Abstract
Perinatal human immunodeficiency virus type 1 (HIV-1) transmission is characterized by acquisition of a homogeneous viral quasispecies, yet the selective factors responsible for this genetic bottleneck are unclear. We examined the role of maternal autologous neutralizing antibody (aNAB) in selective transmission of HIV-1 escape variants to infants. Maternal sera from 38 infected mothers at the time of delivery were assayed for autologous neutralizing antibody activity against maternal time-of-delivery HIV-1 isolates in vitro. Maternal sera were also tested for cross-neutralization of infected-infant-first-positive-time-point viral isolates. Heteroduplex and DNA sequence analyses were then performed to identify the initial infecting virus as a neutralization-sensitive or escape HIV-1 variant. In utero transmitters (n = 14) were significantly less likely to have aNAB to their own HIV-1 strains at delivery than nontransmitting mothers (n = 17, 14.3% versus 76.5%, P = 0.003). Cross-neutralization assays of infected-infant-first-positive-time-point HIV-1 isolates indicated that while 14/21 HIV-1-infected infant first positive time point isolates were resistant to their own mother's aNAB, no infant isolate was inherently resistant to antibody neutralization by all sera tested. Furthermore, both heteroduplex (n = 21) and phylogenetic (n = 9) analyses showed that selective perinatal transmission and/or outgrowth of maternal autologous neutralization escape HIV-1 variants occurs in utero and intrapartum. These data indicate that maternal autologous neutralizing antibody can exert powerful protective and selective effects in perinatal HIV-1 transmission and therefore has important implications for vaccine development.
Acquisition of a homogeneous viral quasispecies is characteristic of perinatal human immunodeficiency virus type 1 (HIV-1) transmission (9, 16, 40), suggesting the presence of selective host pressures. Better understanding of such selective transmission could offer insight into potential protective mechanisms, inform HIV-1 vaccine development, and further the potential use of passive monoclonal antibody prevention regimens. Since maternal antibodies cross the placenta into the fetal bloodstream, perinatal transmission offers the unique opportunity to study potential prophylactic effects of an autologous neutralizing antibody (aNAB) present in both donor and recipient prior to virus exposure. Animal models indicate that antibody can reduce or prevent perinatal transmission of retroviruses (18, 20, 22, 35). The role of maternal neutralizing antibody in prevention of perinatal HIV-1 transmission, however, remains controversial (6, 26, 28, 32, 37). Conflicting reports may be due to limits in definitive data, small sample sizes, inconsistent selection of virus source, differences in HIV-1 gene region analyzed, use of widely disparate maternal and infant sample collection time points, and lack of differentiation of the timing of mother-to-child transmission.
Several small studies have suggested that virus isolates from infants are often resistant to maternal serum, suggesting transmission of maternal aNAB escape variants in some cases (29, 41). In order to better define the potential protective and/or selective roles of maternal HIV-1 aNAB in perinatal transmission, we performed a series of experiments, including measurement of maternal autologous neutralization capacity, along with a genetic analysis of maternal and perinatally transmitted viral strains in a large, prospectively monitored cohort of mother-infant pairs with timing of transmission defined as in utero or intrapartum (7). We also assessed the ability of transmitting mothers to neutralize their own babies' first positive HIV-1 isolate to address the question of whether a transplacentally acquired antibody might have activity against transmitted variants. Cross-neutralization assays were done to assess the breadth of maternal HIV-1 neutralization capacity and the inherent susceptibility/resistance of infant primary HIV-1 isolates at or near the time of delivery. Lastly, to determine if maternal aNAB escape strains are preferentially transmitted in utero and/or intrapartum, HIV-1 envelope gene regions from infected mother-infant pairs at their first positive time point were also compared by heteroduplex assay (11) and sequence analysis. Our results support both preventative and selective effects of maternal aNAB in perinatal transmission and indicate the need for further careful evaluation of antibody-mediated immunity in effective HIV-1 vaccine development.
(Part of this research was presented at the 11th Conference on Retroviruses and Opportunistic Infections, San Francisco, Calif., 2004 [abstract no. 429].)
MATERIALS AND METHODS
Study subjects.
The 38 seropositive mothers studied were monitored as part of a prospective study of maternal-fetal HIV-1 transmission conducted by the Los Angeles Pediatric AIDS Consortium between May 1989 and March 1996 (17). Informed consent and human subjects protocols were approved by the University of California at Los Angeles (UCLA) Institutional Review Board. The mothers were chosen as study participants based on sample availability, including at least one preterm and one time-of-delivery sample. Mothers were also chosen based on availability of samples from their infants from within 48 h of delivery and sufficient clinical follow-up of both the mothers and their infants. Primary HIV-1 culture isolates at the time of delivery from maternal blood samples and at the first positive time point from infant blood samples also had to be available for study participation. All available mother-baby (MB) pairs meeting these criteria were enrolled in our study. Samples were collected from patients with informed consent under the approval of the institutional review boards at each site participating in the study. Four of 21 transmitting mothers received oral zidovudine (ZDV; 500 mg/day) during gestation as part of their own health regimen, one of these mothers also received ZDV infusion during labor (2 mg/kg of body weight loading dose, followed by 1 mg/kg/h), and her infant was treated with ZDV (2 mg/kg/day) during the first 6 weeks following delivery. HIV-1-infected pregnant women received no other antiretrovirals. Two transmitting mothers and 14 nontransmitting mothers treated with ZDV during gestation had to be excluded from the study, because we were unable to culture HIV-1 from their blood at delivery. No other mother-baby pairs with available samples and follow-up were excluded. Samples were collected within 48 h of birth and at 2, 4, 6, 8, 10, and 12 weeks of age from 21 infants that were defined as infected following at least two positive HIV-1 cocultures from peripheral blood at two separate time points and confirmation of seropositive status beyond 15 months of age. According to the current working definition, infected infants with positive HIV-1 cocultures and/or PCRs within 48 h following birth were defined as infected “in utero,” while those with negative coculture/PCR within 48 h of birth and with subsequent positive cocultures/PCRs were defined as infected “intrapartum” (7). No infants were breast fed. Maternal and infant blood samples were collected by venipuncture into EDTA and heparin Vacutainer tubes (Becton Dickinson, Franklin Lakes, NJ).
HIV-1 culture and antibody neutralization assays.
