Abstract
KIF14 is a microtubule motor protein whose elevated expression is associated with poor-prognosis breast cancer. Here we demonstrate KIF14 accumulation in mitotic cells, where it associated with developing spindle poles and spindle microtubules. Cells at later stages of mitosis were characterized by the concentration of KIF14 at the midbody. Time-lapse microscopy revealed that strong RNA interference (RNAi)-mediated silencing of KIF14 induced cytokinesis failure, causing several rounds of endoreduplication and resulting in multinucleated cells. Additionally, less efficacious KIF14-specific short interfering RNAs (siRNAs) induced multiple phenotypes, all of which resulted in acute apoptosis. Our data demonstrate the ability of siRNA-mediated silencing to generate epiallelic hypomorphs associated with KIF14 depletion. Furthermore, the link we observed between siRNA efficacy and phenotypic outcome indicates that distinct stages during cell cycle progression are disrupted by the differential modulation of KIF14 expression.
Kinesins comprise a superfamily of motor proteins that impact a wide array of cellular functions by coupling ATP hydrolysis to the regulated and targeted movement of specific intracellular cargo along microtubule filaments. Kinesins have been functionally linked to various biological phenomena, including, but not limited to, cargo-containing vesicle transport, mitotic spindle formation, chromosome segregation, midbody formation, and cytokinesis completion (16, 22). All kinesins contain a highly conserved motor domain, within which resides the globular catalytic core that contains both microtubule and nucleotide binding sites that function together to generate the microtubule-stimulated ATPase activity (26). In addition, the motor domain contains a highly conserved neck region (adjacent to the catalytic core) that can be subdivided into a neck linker and a neck coiled-coil (CC) region (23). The neck linker serves to determine motor directionality (3), and the neck CC can facilitate oligomer formation (18, 25). The kinesin superfamily has been subdivided into 14 kinesin families (14) and can be further defined by the position of the motor domain at the N terminus (N type), C terminus (C type), or internal region (I type) (23).
KIF14 is a mammalian kinesin classified as an N-type, kinesin-3 family member based upon phylogenetic sequence analysis of its presumptive motor domain (16, 19). Our interest in KIF14 was piqued by the observation that elevated KIF14 expression is associated with poor-prognosis breast cancer (28). However, due to the limited functional annotation for KIF14 and its Drosophila orthologs, it is difficult to speculate on how KIF14 may contribute to breast cancer prognosis. Separate studies characterizing KLP38B, which is the KIF14 ortholog in Drosophila melanogaster, ascribed different functions to this kinesin. One study reported a role for KLP38B in chromosome segregation (17), while another reported a role in cytokinesis (21). In addition, the silencing of KIF14 in human cells by using endoribonuclease-prepared short interfering RNA (esiRNA) was reported to disturb chromosome congression and alignment, resulting in a prolonged delay at the metaphase-to-anaphase transition (33).
In this report, we have characterized the mitotic expression, subcellular localization, and function of human KIF14. We establish that the KIF14 motor domain exhibits microtubule-dependent ATPase activity and that KIF14 manifests the elevated mitotic expression and subcellular localization properties of a mitotic kinesin. In addition, siRNA-mediated silencing of KIF14 produced distinct phenotypes that could be linked to siRNA efficacy. Strong silencing of KIF14 prevented midbody cleavage, resulting in the formation of multinucleated cells. Less efficacious KIF14 siRNAs were found to elicit apoptosis of cells following entry into mitosis, during an attempt at cytokinesis, after binucleation due to cytokinesis failure, or after completion of cell division. Therefore, the data demonstrate that the silencing of KIF14 generates a multitude of mitotic phenotypes in which the efficacy of KIF14 silencing dictates the stage at which cell cycle progression is disrupted.
MATERIALS AND METHODS
Antibodies.
Antibodies used for immunofluorescence are as follows: polyclonal anti-KIF14 antibody (Abcam, Inc.; ab3746) and monoclonal anti-alpha-tubulin antibody (Sigma; clone DM1A0). KIF14 was detected using Alexa Fluor 488 antibody (Invitrogen). Alpha tubulin was detected using Alexa Fluor 594 (Invitrogen) goat anti-mouse immunoglobulin G antibody.
Cell culture and transfections.
