Abstract
Six of seven HLA-A*2402-positive individuals with acute parvovirus B19 infections made vigorous CD8-positive cytotoxic T-cell (CTL) responses to the viral epitope FYTPLADQF. All responders showed highly focused T-cell receptor (TCR) usage, using almost exclusively BV5.1. The BV5.1 TCR dominated the acute response, was maintained over time, and was also used by a remotely infected individual. Nine CTL clones and two oligoclonal lines obtained from three unrelated individuals used BV5.1, BJ2.1, and a conserved TCR CDR3 of nine amino acids. This commonly recognized epitope is likely important in long-term protective immunity and should be included in vaccine design.
We recently described long-lived cytotoxic T-lymphocyte (CTL) expansions in 11 patients with acute parvovirus B19 infections (8). These CD8+ T cells were activated, antigen experienced (as evidenced by the expression of perforin, CD38, and CD57), and increased in frequency over a number of months, despite the rapid resolution of symptoms. In three acutely infected HLA-A*2402-positive adults studied with HLA/B19 tetrameric complexes, the HLA-A*2402 response was numerically dominant. HLA-A*2402 is the commonest HLA class 1 allele in East Asia and has been implicated in several antiviral CTL immune responses, including those to hepatitis B virus, hepatitis C virus (HCV), cytomegalovirus, and human immunodeficiency virus (HIV) (6, 7, 11). Here we characterize the HLA-A*2402-restricted, FYTPLADQF-specific CD8+ T-cell response to parvovirus B19 in these patients, a further four HLA-A*2402-positive patients with acute parvovirus infections, and a seropositive remotely infected donor. All seven responders showed a highly focused T-cell-receptor (TCR) repertoire.
A vigorous HLA-A*2402-restricted FYTPLADQF response is seen in B19-seropositive individuals.
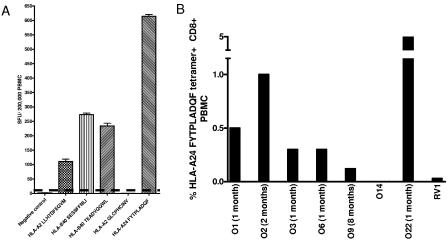
We identified seven HLA-A*2402-positive individuals from a cohort of 22 adult patients with acute B19 infections, as evidenced by arthropathy, rash, fever, and B19 immunoglobulin M (IgM) seropositivity. Ethical approval (CO2.113) and informed patient consent were obtained prior to study initiation and venous sampling. A remotely infected healthy individual was also studied. Three individuals have been identified previously (8). The following epitopes, which were described previously (8), were used in gamma interferon (IFN-γ)-specific enzyme-linked immunospot (ELISPOT) assays and/or to construct B19-specific tetrameric complexes: HLA-A*2402-restricted FYTPLADQF, HLA-B35 QPTRVDQKM, HLA-B35 FPGINADAL, HLA-A*201 LLHTDFEQV, HLA-A*201 GLCPHCINV, HLA-B8 TAKSRVHPL, and HLA-B40 SESSFFNLI and TEADVQQWL). Figure 1A shows that for donor O22, the HLA-A*2402 FYTPLADQF response was numerically dominant by direct ex vivo peripheral blood mononuclear cell (PBMC) IFN-γ ELISPOT assay 1 month after the onset of symptoms. Six of the seven acutely infected individuals showed vigorous responses to peptide FYTPLADQF by IFN-γ ELISPOT assay and/or tetramer staining, with responses ranging from 0.3 to 4.9% of CD8+ T cells (Fig. 1B). In three individuals where responses restricted by other HLA alleles were identified, the A24 responses were immunodominant. Surprisingly, as described previously (8), these responses increased over the first year postinfection, despite the resolution of clinical symptoms and the control of viremia. One HLA-A*2402-positive, remotely infected, IgG-positive, IgM-negative adult with no B19 exposure within 10 years responded to FYTPLADQF by both IFN-γ ELISPOT assay and tetramer staining (20 spot-forming units/million PBMC and 0.03% of CD8+ T cells, respectively [data not shown]). This response was further confirmed by the generation of peptide-specific T-cell lines (data not shown).
