Abstract
The mycovirus cryphonectria hypovirus 1 (CHV1) causes proliferation of vesicles in its host, Cryphonectria parasitica, the causal agent of chestnut blight. These vesicles have previously been shown to contain both CHV1 genomic double-stranded RNA (dsRNA) and RNA polymerase activity. To determine the cellular origins of these virus-induced membrane structures, we compared the fractionation of several cellular and viral markers. Results showed that viral dsRNA, helicase, polymerase, and protease p29 copurify with C. parasitica trans-Golgi network (TGN) markers, suggesting that the virus utilizes the fungal TGN for replication. We also show that the CHV1 protease p29 associates with vesicle membranes and is resistant to treatments that would release peripheral membrane proteins. Thus, p29 behaves as an integral membrane protein of the vesicular fraction derived from the fungal TGN. Protease p29 was also found to be fully susceptible to proteolytic digestion in the absence of detergent and, thus, is wholly or predominantly on the cytoplasmic face of the vesicles. Fractionation analysis of p29 deletion variants showed that sequences in the C terminal of p29 mediate membrane association. In particular, the C-terminal portion of the protein (Met-135-Gly-248) is sufficient for membrane association and is enough to direct p29 to the TGN vesicles in the absence of other viral elements.
Cryphonectria parasitica is the filamentous ascomycete that causes chestnut blight, a disease that resulted in the nearly complete demise of the American chestnut tree (25). Mycovirus cryphonectria hypovirus 1 (CHV1) infection of C. parasitica reduces virulence of the fungus and has been exploited for the biocontrol of chestnut blight in Europe (24). CHV1 is a member of the virus family Hypoviridae, distinguished by the ability to attenuate virulence and alter developmental processes upon infection of the fungal host. Specific symptoms of virus infection of the fungus grown in culture include reduced pigment production, suppressed asexual sporulation, loss of female fertility, and modified expression of specific host genes (27, 29, 39, 40).
CHV1 has a double-stranded RNA (dsRNA) genome and, like other fungal virus dsRNAs, that of CHV1 is not infectious but can be transmitted horizontally by fungal anastomosis (38) and vertically through asexual, but not sexual, spores. Although hypovirus RNA is found in hyphal extracts as dsRNA, the structural characteristics of the dsRNA are reminiscent of a replicative intermediate or replicative form of a single-stranded RNA (ssRNA) virus (53). In CHV1, the 12.7-kb positive, coding strand is polyadenylated and contains two contiguous open reading frames (ORFs A and B) that encode polyproteins that undergo proteolytic processing (6, 7, 54). Domains in both ORF A and ORF B share sequence homology with ssRNA plant potyviruses (30) and the polyadenylated strand is infectious (3), which is consistent with the ssRNA viral replication strategy (11, 44). CHV1 infection of C. parasitica causes proliferation of vesicles (14, 42, 43) that have been previously shown to contain CHV1 dsRNA, RNA-dependent RNA polymerase (RdRp) activity, and the RdRp protein (16, 17). Fahima and coworkers investigated the properties of the RdRp associated with dsRNA-containing vesicles and reported on the nature of the products synthesized in vitro. Evidence suggests that the strategy employed by the hypovirulence-associated dsRNA is that typical of positive-strand RNA viruses (16, 17). Positive-strand RNA virus replication is known to occur in close association with intracellular membranes, and direct evidence for the importance of membranes in virus RNA replication has been obtained in several cases (13, 49-51). Different viruses utilize different types of membranes. Alphaviruses and rubella viruses assemble their replication complexes on endosomal and lysosomal membranes (19, 31), picornaviruses use the endoplasmic reticulum (ER) (1, 52, 56), and arteriviruses use perinuclear ER membranes (46, 62).
The study and characterization of the vesicles associated with dsRNA in C. parasitica are of particular interest not only because of the important role they play in viral replication and transcription, but also because characterization of these vesicles might shed light into the mechanism that triggers the phenotypic effects the virus causes on its fungal host. This study reports the origins of the cytoplasmic membranes associated with the CHV1 infection and replication cycle and describes the integration of a viral protein into these vesicle membranes.
MATERIALS AND METHODS
Strains and growth conditions.
One-liter EP complete liquid cultures (47) were inoculated with fungal strains EP67 (uninfected) or EP802 (isogenic CHV1 infected) that were previously grown for 7 days on PDAmb (potato dextrose agar with 1 mg/liter methionine and biotin) petri dish plates. Solid cultures were homogenized for 1 min in EP complete at full speed in a Waring blender (New Hartford, CT) before being added to liquid medium for growth on an orbital shaker for 3 days. Mycelia were harvested through filtration and immediately processed or frozen and stored at −80°C. The fungal strains used were described previously (60).
Membrane isolation and fractionation.
A protocol adapted from those of Lin et al. and Pearse (35, 45) was used to separate subcellular fractions and for vesicle purification. Homogenization of mycelial pads was carried out as previously described (17, 41) except buffer A (0.1 M morpholineethanesulfonic acid-NaOH, pH 6.5, 0.3 M sorbitol, 1 mM EGTA, 0.5 mM MgCl2, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin) was used. Homogenates were centrifuged in three steps; 5,000 × g for 30 min, 20,000 × g for 30 min in an Avanti J-E centrifuge from Beckman Coulter (Palo Alto, CA), and 90,000 × g for 90 min in an L8-70 ultracentrifuge from Beckman Coulter (Palo Alto, CA) at 4°C. Pellets were washed and resuspended in buffer B (buffer A without sorbitol) and labeled P5,000, P20,000, P90,000, respectively. P90,000 constituted the microsomal fraction. Supernatants S5,000, S20,000, and S90,000 were frozen, lyophilized, and concentrated 10 times. A 0.5-ml aliquot of the microsomal (P90,000) fraction was loaded onto a 10-ml linear gradient of Ficoll-heavy water (9% 2H2O-2% Ficoll to 90% 2H2O-25% Ficoll in buffer B) and centrifuged at 90,000 × g for 16 h in a swinging SW41Ti rotor. One-milliliter fractions were carefully collected, diluted with 6 ml buffer B, and centrifuged for 2 h at 90,000 × g. Pellets were resuspended in buffer B or Kex2 extraction buffer (see the description of the enzyme assay, below), aliquoted, and stored at −80°C prior to further analysis. Whenever Kex2 activity was assayed, dithiothreitol and leupeptin were omitted from buffer A, due to their reported irreversible inhibitory effects on Kex2 activity (20, 21).
Protein determination.
Protein concentrations in each fraction were determined with a Coomassie Plus protein reagent kit or with a BCA reagent kit (Pierce, Rockford, IL). Due to compatibility issues with Kex2 extraction buffer, the Compat-Able protein assay preparation reagent set (Pierce, Rockford, IL) was used prior to protein determination for Kex2 assays. All procedures were carried out according to the manufacturer's recommendations.
Kex2 and AP-1μ subunit cloning.
