Abstract
Lacking an RNA-dependent RNA polymerase, hepatitis delta virus (HDV), which contains a circular RNA of 1.7 kilobases, is nonetheless able to replicate its RNA by use of cellular transcription machineries. Previously, we have shown that the replications of genomic- and antigenomic-strand HDV RNAs have different sensitivities to α-amanitin, suggesting that these two strands are synthesized in different transcription machineries in the cells, but the nature of these transcription machineries is not clear. In this study, we performed metabolic labeling and immunofluorescence staining of newly synthesized HDV RNA with bromouridine after HDV RNA transfection into hepatocytes and confirmed that HDV RNA synthesis had both α-amanitin-sensitive and -resistant components. The antigenomic RNA labeling was α-amanitin resistant and localized to the nucleolus. The genomic RNA labeling was α-amanitin sensitive and more diffusely localized in the nucleoplasm. Most of the genomic RNA labeling appeared to colocalize with the PML nuclear bodies. Furthermore, promyelocytic leukemia protein, RNA polymerase II (Pol II), and the Pol I-associated transcription factor SL1 could be precipitated together with hepatitis delta antigen, suggesting the association of HDV replication complex with the Pol I and Pol II transcription machineries. This conclusion was further confirmed by an in vitro replication assay. These findings provide additional evidence that HDV RNA synthesis occurs in the Pol I and Pol II transcription machineries, thus extending the capability of the cellular DNA-dependent RNA polymerases to utilizing RNA as templates.
Hepatitis delta virus (HDV) causes chronic fulminant hepatitis in human and always associates with hepatitis B virus (HBV) infection. Despite its obligate association with HBV, HDV is able to replicate autonomously in the absence of HBV or any other helper virus. HDV RNA contains a circular single-stranded RNA genome, which encodes a single protein, hepatitis delta antigen (HDAg). HDAg is involved in viral RNA replication and virus assembly; however, it is not a polymerase. Thus, HDV relies exclusively on the mammalian host RNA polymerases for its RNA-templated RNA replication (10, 22, 26, 31, 33, 38). Similar to plant viroids (11), the circular RNA genome of HDV is replicated by a rolling-circle mechanism in the nucleus (22). The genomic RNA strand is first replicated into the full-length antigenomic RNA and is also transcribed into an mRNA (0.8 kb), which encodes HDAg. The antigenomic RNA, in turn, is replicated into genomic RNA by another round of rolling-cycle replication. The mechanisms of genomic RNA, antigenomic RNA, and mRNA synthesis are independent and are probably mediated by different RNA polymerases (17, 20, 22, 26). However, the nature of the polymerases involved in the synthesis of the various HDV RNA species is unclear, inasmuch as the mammalian cells are not known to encode any RNA-dependent RNA polymerase. Based on indirect evidence, cellular DNA-dependent RNA polymerase II (Pol II) has been implicated as the key enzyme for HDV RNA replication (10, 27, 28, 38). Recent studies further showed that the syntheses of HDV genomic and antigenomic RNA strands have different sensitivities to α-amanitin (22, 25, 26), suggesting the involvement of more than one replication machinery. However, the nature of these machineries has been controversial.
Promyelocytic leukemia protein (PML) was originally discovered in acute promyelocytic leukemia patients, associated with a reciprocal chromosomal translocation of chromosomes 15 and 17 (6, 13). PML is a part of a multiple, heterogeneous protein complexes called PML nuclear bodies, nuclear domain 10, or PML oncogenic domains. A large number of proteins are localized to PML nuclear bodies (15, 39). Some of them, namely, sp100, PML, and NDP52, have been reported to be involved in gene expression and replication of many DNA viruses, including simian virus 40, adenovirus 5, human cytomegalovirus, and herpes simplex virus type 1 (14, 23, 24, 32, 34). Overexpression of PML protein enhances the replication of papillomavirus (8). One of the HDAg species, the large form, has been localized to the PML bodies (2).
In light of the unusual ability of mammalian cells to carry out RNA-dependent RNA replication of HDV in the absence of a virus-encoded RNA-dependent RNA polymerase, this virus system offers a unique opportunity to decipher not only the mechanism of HDV RNA replication specifically but also the potential mechanism of RNA amplification in general in mammalian cells.
MATERIALS AND METHODS
Cell culture, cell lines, and RNA transfection.
A human hepatoma cell line, Huh7, was cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. H1δ9 is an HDV cDNA (trimer)-integrated stably transformed cell line, which undergoes active HDV RNA replication (19), and was cultured the same way as Huh7. For BrUTP labeling, 5 × 104 cells were seeded in a well of an eight-well chamber slide and grown until they were 60% confluent. The cells were treated with actinomycin D at 5 μg/ml at 37°C for 45 min to inhibit cellular global transcription. For the indicated experiments, α-amanitin (10 μg/ml) was also added to medium together with actinomycin D. RNA transfection was performed as described previously (26). Briefly, the 1.2× genome-length HDV RNAs were transcribed from plasmids pBSδ 1.2G and pBSδ 1.2AG by using T7 Megascript kits (Ambion) after linearization with restriction enzyme NotI. Plasmids pBSδ 1.2G and pBSδ 1.2AG (templates for in vitro transcription of the 1.2× genome-length genomic and antigenomic HDV RNAs, respectively) have been described elsewhere (22). The capped mRNA for HDAg was transcribed from plasmid pX9-I/II after linearization with restriction enzyme HindIII by using a T7 m-Message m-Machine kit (Ambion) (25). All RNA transfection experiments were performed by use of the DMRIE-C reagent (Gibco BRL) according to the manufacturer's directions.
BrUTP labeling.
BrUTP incorporation assay was performed by using FuGENE 6 transfection reagent as previously described with slight modifications (Roche Applied Biosciences). Briefly, FuGENE 6 was mixed with 0.1 volume of 10 mM BrUTP (Sigma-Aldrich) and incubated for 15 min at room temperature before being used for transfection. H1δ9 cells were washed twice with phosphate-buffered saline (PBS), and 100 μl of the transfection mix was added to each well of a chamber slide and incubated at 4°C for 15 min. The cells were then pulse-chased at 37°C for additional 15 or 45 min with DMEM supplemented with 10% fetal bovine serum. The cells were fixed with 4% paraformaldehyde at room temperature for 20 min and used for immunostaining. For Huh7 cells transfected with HDV gnomic RNA and mRNA, cells were labeled with BrUTP on day 3 after RNA transfection. For BrUTP and HDV RNA cotransfection, DOTAP (Roche) was used. Briefly, 15 μl of DOTAP was diluted to 50 μl with HEPES-buffered saline and then mixed with HDV RNA mixture (1 μg of 1.2× genomic or antigenomic RNA and mRNA) and 2 mM BrUTP in a total volume of 75 μl. The mixture was incubated at room temperature for 15 min and then added to 1 ml OPTI-MEM. The cells were washed twice with OPTI-MEM prior to transfection. The DOTAP-RNA mixture (100 μl) was added to each well of a chamber slide and incubated at room temperature for 15 min. The cells were then pulse-chased for various periods of time and fixed using the same procedures as described above.