FIG. 1.
Vigorous B19 HLA-A24 FYTPLADQF-specific CD8+ T-cell responses are seen in acute and remote B19 infections. (A) IFN-γ ELISPOT responses in acutely B19-infected patient O22 1 month after symptom development. PBMC were stimulated with five B19 CD8+ T-cell epitopes (HLA-A2 LLHTDFEQV, HLA-B40 SESSFFNLI, HLA-B40 TEADVQQWL, HLA-A2 GLCPHCINV, and HLA-A24 FYTPLADQF). Data from triplicate estimations are shown, with positive results defined as the negative control value + 2 standard deviations or two times the negative control (solid horizontal line). SFU, spot-forming units. (B) FYT A24 tetramer staining of PBMC from seven acutely infected HLA-A24-positive individuals and one remotely infected HLA-A24-positive individual (RV1). Months indicate times after symptom development. Patients O1 to O3 and RV1 were described previously (8).
HLA-A*2402 FYTPLADQF-specific T cells use predominantly TCR BV5.1.
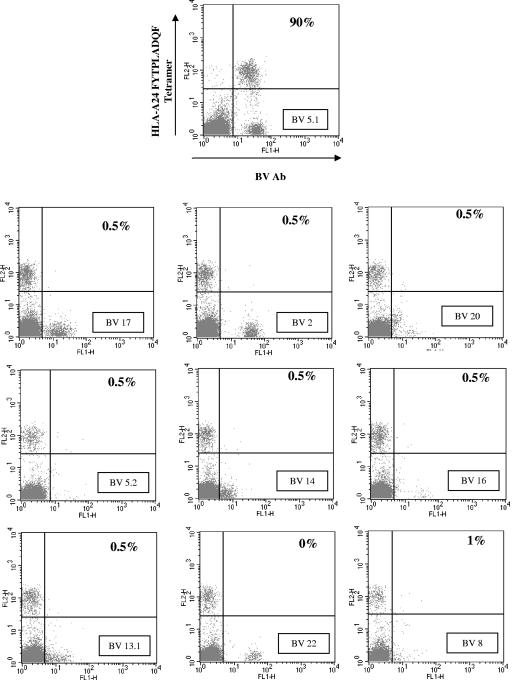
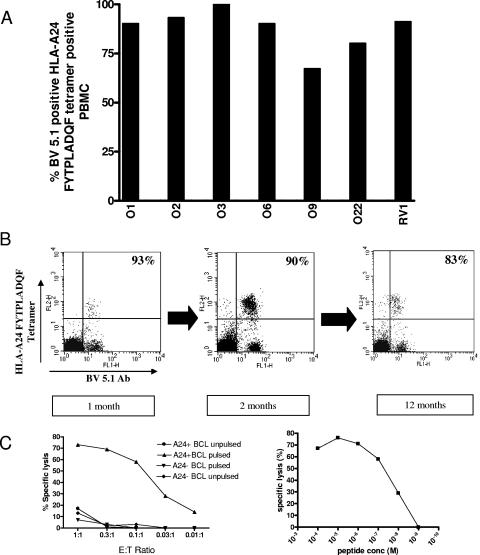
PBMC were costained with anti-CD8 antibody, B19/HLA tetramers (8), and monoclonal antibodies for 11 different TCR V beta (BV) chains (Serotec, United Kingdom). Figure 2 shows that 90% of HLA-A*2402 FYTPLADQF tetramer-positive T cells from patient O1 used TCR BV5.1. This compares to 5.4% of PBMC-derived CD8 T cells in this individual. Figure 3A shows that all seven of the HLA-A*2402 FYTPLADQF responders (six with acute infections and one with remote infection) had predominantly TCR BV5.1 usage (range, 67 to 100%; mean, 87%). Such dramatic restriction was not seen for other B19-specific responses studied ex vivo. No restriction of TCR BV was detected in the HLA-A*0201-restricted LLHTDFEQV response of patient O5, the HLA-B*8-restricted TAKSRVHPL response of O6, or the HLA-B*35-restricted FPGINADAL response of remotely infected individual RV1. HLA-B*35 QPTRVDQKM tetramer-positive PBMC from individual RV1 showed preferential TCR usage, with 32% BV16 and 45% BV8 usage (data not shown).