Cloning of the Ca2+-dependent serine protease of the subtilisin superfamily Kex2 was performed by PCR using degenerate primers created using protein sequence alignments of Neurospora crassa, Saccharomyces cerevisiae, and Aspergillus niger Kex2 with genomic EP67 DNA as template. Derived primer pair sequences were the following: Kex2-360F (5′-GAYGGNTAYACNAAYAGYATHTAYAGYAT-3′) and Kex2-600R (5′-CCARTGNRCNACRCTCATRAANGTCCA-3′); Kex2-270F (5′-TGYGGNGTNGGNGTNGCNTA-3′) and Kex2-450R (5′-ARRTAYTGNAYRTCNCKCCA-3′); and Kex2-450F (5′-TGGMGNGAYRTNCARTAYYT-3′) and Kex2-600R (5′-CCARTGNRCNACRCTCATRAANGTCCA-3′). The compilation of the three PCR fragments derived from these primers allowed the design of two specific primers (60R, 5′-GGCGATGTTGGAGTCGT-3′, and 970R, 5′-GGACAATGAGGCCTC-3′) to start chromosome walking in both directions. In this way several genomic DNA pieces were cloned from which the whole Kex2 sequence was obtained. Kex2 cDNA sequence was assembled from cDNA clones obtained from reverse transcription-PCR (RT-PCR) with specific primers derived from the genomic sequences. A 120-bp PCR fragment of the C. parasitica adaptor protein 1μ subunit (AP-1μ), a coated vesicle adaptor that facilitates cargo selection in the late secretory trans-Golgi network (TGN) and in endosomes (48), was obtained from cDNA prepared from RNA obtained as previously described (60), using primers designed based on the Neurospora crassa sequence (CAD70726): AP50asp50F (5′-TTAATGTCAAATTTGAGATTCCCTAT-3′) and AP50asp160Rev (5′-GACTGCGTGATGTACCGGACCC). The 120-bp fragment was cloned into the PCR vector pGEM-T (Promega, Madison, WI) and used to screen a cosmid genomic EP44 library (8). Positive cosmid genomic clones were analyzed and sequenced, and respective cDNA was obtained by RT-PCR using specific primers designed based on the genomic sequence.
Plasmid constructs.
For the expression of recombinant CHV1-p29 and Kex2 proteins to be used as antigens for the production of polyclonal antibodies, CHV1-p29 (Phe-25 to Ala-229) and Kex2 (Gln-165 to Asn-559) from C. parasitica were expressed by cloning the corresponding cDNA obtained by RT-PCR using Hi-fidelity Taq polymerase (Invitrogen, Carlsbad, CA) and XhoI-EcoRI restriction sites of the His6-tag expression vector pRSET A (Invitrogen, Carlsbad, CA). For the expression of recombinant AP-1μ for the production of polyclonal antibodies, AP-1μ cDNA corresponding to amino acid positions Met-1 to Arg-414 was cloned by RT-PCR into KpnI-HindIII restriction sites of the His6-tag expression vector pRSET C (Invitrogen, Carlsbad, CA). p29-green fluorescent protein (GFP) fusion deletion constructs were cloned into pCPXHY1 (5) KpnI-SacI restriction sites. To generate the plasmids shown below in Fig. 7, DNA spanning the sequences of p29 shown was cloned as a KpnI-XhoI fragment into pBSK (Stratagene, La Jolla, CA) which already contained the XhoI-SacI fragment of GFP. The fusion sequences were then moved as a cassette to pCPXHY1 using KpnI-SacI restriction sites. Oligonucleotides were constructed to contain all required restriction sites for initiation and termination, as necessary. XhoI sites were used for in-frame fusion with the GFP gene. Plasmid pCT74 was used as the source of GFP (36). All DNA fragments generated by PCR were analyzed by DNA sequencing. Deletion constructs were transformed into C. parasitica spheroplasts as previously described (9). For each deletion three independent transformants were grown, harvested, and analyzed.
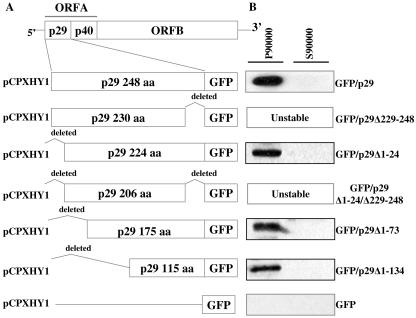
FIG. 7.
(A) Schematic representation of p29 deletion constructs. Deletion constructs of p29 coding sequences were fused to GFP. (B) Western blots using p29 antibodies are shown for the microsomal (P90,000) and soluble (S90,000) fractions for each of the deletions. All analyses were done using three independent transformants; representative results are shown.
Nucleic acid extraction from vesicle preparation and Northern blot analysis.
Nucleic acids were obtained by phenol-chloroform extraction of 100 μl of each fraction. For Northern blot analysis of CHV1 dsRNA, dsRNA was separated in an agarose gel, blotted into Hybond-N+ membrane (Amersham Biosciences, Piscataway, NJ), and hybridized to a CHV1-ORF B probe prepared using a Random Prime DNA labeling kit (Roche, Indianapolis, IN).
Expression of recombinant proteins and antibody generation.
CHV1-p29 (Phe-25 to Ala-229), Kex2 (Gln-165 to Asn-559), and AP-1μ (Met-1 to Arg-414) were expressed in Escherichia coli BL21(DE3)(LysE) cells and purified by using Ni-nitrilotriacetic acid-agarose beads (QIAGEN, Valencia, CA) according to protocols supplied by the manufacturer. Recombinant purified proteins were injected into rabbits for antibody preparation. To remove nonspecific signal, the antibodies were purified against EP67 for p29 and against BL21 cell lysates for Kex2. Lyophilized mycelia (0.1 g) were homogenized using a HandiShear AC homogenizer (Virtis, Gardiner, NY) in 10 ml Tris-buffered saline, 0.2% Tween 20. One hundred microliters of undiluted p29 antibody was added, and the suspension was incubated with gentle agitation for 5 h at room temperature. For preadsorbing of the Kex2 antibodies, an overnight BL21(DE3)(LysE) culture, previously transformed with wild-type pRSET A plasmid, was grown. The bacterial pellet was resuspended in phosphate-buffered saline (PBS), 0.1% Tween 20 and sonicated. Antibodies were added to the suspension in a 1:100 dilution and then incubated with gentle agitation for 5 h at room temperature. Suspensions were then centrifuged for 90 min at 90,000 × g, and the supernatant was removed and stored at −20°C until further use. Polymerase antibodies (16) were prepared through affinity purification against agarose coupled to polymerase antigen using the Amino-link coupling kit (Pierce, Rockford, IL). Helicase antibodies (T. Fahima and N. K. Van Alfen, unpublished data) and AP-1μ antibodies were also affinity purified on columns with their agarose support covalently linked to the original bacterially expressed antigen using the Amino-link coupling kit (Pierce, Rockford, IL).