Immunostaining.
After being fixed with 4% paraformaldehyde, cells were washed twice with PBS and permeabilized with 0.1% Triton X-100. Subsequently, cells were blocked with 5% bovine serum albumin, followed by incubation with mouse anti-PML antibody (Santa Cruz), mouse anti-C23 antibody (Stressgen Biotechnologies Corporation), mouse anti-SC35 antibody (Abcam), or mouse anti-HDAg (3G3). The cells were then further stained with Texas Red- or fluorescein isothiocyanate (FITC)-conjugated donkey anti-mouse antibody. After being washed three times, the cells were stained with Cy2-conjugated (green) or Cy3-conjugated (red) mouse antibromodeoxyuridine (anti-BrdU) antibody at room temperature overnight. The cells were washed three times, mounted, and examined by confocal microscopy. The mouse antibody against anti-BrdU was labeled with Cy2 or Cy3 by using a Cy2-conjugated or Cy3-conjugated monoclonal antibody kit (Amersham Biosciences) according to the manufacturer's instructions. To investigate colocalization of HDAg with PML and SC35, the antibodies against PML and SC35 were conjugated with Cy2 and the antibodies against HDAg were conjugated with Cy3, using the kit described above.
Western blotting.
Western blotting was performed as previously described (26). Immunoblotting was carried out using anti-HDAg (1:200) (3G3), anti-Pol II (1:200) (Abcam), anti-SL1 (1:2500) (a generous gift from Lucio Comai, Department of Molecular Microbiology and Immunology, University of Southern California), anti-coilin (1:500) (BD Biosciences), anti-PML (1:500), anti-NDP52 (1:200) (BD Biosciences), anti-Sp100 (Chemicon), and anti-actin (1:5,000) (Sigma) antibodies. After incubation with the secondary antibody conjugated to horseradish peroxidase, the blots were developed by enhanced chemiluminescence using an ECL Plus blotting detection system kit (Amersham Biosciences).
Immunoprecipitation.
Huh7 cells were transfected with 1.2× HDV genomic RNA and mRNA and harvested at day 3 posttransfection. The cell pellet was resuspended in 1 ml PBS buffer in the presence of protease inhibitor cocktail (protease inhibitor cocktail tablet; Roche) and sonicated to disrupt cellular compartments. The cell lysates were clarified by centrifugation at 12,000 rpm for 10 min, and then 1 μg of monoclonal antibody against HDAg was added to 200 μg of cell lysates in a total volume of 500 μl, which was incubated at 4°C for 18 h with agitation. Subsequently, preclarified protein G-agarose beads were added, and the mixture was further incubated at 4°C for 4 h. The precipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting.
siRNA knockdown.
The small interfering RNAs (siRNAs) targeting PML, NDP52, and Sp100 were purchased from Dharmacon. Nonspecific siRNA was used as a control. Huh7 cells were transfected with the various siRNAs by using Lipofectamine 2000 (Invitrogen). A total of 100 nM of siRNA was used in each well of the six-well plate. At 48 h after transfection, cells were further transfected with HDV 1.2× genomic RNA and mRNA by using DMRIE-C reagent (Invitrogen) as described previously (26). One day after HDV RNA transfection, the cells were transfected with the same siRNA again to ensure sufficient knockdown of the target protein expression. Two more days later, the cells were harvested and analyzed for protein expression by Western blotting and for HDV RNA replication by reverse transcription and real-time PCR, as described above.
In vitro replication assay.
Production and partial purification of HDV virus particles were performed by a modification of the procedure from a previous study (1). Briefly, Huh7 cells were cotransfected with HDAg- and HBsAg-expressing plasmids pCd1.2G (21) and pA3HBV3.8 (36), respectively, using FuGENE 6 (Roche Diagnostics). Cell culture fluids harvested from days 9 to 18 posttransfection were pooled and stored at −20°C. The pooled culture fluids were thawed and clarified by centrifugation, and HDV particles were precipitated from the supernatant by using polyethylene glycol 8000 (OmniPur), redissolved in a volume of serum-free DMEM equivalent to 2.5% of the original volume of the cell culture fluid, filter sterilized, and stored at −80°C. The virus concentration was determined by comparison with known standard quantities of in vitro-transcribed HDV RNA by using Northern blot hybridization. The in vitro replication assay was modified from previous studies (10, 38). Briefly, HeLa nuclear extract (8 units) (Promega) was incubated with α-amanitin (10 μg/ml), 1 μg of anti-SL1 polyclonal antibody, anti-polymerase II, anti-SC35, or anti-HDAg monoclonal antibody at 4°C for 30 min prior to the replication assay. The reaction mixture (50 μl) contained 1.2 × 107 virus particles, HeLa cell extract (8 units; Promega) containing α-amanitin or individual antibody as indicated, 5 mM MgCl2, 40 units of RNasin (Promega), and 20 mM HEPES buffer. The virus particles were disrupted by incubation with 1% NP-40 at 4°C for 30 min prior to the experiment. The reaction mixtures were incubated at 37°C for 60 min. After reaction, RNA was extracted with Tri-reagent (Molecular Research Center, Inc.) and used for reverse transcription to amplify either genomic or antigenomic cDNA by using specific sets of primers. The sequence of the primer for genomic-sense cDNA (G primer) is 5′-CCGGCTACTCTTCTTTCC CTTCTCTCGTC-3′, and that for antigenomic-sense cDNA (AG primer) is 5′-CACCGAAGAAGG AAGGCCCTGGAGAACAA-3′. Reverse transcription was performed by using Superscript III (Invitrogen) according to the manufacturer's instructions.
Real-time PCR.
Real-time PCRs were performed by using the TaqMan PCR reagent kit (Applied Biosystems, Courtab, France). The cDNA (1 μl) from the in vitro reaction was added to a 25-μl PCR mixture containing 3.5 mM MgCl2; 200 μM each of dATP, dCTP and dGTP; 400 μM of dUTP; 300 nM of each primer; and 200 nM of fluorogenic probe. The reaction consisted of an initiating step of 10 min at 95°C, followed by 50 cycles of amplification, each consisting of 15 s at 95°C and 1 min at 60°C. The reactions, data acquisition, and analyses were performed with the ABI PRISM 7000 sequence detection system (Applied Biosystems, Courtab, France).
