the left.
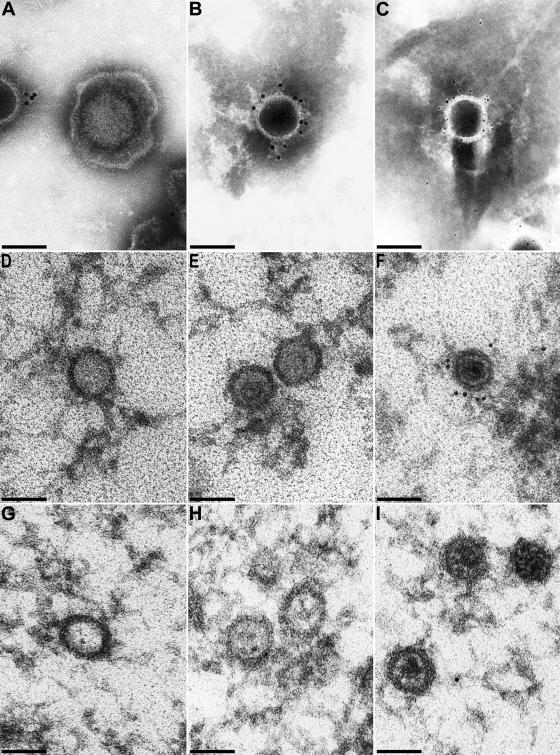
In a second approach, purified virions were investigated by immunoelectron microscopy. As shown in Fig. 8, in purified virion preparations only capsids exhibited specific label (Fig. 8A to C), whereas intact virions were not decorated (Fig. 8A). Analysis of intranuclear capsids in thin sections demonstrated that A (Fig. 8D) and B (Fig. 8E) capsids were devoid of label (30 of 30 A capsids, 15 of 15 B capsids), whereas C capsids were clearly labeled at multiple sites on the capsid (17 of 20 C capsids) (Fig. 8F). For controls, neither A, B, nor C capsids in PrV-ΔUL25F-infected cells exhibited any specific label (Fig. 8G to I).
FIG. 8.
Immunolabeling of purified virions. Purified virion preparations and capsids in ultrathin sections were analyzed by immunoelectron microscopy after incubation with UL25 antiserum and gold-conjugated secondary antibodies. (A to C) Results from analysis of negatively stained purified virion preparations. (D to F) Intranuclear A, B, and C capsids in PrV-Ka-infected RK13 cells. (G to I) Different intranuclear capsid forms in PrV-ΔUL25F-infected cells serving as negative controls to demonstrate the specificity of the antiserum. Secondary antibodies tagged with 5-nm gold particles were used for panel C, whereas 10-nm gold-tagged antibodies were used for all other panels. Bars, 100 μm.
DISCUSSION
Proteins homologous to the UL25 protein of HSV-1 are conserved in all three subfamilies of the Herpesviridae (5, 10, 39). However, their exact role during viral replication is still unclear. The HSV-1 UL25 protein apparently is involved in a late step of the DNA cleavage/encapsidation process (41, 60). Although it is disputed whether it is involved in stably maintaining packaged genomes in the capsid (41) or in the encapsidation process itself (60), it is evident that the protein is not necessary for initiation of the cleavage of concatemeric replication intermediates and their packaging. We studied UL25 function in the alphaherpesvirus PrV. The salient findings of this study are that (i) UL25 is essential for PrV replication in cell culture, (ii) it is not required for cleavage and packaging of viral genomic DNA, and (iii) it is essential for nuclear egress of capsids.
Cleavage of concatemeric DNA and packaging of unit-length herpesvirus genomes into capsids are tightly interconnected processes involving several herpesvirus proteins (reviewed in reference 6). Only the complete set of proteins leads to properly packaged full-length genomes and the release of mature nucleocapsids from the nucleus. Genes encoding proteins that are necessary for these processes are highly conserved throughout the Herpesviridae, indicating the existence of a conserved mechanism. Basic principles of genome replication and packaging are even shared with the large double-stranded DNA-containing bacteriophages which rely on similar mechanisms to replicate and encapsidate their genomes (reviewed in references 6 and 9). Whereas in the bacteriophages the mature capsid serves as the substrate for addition of the tail and tail fibers, the mature herpesvirus capsid proceeds to envelopment at the inner nuclear membrane. So far, however, the molecular requirements for this primary envelopment process and, in particular, for the preferential envelopment of mature C capsids remain unclear.