Ficoll-Hypaque density gradient centrifugation was used to prepare peripheral blood mononuclear cells (PBMC) from heparin-treated samples collected from mothers at the time of delivery and from infants at their first positive time point. Mononuclear cells were washed with normal saline twice and enumerated, and cells not used immediately were stored under liquid nitrogen. HIV-1 cocultures were performed on 1 × 107 maternal or 1 × 106 infant PBMC according to the National Institutes of Health/National Institute of Allergy and Infectious Diseases Clinical Trials Group consensus protocol (1, 13). Supernatant harvested from the initial cocultures were expanded and titrated for viral 50% tissue culture infectious doses (TCID50), as detailed in the National Institutes of Health/National Institute of Allergy and Infectious Diseases Clinical Trials Group virology manual (1). Maternal sera collected at delivery were assayed for autologous neutralizing antibody activity in duplicate against virus cultured from maternal PBMC (MPBMC) collected at the same time point. Serum samples were heat inactivated for 30 min at 56°C prior to testing and serially twofold diluted from 1:10 to 1:2,560 in growth medium (RPMI 1640, 200 mM l-glutamine, 20% fetal calf serum, 10 U/ml interleukin-2, 50 U/ml penicillin, 100 μg/ml streptomycin). Hyperimmune HIV-1 gamma globulin and HIV-1-positive serum with a known neutralizing titer were included as positive controls in each run, with normal pooled HIV-1-negative human serum as a negative control. A volume of 100 μl of each maternal or control serum dilution was combined with an equal volume of growth media containing 100 TCID50 of maternal stock virus in 96-well microtiter plates in duplicate. After a 90-min incubation in a humidified incubator with 5% CO2 at 37°C, 2 × 105 phytohemagglutinin-stimulated PBMC were added to each well, and plates were incubated overnight. The next day the cells were washed twice with growth media and resuspended in 200 μl of the same media. On day 7, supernatants were harvested and diluted 1:10, and p24 antigen levels were determined by the Coulter kinetic assay (Miami, FL). Presence of HIV-1 neutralizing antibody was judged by a decrease in p24 antigen production compared to production in control wells incubated in the absence of maternal serum. The 50% neutralization titer (NT50) was calculated by the methods of Aubert and Montefiori et al. (3, 36). Sera were considered positive for HIV-1 neutralizing antibody if they had a titer greater than 10. Samples undetectable for aNAB were assigned an NT50 titer of 5 in the statistical analysis.
Heteroduplex assays.
PBMC and cultured cell DNA was prepared from maternal and infant samples by using QIAmp blood kits (QIAGEN, Valencia, CA). Heteroduplex mobility analysis was performed on samples as previously described by our group (16). Briefly, an approximately 690-bp fragment spanning the HIV-1 env V3 to V5 regions was amplified by nested PCR using Expand High-Fidelity Polymerase (Hoffman LaRoche, Nutley, NJ). A minimum of 30 HIV-1 proviral DNA copies, as determined by quantitative PCR (13, 15), were amplified in triplicate in the first-round PCR of each maternal and infant sample. First-round PCR products were combined and concentrated using QIAquick PCR columns (QIAGEN, Valencia, CA), and then all of the first round products were transferred to a second-round PCR for further amplification. HIV-1 env gene PCR products were denatured and reannealed, and heteroduplexes were resolved on 5% polyacrylamide gels at 250 V for 2.5 h. Gels were stained with ethidium bromide, UV illuminated, and digitally recorded with a Sony DSC70 Cybershot camera (Tokyo, Japan). For the heteroduplex tracking analysis (HTA), a single-stranded 32P-labeled DNA probe was generated from maternal neutralization culture cell DNA using primers REDE3 and REDE4 as previously described (10). One microliter of maternal neutralization escape probe (≥5,000 cpm/μl) was reannealed with approximately 250 ng of maternal or infant HIV-1 env PCR product generated for the heteroduplex mobility analysis (HMA) assays above by heating the probe-target mixture to 95°C for 1 min and then cooling on ice for up to 30 min. Neutralization escape probe-maternal-infant HIV-1 env DNA heteroduplexes were resolved on polyacrylamide gels as described above. Vacuum-dried gels were used to expose a PhosphorImager plate (Molecular Dynamics, Sunnyvale, CA) for image detection. Images from the digital camera and PhosphorImager were converted to TIFF files and transferred to the graphics program Canvas (Deneba, Miami, FL) for production of figures.
Cloning and sequencing of HIV-1 env gene regions.
Nested PCR products generated in the heteroduplex analyses described above were gel purified using the Geneclean kit (Qbiogene, Carlsbad, CA). V3 to V5 env gene regions were T/A cloned into the pGEM-T vector (Promega, Madison, WI). Ligations were performed according to the manufacturer's instructions and transformed into maximum-efficiency JM109-competent cells (Promega, Madison, WI). Transformed cells were plated onto Luria broth agar plates containing ampicillin (50 μg/ml), and white colonies were screened for inserts by PCR. A small amount of each white colony was picked up using a sterilized toothpick and placed in a tube with 10 μl double-distilled H20. Following lysis of the bacteria, debris was pelleted by microcentrifugation, and the supernatant was added to PCR master mix containing 1.25 mM deoxynucleoside triphosphate, 20 pmol each primers REDE3 and REDE4, 2 mM MgCl2+, GeneAmp Buffer II, and Taq DNA polymerase (Applied Biosystems, Foster City, CA). Samples were amplified in a DNA Thermal Cycler 9600 (Perkin-Elmer, Alameda, CA) for 30 cycles of 98°C for 10 s, 60°C for 10 s, and 72°C for 30 s followed by a final incubation at 72°C for 10 min. PCRs were screened for env gene products by agarose gel electrophoresis. Positive colonies were seeded into 5 ml Luria broth containing 50 μg/ml ampicillin and incubated overnight at 37°C with shaking, and clonal DNA was prepared using the QIAquick plasmid prep kit (QIAGEN, Valencia, CA). Fluorescent DNA sequencing of HIV-1 env gene region clones was performed using the ABI Big Dye Terminator Ready Reaction Cycle Sequencing kit, and sequences were read on an ABI Prism 377XL Automated DNA sequencing machine (Applied Biosystems, Foster City CA).
Phylogenetic analysis. (i) Neighbor-joining tree.
Nucleotide sequences were initially aligned using HMMER (version 2.3.1; Sean Eddy; http://hmmer.wustl.edu/), then codon aligned using GeneCutter (Brian Gaschen; http://www.hiv.lanl.gov/content/hivdb/GENE_CUTTER/cutter.html), and then manually adjusted. After gap stripping, 519 positions remained. Phylogenetic relationships were estimated using the F84 evolutionary model implemented in the Phylip neighbor program (Phylogeny Inference Package; Joe Felsenstein; http://evolution.genetics.washington.edu/phylip.html) with a transition\transversion ratio of 1.3. Two unrelated reference sequences (accession nos. K03455 and AF224507) were used as an outgroup. The reliability of branching orders was assessed by bootstrap analysis with 100 replicates (19).
(ii) Maximum likelihood trees.