RESULTS
BrUTP labeling of HDV RNA in stable HDV RNA-replicating cells.
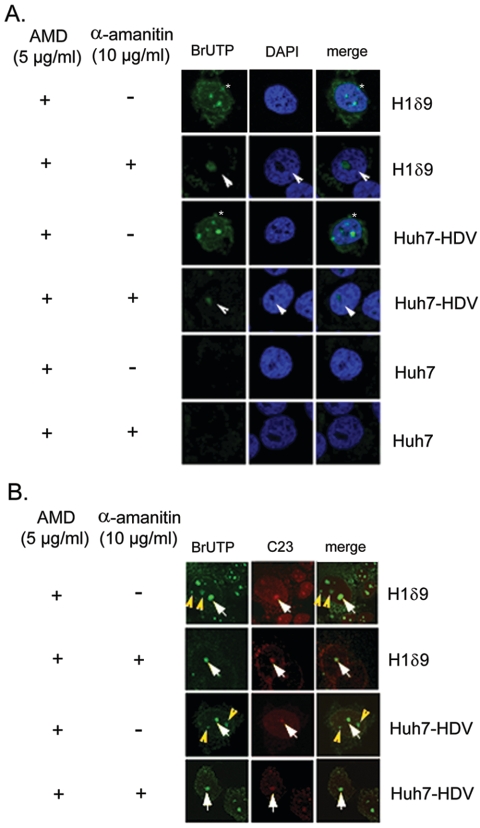
To investigate the nature of the cellular transcription machineries involved in HDV RNA replication, we first set out to study the intranuclear sites of HDV RNA synthesis. We used BrUTP labeling to visualize the newly synthesized RNA initially in a cell line (Η1δ9) that contains a stably integrated HDV cDNA trimer and undergoes active HDV RNA synthesis (19). Short-term BrUTP labeling (15 min) was done in the presence of actinomycin D, which eliminated cellular DNA-dependent transcription. Under such conditions, several distinct and diffuse BrUTP-labeled spots were observed inside the nucleus as well as in the cytoplasm (Fig. 1), whereas no label could be detected in the cells without HDV sequences (Huh7 cells) (Fig. 1A, Huh7), indicating that the BrUTP label represents the newly synthesized HDV RNA. These results are consistent with the previous findings that HDV RNA synthesis occurs in the nucleus and the genomic RNA product is rapidly transported to the cytoplasm (20). After cells were treated with α-amanitin (10 μg/ml), the BrUTP labeling decreased considerably, but several prominent intranuclear spots were still detected (Fig. 1A). These results confirmed the previous finding that part of HDV RNA synthesis, specifically the antigenomic RNA synthesis (using the genomic RNA strand as the template), was resistant to α-amanitin (22, 26). Interestingly, some of the BrUTP labeling colocalized with the nucleolus marker protein C23, whereas some did not (Fig. 1B). After the α-amanitin treatment, all of the remaining BrUTP labeling colocalized with C23 (Fig. 1B). The same results were also obtained for Huh7 cells transfected with HDV RNA by using previously established procedures (25), which led to active RNA replication (Fig. 1A and B, Huh7-HDV). These results provide strong evidence that HDV RNA replication is carried out by two distinct RNA replication machineries at different intranuclear locations.
FIG. 1.
Subcellular distribution of HDV RNA synthesis. BrUTP was incorporated (15-min labeling) into newly synthesized RNA and visualized by immunostaining (Cy2, green staining). (A) BrUTP incorporation in either the stable HDV cDNA-integrated cell line H1δ9 or Huh7 cells transiently transfected with HDV RNA (Huh7-HDV). The cells were treated with actinomycin D (AMD) (+) and with (+) or without (−) α-amanitin prior to BrUTP labeling as designated. The asterisks indicate prominent intranuclear spots, and the arrowheads indicate the BrUTP labeling in the nucleolus-like structure. Untransfected Huh7 cells (Huh7) were used as the negative control. (B) Immunostaining of nucleolin C23 (Texas Red, red staining). The white arrows indicate the colocalization of C23 and BrUTP labeling, and the yellow arrowheads indicate BrUTP labeling outside of nucleolus. The incubation time was 15 min after BrUTP transfection.
BrUTP labeling of separate HDV RNA strands following HDV RNA transfection.
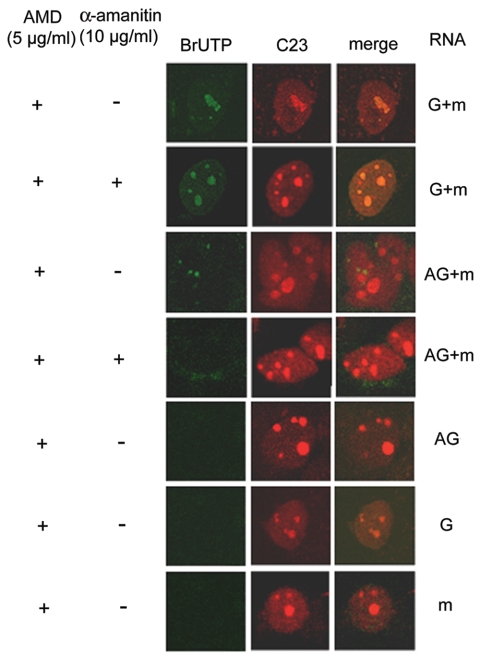
To determine the sites of synthesis of the genomic and antigenomic HDV RNA strands separately, we transfected either genomic or antigenomic RNA (together with the HDAg-encoding mRNA) (25, 26) and BrUTP into HuH7 cells so that BrUTP would be incorporated into the RNA synthesized by using the transfected RNA as the template during a brief period (15 min) following transfection. When the genomic RNA was used for transfection, BrUTP labeling was visualized almost exclusively in the nucleolus (colocalized with C23); the labeling could not be inhibited by α-amanitin (Fig. 2, G +m, with α-amanitin). These results suggested that HDV antigenomic RNA synthesis from the genomic RNA template occurs in the nucleolus and is mediated by RNA polymerase I or an unknown α-amanitin-resistant RNA polymerase. In contrast, when antigenomic HDV RNA was used for transfection, the BrUTP-labeled dots did not colocalize with C23 and were totally inhibited by α-amanitin (Fig. 2, AG+m, with α-amanitin). Thus, genomic RNA synthesis from the antigenomic RNA template is α-amanitin sensitive and occurs outside the nucleolus. When genomic or antigenomic RNA alone (without cotransfection with an HDAg-encoding mRNA) or mRNA alone was used for transfection, no RNA synthesis occurred (25); correspondingly, no BrUTP labeling was detected (Fig. 2).