HeLa-S3, HCT116, and HEK-293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen) at 37°C. For siRNA transfections, cells were seeded in six-well (9.60 cm2) culture dishes at a densities of 60,000 to 90,000 cells/well in 2 ml serum containing growth medium. Twenty-four hours after seeding, cells were transfected with 100 pmol of siRNA using Oligofectamine (Invitrogen) transfection reagent according to the manufacturer's protocol. Cells were incubated with transfection complexes under normal growth conditions until collection for analysis at specific time points posttransfection. All siRNA duplexes (Table 1) were purchased from Dharmacon (Lafayette, CO). Plasmid transfections of HEK-293T cells were performed on cells plated at 2 × 104 cells per well in an eight-well chamber slide (Lab-Tek) and cultured overnight in 10% fetal bovine serum (Invitrogen). Cells were transfected the following day with 1 μg plasmid DNA per well using TransIT 293 reagent (Mirus) in Opti-MEM medium (Invitrogen) according to the manufacturer's protocol and incubated overnight. The colony-forming capacity of siRNA-transfected HeLa-S3 cells was evaluated by plating cells at 100, 500, and 1,000 cells per well 48 h posttransfection followed by 2 weeks of uninterrupted culture before staining with crystal violet.
TABLE 1.
Sense sequence of siRNA duplexes
| Gene | Sense sequence |
|---|---|
| Luciferase | CGUACGCGGAAUACUUCGATT |
| KIF14 M-1 | AAACUGGGAGGCUACUUACTT |
| KIF14 M-2 | GUGAGUAUUAUCCCAGUUGTT |
| KIF14 S-1 | GUUGGCUAGAAUUGGGAAATT |
| KIF14 S-2 | CAGGGAUGCUGUUUGGAUATT |
| KIF14 S-3 | GGGAUUGACGGCAGUAAGATT |
EGFP-tagged KIF14 (KIF14-EGFP) cloning.
The KIF14 open reading frame was PCR amplified from cDNA clone KIAA0042 (purchased from Kazusa DNA Research Institute) by using the following primers: KIF14-forward (5′-CTAGAAACCGTCCGGAATGTCATTACACAGTACTCAT-3′) and KIF14-reverse (5′-CTAGAAACGCGGATCCCACCCACTGAATCCTACTGG-3′). The PCR product was restriction digested and ligated into BspE1/BamHI-digested enhanced green fluorescent protein (EGFP) fusion vector pEGFP-C1 (BD Biosciences).
Immunoblot analysis of KIF14 expression.
Transfected cells were trypsinized, collected by centrifugation (5 min at 300 × g), and washed with phosphate-buffered saline. Cell pellets were resuspended in lysis buffer (20 mM Tris HCl [pH 7.6], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) containing 1× protease inhibitor mix (Roche Complete) and incubated for 10 min on ice. Following centrifugation (10 min at 10,000 × g), the supernatant was collected and the protein concentration was determined by using the Bio-Rad DC protein assay kit. Equivalent protein (25 μg/sample) was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on Bio-Rad ready gels (7.5% acrylamide or 4 to 15% acrylamide gradient). Proteins were transferred to a nitrocellulose membrane and blocked in TBST buffer (150 mM NaCl, 10 mM Tris-HCl [pH 7.6], 0.1% Tween 20) containing 5% nonfat dry milk (blocking buffer) for 30 min at room temperature. The membrane was then incubated with anti-KIF14 antibody (Abcam, Inc.; ab3746) diluted 1:1,000 in blocking buffer for 90 min at room temperature. After three washes in TBST, the membrane was incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G antibody (Zymed) diluted 1:10,000 in blocking buffer for 45 min at room temperature. Following three washes in TBST, the membrane was incubated in chemiluminescence detection reagents (ECL Plus, Amersham) and the image was captured using a charge-coupled device camera (Kodak Image Station 440CF).
Immunofluorescence microscopy.
HeLa-S3 and 293T cells cultured on glass chamber well slides were fixed and permeabilized as described previously (12) for 15 min in immunohistochemical buffer. Following fixation, cells were washed with TBST and incubated with primary antibody for 90 min at 37°C. Cells were washed in TBST prior to secondary antibody addition and incubation at room temperature for 1 h followed by washes in TBST supplemented with 10 μg/ml Hoechst in order to stain DNA. Samples were mounted with Fluoromount G (Southern Biotech) and visualized directly on a DeltaVision (version 3.5) deconvolution microscope (Applied Precision, Inc.) or an InCell Analyzer 1000 (GE Healthcare) by using DAPI (4′,6′-diamidino-2-phenylindole), fluorescein isothiocyanate, and rhodamine-Texas Red-phycoerythrin filter sets for blue, green, and red, respectively.