We show here that the PrV UL25 protein is required for primary envelopment but is not essential for cleavage and encapsidation of genomic DNA. Thus, in the absence of UL25, all three described intranuclear capsid forms of PrV (44)—empty A capsids, scaffold-containing B capsids, and genome-containing C capsids—were observed, often in intimate contact with the inner nuclear membrane. However, primary envelopment was never seen, indicating that even apparently “mature” C capsids are not competent for budding at the inner nuclear membrane in the absence of UL25.
This defect in primary envelopment parallels the requirement for UL25 for productive herpesvirus replication. PrV UL25, like its homologs in HSV-1 (3) and bovine herpesvirus type 1 (BHV-1) (14), is essential for productive replication in cell culture. Only very little infectious progeny was detectable after replication on noncomplementing cells at levels reduced ca. 10,000-fold from those obtained from complementing cells. Since the virus stocks used for infection had to be propagated on complementing cells, this residual infectivity may have been due to input UL25 molecules and/or rescue of UL25.
The HSV-1 UL25 protein has been proposed to be essential for cleavage and/or packaging of replicated viral genomic DNA. In its absence, there was a marked increase in the number of immature A and B capsids in the nucleus and a concomitant lack of C capsids (41). The same phenomenon has been observed in a UL25-deficient BHV-1 mutant (14). However, it was also shown that neither HSV-1 nor BHV-1 UL25 is essential for cleavage and packaging per se. A low level of stably packaged unit-length viral genomes were detected even in the absence of UL25, and engineered amplicons with a size smaller than the viral genome were packaged and encapsidated with wild-type-like efficiency (60), whereas cleavage appeared unimpaired by the absence of UL25 in BHV-1 (14). In PrV-ΔUL25F-infected cells, C capsids were visible in electron microscopic images, indicating that DNA can be packaged in the absence of UL25. Surprisingly, the numbers of B capsids did not differ in the absence or presence of UL25 protein. Moreover, particles exhibiting an intermediate appearance, i.e., a partial degradation of scaffold but no completed DNA packaging, were detected more frequently in the absence of UL25. All of these data support the notion that UL25 is not required for cleavage and encapsidation but enhances its efficiency.
Cleavage of concatemeric viral DNA and encapsidation of unit-length genomes into preformed capsids are tightly linked processes. Each unit-length genome is the result of two cleavage steps, generating the free UL and US ends, respectively. Published data indicate that the second cleavage, which is necessary for release of unit-length genomes from the concatemer, occurs only if the genome is properly packaged (reviewed in reference 6). Although directionality of genome packaging has not been formally proven, it is assumed that the genome is inserted with the UL terminus first, while the second cut generates the mature US terminus (reviewed in reference 6). In PrV, cis-acting signals that play a role in cleavage/encapsidation are present at both ends of the linear genome (64); these are unique, with no terminal redundancy (13), and are separated by the cleavage site in the newly replicated concatemeric DNA. In our Southern blot analyses, the UL-terminal BamHI fragment 14′ was present in similar amounts in the presence or absence of UL25, indicating that the UL terminus, which is thought to be generated by the first cleavage event, is efficiently formed in the absence of UL25. However, the US terminus was strikingly underrepresented in DNA of PrV-ΔUL25F-infected RK13 cells, while the signal for the junction fragment was not enhanced. This implies that efficient initiation of packaging occurs in the absence of UL25, leading to free UL ends, while completion of packaging proceeds only inefficiently. It has been suggested that the US end of the preceding genome, which is formed simultaneously with the cleavage that generates the mature left end of the following molecule, may be degraded (6). These results and others (60) argue for a role of herpesvirus UL25 homologs in efficient packaging of full-length genomes but concur with the findings that UL25 is not required for cleavage.