FIG. 2.
Subcellular localization of sites of synthesis of individual HDV RNA strands. Huh7 cells were cotransfected with BrUTP, HDAg mRNA and either genomic RNA (G+m) or antigenomic RNA (AG+m) as previously described (25, 26). Cells were treated with actinomycin D (AMD) (+) and with (+) or without (−) α-amanitin before transfection. Transfection of genomic (G), antigenomic (AG), or mRNA (m) alone, none of which alone lead to RNA replication, served as negative controls. At 15 min after RNA transfection, BrUTP-labeled RNA was detected by staining with BrdU-specific antibody. BrUTP labeling, Cy2 (green); C23, Texas Red (red).
Colocalization of genomic RNA synthesis with PML bodies and of antigenomic RNA synthesis with the nucleolus.
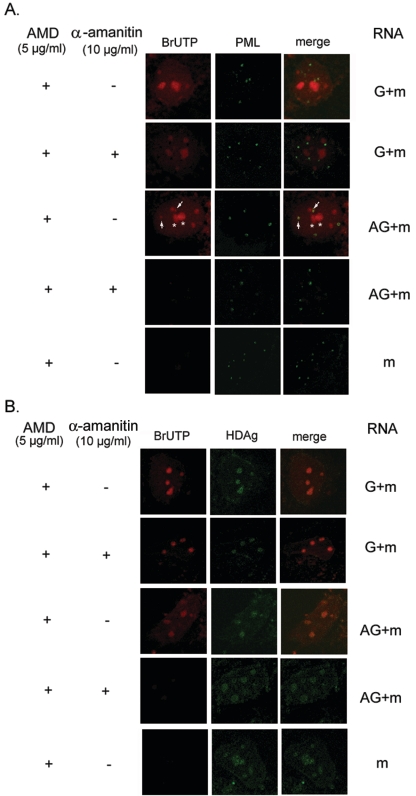
We subsequently explored which nuclear body structure was associated with genomic RNA synthesis. A previous study has demonstrated that the large form of HDAg was colocalized with the PML bodies, although the biological significance of such colocalization was not clear (2). We therefore proceeded to study whether the genomic RNA synthesis occurs in the PML bodies. The structure and localization of PML bodies are known to be dynamic in the nucleoplasm at different stages of cell cycle or during virus infection (9, 14). Therefore, we labeled the cells with BrUTP for a longer period of time (60 min) after HDV RNA transfection so that PML bodies could be more clearly identified. When the genomic RNA was used for transfection, BrUTP label did not colocalize with PML, and most of the label was resistant to α-amanitin treatment (Fig. 3A, G +m, with or without α-amanitin). However, when the antigenomic RNA was transfected, some of the BrUTP-labeled RNA appeared to colocalize with, or was very near, PML bodies (Fig. 3A). In contrast to what is seen in Fig. 2, where BrUTP labeling was carried out for only 15 min, the BrUTP label was also detected in the nucleolus-like structures when BrUTP was labeled for 60 min (Fig. 3A). All of the labels, including the nucleolus-like labels, disappeared when the cells were treated with α-amanitin (Fig. 3, AG+m, with α-amanitin). These findings are consistent with the interpretation that the BrUTP label represented the α-amanitin-sensitive genomic RNA synthesis and the subsequent transport of the RNA products to other sites for storage or to serve as a template for the ensuing antigenomic RNA synthesis. Taken together, these data suggested that the genomic RNA synthesis occurs in a structure associated with the PML body and is mediated by host RNA polymerase II. In contrast, antigenomic RNA synthesis occurs in the nucleolus.
FIG. 3.
(A) HDV genomic RNA synthesis is colocalized with the PML body. The BrUTP-labeled RNA and PML body are identified by red (Cy3) and green (FITC) staining, respectively. The arrows indicate the convergence of PML body and BrUTP labeling, and the asterisks indicate the BrUTP labeling in the nucleolus-like structures. (B) HDAg is colocalized with HDV genomic and antigenomic RNA synthesis. The BrUTP labeling and HDAg are identified as red (Cy3) and green (FITC), respectively. The RNA transfection and drug treatments were the same as described for Fig. 2. The incubation time was 60 min after RNA transfection. AMD, actinomycin D.
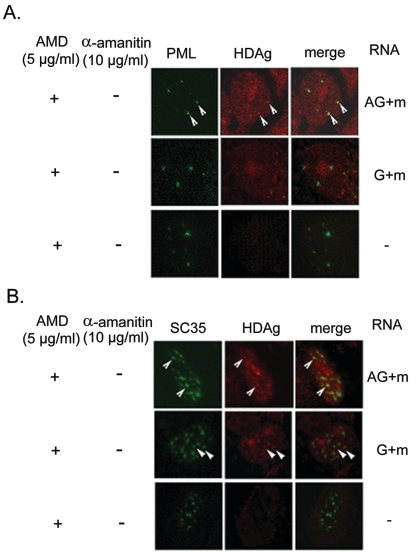
Since HDAg is required for HDV RNA synthesis, the HDAg that is newly synthesized from the transfected mRNA likely will be accumulated at the site of active RNA synthesis. We therefore studied the distribution of HDAg immediately after HDV RNA transfection. All of the HDAg made within 60 min following the RNA transfection coincided with BrUTP labeling in either the genomic or the antigenomic RNA-transfected cells (Fig. 3B). The distribution of HDAg was not affected by the α-amanitin treatment. Furthermore, we found that HDAg colocalized with PML when antigenomic RNA, but not when genomic RNA, was transfected (Fig. 4A). These results suggested that the HDV antigenomic RNA and HDAg are recruited into the PML bodies. Some of HDAg was also colocalized with nuclear SC35 speckles, when either genomic or antigenomic RNA was used for transfection (Fig. 4B). The nuclear SC35 speckle has been reported to interact with HDAg and is presumed to be involved in HDV RNA replication (3, 4).
FIG. 4.
Colocalization of HDAg and PML (A) or SC35 (B). Huh7 cells were cotransfected with HDAg-encoding mRNA and either genomic RNA (G+m) or antigenomic RNA (AG+m) as indicated. At 60 min after RNA transfection, the cells were examined for the distribution of HDAg and PML or SC35 by immunostaining. Untransfected Huh7 cells served as negative control (−). The cells were treated with actinomycin D (AMD) at 5 μg/ml prior to transfection. PML and SC35 are identified by Cy2 (green), and HDAg is identified by Cy3 (red). The arrowheads show the colocalization of HDAg and PML or SC35.
Coimmunoprecipitation of Pol II and a Pol I transcription factor, SL1, with HDAg.