Gel electrophoresis and immunoblotting.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using either 12%, 15%, or 4 to 20% gradient acrylamide gels as required and blotted onto nitrocellulose membranes. Western blot detection by chemiluminescent ECL, with SuperSignal West Pico substrate from Pierce (Rockford, IL), was carried out according to the manufacturer's recommendations. Anti-KDEL mouse monoclonal and secondary goat anti-mouse antibody-horseradish peroxidase conjugates were purchased from Stressgen Biotechnologies (Victoria, Canada) and used as specified by the manufacturer. Anti-KDEL was used at 4 μg/ml in PBS, 0.05% Tween 20, 3% nonfat dried milk, and secondary goat anti-mouse was at 1:1,000 in PBS, 0.1% Tween 20, and 5% milk. Secondary anti-rabbit-horseradish peroxidase conjugate and primary anti-β-COP clone M3A5 were purchased from Sigma (St. Louis, MO) and were used at 1:5,000 and 1:500 dilutions in PBS, 0.3% Tween 20, 7% milk, respectively. Anti-P29, anti-Kex2, anti-helicase, anti-polymerase, and anti-AP-1μ were rabbit polyclonal antibodies self-prepared as described and used in a 1:4,000, 1:8,000, 1:2,000, 1:100, and 1:200 dilution in PBS, 0.3% Tween 20, 7% milk, respectively. Rabbit polyclonal glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antiserum was kindly provided by M. L. Delgado (12) and used in a 1:3,000 dilution in PBS, 0.1% Tween 20, 7% milk.
Kex2 enzymatic assay.
Kex2 protease was solubilized from the crude microsomal membranes or from microsomal fractions by resuspending pellets in Kex2 extraction buffer (50 mM sodium HEPES [pH 7.6], 1 mM EDTA, 50 mM NaCl, 2% sodium deoxycholate, 20% glycerol) and incubated for 60 min at 4°C. The resulting fraction was stored at −20°C until assayed for activity. The reaction mixture (200 μl) contained 200 mM sodium HEPES (pH 7), 1.5 mM CaCl2, 0.1 mM l-1-tosylamido-2-phenylethyl-CK, and 100 μM N-t-BOC-GLN-ARG-ARG 7-amido-4-methylcoumarine from Sigma Chemical Co. (St. Louis, Mo.) as substrate. After substrate addition, reaction mixtures were incubated at 37°C for 30 min. Reactions were terminated by addition of 1.6 ml of 125 mM ZnSO4 and 0.2 ml of saturated Ba(OH)2. The precipitate was removed by centrifugation for 5 min at 20,000 × g in a microcentrifuge, and the amount of 7-amino-4-methylcoumarin liberated was determined fluorimetrically at an excitation wavelength of 370 nm and an emission wavelength of 440 nm (28).
Protease susceptibility assay.
Subcellular fractions were prepared as previously described but with HEPES-based lysis buffer (HEPES pH 6.8, 150 mM potassium acetate, 250 mM sorbitol, 1 mM magnesium acetate) to prevent permeabilization of membrane compartments. No protease inhibitors were included during the procedure. The P90,000 fraction was resuspended in Feldheim's lysis buffer (18), and proteinase K (Boehringer Manheim, Germany) was added to the mixtures to achieve a final concentration of 0, 4, 8, 10, 15, 20, and 30 μg of proteinase K/ml in the presence or absence of 0.1% Triton X-100. Assays were incubated for 10 min on ice. Proteolytic activity was stopped by adding 4× SDS-PAGE loading buffer containing 5 mM phenylmethylsulfonyl fluoride, and samples were immediately boiled for 10 min and analyzed by SDS-PAGE and subsequent Western blotting.
Biochemical treatments of membranes.
Subcellular fractions were prepared as previously described. The microsomal fraction, P90,000, was resuspended in buffer B and treated with 0.1 volume of 1 M Na2CO3 (pH 11) or with 4 M urea or 2 M NaCl in a 1:1 dilution, incubated at 4°C with shaking for 30 min, and centrifuged at 90,000 × g. Pelleted and lyophilized soluble fractions were resuspended in buffer B and analyzed by SDS-PAGE. For the separation of integral membrane proteins in Triton X-114 solution, the P90,000 fraction was diluted 1:1 in 20 mM Tris-HCl, pH 7.4, 300 mM NaCl, and 2% Triton X-114 at 0°C. The clear protein sample was then overlaid on a sucrose cushion of 6% (wt/vol) sucrose, 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 0.06% Triton X-114 which was previously placed at the bottom of a microcentrifuge tube; the tube was incubated for 3 min at 30°C. Clouding of the solution occurred, and so the tube was centrifuged for 3 min at 300 × g at room temperature. The detergent phase was found as an oily droplet at the bottom of the tube. The upper aqueous phase was removed from the tube and was rinsed with 2% Triton X-114 in a separate tube without sucrose cushion and spun as before. The detergent phase of this last condensation was discarded, and the aqueous phase was analyzed by SDS-PAGE together with the detergent phase previously obtained (2).
Nucleotide sequence accession numbers.
Accession numbers for C. parasitica Kex2 and AP-1μ are DQ219470 and DQ218057, respectively.
RESULTS
Subcellular fractionation of virus-infected C. parasitica.
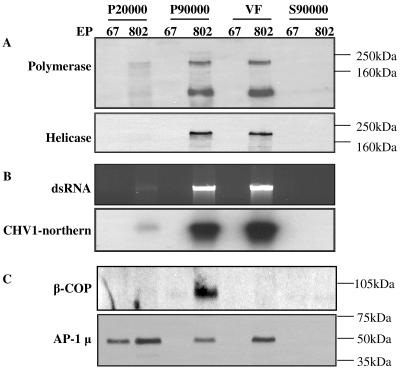
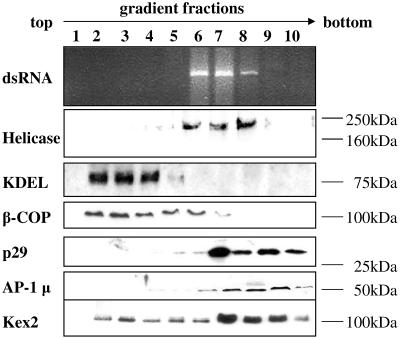
Given the precedents pointing to dsRNA accumulation in vesicle fractions, we used a differential centrifugation and Ficoll-2H2O gradient fractionation protocol widely used for the purification of eukaryotic vesicles to identify the nature of the virus-associated vesicle fraction (15, 23). We monitored dsRNA accumulation and the presence of CHV1 polymerase and helicase proteins in each fraction (Fig. 1A and B). Double-stranded RNA was found to accumulate in the microsomal fraction (P90,000) and, after P90,000 fractionation in a Ficoll-2H2O gradient, in fractions 6, 7, and 8. These fractions were located close to the bottom of the gradient and are referred to as the vesicle fraction (VF) (Fig. 1B, top panel). Together with the dsRNA, the microsomal fraction and VF contained viral polymerase and helicase proteins (Fig. 1A, top and bottom panels, respectively). The dsRNA was verified to be CHV1 specific through Northern blot analysis (Fig. 1B, bottom panel). The same fractions from uninfected strain EP67 did not show the presence of either CHV1 dsRNA or helicase and polymerase proteins. In an attempt to better understand the localization of different vesicle structural components in the fractionation procedure, we looked for β-COP and the AP-1μ subunit in the same fractions (Fig. 1C). At the concentration used, β-COP could only be detected in the microsomal fraction of the virus-containing strain while AP-1μ was present in the P20,000 fraction in both isolates and in the microsomal fraction and VF of the viral isolate alone. These results show that there is greater membrane proliferation in the viral strain, as shown by the abilities of the antibodies to detect β-COP and AP-1μ antigen in the microsomal fraction of EP802 while not in the microsomal fraction of EP67. Also evident from this blot is that β-COP is not detected in subfraction VF, while AP-1μ is only detected in the VF of EP802.