A maximum likelihood tree was constructed for each mother-infant pair. After gap stripping, the alignments for each mother-infant pair were subjected to maximum likelihood analysis by the method described in Korber et al. (30). The same two reference sequences were used as outgroups (accession nos. K03455 and AF224507) for all trees. BranchLength.pl (B. Korber; www.santafe.edu/∼btk/sciencepaper/bette.html) was used to calculate the branch length to the infant's ancestral node for each maternal sequence.
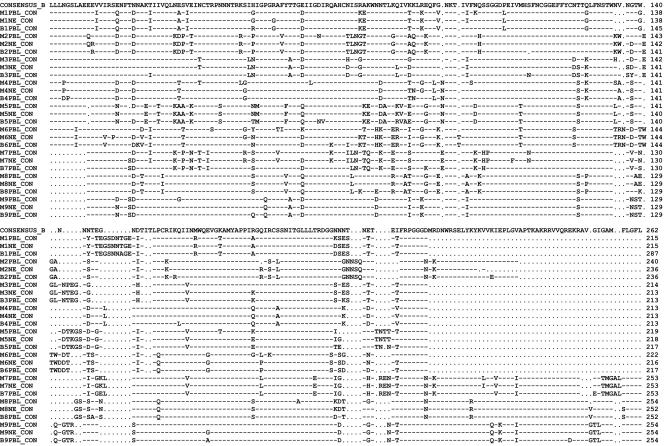
(iii) Consensus protein alignment of HIV-1 gp120 V3 to V5 env gene regions.
Nucleotide sequences were initially aligned using HMMER (version 2.3.1; Sean Eddy; http://hmmer.wustl.edu/), then codon aligned using GeneCutter (Brian Gaschen; http://www.hiv.lanl.gov/content/hiv-db/GENE_CUTTER/cutter.html), and then hand aligned to optimize. Shown is a comparison of V3 to V5 regions of a subtype B consensus sequence (top), consensus sequences for mother's time-of-delivery PBMC (MPBL), mother's neutralization escape (MNE), and infant's first-positive-time-point PBMC (BPBL) for each of the nine MB pairs analyzed.
Statistical analysis.
Descriptive statistics are provided as medians (25th to 75th percentile). Data were compared using the Mann-Whitney U and Fisher's exact tests as well as by logistic regression analysis. Branch length distances from maternal neutralization escape variants (MNE) and maternal PBMC (MPBMC) variants to their infant's ancestral nodes were compared by nonparametric Mann-Whitney matched pairs tests. The test included the average of the two distances to the different baby nodes in mother-baby pair 3. P values of less than or equal to 0.05 were considered significant.
Nucleotide sequence accession numbers.
Sequences determined in the course of this work were deposited in GenBank under accession numbers DQ526029 to DQ526372.
RESULTS
Study cohort.
Thirty-eight mother-infant pairs monitored in the UCLA cohort of the Los Angeles Pediatric AIDS Consortium between 1989 and 1996 were studied as part of a prospective study on maternal-fetal HIV-1 transmission (5, 17). All deliveries occurred prior to the routine use of zidovudine (ZDV) prophylaxis for the prevention of perinatal HIV-1 transmission (last delivery, March 1994). Mother-infant pairs were included in this study based on sufficient sample availability and follow-up during gestation, at delivery, and through 2 years after birth. Mother-infant pairs were excluded from the study based on a lack of sufficient samples (n = 28), loss of mother and/or infant to follow-up (n = 6), and inability to coculture HIV-1 from maternal PBMC at delivery (n = 16). All other eligible mother-infant pairs were enrolled in this study, including 21 transmitting and 17 nontransmitting pairs. Fourteen infected infants acquired HIV-1 in utero, and 7 acquired HIV-1 intrapartum (7).
Transmitting mothers had significantly higher plasma HIV-1 RNA levels at delivery than nontransmitting mothers (median [25th to 75th percentile] HIV-1 RNA copies/ml of 81,962 [28,711 to 226,305] versus 7,400 [2,898 to 17,254]; P < 0.0001). Mothers who transmitted HIV-1 to their infants also had significantly lower CD4+ T-cell counts at delivery (CD4+ cells/mm3, 368 [210 to 672] versus 667 [523 to 904]; P = 0.0007).
Maternal neutralization of autologous and infant HIV-1 isolates.
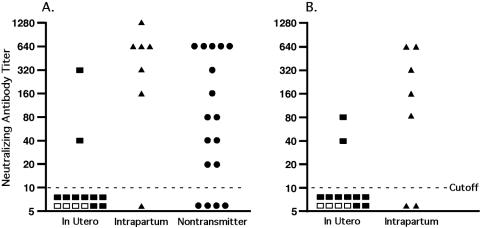
As seen in Fig. 1A, the ability of HIV-1-infected mothers to neutralize their own virus from the time of delivery was measured in vitro. In utero transmitters were significantly less likely to have aNAB to their own HIV-1 strains at delivery than nontransmitting mothers (14.3% versus 76.5%; P = 0.003). Intrapartum transmitters and nontransmitters did not differ in the rate of autologous neutralization (86% versus 76.5%; P = 0.61). The median (25th to 75th percentile) aNAB titer among the seven intrapartum transmitters (640 [160 to 640]) was also not significantly different from that of the 17 nontransmitters (80 [20 to 640]; P = 0.10). The median aNAB titer among in utero transmitters (5 [5 to 5]), however, was significantly lower than that of both intrapartum transmitters (P = 0.009) and nontransmitters (P = 0.007). Thus, women with low or undetectable aNAB are at a higher risk of transmitting HIV-1 to their infants in utero, suggesting that antibody may provide protective and/or selective effects in perinatal transmission.
FIG. 1.
Maternal HIV-1 neutralizing antibody titer at delivery. Maternal neutralizing antibody titer to autologous virus from the time of delivery (A) and maternal neutralizing antibody to the mother's infant's first-positive-time-point HIV-1 isolate (B) are shown. □, Mothers treated with ZDV.
The ability of each transmitting mother's serum to neutralize her own baby's HIV-1 isolate from the first positive time point was then measured in vitro. As seen in Fig. 1B, only 2 of 14 in utero transmitters were able to neutralize their own baby's HIV-1 isolate, versus 5 of 7 intrapartum transmitters (P = 0.017). Of the eight transmitting mothers in which aNAB was detectable, seven were also able to neutralize their own infant's first HIV-1 isolate. In five of these seven mothers, the neutralizing antibody titer to the infant's virus was lower than the mother's titer to her own isolate.
Breadth of maternal antibody neutralization of primary infant HIV-1 isolates.