Time-lapse microscopy.
Live cells were monitored in eight-well coverslip bottom chamber slides beginning at 20 or 40 h posttransfection via time-lapse image capture using n a DeltaVision version 3.5 deconvolution microscope (Applied Precision, Inc.). The microscope was fitted with a humidified 37°C/5% CO2 incubation chamber (Solent Scientific) to maintain a suitable environment for sustained image collection. Bright-field images of 1,024 by 1,024 or 512 by 512 pixels (see video S3 in the supplemental material) were collected at 5-min time intervals. Multiple coordinates per treatment from the chamber wells were collected through a 20× lens objective, magnified to ×30 with a ×1.5 magnifier, for an overall duration ranging from 20 to 40 h. Select time intervals were linked to generate MPEG movies using SoftWoRx software (Applied Precision, Inc.).
Cell synchronization by thymidine block.
A double-thymidine block was used to synchronize HCT116 cells seeded in 10-cm plates at 1.5 × 106 cells per plate and grown overnight as previously described (29). After the second thymidine block, cells were washed and put into deoxycytidine medium; this process was designated as time zero. Samples were collected for flow cytometry, and RNA extraction was performed at 2-h intervals from time zero to 24 and 36 h.
Molecular profiling of synchronized HCT116.
Cell lysates were homogenized using QIAshredder spin columns and total cellular RNA was isolated using the RNeasy mini kit (QIAGEN). RNA amplification, labeling, and hybridization to hu25K ink-jet DNA microarrays was carried out as previously described (10, 28).
Cell cycle analysis.
To analyze cell cycle profiles, approximately 1 × 105 to 5 × 105 cells were harvested along with their accompanying medium and pelleted by centrifugation in order to obtain both adherent and detached cells for subsequent analysis. Ethanol-fixed cells were prepared for flow cytometry as previously described (24). For each sample, 10,000 events were collected using a FACSCalibur flow cytometer (Becton Dickinson) and incorporation of propidium iodide was used as a marker for DNA content. Aggregated cells were gated out, and cell cycle profiles were analyzed using FlowJo cytometry analysis software (version 4.0.2).
Expression and purification of recombinant KIF14 motor domain.
A KIF14 gene fragment encoding amino acids V342 through to K720 was PCR amplified from cDNA clone KIAA0042 and cloned into pET22b (Novagen) using NdeI and XhoI sites engineered into the PCR primers. Escherichia coli BL21 (DE3) was transformed and cultured for 50 h at 18°C in LB broth containing 2 mM MgCl2 and 50 μg/ml carbenicillin. Harvested cells (20 g) were suspended in buffer (20 mM Tris-Cl [pH 8.0], 300 mM NaCl, 0.1% Tween, 10 mM imidazole, 2 mM MgCl2, 5 mM β-mercaptoethanol) and lysed by French press. The sample was clarified and batch bound to 0.3 ml nickel-nitrilotriacetic acid (QIAGEN). Bound proteins were eluted by applying a step gradient of lysis buffer containing imidazole and analyzed by SDS-PAGE. Fractions were pooled, diluted 10-fold with cation exchange buffer (50 mM HEPES [pH 6.8], 1 mM MgCl2, 1 mM EDTA, 10 μM ATP, 1 mM dithiothreitol), and applied to a 1.0-ml HiTrap SP HP (Amersham Biosciences). Protein was eluted with a linear gradient to 750 mM KCl and analyzed by SDS-PAGE. Pooled fractions were diluted with buffer to 200 mM KCl, concentrated in a Centricon-30 concentrator, and spiked with glycerol to 10% vol/vol for storage at −70°C.
ATPase activity assay.
Taxol-stabilized microtubules were prepared as described previously (13) and stored at 20 μM at room temperature. ATPase activity reactions (50 μl) contained 50 mM (PIPES) piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 7.0), 1 mM EGTA, 1 mM dithiothreitol, 100 μg/ml bovine serum albumin, 2 mM MgCl2, 1 mM NaATP, 150 nM KIF14 motor domain, and differing amounts of taxol-stabilized microtubules. Reactions were incubated for 0 to 20 min at room temperature and quenched with 50 μl stop solution (1.8 M KCl, 50 mM EDTA). Propidium iodide was detected by addition of Quinaldine Red-ammonium molybdate.
RESULTS
KIF14 contains an internally positioned N-type motor that exhibits microtubule-dependent ATPase activity.