FIG. 2.
The B19 HLA-A24 FYTPLADQF tetramer-specific T-cell response in patient O1 predominantly uses TCR gene segment BV5.1. PBMC from patient O1 (2 months after symptom development) were stained with tetramer and BV monoclonal antibodies. Percentages shown are percentages of tetramer-positive PBMC.
FIG. 3.
B19 HLA-A24 FYTPLADQF-specific T-cell responses show striking BV5.1 TCR usage and efficient cytolysis. (A) BV5.1 usage of HLA-A24 FYTPLADQF tetramer-positive T cells from six acute and one remote responder. (B) Longitudinal BV5.1 monoclonal antibody staining of PBMC from acutely infected patient O1. (C) Chromium release cytolysis (using HLA-A24-positive or -negative B-cell targets) of HLA-A24 FYTPLADQF-specific CD8+ T-cell clone O22 clone 1. The peptide concentration for the left panel was 10−6 M, and the E:T ratio in the right panel was 1:1.
Figure 3B shows that restricted TCR BV5.1 usage was observed in patient O1 from the very first sample time (1 month after symptom development). Indeed, although a huge increase in tetramer-positive T cells was observed subsequently, the percentage of BV5.1 fell slightly.
HLA-A*2402 FYTPLADQF-specific T-cell clones and clonal lines from unrelated individuals use BV5.1 and BJ2.1, with a conserved CDR3 length and amino acid usage.
T-cell lines were derived by stimulation with FYTPLADQF for 10 days ex vivo (8); CTL clones were derived from patients O9 and O22 by limiting dilution, and long-term CTL lines were derived from patient O9 and remotely infected donor OR4 by repeated ex vivo stimulation with peptide-pulsed irradiated autologous B cells (2). CTL clones were CD8+ FYT/HLA-A*2402 tetramer positive, and BV5.1 antibody positive (data not shown). CTL clones killed at low effector/target (E:T) ratios and at nM peptide concentrations, as shown for O22 clone 4 in Fig. 3C. RNA was extracted from either cell-sorted or magnetic bead-enriched cell populations (to exclude feeder cell contamination) using an RNeasy mini kit (QIAGEN, United Kingdom). Reverse transcription was carried out using the Moloney murine leukemia virus reverse transcriptase (Stratagene, Amsterdam, The Netherlands) and oligo(dT)(12-18) primers (Life Technologies, Heidelberg, Germany) according to the manufacturer's instructions. PCR was carried out using Pfu polymerase (Stratagene, Amsterdam, The Netherlands) and BV5.1 (5′ATACTTCAGTGAGACACAGAGAAAC3′) and Cβ (5′TTCTGATGGCTCAAACAC3′) primers. PCR products were gel purified, and DNAs were extracted using a QIAGEN gel extraction kit before either bulk sequencing or subcloning into Topo (Invitrogen) and clone sequencing. All sequences were confirmed by reverse sequencing.