In order to assess the breadth of maternal HIV-1 neutralization capacity and to study the inherent neutralization susceptibility/resistance of infant primary viral isolates, cross-neutralization assays were performed. Sufficient maternal time-of-delivery sera were available from 10 study mothers to perform cross-neutralization experiments on 20 of the study infant HIV-1 isolates. We were unable to generate a sufficient titer viral stock from one intrapartum-infected infant, excluding cross-neutralization analysis of this isolate. The cross-neutralization experiments were performed on infant first-positive-time-point HIV-1 isolates using maternal time-of-delivery sera from four in utero transmitters, two intrapartum transmitters, and four nontransmitters. The nontransmitting mothers showed broader neutralization capacity than the transmitting mothers (average of 95% of isolates neutralized versus 55%; P = 0.069). Nontransmitters also showed a higher median cross-neutralization titer than transmitters, although this trend did not reach significance (160 [40 to 320] versus 40 [20 to 160]; P = 0.063). Maternal cross-neutralization titers did not correlate significantly with maternal aNAB titers; however, the number of mothers with sufficient sera available to perform this analysis was small. All of the infant isolates tested were cross-neutralized by one or more maternal panel sera, indicating that none of the strains were inherently resistant to HIV-1 antibody neutralization.
Maternal HIV-1 env gene diversity in vivo and in vitro.
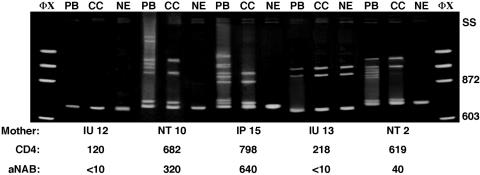
Heteroduplex mobility analysis (HMA) was first performed on all study mothers to determine if sufficient viral diversity is present in vivo in maternal PBMC and in vitro after culture to allow us to molecularly differentiate among aNAB-susceptible and -resistant HIV-1 strains. DNA was extracted from maternal time-of-delivery PBMC as well as from maternal PBMC coculture cells incubated in the presence and absence of autologous time-of-delivery serum. A 690-base sequence of the HIV-1 env gp120 gene, including the neutralizing domain of the V3 loop (23) through the V5 region, was PCR amplified from each DNA source and denatured, and HMA banding patterns were visualized following polyacrylamide gel electrophoresis (10, 16). The majority (87%) of HIV-1-infected mothers studied showed multiple, slowly migrating heteroduplexes formed from amplified PBMC and PBMC coculture DNA at delivery (Fig. 2), indicating a heterogeneous HIV-1 quasispecies. Both transmitting and nontransmitting mothers with detectable aNAB showed greater HIV-1 env gene diversity in PBMC and PBMC coculture-derived virus PCR products at delivery than mothers without detectable aNAB.
FIG. 2.
Heteroduplex mobility analysis of maternal HIV-1 env gene diversity. ΦX, DNA molecular weight markers. SS, single-stranded DNA. PB, maternal PBMC derived viral DNA PCR products; CC, maternal PBMC coculture cell-derived DNA PCR products; NE, maternal neutralization assay cell-derived DNA PCR products. The size (in base pairs) of the molecular weight markers is located to the right of the gel. IU, in utero transmitter; IP, intrapartum transmitter; NT, nontransmitter. CD4, maternal CD4 cell count. aNAB values are 50% neutralizing titers.
HMA of DNA extracted from maternal PBMC coculture cells indicated a reduction of viral diversity in vitro compared to that of uncultured PBMC, most likely due to the loss of replication-incompetent variants (Fig. 2). In vitro replication-competent HIV-1 strains were then tested in aNAB assays, and HMA was used to determine the env gene diversity of escape variants. Following incubation with autologous maternal time-of-delivery serum, a further loss in env gene diversity was observed. Thirty-three of 38 (87%) maternal aNAB cultures produced a single aNAB escape variant, i.e., a single, rapidly migrating homoduplex band, on an HMA gel. Four in utero transmitters and one intrapartum transmitter with no detectable aNAB had multiple aNAB escape variants detected in vitro by HMA. We could amplify maternal HIV-1 env gene PCR products from aNAB assay cells on all 38 mothers studied, indicating the lack of complete in vitro viral neutralization even in women with relatively high aNAB titers. Thus, HMA showed that mothers with a diverse HIV-1 quasispecies are infected with a combination of aNAB escape and -sensitive variants which can be differentiated by genetic analysis.
Selective transmission of maternal aNAB escape variants.
To determine if maternal aNAB escape variants are selectively transmitted to infants in utero and/or intrapartum, a genetic comparison of maternal time-of-delivery and infant first-positive-time-point HIV-1 strains was conducted. We analyzed the genetic relationship between maternal aNAB escape and paired infant HIV-1 variants amplified from PBMC DNA by single-stranded heteroduplex tracking analysis (HTA) (16, 38) on all transmitting mother-infant pairs in which the mother had a single HIV-1 aNAB escape variant (n = 16). In five cases where the mother had multiple aNAB escape variants, a single aNAB escape HTA probe could not be derived, so maternal and infant samples were compared by HMA. Results of both HTA and HMA indicated that the majority of both the in utero (10/14)- and intrapartum (5/7)-infected infants had a single identifiable HIV-1 variant at their first positive time points. As the majority of the transmitters had multiple env variants at delivery, these data indicate that a transmission bottleneck exists in both in utero and intrapartum perinatal transmission.
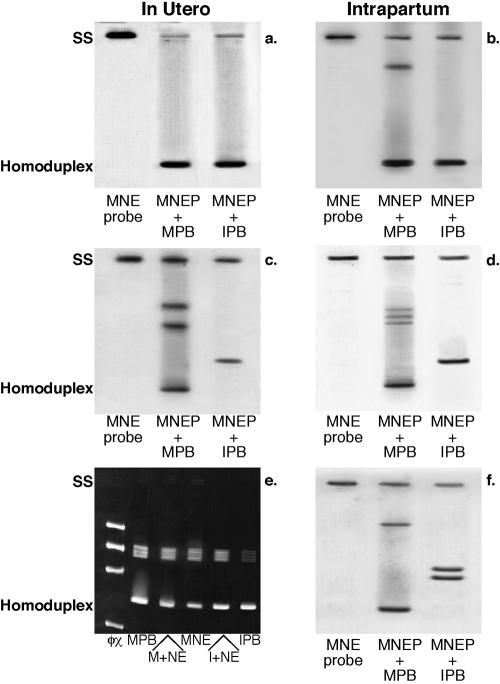
For the HTA analysis, single-stranded env gene PCR products were amplified from maternal aNAB assay culture cell DNA and labeled with 32P. The HTA assay results in the formation of a single rapidly migrating homoduplex when maternal aNAB escape probe and infant HIV-1 env DNA PCR products are ≥97% identical (10). Lack of close genetic identity between PCR products combined and analyzed by single-stranded HTA or HMA is indicated by the formation of new, unique slowly migrating heteroduplexes. Figure 3 contains representative examples of heteroduplex analyses from 6 of the 21 infected mother-infant pairs. The aNAB escape variants from 12 of 14 in utero transmitting mothers formed rapidly migrating homoduplexes and/or did not form any novel heteroduplexes when paired maternal and infant DNAs were denatured and allowed to reanneal in the heteroduplex analysis (Fig. 3a and e). None of these 12 in utero aNAB escape variant transmitting mothers had measurable aNAB to their own isolate at delivery. In contrast to the in utero transmitters, only one of seven intrapartum transmitters infected her infant with an HIV-1 variant which formed a rapidly migrating homoduplex with the maternal aNAB escape variant (Fig. 3b). Interestingly, this intrapartum transmitter was also the only one to lack detectable aNAB to her own virus isolate from the time of delivery. In six of seven intrapartum (Fig. 3d and f) and two in utero infections (Fig. 3c), the HIV-1 env sequences from the infants' first positive time point formed unique heteroduplex bands upon annealing with maternal aNAB escape variant probes.