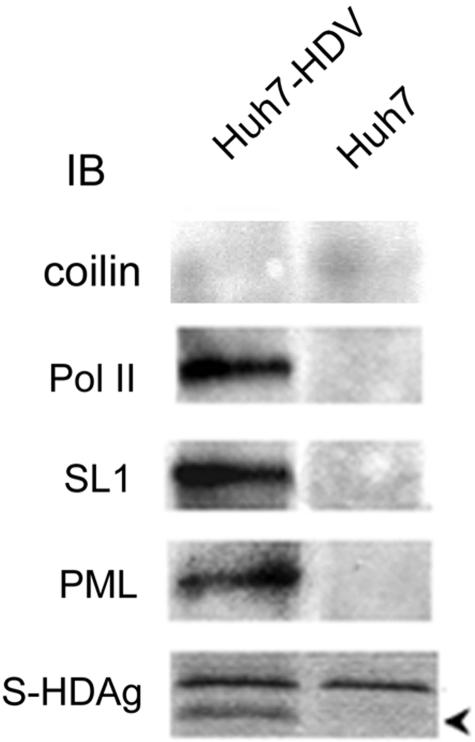
Since HDAg acts as a transcription activator and supports HDV RNA replication (5, 37), it has been presumed to be a component of the HDV RNA replication complex. Therefore, we studied whether HDAg is associated with components of the cellular transcription machineries in Huh7 cells transfected with HDV RNA. We performed immunoprecipitation of cellular lysates with antibodies against HDAg and determined whether any cellular transcription factors could be coprecipitated. The results showed that in the HDV RNA-transfected cells, but not in the untransfected cells, SL1, PML, and RNA polymerase II (38) could be coprecipitated with HDAg by the HDAg-specific antibodies, indicating that they were present together with HDAg in a complex (Fig. 5). The coprecipitated proteins were not recognized by anticoilin antibody, indicating that HDAg specifically binds to the particular nuclear proteins. These results further suggest that the components of both Pol I and Pol II transcription machineries are associated with HDV RNA replication. In addition, the PML body likely participates in HDV RNA replication.
FIG. 5.
Immunoprecipitation of cellular proteins in the HDAg-containing complex. Cellular extracts of Huh7 cells that had been transfected with HDV RNA (Huh7-HDV) or untransfected Huh7 cells were immunoprecipitated by antibody specific for HDAg and subsequently immunoblotted (IB) with antibodies against coilin, RNA polymerase II, SL1, PML, or HDAg. The arrow indicates HDAg.
Effects of knockdown of PML nuclear body proteins on HDV RNA replication.
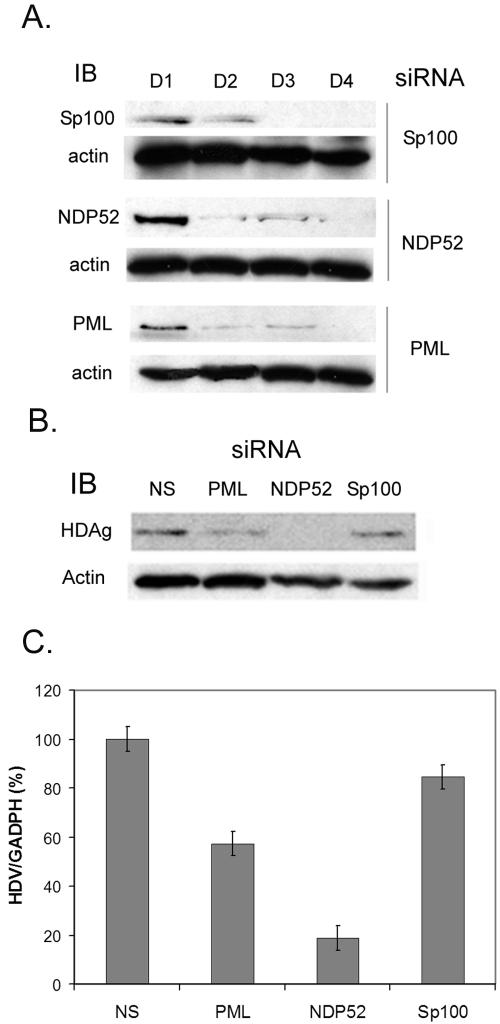
To further assess the functional importance of the PML nuclear body in the HDV life cycle, we used the siRNA approach to knock down the proteins in or associated with the PML nuclear body, including PML, sp100, and NDP52 (Fig. 6A), all of which have been reported to be involved in the replication of some viruses (15, 16, 30). All three siRNAs specifically knocked down the protein expression levels of the respective proteins by day 2 or 3 posttransfection, indicating efficient siRNA knockdown under our assay condition (Fig. 6A).
FIG. 6.
(A) siRNA knockdown of PML nuclear body-associated proteins. The siRNAs of Sp100, NDP52, or PML were transfected into Huh7 cells to knock down the endogenous protein expression. Two days later, siRNA transfection was repeated to enhance the extent of knockdown. The proteins were identified by immunoblotting (IB) using individual antibodies on different days after siRNA transfection. (B and C) Effects of NDP52, Sp100, and PML siRNA knockdown on HDV RNA synthesis (C) and HDAg production (B). The siRNA of PML, NDP52, or Sp100 or nonspecific (NS) siRNA was transfected into HuH7 cells. Two days after siRNA transfection, cells were transfected with HDV RNA and mRNA, followed by retransfection with the same siRNA 24 h later to maintain the protein knockdown. The relative levels of HDAg (B) and HDV RNA (C) in each reaction were determined at 48 h after the second siRNA transfection (see Materials and Methods). The proteins were detected by immunoblotting using specific antibodies (B). Actin was used as a protein sample loading control. The HDV RNA quantitative analysis (C) was performed by specific reverse transcription and real-time PCR as described in Materials and Methods. Error bars indicate standard deviations.
To examine which PML-associated protein is essential for HDV replication, the cells were first transfected with these siRNAs and then transfected with HDV RNA to initiate HDV RNA replication. Among these three proteins, NDP52 knockdown caused the most dramatic reduction of HDV RNA replication (Fig. 6C) and, correspondingly, HDAg production (Fig. 6B). The PML-specific siRNA also caused a nearly 50% reduction of HDV RNA replication and HDAg production (Fig. 6B and C). In contrast, Sp100-specific siRNA had very little effect on HDV RNA replication or HDAg production. These results further suggested that at least some of the PML body-associated proteins, including PML and NDP52, are involved in HDV RNA replication. NDP52 has been shown to be involved in maintaining the PML nuclear body stability and viral RNA transcription (15). In contrast, PML and Sp100 are dynamic components of the PML body which do not play a strict structural role in this body (35). NDP52 may be involved in HDV RNA replication either directly or indirectly through the stabilization of PML protein and the PML nuclear body. Another possibility is that NDP52 mediates the interaction between RNA polymerase II and HDAg or recruits RNA polymerase to the HDV RNA replication complex.