Although C capsids were formed readily in PrV-infected cells even in the absence of UL25, and were often found close to and in contact with the nuclear membrane (Fig. 5), neither budding at the inner nuclear membrane nor translocation of capsids into the cytoplasm was detectable, indicating that either the UL25 protein itself or a protein dependent on the presence of UL25 is necessary for primary envelopment. Egress from the nucleus appears to be highly selective, and predominantly C capsids are translocated while budding B capsids or nuclear “L particles” have been observed only very infrequently in electron microscopic examinations (2, 22, 23). The molecular basis for this selective envelopment remains enigmatic. Stow (60) hypothesized that the HSV-1 UL25 protein may act in a manner similar to that of the head decoration protein gpD in bacteriophage λ, which stabilizes the head shell following the rearrangement of the gpE subunits of the head shell lattice that accompanies expansion of the head. Indeed, parallels between the phenotypes of gpD and UL25 deletion mutants are startling. In phage λ, gpD is added to the head after DNA packaging has been initiated (17). In HSV-1 and PrV, UL25 (27, 61) was found in larger amounts on mature C capsids than on immature capsid forms (27, 50, 61), indicating that UL25 may be added to the capsid concomitantly with DNA packaging. Recently, an interaction between UL17 and UL25 has been proposed to mediate UL25 binding to capsids (62). Our immunoelectron microscopic analyses failed to detect UL25 on defective A and immature B capsids of wild-type PrV, whereas C capsids were heavily decorated by the antiserum. Whether this indicates that immature PrV capsids are devoid of UL25, or whether the amount associated with them is below the detection level of our method, is unclear. However, it is important to emphasize that a clear association between immunoreactivity and a specific capsid form can be made only by concomitant visualization of both the capsid and the immune reaction. Routinely, the different intranuclear capsid forms are prepared by differential centrifugation followed by analysis in immunoblot experiments, with no visual control of the purity of the preparations (27, 50, 58).
Encapsidation is thought to be accompanied by structural changes of the capsid (24), which may be required for or be the result of addition of UL25. In our immunolabeling studies, we found UL25-specific label on multiple sites on intranuclear C capsids, as well as capsids released from mature extracellular virions (Fig. 8), which further supports the idea that UL25 may decorate the mature capsid. A tight interaction between capsid and the UL25 protein can also be deduced from the fractionation experiments (Fig. 7). The relative abundance of the UL25 protein, which had previously been designated a “minor” capsid protein (27), is also reflected by its identification as a rather prominent protein in purified virus particles (45).
In bacteriophage λ, two other proteins are added after gpD has been deposited on the head (17). These are, in turn, necessary for attachment of the tail. It is also conceivable that onto the UL25-decorated herpesvirus capsids, one or more proteins have to be added, which then form the trigger for primary envelopment.
In summary, we show here that PrV UL25 is essential for virus replication. It is not required for DNA cleavage or packaging but is involved in a later step in virion morphogenesis, forming the trigger either for primary envelopment itself or for addition of other proteins which may then prompt nuclear egress of capsids.
Acknowledgments
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG Me 854/5-2).
We thank Petra Meyer, Diana Werner, Kathrin Müller, and Katrin Giesow for expert technical assistance; Mandy Jörn for photographic help; and Egbert Mundt for help with preparation of the antiserum.