FIG. 3.
Heteroduplex comparison analysis of maternal neutralization escape and infant PBMC HIV-1 DNA. SS, single-stranded DNA. MNE, maternal aNAB escape; MPB, maternal PBMC-derived HIV-1 DNA PCR products; IPB, infant PBMC-derived HIV-1 DNA PCR products. (a) MB pair 10, aNAB < 10; (b) MB pair 4, aNAB < 10; (c) MB pair 8, aNAB = 40; (d) MB pair 11, aNAB = 640; (e) MB pair 14, aNAB < 10; (f) MB pair 16, aNAB = 160. MNEP, maternal neutralization escape probe; M+NE, maternal PBMC HIV-1 DNA mixed with maternal NE probe; I+NE, infant PBMC HIV-1 DNA mixed with maternal NE probe.
In total, heteroduplex analysis confirmed transmission of one or more maternal aNAB escape variant strains in 13 of 21 infected mother-infant pairs studied in which the mother lacked detectable aNAB at delivery. The identity of the transmitted HIV-1 variant in the eight study pairs in which the mothers had detectable aNAB at delivery was less clear following heteroduplex analysis. HMA/HTA banding patterns did not identify the transmitted variant(s) in these cases as a ≥97% match with either maternal aNAB escape or aNAB-sensitive variants. The lack of a close match between the infants' first HIV-1 env variant and any of the mothers' PBMC and/or aNAB escape variants noted in several pairs suggests that transmission of a minor maternal variant and/or transmission of a variant arising from a source other than maternal PBMC may have occurred. An alternative hypothesis to this is that HIV-1 env gene evolution occurring in the mothers and/or infected infants soon after transmission resulted in viral diversification to the point that a ≥97% identical genetic maternal viral env gene match could not be found by heteroduplex analysis. As the lack of a good maternal-infant HIV-1 env gene match was noted only in MB pairs where the mother possessed detectable aNAB (primarily intrapartum transmitters), our data suggest that maternally derived aNAB may promote viral evolution in infected infants in utero or soon after birth. In order to more carefully characterize the genetic relationship between maternal and infant HIV-1 variants, a more detailed viral phylogenetic sequence analysis was undertaken for nine mother-infant pairs with transmission patterns representative of all of those observed by heteroduplex analysis. Five in utero and four intrapartum transmitting pairs were chosen, including five pairs in which the mother had aNAB at delivery (two in utero and three intrapartum) and four pairs (three in utero and one intrapartum) in which maternal aNAB was not detected.
Perinatal transmission of maternal aNAB HIV-1 variants.
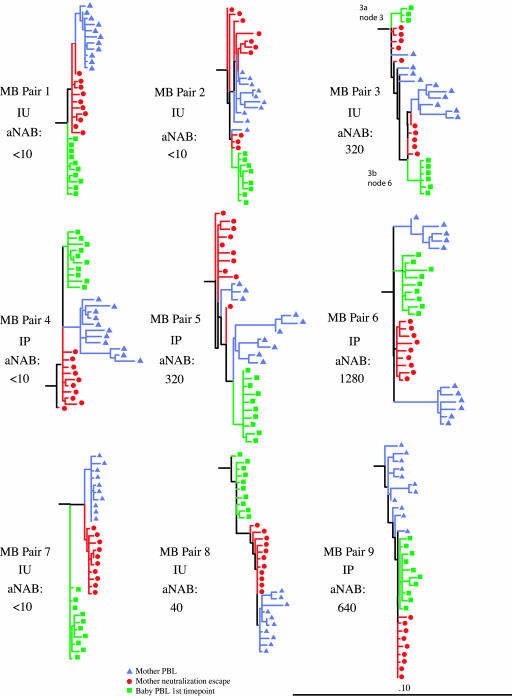
HIV-1 env gene DNAs generated by PCR of maternal PBMC, maternal aNAB escape viruses, and infant PBMC for the heteroduplex analyses were cloned into pGEMT and sequenced. Sequences were compiled, and a single neighbor-joining tree was constructed to check for sequence overlap. The neighbor-joining tree and bootstrap values verified separate clustering of the nine mother-infant pairs (data not shown). Thus, no evidence of sequence contamination was found. Clustering within each pair was also evident. Maximum likelihood trees were then constructed for each of the nine mother-infant pairs for use in statistical data analyses (Fig. 4). For each maximum likelihood tree constructed, the branch length distances from the infant node of origin to all maternal env sequences were determined and compared. The shortest distance to the infant node was found to be from one of the mother's aNAB escape variants in all nine of the nine pairs analyzed. Comparison of these distances in a Mann-Whitney matched pairs test confirmed statistical significance (P = 0.008) (Table 1). In addition, a comparison of the median distance from aNAB escape or -sensitive variant maternal sequences to their infant's node of origin indicated aNAB escape variant sequences were generally more closely related to the infants' in eight of nine pairs (P = 0.012) (Table 1). Although reported by others (33, 39), we did not find a correlation between N-linked glycosylation site number acquisition or loss with either neutralization escape or transmission (data not shown).
FIG. 4.
Maximum likelihood trees. Nucleotide sequences from maternal time-of-delivery PBMC (blue), maternal time-of-delivery aNAB culture cells (red), and infant first-positive-time-point PBMC (green). Horizontal branch lengths are drawn to scale, with the black bar representing 10% divergence. IU, in utero; IP, intrapartum.
TABLE 1.