FIG. 1.
Fractions of uninfected (EP67) and CHV1-infected (EP802) strains of C. parasitica, obtained by differential centrifugation. Cell lysates were centrifuged at 5,000 × g, 20,000 × g, and 90,000 × g. Pellets (P) and supernatants (S) were collected and analyzed. The microsomal fraction, P90,000, was further fractionated on a Ficoll-2H2O gradient. The fractions from EP802 that contained viral elements were pooled together and labeled as VF. Corresponding fractions from strain EP67 were also pooled and labeled as such. (A) Western blots of fractions using viral polymerase and helicase antibodies. (B) Nucleic acid extraction from the fractions, with the top panel showing ethidium bromide staining of dsRNA. The bottom panel shows Northern hybridization of the same samples probed with radioactively labeled CHV1-ORF B. (C) Western blots of the same fractions using β-COP and AP-1μ subunit antibodies. Equal total protein amounts were loaded.
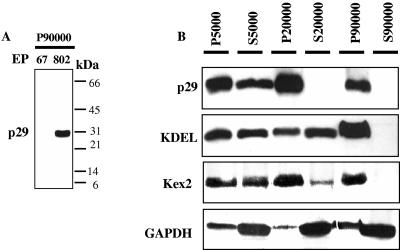
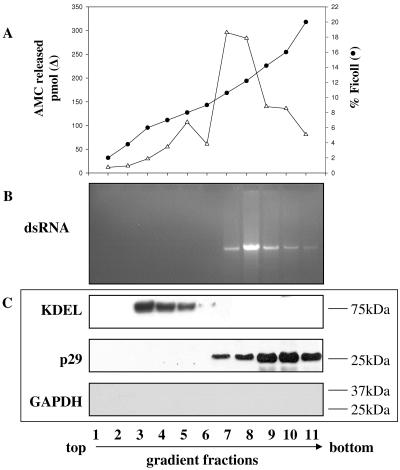
To further characterize the kind of vesicles utilized by the virus, we monitored dsRNA accumulation and other specific CHV1 markers in each fraction together with C. parasitica organelle-specific markers. The viral p29 gene was cloned and expressed, and the recombinant protein was purified and used to produce polyclonal antibodies. Antibodies raised against p29 reacted specifically on Western immunoblot assays with a 29-kDa polypeptide from the P90,000 fraction of EP802, but not with any fraction of EP67 (Fig. 2A). To identify the various fractions of the isolation procedure, the viral protease p29, the ER marker KDEL, the trans-Golgi Kex2 protease, and the cytosolic protein GAPDH were sought in each fraction (Fig. 2B). All antibodies used reacted specifically with bands of the expected molecular weight. As expected for soluble proteins, cytosolic GAPDH could be found in all fractions and was predominantly recovered in the S90,000 fraction. p29, KDEL, and Kex2 fractionated into the microsomal fraction (P90,000) and were undetectable in S90,000, reflecting localization within vesicles or their membranes. Figure 3 shows the Ficoll-2H2O gradient fractionation profile of the P90,000 microsomal fraction. Gradient fractions were assayed for Kex2 activity, for the presence of KDEL and GAPDH, and for viral dsRNA and p29. Viral dsRNA and p29 were always associated with fractions near the bottom of the gradient from strain EP802 (Fig. 3B and C). The viral elements were not detected in analogous fractions extracted from EP67 (data not shown). Fractions containing the viral elements also contained the peak in Kex2 activity (Fig. 3A), indicating that the viral elements cofractionated with fungal TGN. ER marker KDEL was localized in upper fractions, and GAPDH was not detected in the gradient fractions. Due to the fact that dsRNA gels were loaded using equal volumes while Westerns blots were loaded based on equal protein amounts (see Materials and Methods), intensities on the Western blot signal and dsRNA gels cannot be quantitatively compared.
FIG. 2.
(A) Western blot of the microsomal fractions of uninfected (EP67) and CHV1-infected (EP802) cells using CHV1-encoded p29 antibodies. (B) Western blot showing subcellular fractionation of cell lysates from CHV1-infected strain EP802 separated by differential centrifugation. Lysates were centrifuged at 5,000 × g, 20,000 × g, and 90,000 × g. Pellets (P) and supernatants (S) were collected and treated with p29, KDEL, Kex2, or GAPDH antibodies. Equal total protein amounts were loaded.
FIG. 3.
Ficoll-2H2O gradient fractions of the subcellular microsomal fraction, P90,000, of CHV1-infected strain EP802. (A) ▵, Kex2 activity expressed as pmol 7-amino-4-methylcoumarin (AMC) released; •, activity expressed as % Ficoll. (B) Nucleic acid extraction of 100 μl from each gradient fraction and staining with ethidium bromide. (C) Western blot analysis of gradient fractions using antibodies to KDEL, p29, and GAPDH. Fractions were labeled 1 to 11, from top to bottom. For Western assays, equal protein amounts were loaded for all fractions except fraction 1, which did not have enough protein and so the maximal volume was loaded. The percent Ficoll was determined by refractometry.
For a more detailed characterization of the vesicles of EP802 separated on the gradient, three more markers were used to better identify the fractions: the viral helicase, β-COP, required for the retrieval of proteins from an early Golgi compartment to the ER (15), and the AP-1μ subunit adaptor protein, found specifically on the TGN and endosomes (48) (Fig. 4). The same fractionation pattern as in Fig. 3 could be observed, with viral elements that associated with the bottom fractions and cofractionating with Kex2 and AP-1μ, indicating that the viral elements are associated with the TGN. In contrast, KDEL and β-COP migrated to the upper fractions, showing clear distinction between the ER and the intermediate compartment, and the viral vesicles associated with the TGN.
FIG. 4.
Agarose gels showing Ficoll-2H2O gradient fractions of the CHV1-infected strain EP802 microsomal fraction, P90,000. The presence of CHV1 dsRNA, p29, CHV1-helicase, KDEL (ER marker), Kex2 (TGN marker), β-COP (intermediate compartment, ER to Golgi marker) and AP-1μ (TGN and endosome marker) in the various fractions were detected using ethidium bromide staining for nucleic acids (dsRNA) and Western blot assays using antibodies to the proteins. The profile of GAPDH is not shown since it was below the detection limit in the gradient fractions.
p29 is an integral membrane protein.