Requirement of nuclear factors in in vitro HDV RNA replication assays.
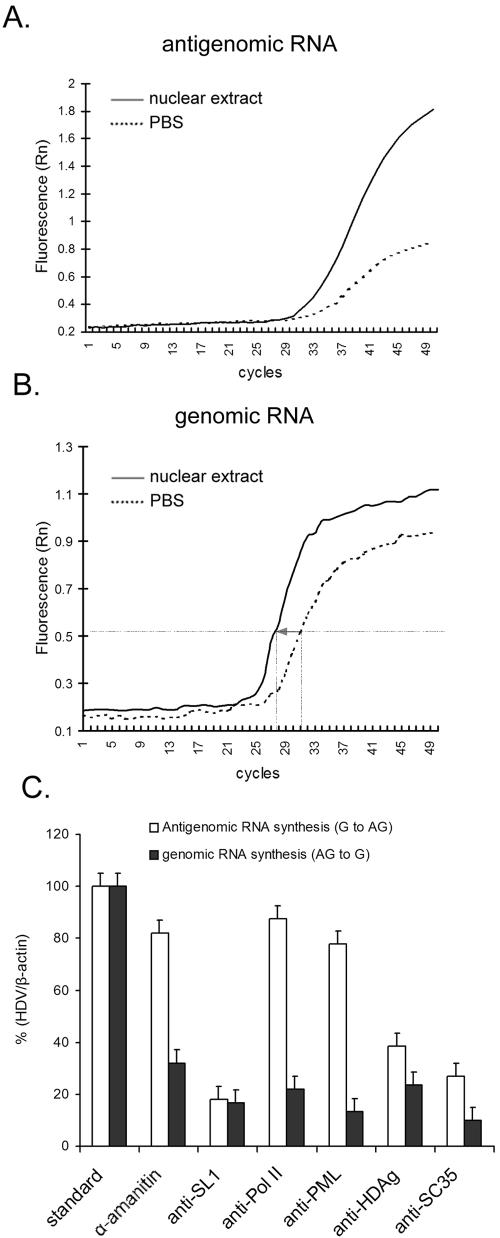
The BrUTP labeling data thus suggested that the syntheses of genomic and antigenomic HDV RNAs were carried out by two distinct transcription machineries. To further support this conclusion, we studied the requirement for nuclear factors in HDV RNA replication in a novel in vitro replication system. We used purified HDV particles produced from Huh7 cells expressing both HDAg and HBsAg (1) to perform in vitro RNA replication using HeLa nuclear extracts (Fig. 7A). The HDV virus particles contain the circular HDV genomic RNA complexed with HDAg, thus representing the authentic viral RNA template during virus infection. HeLa nuclear extract was treated with antibodies against various nuclear factors to block their biological functions prior to the in vitro replication assay. The antigenomic RNA products were detected by reverse transcription and real-time PCR using specific primers (Fig. 7A). Approximately 1.1 × 108 copies of antigenomic RNA products could be obtained from a typical reaction mix, as determined by quantitative PCR, after 60 min of incubation (Fig. 7A). Significantly, the reaction was almost totally refractory to inhibition by α-amanitin (10 μg/ml) (Fig. 7C). Pretreatment with the antibody against SL1, which is the specific initiation factor for RNA polymerase I-mediated transcription (7, 12), reduced HDV antigenomic RNA synthesis by more than 80%. In contrast, neither anti-Pol II antibody nor anti-PML antibody affected antigenomic RNA synthesis. Anti-SC35 also significantly reduced antigenomic RNA synthesis. The antibody against HDAg inhibited the antigenomic RNA synthesis by more than 60% (Fig. 7C). We also detected genomic RNA products in the in vitro reaction by following the kinetics of the increase of genomic RNA by using quantitative PCR (Fig. 7C); this RNA product would represent the products from two cycles of reaction (from genomic to antigenomic and back to genomic RNA). Under the assay conditions used, the amounts of genomic RNA increased by approximately 16-fold after a 60-minute reaction (Fig. 7B), suggesting that the in vitro reaction was capable of two cycles of RNA synthesis. Interestingly, the genomic RNA synthesis was inhibited by α-amanitin and by all of the antibodies tested, notably anti-Pol II and anti-PML antibodies (Fig. 7C). Because anti-SL1 antibody already inhibited the antigenomic RNA synthesis, we could not determine whether it also inhibited genomic RNA synthesis in this in vitro assay. These results further support the conclusion that genomic RNA synthesis is carried out by Pol II in a nuclear compartment associated with the PML bodies, whereas antigenomic RNA synthesis involves components of the Pol I transcription machinery.
FIG. 7.
In vitro replication assay. (A and B) Quantification of HDV antigenomic and genomic RNAs after in vitro replication. In vitro replication was performed using genomic RNA from the disrupted HDV particles as a template in the HeLa nuclear lysates. After reaction, cDNAs of antigenomic or genomic RNA (after reverse transcription using primers specific for either antigenomic RNA [A] or genomic RNA [B]) were amplified and detected by real-time quantitative PCR. The RNA was normalized with β-actin mRNA. The same amount of viral particles was incubated with either HeLa nuclear extract or PBS (as a negative control) to carry out the in vitro replication assay. The increase of the HDV RNA amounts after the reaction was determined by the reduction of the PCR cycle numbers as indicated by the arrow between the solid and dotted lines. (C) Effects of α-amanitin or antibodies on in vitro RNA synthesis. The in vitro reactions were carried out using purified HDV particles and HeLa nuclear lysates as in panels A and B. α-Amanitin and antibodies against SL1, polymerase II, PML, SC35, or HDAg were added in the nuclear extracts as indicated and incubated as described above. The amounts of antigenomic and genomic RNA products in each reaction were determined by reverse transcription-PCR as shown in panels A and B. The relative amounts of RNA products in each reaction are presented as the ratios of the products relative to those in the standard reactions. Error bars indicate standard deviations. RNA [B]) were amplified and detected by real-time quantitative PCR. The RNA was normalized with β-actin mRNA. The same amount of viral particles was incubated with either HeLa nuclear extract or PBS (as a negative control) to carry out the in vitro replication assay. The increase of the HDV RNA amounts after the reaction was determined by the reduction of the PCR cycle numbers as indicated by the arrow between the solid and dotted lines. (C) Effects of α-amanitin or antibodies on in vitro RNA synthesis. The in vitro reactions were carried out using purified HDV particles and HeLa nuclear lysates as in panels A and B. α-Amanitin and antibodies against SL1, polymerase II, PML, SC35, or HDAg were added in the nuclear extracts as indicated and incubated as described above. The amounts of antigenomic and genomic RNA products in each reaction were determined by reverse transcription-PCR as shown in panels A and B. The relative amounts of RNA products in each reaction are presented as the ratios of the products relative to those in the standard reactions. Error bars indicate standard deviations.