Branch length distances from maternal HIV-1 sequences to baby ancestral nodes
| MB pair no. | Branch length distance
|
|||
|---|---|---|---|---|
| Minimum
|
Median
|
|||
| MNEa | MPBMCb | MNEa | MPBMCb | |
| 1 | 0.00213 | 0.00642 | 0.00428 | 0.00845 |
| 2 | 0.00395 | 0.00689 | 0.00692 | 0.01233 |
| 3 (node a) | 0.00571 | 0.01196 | 0.00951 | 0.02250 |
| 3 (node b) | 0.00590 | 0.01302 | 0.01076 | 0.01836 |
| 4 | 0.00891 | 0.00993 | 0.01091 | 0.01140 |
| 5 | 0.00769 | 0.01399 | 0.01590 | 0.01839 |
| 6 | 0.00805 | 0.01234 | 0.00943 | 0.02664 |
| 7 | 0.00891 | 0.00993 | 0.01091 | 0.00993 |
| 8 | 0.00924 | 0.01265 | 0.01192 | 0.01560 |
| 9 | 0.00108 | 0.00328 | 0.00218 | 0.00774 |
HIV-1 env sequences of maternal neutralization escape variants (MNE).
HIV-1 env sequences of maternal PBMC (MPBMC) variants.
The data also showed that the HIV-1 envelope gene gp120 regions from some mother-baby pairs were more closely related than others. In several cases where mothers possessed aNAB to their own virus at delivery, both heteroduplex and sequence analyses found greater genetic distance between mother and infant virus strains (Fig. 3c, and d, and 4, MB pair 5). Figure 5 contains consensus sequence alignments for all nine MB pairs sequenced in this study. The sequences for the intrapartum transmitting MB pair 5 indicate that the 5′ end of the virus genome containing the V3 loop of the baby's virus at 4 weeks of age matches the mother's aNAB escape variant at delivery closely. However, within the V4 to V5 region at the 3′ end of the virus, the infants' sequences match those of a single, minor maternal PBMC, non-aNAB escape variant sequence, a pattern suggestive of recombination (data not shown). In addition, mutations unique to infant virus strains were found in the env sequences from baby 5 and, to a lesser extent, in all infants analyzed at the first positive time point (Fig. 5). Thus, minor maternal variants undetected due to compartmentalization or sampling limitations can be transmitted, and/or viral evolution occurs in infected infants immediately after HIV-1 acquisition. The fact that more variation was found at the first positive time point in infected infants born to aNAB-positive women suggests that maternal aNAB promotes HIV-1 evolution very early in infection. It remains unclear, therefore, if mutations associated with aNAB escape were preselected in the initial infecting virus and/or if maternally derived aNAB drove acquisition of these mutations in the infant immediately after infection.
FIG. 5.
Consensus protein alignment of HIV-1 gp120 V3 to V5 env gene regions. Shown is a comparison of regions V3 through V5 of a subtype B consensus sequence (top) and consensus sequences for mother time-of-delivery PBMC (MPBMC), mother neutralization escape (MNE), and infant first-positive-time-point PBMC (BPBL) for each of the nine MB pairs analyzed. Dashes indicate conserved residues relative to the subtype B consensus; dots indicate deleted residues; substitutions are indicated by the amino acid letter. ConB, clade B consensus sequence.
DISCUSSION
Our analysis of the largest cohort of perinatally infected mother-infant pairs with defined timing of transmission indicated the presence of selective pressures resulting in a transmission bottleneck both in utero and intrapartum. Genetic comparison of maternal and infant HIV-1 gp120 envelope gene regions by heteroduplex and/or sequence analyses strongly suggests a role for maternal aNAB in the selective transmission and/or outgrowth of escape variants. These data do not rule out other selective factors in perinatal HIV-1 transmission, such as CD8+ T-cell responses (2, 24, 25). Indeed, we noted that among five transmitting mothers with multiple aNAB escape variants present at delivery, only one transmitted all of her identifiable escape variants to her infant.
Our results do not indicate the exact mechanism(s) by which maternal aNAB prevents infection with sensitive variants in the infant. Maternal aNAB may act to prevent exposure of the infant to sensitive variants in utero. However, as maternal antibody crosses the placenta beginning at 18 weeks of gestation, peaks at delivery, and persists in the infant for up to 18 months of age, it may also act to prevent or modify HIV-1 infection in the infant. Previous studies in neonatal macaques challenged with simian HIV (SHIV) have shown that neutralizing monoclonal antibodies given to the pregnant dams prior to delivery and/or to the newborn monkeys can provide partial protection against subsequent oral challenge with pathogenic SHIV 89.6P (4, 27). Complete protection by monoclonal neutralizing antibody postexposure prophylaxis against SHIV 89.6P challenge in neonatal macaques has also been reported in some cases (20). Thus, maternal aNABs may have dual protective effects that help to decrease the risk of perinatal transmission both in utero and intrapartum.
In our study, lack of maternal aNAB was primarily associated with in utero perinatal HIV-1 transmission. Intrapartum transmission may be influenced by delivery factors such as prolonged labor, duration of ruptured membranes, and infant exposure to maternal blood (5, 31, 34), which may overcome the protective effect of maternal aNAB and allow transmission to occur. In addition, cell-to-cell transmission of virus could occur despite the presence of maternal aNAB. While the majority of intrapartum transmitting mothers in our study could neutralize their own and their baby's HIV-1 isolates, phylogenetic analysis indicated a close degree of relatedness of the transmitted variant to the mother's aNAB escape env variant at delivery. Sequence analysis of MB pairs transmitting in the presence of maternal aNAB showed that the HIV-1 variant transmitted had features of both maternal aNAB-sensitive strains and aNAB escape strains, suggesting the potential transmission of a rare maternal variant and/or a recombinant virus. A more likely explanation, however, is that these mothers transmitted an aNAB-sensitive variant which quickly evolved to acquire aNAB escape mutations similar to those of the mother's virus under pressure from transplacentally acquired maternal aNAB. Transplacental antibody with any neutralizing activity against the infecting strain could still modify primary viremia and disease progression in the infant. There are data to support this from previous studies of this cohort showing that intrapartum-infected infants have significantly slower disease progression compared to that of in utero-infected infants (13, 14). Thus, our results indicate that maternal aNAB can have both protective and selective effects, resulting in prevention of transmission or slowing of disease progression in infected infants.