Tables 1 and 2 display the nucleotide and predicted amino acid sequences of the TCR BV CDR3 region sequences of HLA-A*2402-restricted FYTPLADQF-specific T-cell lines and clones. For the O9 lines, both a pool and 30 molecular clone sequences were obtained. Even though molecular cloning showed the line to be polyclonal, the CDR3 length was absolutely conserved even at this stage, and 29/30 clones used the BJ2.1 segment. Seven clones from individual O22 were sister clones with the same nucleotide sequence. The RV1 line amino acid sequence was identical to that obtained from the O22 clones, despite differences at the nucleotide level. This both excludes PCR contamination and suggests antigen-driven selection. All sequences from all three individuals had CDR3 regions of the same length (nine amino acids), and all but one used JB2.1. This region is critical in the recognition of bound peptide in several TCR/major histocompatibility complexes (reviewed in reference 5).
TABLE 1.
TCR beta chain CDR3 region nucleotide sequences of FYT A24 CD8+ cell lines and clones from individuals O9, O22, and RV1
| Cell line or clone | Sequencea |
|---|---|
| O9 cell line pool | ACCCCTAGCGGGGGATACAATGAGCAG |
| O9 cell lines | |
| A 14/30 | ACCCCTAGCGGGGGATACAATGAGCAG |
| B 4/30 | AGCAGTGCCGGGGGGTACAATGAGCAG |
| C 3/30 | AGCTTGTCAGGGGGCTACAATGAGCAG |
| D 1/30 | AGCTTGTCAGGGGGCTACAATGAGCAG |
| E 1/30 | AGCTTAAGTGGGGGCTACAATGAGCAG |
| F 1/30 | AGCTTTGCCGGGGGGTACAATGAGCAG |
| G 1/30 | AGCTTTAGCGGGGGTTACAATGAGCAG |
| H 1/30 | ACCCCTGGCGGGGGATACAATGAGCAG |
| I 1/30 | AATCTAGCGGGAGGGTACAATGAGCAG |
| J 1/30 | AGCTTGGCCGGAGGGTTCACTGAAGCT |
| K 1/30 | ACCTTTAGCGGGGGTTACAATGAGCAG |
| L 1/30 | AGCTCGGGGGGAGGGTACAATGAGCAG |
| O9 clone 1 | AGCTCGGGGGGAGGGTACAATGAGCAG |
| O9 clone 2 | ACCTTGGGTGGTGCCTACAATGAGCAG |
| O22 clone 1 | AGCTCCGCTGGGGGTTACAATGAGCAG |
| O22 clone 2 | AGCTCCGCTGGGGGTTACAATGAGCAG |
| O22 clone 3 | AGCTCCGCTGGGGGTTACAATGAGCAG |
| O22 clone 4 | AGCTCCGCTGGGGGTTACAATGAGCAG |
| O22 clone 5 | AGCTCCGCTGGGGGTTACAATGAGCAG |
| O22 clone 6 | AGCTCCGCTGGGGGTTACAATGAGCAG |
| O22 clone 7 | AGCTCCGCTGGGGGTTACAATGAGCAG |
| RV1 cell line pool | AGCTCGGCGGGGGGGTACAATGAGCAG |
For the O9 cell line, both a pool sequence and molecular clones were obtained. Letters in bold indicate nucleotides that are probably contributed by the BV5.1 gene segment, and underlining indicates nucleotides that are probably contributed by the BJ2.1 gene segment.
TABLE 2.