DISCUSSION
The data presented in this report altogether suggest that HDV antigenomic RNA synthesis occurs in the nucleolus, whereas the genomic RNA synthesis likely occurs in the PML body-associated structure. Furthermore, the genomic RNA synthesis is carried out by Pol II and involves PML and NDP52 proteins. In contrast, the antigenomic RNA synthesis is carried out by Pol I or a Pol I-like polymerase in the nucleolus and involves the Pol I-specific transcription factor SL1. These findings, plus the fact that the genomic RNA is exported to the cytoplasm immediately after its synthesis, whereas antigenomic RNA is not (20), suggest that these two RNA strands are synthesized by two different transcription machineries coupled to different RNA export pathways. Our studies did not resolve clearly where the transcription of the HDAg mRNA takes place. However, when the genomic RNA was used for transfection, BrUTP labeling was observed not only in the nucleolus but also in the nucleoplasm; the nucleoplasmic labeling was sensitive to the α-amanitin treatment (Fig. 2). Since the genomic RNA is used as the template for the synthesis of both antigenomic RNA and the HDAg-encoding mRNA, it is reasonable to conclude that the mRNA transcription is carried out by Pol II in the nucleoplasm. However, the extranucleolar BrUTP labeling did not colocalize with PML; thus, the mRNA transcription may occur at a site different from that of the genomic RNA synthesis, which colocalizes with PML. These results further support the replication model recently postulated for HDV RNA (17).
The role of Pol II in HDV RNA replication is now more clearly defined from the in vitro replication assay, in which the antibody against Pol II inhibited genomic RNA synthesis. This finding, coupled with the experiment showing that the introduction of an α-amanitin-resistant Pol II conferred resistance of HDV RNA synthesis to the drug (26), established the role of Pol II in HDV RNA replication. However, whether HDV genomic RNA synthesis takes place exclusively in the PML body remains to be confirmed. PML is a dynamic nuclear body that is often involved in DNA virus replication. Its involvement in HDV RNA replication will be consistent with its physiological nature. On the other hand, the role of Pol I in HDV antigenomic RNA synthesis is still equivocal. Because of the lack of a suitable antibody, we have so far not been able to demonstrate directly the participation of Pol I in HDV RNA replication. However, in this study, we have demonstrated the indispensable role of the Pol I-specific factor SL1 in the in vitro replication assay and also its coprecipitation with HDAg in HDV-replicating cells. The Pol I transcription machinery is presumed to be involved in HDV antigenomic RNA synthesis; nevertheless, we cannot rigorously rule out the possibility that an unknown α-amanitin-resistant and SL1-associated RNA polymerase, or even Pol III, carries out HDV antigenomic RNA synthesis.
Why does HDV, a small satellite RNA virus, need such a complex RNA replication mechanism? HDV RNA replication occurs via a rolling-circle mechanism, generating genomic and antigenomic strands asymmetrically. In addition, it has to produce a subgenomic mRNA species. The virus encodes only one protein, which is involved in all aspects of RNA replication and mRNA transcription. The separation of these various replication functions will be difficult to achieve entirely through regulation by this single viral protein. The spatial separation of these transcription and replication functions will allow these different steps of RNA synthesis to be independently regulated.
These findings also imply that the genomic and antigenomic RNA strands are directed to different sites in the nucleus. Two possibilities can be considered: first, the fate of HDV RNA strands is determined by their primary sequences or structures; second, the different RNA strands are carried by the different modified forms of HDAg (e.g., phosphorylated, acetylated, or methylated) to the different nuclear bodies (18, 29).
Our in vitro HDV RNA replication system represents a novel system for studying HDV RNA synthesis. It uses the authentic HDV genomic RNA template, which can establish infection in Huh7 cells. For the first time, the essential role of HDAg in HDV RNA replication can be demonstrated in an in vitro reaction. Although the efficiency of this in vitro replication assay system is still low, it allows the demonstration of the essential factors involved in HDV RNA synthesis. Further improvement of this system will be very valuable for dissecting the mechanism of HDV RNA synthesis.
This HDV RNA replication mechanism may prove to be a new aspect of cellular transcription machineries. The entire process of HDV RNA replication is carried out by the host DNA-dependent RNA polymerases. Our studies suggest that not only RNA polymerase II but also RNA polymerase I or a closely related, nucleolus-associated polymerase are involved in HDV RNA replication. These mechanisms may also apply to some cellular RNA molecules as well.