Our results of selective perinatal HIV-1 transmission of aNAB escape variants contrast with a report of heterosexual transmission in infected adults in Zambia, which indicated preferential transmission of aNAB-hypersensitive variants (12). A recent study of sexual HIV-1 transmission among men having sex with men, however, reported transmission of variants with enhanced neutralization sensitivity in only two of eight pairs (21). In addition, previous studies evaluating the ability of mothers to neutralize their baby's early HIV-1 isolates have suggested preferential transmission of antibody escape strains (29, 37), and a recent molecular study of intrapartum transmission of non-clade B strains also indicated selective transmission of aNAB escape strains (41). Unlike adults with primary infection, perinatally exposed infants acquire maternal antibody prior to birth, providing an opportunity for HIV-1 aNAB to exert preventative/selective pressures immediately upon infant viral exposure. Thus, perinatal HIV-1 transmission is more similar to that of vaccinated individuals with neutralizing antibody present prior to viral challenge which may be protective or, if failing to protect, may modify the course of disease progression. Sequence data from our study indicated preferential infection by aNAB escape variants but also showed that intrapartum-infected infants' first viral isolates shared some sequence characteristics with maternal aNAB-sensitive strains. These data suggest that mechanisms of intrapartum viral transfer may differ from those in utero. Thus, viral characteristics may differ by the mode of transmission, suggesting that vaccine/passive antibody trials focusing on prevention of acute infection should treat HIV-1 sequences derived from acutely infected infants and adults as separate categories for both vaccine design and neutralizing antibody assay reagent selection. Our observations that mothers with detectable aNAB titers were less likely to transmit HIV-1 to their infants in utero and that the infecting strain was more closely related to maternal aNAB escape variants suggest that neutralizing antibody is an important immune correlate of protection from HIV-1 infection. In addition, while we found clear evidence of selective perinatal transmission of aNAB escape variants, no infant first viral isolate tested was found to be innately resistant to antibody neutralization in vitro (8). This observation is critical for the proposed combined vaccine/passive monoclonal antibody trials for prevention of HIV-1 transmission by breast feeding. Our results underscore the urgent need for further research into the protective role of neutralizing antibody in all forms of HIV-1 transmission, providing hope for the development of successful vaccine and passive antibody approaches to HIV-1.
Acknowledgments
This research was supported in part by grants HD30629 and HD26621 from the National Institute of Child Health and Development, by grant ACTG AI32440 from the National Institute of Allergy and Infectious Diseases, by Universitywide AIDS Research Program grants K97-LA-101 and R99-LA-042, by Clinical Research Center grant RR-00-865, and by the Pediatric AIDS Foundation, Santa Monica, Calif.
We thank Steve Wolinsky and Brian Gaschen for their advice and technical assistance. We also thank Mary Ann Dillon, Audra Deveikis, Margaret Keller, Karin Nielsen, and the patients who participated in the study.
REFERENCES
- 1.AIDS Clinical Trials Group Virology Technical Advisory Committee. 1994. ACTG virology manual for HIV laboratories. Division of AIDS, National Institute of Allergy and Infectious Diseases, Nationl Institutes of Health, Bethesda, Md.
- 2.Altfeld, M., T. M. Allen, X. G. Yu, M. N. Johnston, D. Agrawal, B. T. Korber, D. C. Montefiori, D. H. O'Connor, B. T. Davis, P. K. Lee, E. L. Maier, J. Harlow, P. J. Goulder, C. Brander, E. S. Rosenberg, and B. D. Walker. 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature 420:434-439. [DOI] [PubMed] [Google Scholar]
- 3.Aubert, M. F. 1996. Laboratory techniques in rabies. World Health Organization, Geneva, Switzerland.
- 4.Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, P. J., M. Dillon, M. Navaie, A. Deveikis, M. Keller, S. O'Rourke, and Y. J. Bryson. 1994. Factors predictive of maternal-fetal transmission of HIV-1. Preliminary analysis of zidovudine given during pregnancy and/or delivery. JAMA 271:1925-1930. [PubMed] [Google Scholar]
- 6.Bryson, Y. J., D. Lehman, E. Garratty, R. Dickover, S. Plaeger-Marshall, and S. O'Rourke. 1993. The role of maternal autologous neutralizing antibody in prevention of maternal fetal HIV-1 transmission. J. Cell Biochem. Suppl. 17E:95. [Google Scholar]
- 7.Bryson, Y. J., K. Luzuriaga, J. L. Sullivan, and D. W. Wara. 1992. Proposed definitions for in utero versus intrapartum transmission of HIV-1. N. Engl. J. Med. 327:1246-1247. [DOI] [PubMed] [Google Scholar]
- 8.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 9.Contag, C. H., A. Ehrnst, J. Duda, A. B. Bohlin, S. Lindgren, G. H. Learn, and J. I. Mullins. 1997. Mother to infant transmission of human immunodeficiency virus type 1 involving five envelope sequence subtypes. J. Virol. 71:1292-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delwart, E. L., H. Pan, H. W. Sheppard, D. Wolpert, A. U. Neumann, B. Korber, and J. I. Mullins. 1997. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J. Virol. 71:7498-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delwart, E. L., E. G. Shpaer, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 12.Derdeyn, C. A., J. M. Decker, F. Bibollet-Ruche, J. L. Mokili, M. Muldoon, S. A. Denham, M. L. Heil, F. Kasolo, R. Musonda, B. H. Hahn, G. M. Shaw, B. T. Korber, S. Allen, and E. Hunter. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019-2022. [DOI] [PubMed] [Google Scholar]
- 13.Dickover, R. E., M. Dillon, S. Gillette, A. Deveikis, M. Keller, S. Plaeger-Marshall, I. Chen, A. Diagne, and Y. J. Bryson. 1994. Rapid increases in HIV-1 load correlate with early disease progression and loss of CD4 cells in vertically infected infants. J. Infect. Dis. 170:1279-1284. [DOI] [PubMed] [Google Scholar]
- 14.Dickover, R. E., M. Dillon, K. M. Leung, P. Krogstad, S. Plaeger, S. Kwok, C. Christopherson, A. Deveikis, M. Keller, E. R. Stiehm, and Y. J. Bryson. 1998. Early prognostic indicators in primary perinatal human immunodeficiency virus type 1 infection: importance of viral RNA and the timing of transmission on long-term outcome. J. Infect. Dis. 178:375-387. [DOI] [PubMed] [Google Scholar]