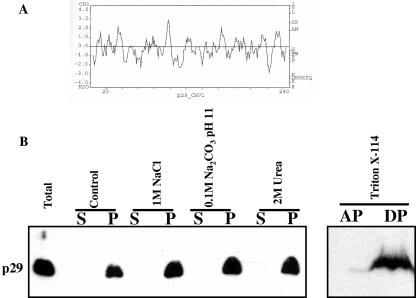
Analysis of the primary sequence of p29 does not reveal regions that are typically found in proteins associated with membranes, i.e., the hydrophobicity profile of p29 does not show any regions of sufficient length and hydrophobicity that could qualify as transmembrane domains (Fig. 5A). To determine if p29 is an integral or peripheral membrane protein, biochemical analyses were performed using the microsomal fraction from EP802. p29 remained associated with the membrane fraction after extraction with 1 M NaCl, 0.1 M Na2CO3 (pH 11), or 2 M urea (Fig. 5B, left panel). These treatments would be expected to dislodge proteins that were weakly or peripherally associated with membranes. A Triton X-114 phase partition analysis was also carried out. In this experiment the p29 in the P90,000 fraction partitioned mainly to the detergent phase (Fig. 5B, right panel). Thus, despite the absence of obvious membrane-spanning regions, p29 displayed high affinity for membranes.
FIG. 5.
(A) CHV1-p29 hydrophobicity plot. Hydrophobicity was calculated by using the algorithm of Kyte-Doolittle with a window size of 7 amino acids using the SDSC Biology workbench web-based tool (34). (B) Extraction and Western blot analysis of p29 in CHVI-infected strain EP802. The P90,000 fraction was extracted with either 1 M NaCl, 0.1 M Na2CO3 (pH 11.5), or 2 M urea and subjected to centrifugation at 90,000 × g, yielding the soluble fraction (S) and pellet (P). The total P90,000 fraction was also fractionated into aqueous (AP) and detergent-soluble (DP) phases after treatment with 1% Triton X-114. Samples were analyzed by Western blotting using p29 antibodies.
Protease susceptibility of membrane-associated p29.
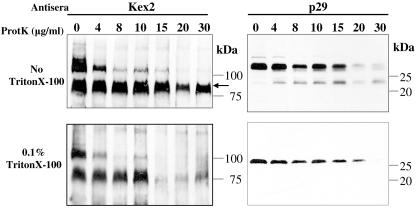
To investigate the distribution of p29 with respect to the cytosolic and lumenal faces of the TGN vesicles, we assayed the susceptibility of p29 to proteolysis. Parts of p29 that localize to the cytoplasmic side of the vesicles will be accessible to the protease in a detergent-independent fashion, while any parts of p29 that protrude into the lumen of the vesicle should only be degradable after solubilization of the vesicle membrane with detergent. The P90,000 membrane fractions of strain EP802 were prepared and subjected to increasing amounts of proteinase K (Fig. 6). As a control we used Kex2 protease, an enzyme that resides in the lumen of the late Golgi and has a transmembrane domain and a cytoplasmic tail (21, 22). The Kex2 cytoplasmic tail was degraded due to proteolytic activity with increasing concentrations of proteinase K, while a 70-kDa fragment was protected from proteolytic activity in the absence of detergent even at the highest protease concentrations. Addition of Triton X-100 rendered the Kex2 70-kDa lumenal fragment susceptible to degradation. The sensitivity of p29 to proteinase K was analyzed in parallel to that of Kex2. In contrast to Kex2, p29 was equally susceptible to proteolysis in the absence or presence of Triton X-100. Degradation intermediates that accumulated and were degraded were observed and had increased susceptibility to protease upon addition of Triton X-100; however, a slight increase in susceptibility to protease upon addition of Triton X-100 was also observed in the cytoplasmic tail of Kex2. When comparing Kex2 and p29 in the absence of detergent, p29 and its degradation intermediate were both almost totally degraded at 20 and 30 μg/ml proteinase K, while the lumenal part of Kex2 was completely protected at these concentrations (Fig. 6, top panel).
FIG. 6.
Protease susceptibility of CHV1 p29. Aliquots of the P90,000 microsomal fraction of CHV1-infected strain EP802 were incubated with increasing amounts of proteinase K. Assays were carried out in the absence (top panel) or presence (bottom panel) of 0.1% Triton X-100. Western blots were generated using p29 antibodies and as a control using antibodies against Kex2. Kex2 is a lumenal (arrow) TGN resident that has a transmembrane domain and a cytoplasmic tail.
Deletion analysis of p29 membrane association.
To characterize the p29 membrane interaction in vitro and in vivo, plasmids were constructed that expressed p29 derivatives with C-terminal or N-terminal truncations of various lengths fused to GFP (Fig. 7A). The microsomal and soluble fractions, P90,000 and S90,000, were analyzed (Fig. 7B). In these assays, p29 was recovered in all cases in the P90,000 fraction and none could be detected in the soluble fraction.
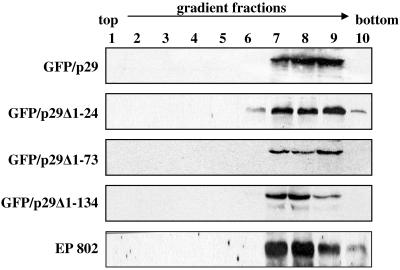
Deletion of the C-terminal region in constructs GFP/p29Δ229-248 and GFP/p29Δ(1-24) (229-248) resulted in unstable proteins that could not be detected by Western blotting or UV fluorescence microscopy; plasmid presence was verified by Southern blot analysis (data not shown). Constructs GFP/p29Δ1-24, GFP/p29Δ1-73, and GFP/p29Δ1-134 with N-terminal deletions did not alter membrane association patterns of p29 under the experimental conditions used. Thus, the p29 membrane association domain could be mapped to its C-terminal region, between amino acids Met-135 and Gly-248. Although the C-terminal region alone retained high levels of membrane association, we cannot rule out a possible accessory role of the N-terminal region of p29 in membrane association. Attempts to visualize and characterize the GFP fusions in vivo were unsuccessful due to high background levels of free GFP, likely derived from self-cleavage of the fusion protein. When analyzing constructs GFP/p29 and GFP/p29Δ1-24 on a 15% SDS-PAGE, no significant molecular mass differences could be observed between these two constructs and EP802 p29. Constructs GFP/p29Δ1-73 and GFP/p29Δ1-134, however, produced bands of approximately 22 and 23 kDa, respectively, that reacted with both p29 and GFP antibodies in Western blot analysis. To assess the nature of the membrane to which each of the constructs attached, a Ficoll-2H2O gradient was used to fractionate the P90,000 extract of each independent construct (Fig. 8). All N-terminal deletions were found in the same fractions as virus-containing vesicles of strain EP802. These results indicate that the C-terminal region of p29 (Met-135-Gly-248) not only mediates membrane association but also is able to direct its association to the same membranes as the native p29. Moreover, it is able to do so in the absence of other viral components.
FIG. 8.
Western blot analysis showing gradient fractions for each of the deletion constructs transformed into strain EP67 and CHV1-infected strain EP802 analyzed by Western blotting using p29 antibodies. Deletion construct nomenclature is the same as for Fig. 7, and gradient fractions are the same as in Fig. 3 and 4. Analyses were carried out using three independent transformants for each construct; representative results are shown.