Predicted TCR beta chain CDR3 region amino acid sequences of FYT A24 CD8+ cell lines and clones from individuals O9, O22, and RV1
| Gene segment | Amino acid at CDR3 positiona:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 95 | 96 | 97 | 98 | 99 | 100 | 101 | 102 | 103 | |
| O9 cell line pool | T | P | S | G | G | Y | N | E | Q |
| O9 cell lines | |||||||||
| A 14/30 | T | P | S | G | G | Y | N | E | Q |
| B 4/30 | S | S | A | G | G | Y | N | E | Q |
| C 3/30 | S | L | S | G | A | Y | N | E | Q |
| D 1/30 | S | L | S | G | G | Y | N | E | Q |
| E 1/30 | S | L | S | G | G | Y | N | E | Q |
| F 1/30 | S | F | A | G | G | Y | N | E | Q |
| G 1/30 | S | F | S | G | G | Y | N | E | Q |
| H 1/30 | T | P | G | G | G | Y | N | E | Q |
| I 1/30 | N | L | A | G | G | Y | N | E | Q |
| J 1/30 | S | L | A | G | G | F | T | E | A |
| K 1/30 | T | F | S | G | A | Y | N | E | Q |
| L 1/30 | S | S | G | G | G | Y | N | E | Q |
| O9 clone 1 | S | S | G | G | G | Y | N | E | Q |
| O9 clone 2 | T | L | G | G | A | Y | N | E | Q |
| O22 clone 1 | S | S | A | G | G | Y | N | E | Q |
| O22 clone 2 | S | S | A | G | G | Y | N | E | Q |
| O22 clone 3 | S | S | A | G | G | Y | N | E | Q |
| O22 clone 4 | S | S | A | G | G | Y | N | E | Q |
| O22 clone 5 | S | S | A | G | G | Y | N | E | Q |
| O22 clone 6 | S | S | A | G | G | Y | N | E | Q |
| O22 clone 7 | S | S | A | G | G | Y | N | E | Q |
| RV1 cell line pool | S | S | A | G | G | Y | N | E | Q |
For the O9 cell line, both a pool sequence and molecular clones were obtained. Letters in bold indicate amino acids that are probably contributed by the BV5.1 gene segment, and underlining indicates amino acids that are probably contributed by the BJ2.1 gene segment. The TCR beta CDR3 loop is predicted to span amino acids 95 to 106, with a framework cysteine at position 92.
This striking TCR conservation is not a universal property of B19 infection, since other epitopes did not show TCR conservation, nor is it unique to B19 infection. However, although previous studies of other infections have identified striking oligoclonality, these have largely been in situations of long-term repetitive stimulation (cytomegalovirus and Epstein-Barr virus) or after recurrent challenge (influenza virus) (1, 9, 13). In the patients studied here, although ongoing antigenic stimulation appears to influence the magnitude and phenotype of the HLA-A*2402-restricted T-cell response over time, the BV usage was markedly restricted even during acute disease. This suggests that the selection of specific TCRs for this HLA-A*2402-restricted epitope occurs at a very early stage and also shows that the phenotypic changes observed do not result from the emergence of new T-cell clones. Potential mechanisms include structural constraints induced by the peptide-major histocompatibility complex combination, thymic constraints imposed by the self-peptide repertoire, and cross-reactivities with other immunogens (2, 4, 12). The functional avidity of this response has not been compared with those of other HLA-A*24-restricted responses but would appear to be in the upper normal range, as evidenced by killing at E:T ratios of 0.03:1. In our patient cohort, the tight restriction of TCR usage had no apparent deleterious effects, perhaps because B19 exhibits little sequence variation. However, narrow TCR repertoires may predispose individuals to viral escape, as described for chronic HCV infection (10).
In summary, seven of eight HLA-A*2402-positive individuals made vigorous responses to the HLA-A*2402-restricted epitope FYTPLADQF that were dominated by a conserved BV5.1 TCR. A highly focused T-cell response may aid in the rapid identification and elimination of even low levels of virus. This has important implications for vaccine development, particularly because HLA-A*2402 is not only the commonest East Asian HLA type but is likely part of a cross-presenting “supertype” that also includes A*2301 and A*3001 (3).