- 15.Dickover, R. E., R. M. Donovan, E. Goldstein, S. Dandekar, C. E. Bush, and J. R. Carlson. 1990. Quantitation of human immunodeficiency virus DNA by using the polymerase chain reaction. J. Clin. Microbiol. 28:2130-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickover, R. E., E. M. Garratty, and Y. J. Bryson. 2001. Perinatal transmission of major, minor and multiple maternal human immunodeficiency virus type 1 variants in utero and intrapartum. J. Virol. 75:2194-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickover, R. E., E. M. Garratty, S. A. Herman, M. S. Sim, S. Plaeger, P. J. Boyer, M. Keller, A. Deveikis, E. R. Stiehm, and Y. J. Bryson. 1996. Identification of levels of maternal HIV-1 RNA associated with risk of perinatal transmission: effect of maternal zidovudine treatment on viral load. JAMA 275:599-605. [PubMed] [Google Scholar]
- 18.Emini, E. A., W. A. Schleif, J. H. Nunberg, A. J. Conley, Y. Eda, S. Tokiyoshi, S. D. Putney, S. Matsushita, K. E. Cobb, C. M. Jett, et al. 1992. Prevention of HIV-1 infection in chimpanzees by gp120 V3 domain-specific monoclonal antibody. Nature 355:728-730. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein, J. 1992. Estimating effective population size from samples of sequences: a bootstrap Monte Carlo integration method. Genet. Res. 60:209-220. [DOI] [PubMed] [Google Scholar]
- 20.Ferrantelli, F., R. A. Rasmussen, K. A. Buckley, P. L. Li, T. Wang, D. C. Montefiori, H. Katinger, G. Stiegler, D. C. Anderson, H. M. McClure, and R. M. Ruprecht. 2004. Complete protection of neonatal rhesus macaques against oral exposure to pathogenic simian-human immunodeficiency virus by human anti-HIV monoclonal antibodies. J. Infect. Dis. 189:2167-2173. [DOI] [PubMed] [Google Scholar]
- 21.Frost, S. D., Y. Liu, S. L. Pond, C. Chappey, T. Wrin, C. J. Petropoulos, S. J. Little, and D. D. Richman. 2005. Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J. Virol. 79:6523-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Girard, M., B. Meignier, F. Barre-Sinoussi, M. P. Kieny, T. Matthews, E. Muchmore, P. L. Nara, Q. Wei, L. Rimsky, K. Weinhold, et al. 1995. Vaccine-induced protection of chimpanzees against infection by a heterologous human immunodeficiency virus type 1. J. Virol. 69:6239-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorny, M. K., K. Revesz, C. Williams, B. Volsky, M. K. Louder, C. A. Anyangwe, C. Krachmarov, S. C. Kayman, A. Pinter, A. Nadas, P. N. Nyambi, J. R. Mascola, and S. Zolla-Pazner. 2004. The v3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J. Virol. 78:2394-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 25.Goulder, P. J., C. Pasquier, E. C. Holmes, B. Liang, Y. Tang, J. Izopet, K. Saune, E. S. Rosenberg, S. K. Burchett, K. McIntosh, M. Barnardo, M. Bunce, B. D. Walker, C. Brander, and R. E. Phillips. 2001. Mother-to-child transmission of HIV infection and CTL escape through HLA-A2-SLYNTVATL epitope sequence variation. Immunol. Lett. 79:109-116. [DOI] [PubMed] [Google Scholar]
- 26.Hengel, R. L., M. S. Kennedy, R. W. Steketee, D. M. Thea, E. J. Abrams, G. Lambert, J. S. McDougal, et al. 1998. Neutralizing antibody and perinatal transmission of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 14:475-481. [DOI] [PubMed] [Google Scholar]
- 27.Hofmann-Lehmann, R., J. Vlasak, R. A. Rasmussen, B. A. Smith, T. W. Baba, V. Liska, F. Ferrantelli, D. C. Montefiori, H. M. McClure, D. C. Anderson, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, H. Katinger, G. Stiegler, L. A. Cavacini, M. R. Posner, T. C. Chou, J. Andersen, and R. M. Ruprecht. 2001. Postnatal passive immunization of neonatal macaques with a triple combination of human monoclonal antibodies against oral simian-human immunodeficiency virus challenge. J. Virol. 75:7470-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Husson, R. N., Y. Lan, E. Kojima, D. Venzon, H. Mitsuya, and K. McIntosh. 1995. Vertical transmission of human immunodeficiency virus type 1: autologous neutralizing antibody, virus load, and virus phenotype. J. Pediatr. 126:865-871. [DOI] [PubMed] [Google Scholar]
- 29.Kliks, S. C., D. W. Wara, D. V. Landers, and J. A. Levy. 1994. Features of HIV-1 that could influence maternal-child transmission. JAMA 272:467-474. [PubMed] [Google Scholar]
- 30.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 31.Kuhn, L., R. W. Steketee, J. Weedon, E. J. Abrams, G. Lambert, M. Bamji, E. Schoenbaum, J. Farley, S. R. Nesheim, P. Palumbo, R. J. Simonds, and D. M. Thea. 1999. Distinct risk factors for intrauterine and intrapartum human immunodeficiency virus transmission and consequences for disease progression in infected children. Perinatal AIDS collaborative transmission study. J. Infect. Dis. 179:52-58. [DOI] [PubMed] [Google Scholar]
- 32.Lathey, J. L., J. Tsou, K. Brinker, K. Hsia, W. A. Meyer III, and S. A. Spector. 1999. Lack of autologous neutralizing antibody to human immunodeficiency virus type 1 (HIV-1) and macrophage tropism are associated with mother-to-infant transmission. J. Infect. Dis. 180:344-350. [DOI] [PubMed] [Google Scholar]
- 33.Lue, J., M. Hsu, D. Yang, P. Marx, Z. Chen, and C. Cheng-Mayer. 2002. Addition of a single gp120 glycan confers increased binding to dendritic cell-specific ICAM-3-grabbing nonintegrin and neutralization escape to human immunodeficiency virus type 1. J. Virol. 76:10299-10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandelbrot, L., M. J. Mayaux, A. Bongain, A. Berrebi, Y. Moudoub-Jeanpetit, J. L. Benifla, N. Ciraru-Vigneron, J. Le Chenadec, S. Blanche, J. F. Delfraissy, et al. 1996. Obstetric factors and mother-to-child transmission of human immunodeficiency virus type 1: the French perinatal cohorts. Am. J. Obstet. Gynecol. 175:661-667. [DOI] [PubMed] [Google Scholar]
- 35.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 36.Montefiori, D. C., T. S. Hill, H. T. Vo, B. D. Walker, and E. S. Rosenberg. 2001. Neutralizing antibodies associated with viremia control in a subset of individuals after treatment of acute human immunodeficiency virus type 1 infection. J. Virol. 75:10200-10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scarlatti, G., T. Leitner, V. Hodara, E. Halapi, P. Rossi, J. Albert, and E. M. Fenyo. 1993. Neutralizing antibodies and viral characteristics in mother-to-child transmission of HIV-1. AIDS 7(Suppl. 2):S45-S48. [DOI] [PubMed] [Google Scholar]
- 38.Sodora, D. L., F. Lee, P. J. Dailey, and P. A. Marx. 1998. A genetic and viral load analysis of the simian immunodeficiency virus during the acute phase in macaques inoculated by the vaginal route. AIDS Res. Hum. Retrovir. 14:171-181. [DOI] [PubMed] [Google Scholar]
- 39.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 40.Wolinsky, S. M., C. M. Wike, B. T. Korber, C. Hutto, W. P. Parks, L. L. Rosenblum, K. J. Kunstman, M. R. Furtado, and J. L. Munoz. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134-1137. [DOI] [PubMed] [Google Scholar]
- 41.Wu, X., A. B. Parast, B. A. Richardson, R. Nduati, G. John-Stewart, D. Mbori-Ngacha, S. M. Rainwater, and J. Overbaugh. 2006. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol. 80:835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]