REFERENCES
- 1.Barrera, A., and R. E. Lanford. 2004. Infection of primary chimpanzee hepatocytes with recombinant hepatitis D virus particles: a surrogate model for hepatitis B virus. Methods Mol. Med. 96:131-142. [DOI] [PubMed] [Google Scholar]
- 2.Bell, P., R. Brazas, D. Ganem, and G. G. Maul. 2000. Hepatitis delta virus replication generates complexes of large hepatitis delta antigen and antigenomic RNA that affiliate with and alter nuclear domain 10. J. Virol. 74:5329-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bichko, V. V., and J. M. Taylor. 1996. Redistribution of the delta antigens in cells replicating the genome of hepatitis delta virus. J. Virol. 70:8064-8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, J., S. O. Gudima, C. Tarn, X. Nie, and J. M. Taylor. 2005. Development of a novel system to study hepatitis delta virus genome replication. J. Virol. 79:8182-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao, M., S. Y. Hsieh, and J. Taylor. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 64:5066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Z., and S. J. Chen. 1992. RARA and PML genes in acute promyelocytic leukemia. Leuk Lymphoma 8:253-260. [DOI] [PubMed] [Google Scholar]
- 7.Comai, L., N. Tanese, and R. Tjian. 1992. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 68:965-976. [DOI] [PubMed] [Google Scholar]
- 8.Day, P. M., C. C. Baker, D. R. Lowy, and J. T. Schiller. 2004. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc. Natl. Acad. Sci. USA 101:14252-14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett, R. D., and J. Murray. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filipovska, J., and M. M. Konarska. 2000. Specific HDV RNA-templated transcription by pol II in vitro. RNA 6:41-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa, M., T. Meshi, T. Ohno, Y. Okada, T. Sano, I. Ueda, and E. Shikata. 1984. A revised replication cycle for viroids: the role of longer than unit length RNA in viroid replication. Mol. Gen. Genet. 196:421-428. [DOI] [PubMed] [Google Scholar]
- 12.Jordan, P., M. Mannervik, L. Tora, and M. Carmo-Fonseca. 1996. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J. Cell Biol. 133:225-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kastner, P., A. Perez, Y. Lutz, C. Rochette-Egly, M. P. Gaub, B. Durand, M. Lanotte, R. Berger, and P. Chambon. 1992. Structure, localization and transcriptional properties of two classes of retinoic acid receptor alpha fusion proteins in acute promyelocytic leukemia (APL): structural similarities with a new family of oncoproteins. EMBO J. 11:629-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly, C., R. Van Driel, and G. W. Wilkinson. 1995. Disruption of PML-associated nuclear bodies during human cytomegalovirus infection. J Gen. Virol. 76:2887-2893. [DOI] [PubMed] [Google Scholar]
- 15.Korioth, F., C. Gieffers, G. G. Maul, and J. Frey. 1995. Molecular characterization of NDP52, a novel protein of the nuclear domain 10, which is redistributed upon virus infection and interferon treatment. J. Cell Biol. 130:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korioth, F., G. G. Maul, B. Plachter, T. Stamminger, and J. Frey. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155-158. [DOI] [PubMed] [Google Scholar]
- 17.Lai, M. M. 2005. RNA replication without RNA-dependent RNA polymerase: surprises from hepatitis delta virus. J. Virol. 79:7951-7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Y. J., M. R. Stallcup, and M. M. Lai. 2004. Hepatitis delta virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J. Virol. 78:13325-13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macnaughton, T. B., E. J. Gowans, A. R. Jilbert, and C. J. Burrell. 1990. Hepatitis delta virus RNA, protein synthesis and associated cytotoxicity in a stably transfected cell line. Virology 177:692-698. [DOI] [PubMed] [Google Scholar]
- 20.Macnaughton, T. B., and M. M. Lai. 2002. Genomic but not antigenomic hepatitis delta virus RNA is preferentially exported from the nucleus immediately after synthesis and processing. J. Virol. 76:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macnaughton, T. B., and M. M. Lai. 2002. Large hepatitis delta antigen is not a suppressor of hepatitis delta virus RNA synthesis once RNA replication is established. J. Virol. 76:9910-9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macnaughton, T. B., S. T. Shi, L. E. Modahl, and M. M. Lai. 2002. Rolling-circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J. Virol. 76:3920-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin, M. A., L. D. Gelb, C. Garon, K. K. Takemoto, T. N. Lee, G. H. Sack, Jr., and D. Nathans. 1974. Characterization of “heavy” and “light” SV40-like particles from a patient with PML. Virology 59:179-189. [DOI] [PubMed] [Google Scholar]
- 24.Maul, G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660-667. [DOI] [PubMed] [Google Scholar]
- 25.Modahl, L. E., and M. M. Lai. 1998. Transcription of hepatitis delta antigen mRNA continues throughout hepatitis delta virus (HDV) replication: a new model of HDV RNA transcription and replication. J. Virol. 72:5449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modahl, L. E., T. B. Macnaughton, N. Zhu, D. L. Johnson, and M. M. Lai. 2000. RNA-dependent replication and transcription of hepatitis delta virus RNA involve distinct cellular RNA polymerases. Mol. Cell. Biol. 20:6030-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monjardino, J. 1996. Replication of hepatitis delta virus. J. Viral Hepat. 3:163-166. [DOI] [PubMed] [Google Scholar]
- 28.Moraleda, G., and J. Taylor. 2001. Host RNA polymerase requirements for transcription of the human hepatitis delta virus genome. J. Virol. 75:10161-10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mu, J. J., Y. G. Tsay, L. J. Juan, T. F. Fu, W. H. Huang, D. S. Chen, and P. J. Chen. 2004. The small delta antigen of hepatitis delta virus is an acetylated protein and acetylation of lysine 72 may influence its cellular localization and viral RNA synthesis. Virology 319:60-70. [DOI] [PubMed] [Google Scholar]
- 30.Nicewonger, J., G. Suck, D. Bloch, and S. Swaminathan. 2004. Epstein-Barr virus (EBV) SM protein induces and recruits cellular Sp110b to stabilize mRNAs and enhance EBV lytic gene expression. J. Virol. 78:9412-9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid, C. E., and D. W. Lazinski. 2000. A host-specific function is required for ligation of a wide variety of ribozyme-processed RNAs. Proc. Natl. Acad. Sci. USA 97:424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosa-Calatrava, M., F. Puvion-Dutilleul, P. Lutz, D. Dreyer, H. de The, B. Chatton, and C. Kedinger. 2003. Adenovirus protein IX sequesters host-cell promyelocytic leukaemia protein and contributes to efficient viral proliferation. EMBO Rep. 4:969-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Symons, R. H., C. J. Hutchins, A. C. Forster, P. D. Rathjen, P. Keese, and J. E. Visvader. 1987. Self-cleavage of RNA in the replication of viroids and virusoids. J. Cell Sci. Suppl. 7:303-318. [DOI] [PubMed] [Google Scholar]
- 34.Tang, Q., P. Bell, P. Tegtmeyer, and G. G. Maul. 2000. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J. Virol. 74:9694-9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiesmeijer, K., C. Molenaar, I. M. Bekeer, H. J. Tanke, and R. W. Dirks. 2002. Mobile foci of Sp100 do not contain PML: PML bodies are immobile but PML and Sp100 proteins are not. J. Struct. Biol. 140:180-188. [DOI] [PubMed] [Google Scholar]
- 36.Wu, J. C., P. J. Chen, M. Y. Kuo, S. D. Lee, D. S. Chen, and L. P. Ting. 1991. Production of hepatitis delta virus and suppression of helper hepatitis B virus in a human hepatoma cell line. J. Virol. 65:1099-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia, Y. P., and M. M. Lai. 1992. Oligomerization of hepatitis delta antigen is required for both the trans-activating and trans-dominant inhibitory activities of the delta antigen. J. Virol. 66:6641-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamaguchi, Y., J. Filipovska, K. Yano, A. Furuya, N. Inukai, T. Narita, T. Wada, S. Sugimoto, M. M. Konarska, and H. Handa. 2001. Stimulation of RNA polymerase II elongation by hepatitis delta antigen. Science 293:124-127. [DOI] [PubMed] [Google Scholar]
- 39.Zuber, M., T. S. Heyden, and A. M. Lajous-Petter. 1995. A human autoantibody recognizing nuclear matrix-associated nuclear protein localized in dot structures. Biol. Cell 85:77-86. [PubMed] [Google Scholar]