DISCUSSION
Interest in CHV1 stems both from its potential as a biological control agent for chestnut blight and as a tool to study fungal development, especially virulence expression. Since CHV1 affects development without affecting vegetative growth of the fungus, it is likely that the effect of the virus on fungal development is specific rather than a result of general debilitation, making this an excellent model system. There has been progress in the identification of virus-encoded hypovirulence symptom determinants (57, 58), the identification of specific host genes and proteins affected by CHV1 (29, 41) and, with this report, an understanding of the nature of the vesicles used by the virus for replication.
Current evolutionary evidence indicates a common ancestry of the chestnut blight hypovirulence-associated dsRNA with the potyviruses (30), a group of positive-strand RNA plant viruses. The majority of CHV1 RdRp in vitro products were found to be plus stranded (16), and the positive strand of CHV1 was found to encode two ORFs and to be infectious (3, 6, 7, 53). These data are consistent with an ssRNA viral replication strategy (11, 44), and it has been proposed that the dsRNA is a replicative form of an ssRNA virus (53). A universal feature of the replication complexes of positive-stranded RNA viruses is their association with host membranes (49, 55, 64). CHV1 dsRNA has previously being shown to associate with vesicles and to cause vesicle accumulation in C. parasitica (14, 23, 42, 43). In this study we have confirmed these findings and have further characterized the intracellular membranes targeted by CHV1. We show that CHV1 elements colocalize with membranes derived from the TGN of C. parasitica (Fig. 3 and 4). In contrast, ER markers were not associated with elements of the virus (Fig. 3 and 4). This is important not only because it rules out the presence of mixed membranes in the fractions of interest, but also because most potyviruses are known to assemble their replication complexes on ER-derived membranes. Therefore, the CHV1 replication complex behaves differently from its ancestors. The finding that CHV1 elements colocalize with the TGN markers is consistent with the ultrastructural studies of infected C. parasitica cells, which describe membrane-bound virus-like particles, measuring 50 to 90 nm in diameter, located behind the hyphal tip and found in association with a unique type of Golgi cisterna (42, 43). Our finding that CHV1 targets the TGN would also explain the presence of the virus-like particles behind the hyphal tip, as found by Newhouse and coworkers, a region that is usually free of mycovirus particles in other infected fungi (63).
Most picornaviruses, including potyviruses, cause a significant proliferation of various host membranes, but they assemble their replication complexes on ER-derived membranes (1, 52, 56). Deletion analysis of potyvirus tobacco etch virus peptide 6K2 has demonstrated its association with large vesicular compartments derived from the ER, and it has consequently been proposed that the 6K2 peptide anchors the replication apparatus to ER-like membranes (51, 61). The hijacking of membranes of the late secretory pathway, however, is not uncommon. Some alphaviruses, for example, assemble their replication complexes in the endo-lysosomal compartment (19, 31), and recently it was found that Kunjin virus, a member of the flavivirus superfamily, assembles its replication complex at TGN membranes (37). A unique aspect of the host-virus interactions of CHV1 is the general lack of adverse effects of infection on vegetative growth. The TGN vesicles that proliferate as a result of virus infection may not, therefore, be important for vegetative growth but may be specific to developmental processes. If true, the virus targets a subset of the TGN vesicles.
Similarities between p29 and the multifunctional potyvirus protease HC-Pro have been noted (10, 57). Plant potyvirus HC-Pro proteins consist of three possible domains: (i) a cysteine-rich, N-terminal region, (ii) a central domain, and (iii) a C-terminal proteolytic domain (61). p29 of CHV1 contains the cysteine-rich and protease domains but lacks the central domain (30). In addition to the similarities regarding the autocatalytic cleavage at glycine dipeptides and the C-terminal protease domains, the N-terminal domains of p29 and HC-Pro contain four conserved cysteine residues. These conserved residues reside within the p29 symptom determinant domain. Similarities between HC-Pro and p29 also relate to their multifunctional nature. HC-Pro has been reported to facilitate aphid transmission, to promote potyvirus genome amplification, to catalyze polyprotein processing, to stimulate long distance movement, and to suppress RNA silencing (61). Functions assigned to p29 include autoproteolysis and suppression of host lacasse expression, pigment production, and asexual sporulation (10). p29 also enhances viral dsRNA accumulation and vertical virus transmission through asexual spores (58).
p29 membrane affinity and topology.
Despite the lack of regions with sufficient hydrophobicity to serve as membrane-spanning domains, p29 was always found associated with membranes. Treatments known to dissociate peripheral proteins from membranes, such as treatment with high salt and high pH, did not release p29 from the membrane fraction (Fig. 5B). The behavior of p29 protein during these treatments strongly suggests that it associates with membranes as an integral protein. Experiments with deletion derivatives fused to GFP showed that the necessary sequence for membrane association is comprised at the C-terminal domain of p29, in particular between amino acids 135 and 248, and that this region is sufficient to direct p29 to the TGN. No other viral sequences were necessary for the direction and integration of p29 into the TGN membranes. Constructs lacking the p29 C-terminal domain were unstable and, thus, we were unable to determine if this deletion renders the protein soluble. It would be interesting to know if the p29 of CHV2-NB, another member of the hypoviridiae, that lacks the C-terminal region of p29 (26) retains a membrane-anchoring function.
The possibility that portions of the membrane-associated p29 protein reside within the lumens of these vesicles was tested using protease susceptibility analyses. Results showed that the viral protease p29 is predominantly on the cytoplasmic face of the vesicles. Our results do not rule out that there could be one or more lumenal regions of p29 that lack epitopes for the polyclonal anti-p29 antisera used. Nevertheless, they show that substantial portions of p29 are protease accessible in the absence of detergent, indicating that p29 is predominantly associated with the cytoplasmic face of the vesicles (Fig. 6). This is consistent with its lack of highly hydrophobic and potential transmembrane domains (Fig. 5A). The basis of the high affinity of p29 to the TGN membranes is not known. Protease p29 may belong to a “nonconformist” class of integral membrane proteins containing C-terminal or C-terminal-proximal anchor sequences that insert by a posttranslational signal recognition particle-independent pathway (33). These tail-anchored proteins, which include cytochrome b5 and synaptobrevin, face the cytoplasm with their C-terminal insertion sequence (typically 16 to 20 amino acids) embedded within the membrane (32, 33, 51). Alternatively, monotopic binding by amphipathic α-helix has been suggested for several viral proteins which lack continuous hydrophobic anchor sequences. The amphipathic α-helix strategy has been proposed for components of picornavirus and several plant virus replication complexes (49). Poliovirus protein 3A localizes to the ER when expressed in isolation, and its precursor behaves as an integral membrane protein, the binding region being mapped to a C-terminal hydrophobic region of only 7 amino acids. However, the exact binding mechanism is not known (4, 59). Similar to p29, 3A also shows a cytosolic topology. Interestingly, CHV1-p29 protein sequence predicts an amphipathic α-helix at its C-terminal end (data not shown).