REFERENCES
- 1.Addison, C., F. J. Rixon, J. W. Palfreyman, M. O′Hara, and V. G. Preston. 1984. Characterization of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology 138:246-259. [DOI] [PubMed] [Google Scholar]
- 2.Aleman, N., M. Quiroga, M. Lopez-Pena, S. Vazquez, F. Guerriero, and J. Nieto. 2003. L-particle production during primary replication of pseudorabies virus in the nasal mucosa of swine. J. Virol. 77:5657-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali, M. A., B. Forghiani, and E. M. Cantin. 1996. Characterization of an essential HSV-1 protein encoded by the UL25 gene reported to be involved in virus penetration and capsid assembly. Virology 216:278-283. [DOI] [PubMed] [Google Scholar]
- 4.Al-Kobaisi, M. F., F. J. Rixon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology 180:380-388. [DOI] [PubMed] [Google Scholar]
- 5.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. F. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 6.Baines, J. D., and S. K. Weller. 2005. Cleavage and packaging of herpes simplex virus 1 DNA, p. 135-150. In C. E. Catalano (ed.), Viral genome packaging machines: genetics, structures, and mechanism. Landes Biosciences, Georgetown, Tex.
- 7.Baines, J. D., C. Cunningham, D. Nalwanga, and A. J. Davison. 1997. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J. Virol. 71:2666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker, M. L., W. Jiang, F. J. Rixon, and W. Chiu. 2005. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 79:14967-14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catalano, C. E. 2005. Viral genome packaging machines: an overview, p. 1-4. In C. E. Catalano (ed.), Viral genome packaging machines: genetics, structures, and mechanism. Landes Biosciences, Georgetown, Tex.
- 10.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchinson, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 11.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeMarchi, J. M., Z. Lu, G. Rall, S. Kupershmidt, and T. Ben-Porat. 1990. Structural organization of the termini of the L and S components of the genome of pseudorabies virus. J. Virol. 64:4968-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desloges, N., and C. Simard. 2003. Implication of the product of bovine herpesvirus type 1 UL25 gene in capsid assembly. J. Gen. Virol. 84:2485-2490. [DOI] [PubMed] [Google Scholar]
- 15.Dezelee, S., F. Bras, P. Vende, B. Simonet, X. Nguyen, A. Flamand, and M. J. Masse. 1996. The BamHI fragment 9 of pseudorabies virus contains genes homologous to the UL24, UL25, UL26, and UL 26.5 genes of herpes simplex virus type 1. Virus Res. 42:27-39. [DOI] [PubMed] [Google Scholar]
- 16.Donate, L. E., L. Herranz, J. P. Secilla, J. M. Carazo, H. Fujisawa, and J. L. Carrascosa. 1988. Bacteriophage T3 connector: three-dimensional structure and comparison with other viral head-tail connecting regions. J. Mol. Biol. 201:91-100. [DOI] [PubMed] [Google Scholar]
- 17.Feiss, M., and C. E. Catalano. 2005. Bacteriophage lambda terminase and the mechanism of viral DNA packaging, p. 5-39. In C. E. Catalano (ed.), Viral genome packaging machines: genetics, structures, and mechanism. Landes Biosciences, Georgetown, Tex.
- 18.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson, W., and B. Roizman. 1972. Proteins specified by the herpes simplex virus. VIII. Characterization and composition of multiple capsid-forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 21.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold labeling study. J. Virol. 79:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Granzow, H., F. Weiland, A. Jöns, B. G. Klupp, A. Karger, and T. C. Mettenleiter. 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 71:2072-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heymann, J. B., N. Cheng, W. W. Newcomb, B. L. Trus, J. C. Brown, and A. C. Steven. 2003. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat. Struct. Biol. 10:334-341. [DOI] [PubMed] [Google Scholar]
- 25.Hohn, T., and B. Hohn. 1973. A minor pathway leading to plaque-forming particles in bacteriophage lambda. Studies on the function of gene D. J. Mol. Biol. 79:649-662. [DOI] [PubMed] [Google Scholar]