REFERENCES
- 1.Annels, N. E., M. F. Callan, L. Tan, and A. B. Rickinson. 2000. Changing patterns of dominant TCR usage with maturation of an EBV-specific cytotoxic T cell response. J. Immunol. 165:4831-4841. [DOI] [PubMed] [Google Scholar]
- 2.Bowness, P., P. A. Moss, S. Rowland-Jones, J. I. Bell, and A. J. McMichael. 1993. Conservation of T cell receptor usage by HLA B27-restricted influenza-specific cytotoxic T lymphocytes suggests a general pattern for antigen-specific major histocompatibility complex class I-restricted responses. Eur. J. Immunol. 23:1417-1421. [DOI] [PubMed] [Google Scholar]
- 3.Burrows, S. R., R. A. Elkington, J. J. Miles, K. J. Green, S. Walker, S. M. Harvana, D. J. Moss, H. Dunckley, J. M. Burrows, and R. Khanna. 2003. Promiscuous CTL recognition of viral epitopes on multiple human leukocyte antigens: biological validation of the proposed HLA A24 supertype. J. Immunol. 171:1407-1412. [DOI] [PubMed] [Google Scholar]
- 4.Burrows, S. R., R. Khanna, J. M. Burrows, and D. J. Moss. 1994. An alloresponse in humans is dominated by cytotoxic T lymphocytes (CTL) cross-reactive with a single Epstein-Barr virus CTL epitope: implications for graft-versus-host disease. J. Exp. Med. 179:1155-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia, K. C., L. Teyton, and I. A. Wilson. 1999. Structural basis of T cell recognition. Annu. Rev. Immunol. 17:369-397. [DOI] [PubMed] [Google Scholar]
- 6.Goulder, P. J., A. Edwards, R. E. Phillips, and A. J. McMichael. 1997. Identification of a novel HLA-A24-restricted cytotoxic T-lymphocyte epitope within HIV-1 Nef. AIDS 11:1883-1884. [DOI] [PubMed] [Google Scholar]
- 7.Ikeda-Moore, Y., H. Tomiyama, K. Miwa, S. Oka, A. Iwamoto, Y. Kaneko, and M. Takiguchi. 1997. Identification and characterization of multiple HLA-A24-restricted HIV-1 CTL epitopes: strong epitopes are derived from V regions of HIV-1. J. Immunol. 159:6242-6252. [PubMed] [Google Scholar]
- 8.Isa, A., V. Kasprowicz, O. Norbeck, A. Loughry, K. Jeffery, K. Broliden, P. Klenerman, T. Tolfvenstam, and P. Bowness. 2005. Prolonged activation of virus-specific CD8(+) T cells after acute B19 infection. PLoS Med. 2:e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawson, T. M., S. Man, S. Williams, A. C. Boon, M. Zambon, and L. K. Borysiewicz. 2001. Influenza A antigen exposure selects dominant Vbeta17+ TCR in human CD8+ cytotoxic T cell responses. Int. Immunol. 13:1373-1381. [DOI] [PubMed] [Google Scholar]
- 10.Meyer-Olson, D., N. H. Shoukry, K. W. Brady, H. Kim, D. P. Olson, K. Hartman, A. K. Shintani, C. M. Walker, and S. A. Kalams. 2004. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J. Exp. Med. 200:307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobao, Y., K. Sugi, H. Tomiyama, S. Saito, S. Fujiyama, M. Morimoto, S. Hasuike, H. Tsubouchi, K. Tanaka, and M. Takiguchi. 2001. Identification of hepatitis B virus-specific CTL epitopes presented by HLA-A*2402, the most common HLA class I allele in East Asia. J. Hepatol. 34:922-929. [DOI] [PubMed] [Google Scholar]
- 12.Stewart-Jones, G. B., A. J. McMichael, J. I. Bell, D. I. Stuart, and E. Y. Jones. 2003. A structural basis for immunodominant human T cell receptor recognition. Nat. Immunol. 4:657-663. [DOI] [PubMed] [Google Scholar]
- 13.Wang, E. C., P. A. Moss, P. Frodsham, P. J. Lehner, J. I. Bell, and L. K. Borysiewicz. 1995. CD8highCD57+ T lymphocytes in normal, healthy individuals are oligoclonal and respond to human cytomegalovirus. J. Immunol. 155:5046-5056. [PubMed] [Google Scholar]