Intracellular localization of deletion derivatives.
Previous deletion analyses of CHV1 infectious cDNA clones used to identify virus-encoded symptom determinants concluded that, while nonessential for viral replication or hypovirulence, p29 does contribute to specific phenotypic changes in CHV1-infected fungal strains, including reduction of host pigmentation and significant reduction of asexual sporulation (10). Moreover, Suzuki and coworkers (57) mapped the p29 symptom determinant domain within the N-terminal region between Phe-25 and Gln-73. Mutations of the CHV1-EP713 infectious cDNA involving cysteine-to-glycine substitutions identified conserved p29 Cys-70 and Cys-72 as crucial in p29-mediated phenotypic modulations (57).
In this study we have shown that p29 membrane association is independent of its symptom determinant domain. Transformation of uninfected C. parasitica with full-length p29 and p29Δ1-24 resulted in suppression of asexual sporulation and pigment formation (data not shown). Deletions p29Δ1-73 and p29Δ1-134, as well as GFP alone, resulted in no effect on the fungal phenotype, yet all deletions integrated into the membrane (Fig. 8). These findings are consistent with the report (10) that p29-mediated symptom expression is independent of its protease activity. Symptom expression was mapped to the N-terminal region of p29, while its proteolytic activity was found in its C-terminal domain. Our results are consistent with previous reports (9, 54) that p29 causes loss of host asexual sporulation and colony pigmentation without the need for other viral proteins.
Our results show that the CHV1 replication complex associates with membranes derived from the TGN of C. parasitica and that the CHV1 p29 protease is an integral membrane protein, with most of the protein exposed to the cytoplasmic face of the TGN vesicles. Membrane association determinants were shown to be within the C-terminal domain of p29 (Met-135-Gly-248) and N-terminal deletions of p29 until Met-135 did not affect its intracellular localization in EP67, suggesting that the C-terminal region is enough to direct p29 to the TGN vesicles in the absence of other viral elements. However, a possible accessory contribution of the N-terminal region to this membrane association cannot be ruled out. The finding that most of the p29 protein is localized to the cytoplasmic face of TGN vesicles may provide clues as to how this protein is involved in causing some of the virus-induced host symptoms. With its cytoplasmic orientation, p29 is available for interaction not only with other viral elements but also with fungal elements as well. These interactions or abolishment of specific fungal interactions by p29 could be the cause of the fungal phenotypic changes observed and deserves further examination.
Acknowledgments
We thank Laura Kong for excellent technical assistance. We also thank Bryce Falk and Pam Kazmierczak for critical reading of the manuscript and for helpful discussions. We also gratefully acknowledge the receipt of plasmids from the laboratories of Linda Ciufetti, Oregon State University, and Donald Nuss, University of Maryland.
REFERENCES
- 1.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 2.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 3.Chen, B., G. H. Choi, and D. L. Nuss. 1994. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 264:1762-1764. [DOI] [PubMed] [Google Scholar]
- 4.Choe, S. S., and K. Kirkegaard. 2004. Intracellular topology and epitope shielding of poliovirus. J. Virol. 78:5973-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, G. H., and D. L. Nuss. 1992. A viral gene confers hypovirulence-associated traits to the chestnut blight fungus. EMBO J. 11:473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, G. H., D. M. Pawlyk, and D. L. Nuss. 1991. The autocatalytic protease P29 encoded by a hypovirulence-associated virus of the chestnut blight fungus resembles the potyvirus-encoded protease Hc-Pro. Virology 183:747-752. [DOI] [PubMed] [Google Scholar]
- 7.Choi, G. H., R. Shapira, and D. L. Nuss. 1991. Co-translational autoproteolysis involved in gene expression from a double-stranded RNA genetic element associated with hypovirulence of the chestnut blight fungus. Proc. Natl. Acad. Sci. USA 88:1167-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Churchill, A. C. L. 1993. Transformation and gene isolation by functional complementation in Cryphonectria parasitica, the chestnut blight fungus. Ph.D. thesis. Utah State University, Logan.
- 9.Churchill, A. C. L., L. M. Ciuffetti, D. R. Hansen, H. D. Van Etten, and N. K. Van Alfen. 1990. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 17:25-32. [Google Scholar]
- 10.Craven, M. G., D. M. Pawlyk, G. H. Choi, and D. L. Nuss. 1993. Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J. Virol. 67:6513-6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawe, A. L., and D. L. Nuss. 2001. Hypoviruses and chestnut blight: exploiting viruses to understand and modulate fungal pathogenesis. Annu. Rev. Genet. 35:1-29. [DOI] [PubMed] [Google Scholar]
- 12.Delgado, M. L., J. E. O'Connor, I. Azorin, J. Renau-Piqueras, M. L. Gil, and D. Gozalbo. 2001. The glyceraldehyde-3-phosphate dehydrogenase polypeptides encoded by the Saccharomyces cerevisiae TDH1, TDH2 and TDH3 genes are also cell wall proteins. Microbiology 147:411-417. [DOI] [PubMed] [Google Scholar]
- 13.den Boon, J. A., J. Chen, and P. Ahlquist. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75:12370-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dodds, J. A. 1980. Association of type 1 viral-like double stranded RNA with club shaped particles in hypovirulent strains of Endothia parasitica. Virology 107:1-12. [DOI] [PubMed] [Google Scholar]
- 15.Eugster, A., G. Frigerio, M. Dale, and R. Duden. 2004. The α- and β′-COP WD40 domains mediate cargo-selective interactions with distinct di-lysine motifs. Mol. Biol. Cell 15:1011-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fahima, T., P. Kazmierczak, D. R. Hansen, P. Pfeiffer, and N. K. Van Alfen. 1993. Membrane-associated replication of an unencapsidated double-strand RNA of the fungus, Cryphonectria parasitica. Virology 195:81-89. [DOI] [PubMed] [Google Scholar]
- 17.Fahima, T., Y. Wu, L. Zhang, and N. K. Van Alfen. 1994. Identification of the putative RNA polymerase of Cryphonectria hypovirus in a solubilized replication complex. J. Virol. 68:6116-6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldheim, D., J. Rothblatt, and R. Schekman. 1992. Topology and functional domains of Sec63p an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol. Cell. Biol. 12:3288-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuller, R. S., A. Brake, and J. Thorner. 1989. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc. Natl. Acad. Sci. USA 86:1434-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuller, R. S., A. J. Brake, and J. Thorner. 1989. Intracellular targeting and structural conservation of a prohormone-processing endoprotease. Science 246:482-486. [DOI] [PubMed] [Google Scholar]