- 26.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 27.Kaelin, K., S. Dezelee, M. J. Masse, F. Bras, and A. Flamand. 2000. The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and to microtubules. J. Virol. 74:474-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 29.Klupp, B. G., and T. C. Mettenleiter. 1999. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J. Virol. 73:3014-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klupp, B. G., A. Karger, H. Granzow, and T. C. Mettenleiter. 2005. Identification, subviral localization, and functional characterization of the pseudorabies virus UL17 protein. J. Virol. 79:13442-13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klupp, B. G., C. J. Hengartner, T. C. Mettenleiter, and L. W. Enquist. 2004. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78:424-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 34.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klupp, B. G., H. Granzow, W. Fuchs, E. Mundt, and T. C. Mettenleiter. 2004. Pseudorabies virus UL3 gene codes for a nuclear protein which is dispensable for viral replication. J. Virol. 78:464-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 38.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGeoch, D. J., C. Cunningham, G. McIntyre, and A. Dolan. 1991. Comparative analysis of the long repeat regions and adjoining parts of the long unique regions in the genomes of herpes simplex viruses type 1 and 2. J. Gen. Virol. 72:3057-3075. [DOI] [PubMed] [Google Scholar]
- 40.McLauchlan, J., and F. J. Rixon. 1992. Characterization of enveloped tegument structures (L-particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J. Gen. Virol. 73:269-276. [DOI] [PubMed] [Google Scholar]
- 41.McNab, A. R., P. Desai, S. Person, L. Roof, D. R. Thompson, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167-180. [DOI] [PubMed] [Google Scholar]
- 44.Mettenleiter, T. C., A. Saalmüller, and F. Weiland. 1993. Pseudorabies virus protein homologous to herpes simplex virus type 1 ICP18.5 is necessary for capsid maturation. J. Virol. 67:1236-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michael, K., B. G. Klupp, T. C. Mettenleiter, and A. Karger. 2006. Composition of virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J. Virol. 80:1332-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 47.Newcomb, W. W., F. L. Homa, D. R. Thomsen, Z. Ye, and J. C. Brown. 1994. Cell-free assembly of the herpes simplex virus capsid. J. Virol. 68:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpesvirus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 74:7137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into herpes simplex virus capsids: location of the UL25 product in the capsids and demonstration that it binds DNA. J. Virol. 75:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel, A. H., F. J. Rixon, C. Cunningham, and A. J. Davison. 1996. Isolation and characterization of herpes simplex type 1 mutants defective in the UL6 gene. Virology 217:111-123. [DOI] [PubMed] [Google Scholar]
- 52.Perdue, M. L., J. C. Cohen, C. C. Randall, and D. J. O'Callaghan. 1976. Biochemical studies of the maturation of herpesvirus nucleocapsid species. Virology 74:194-208. [PubMed] [Google Scholar]
- 53.Perucchetti, R., W. Parris, A. Becker, and M. Gold. 1988. Late stages in bacteriophage lambda head morphogenesis: in vitro studies on the action of the bacteriophage lambda D-gene and W-gene products. Virology 165:103-114. [DOI] [PubMed] [Google Scholar]
- 54.Poon, A. P., and B. Roizman. 1993. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J. Virol. 67:4497-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rixon, F. J., and D. McNab. 1999. Packaging-competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J. Virol. 73:5714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roizman, B., and D. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 57.Salmon, B., C. Cunningham, A. Davison, W. Harris, and J. D. Baines. 1998. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex DNA cleavage and packaging proteins associated with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sternberg, N., and R. Weisberg. 1977. Packaging of coliphage lambda DNA. II. The role of the gene D protein. J. Mol. Biol. 117:733-759. [DOI] [PubMed] [Google Scholar]
- 60.Stow, N. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thurlow, J. K., F. J. Rixon, M. Murphy, P. Targett-Adams, M. Hughes, and V. G. Preston. 2005. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J. Virol. 79:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thurlow, J. K., M. Murphy, N. D. Stow, and V. G. Preston. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 80:2118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wing, B. A., G. C. Y. Lee, and E. S. Huang. 1996. The human cytomegalovirus UL94 open reading frame encodes a conserved herpesvirus capsid/tegument-associated virion protein that is expressed with true late kinetics. J. Virol. 70:3339-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu, C. A., L. Harper, and T. Ben-Porat. 1986. cis functions involved in replication and cleavage-encapsidation of pseudorabies virus. J. Virol. 59:318-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu, D., and S. K. Weller. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]