- 22.Fuller, R. S., R. E. Sterne, and J. Thorner. 1988. Enzymes required for yeast prohormone processing. Annu. Rev. Physiol. 50:345-362. [DOI] [PubMed] [Google Scholar]
- 23.Hansen, D. R., N. K. Van Alfen, K. Gillies, and W. A. Powell. 1985. Naked double-stranded RNA associated with hypovirulence of Endothia parasitica is packaged in fungal vesicles. J. Gen. Virol. 66:2605-2614. [Google Scholar]
- 24.Heiniger, U. R., D. 1994. Biological control of chestnut blight in Europe. Annu. Rev. Phytopathol. 32:581-599. [Google Scholar]
- 25.Hepting, G. H. 1974. Death of the American chestnut. J. For. Hist. 18:60-67. [Google Scholar]
- 26.Hillman, B. I., B. Halpern, and M. P. Brown. 1994. A viral dsRNA element of the chestnut blight fungus with a distinct genetic organization. Virology 201:241-250. [DOI] [PubMed] [Google Scholar]
- 27.Hillman, B. I., R. Shapira, and D. L. Nuss. 1990. Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80:950-956. [Google Scholar]
- 28.Jalving, R., P. J. I. van de Vondervoort, J. Visser, and P. J. Schaap. 2000. Characterization of the kexin-like maturase of Aspergillus niger. Appl. Environ. Microbiol. 66:363-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazmierczak, P., P. Pfeiffer, L. Zhang, and N. K. Van Alfen. 1996. Transcriptional repression of specific host genes by the mycovirus Cryphonectria hypovirus 1. J. Virol. 70:1137-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koonin, E. V., G. H. Choi, D. L. Nuss, R. Shapira, and J. C. Carrington. 1991. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl. Acad. Sci. USA 88:10647-10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kujala, P., T. Ahola, N. Ehsani, P. Auvinen, H. Vihinen, and L. Kaariainen. 1999. Intracellular distribution of rubella virus nonstructural protein P150. J. Virol. 73:7805-7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kutay, U., G. Ahnert-Hilger, E. Hartmann, B. Wiedenmann, and T. A. Rapoport. 1995. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 14:217-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kutay, U., E. Hartmann, and T. A. Rapoport. 1993. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 3:72-75. [DOI] [PubMed] [Google Scholar]
- 34.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 35.Lin, H.-B., S. M. Harley, J. M. Butler, and L. Beevers. 1992. Multiplicity of clathrin light-chain-like polypeptides from developing pea (Pisum sativum L.) cotyledons. J. Cell Sci. 103:1127-1137. [Google Scholar]
- 36.Lorang, J. M., R. P. Tuori, J. P. Martinez, T. L. Sawyer, R. S. Redman, J. A. Rollins, T. J. Wolpert, K. B. Johnson, R. J. Rodriguez, M. B. Dickman, and L. M. Ciuffetti. 2001. Green fluorescent protein is lighting up fungal biology. Appl. Environ. Microbiol. 67:1987-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackenzie, J. M., M. K. Jones, and E. G. Westaway. 1999. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 73:9555-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin, R. M., and N. K. Van Alfen. 1991. The movement of viral-like RNA between colonies of Cryphonectria parasitica. Mol. Plant-Microbe Interact. 4:507-511. [Google Scholar]
- 39.McCabe, P. M., P. Pfeiffer, and N. K. Van Alfen. 1999. The influence of dsRNA viruses on the biology of plant pathogenic fungi. Trends Microbiol. 7:377-381. [DOI] [PubMed] [Google Scholar]
- 40.McCabe, P. M., and N. K. Van Alfen. 2001. Molecular basis of symptom expression by the Cryphonectria hypovirus, p. 125-144. In S. M. Tavantzis (ed.), ds RNA genetic elements: concepts and applications in agriculture, forestry and medicine. CRC Press, Boca Raton, Calif.
- 41.McCabe, P. M., and N. K. Van Alfen. 1999. Secretion of cryparin, a fungal hydrophobin. Appl. Environ. Microbiol. 65:5431-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newhouse, J. R., H. C. Hoch, and W. L. Macdonald. 1983. The ultrastructure of Endothia parasitica comparison of a virulent with a hypovirulent isolate. Can. J. Bot. 61:389-399. [Google Scholar]
- 43.Newhouse, J. R., W. L. Macdonald, and H. C. Hoch. 1990. Virus-like particles in hyphae and conidia of european hypovirulent dsRNA-containing strains of Cryphonectria parasitica. Can. J. Bot. 68:90-101. [Google Scholar]
- 44.Nuss, D. L. 2005. Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3:632-642. [DOI] [PubMed] [Google Scholar]
- 45.Pearse, B. M. F. 1983. Isolation of coated vesicles. Methods Enzymol. 98:320-326. [DOI] [PubMed] [Google Scholar]
- 46.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endo-plasmic reticulum-derived double-membranes vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puhalla, J. E., and S. L. Anagnostakis. 1971. Genetics and nutritional requirements of Endothia-Parasitica. Phytopathology 61:169-173. [Google Scholar]
- 48.Robinson, M. S. 2004. Adaptable adaptors for coated vesicles. Trends Cell Biol. 14:167-174. [DOI] [PubMed] [Google Scholar]
- 49.Salonen, A., T. Ahola, and L. Karriainen. 2004. Viral RNA replication in association with cellular membranes, p. 139-173. In M. Marsh (ed.), Membrane trafficking in viral replication. Springer, Berlin, Germany. [DOI] [PMC free article] [PubMed]
- 50.Salonen, A., L. Vasiljeva, A. Merits, J. Magden, E. Jokitalo, and L. Kaarianen. 2003. Properly folded nonstructural polyprotein directs the Semliki Forest virus replication complex to endosomal compartment. J. Virol. 77:4679-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shapira, R., G. H. Choi, and D. L. Nuss. 1991. Virus-like genetic organization and expression strategy for a double-stranded RNA genetic element associated with biological control of chestnut blight. EMBO J. 10:731-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shapira, R., and D. L. Nuss. 1991. Gene expression by a hypovirulence-associated virus of the chestnut blight fungus involves two papain-like protease activities. J. Biol. Chem. 266:19419-19425. [PubMed] [Google Scholar]
- 55.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki, N., B. Chen, and D. L. Nuss. 1999. Mapping of a hypovirus p29 protease symptom determinant domain with sequence similarity to potyvirus HC-Pro protease. J. Virol. 73:9478-9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki, N., K. Maruyama, M. Moriyama, and D. L. Nuss. 2003. Hypovirus papain-like protease p29 functions in trans to enhance viral double-stranded RNA accumulation and vertical transmission. J. Virol. 77:11697-11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810-26818. [DOI] [PubMed] [Google Scholar]
- 60.Turina, M., A. Prodi, and N. K. Van Alfen. 2003. Role of the Mf1-1 pheromone precursor gene of the filamentous ascomycete Cryphonectria parasitica. Fungal Genet. Biol. 40:242-251. [DOI] [PubMed] [Google Scholar]
- 61.Urcuqui-Inchima, S., A. Haenni, and F. Bernardi. 2001. Potyvirus proteins: a wealth of functions. Virus Res. 74:157-175. [DOI] [PubMed] [Google Scholar]
- 62.van der Meer, Y., H. van Tol, J. Krijnse-Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weber, H. 1979. Ultrastructural evidence for viruses in ascomycetes and fungi imperfecti, p. 363-404. In P. A. Lemke (ed.), Viruses and plasmids in fungi. Marcel Dekker, Inc., New York, N.Y.
- 64.Wimmer, E., C. U. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]