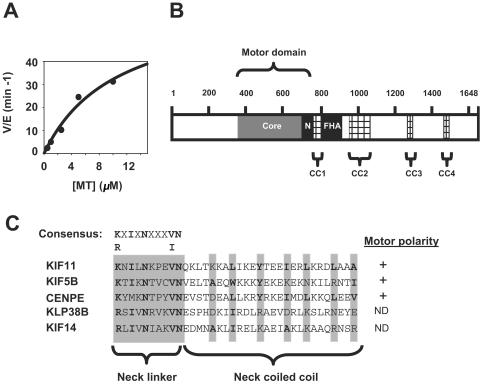
A defining characteristic of kinesin motor domains is their microtubule-dependent ATPase activity. We assayed the ATPase activity of KIF14 in vitro by using recombinant KIF14 motor domain. As shown in Fig. 1A, the intrinsic ATPase activity of the KIF14 motor is enhanced upon the addition of taxol-stabilized microtubules. Notably, the internal location within the polypeptide of the KIF14 motor, 5-prime of four coiled-coil domains and a forkhead-associated domain as detailed in Fig. 1B, indicates an I-type classification for this kinesin. However, the KIF14 neck sequence is carboxy terminal and contains conserved plus-end-directed determinants (26), explaining its phylogenetic classification as an N-type kinesin (Fig. 1C) (16).
FIG. 1.
KIF14 contains an internal motor domain with plus-end-directed determinants and microtubule-dependent ATPase activity. (A) The ATPase activity of a fragment corresponding to the KIF14 motor domain was assayed as described in Materials and Methods. The ATPase activity of the KIF14 motor (rate [V] per enzyme [E])/min is graphed as a function of microtubule (MT) concentration. (B) The polypeptide chain of KIF14 contains several distinct domains that are likely to be involved in regulating aspects of KIF14 function in vivo. The internal motor domain contains the highly conserved catalytic core (shown in dark gray) and the N-type conserved neck region (N). In addition to the coiled-coil region predicted to exist within the neck, KIF14 contains four additional regions predicted to form coiled-coil regions, CC1 to CC4 (hatched lines). The KIF14 forkhead-associated domain is bordered by CC1 and CC2. (C) The KIF14 neck linker contains the consensus N-type motor sequence K/RXIXNXXXV/IN found in other N-type kinesins (KIF11, KIF5B, and CENPE). In addition, KIF14 contains the N-type neck coiled-coil sequence φ-XX(X)-φ-XXX-φ-XX-φ, where φ denotes a highly conserved hydrophobic residue (shaded in gray and shown in bold type). The plus-end-directed classifications of both KIF14 and KLP38B are putative and have not been biochemically confirmed. +, positive motor polarity; ND, not determined.
KIF14 expression is elevated in mitotic cells.
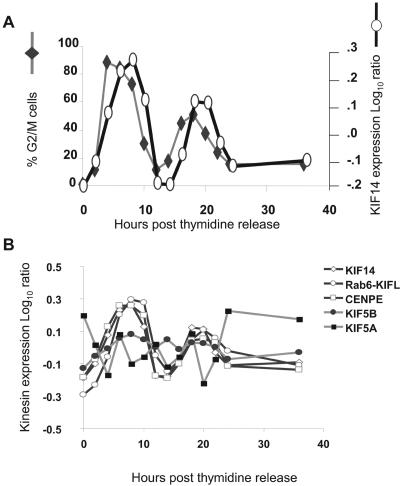
The annotation for KIF14 and its orthologs, although limited, is consistent with functions ascribed to mitotic kinesins (17, 21, 33). As a first step in further defining KIF14 as a mitotic kinesin, we examined whether KIF14 expression is elevated during mitosis as has been reported for several known mitotic kinesins (1, 5, 31). We used microarray profiling to analyze the expression of KIF14 mRNA in synchronized HCT116 cells (Fig. 2A). Since our human 25K gene chip contains probes corresponding to a number of mitotic and transport kinesins, we were able to compare the regulation of KIF14 expression to that of well-annotated kinesins. A double-thymidine block was used to synchronize HCT116 at the G1/S transition. Arrested cells were released by the removal of thymidine, and cells were harvested for microarray hybridization as described in Materials and Methods. In addition, cells were also collected for cell cycle analysis by flow cytometry in order to quantitate the percentage of cells at each stage of the cell cycle. As shown in Fig. 2A, KIF14 transcripts accumulated in cells progressing through G2/M and peak levels were achieved approximately 2 h after the detection of maximal DNA content. This cell cycle-dependent regulation more closely mirrored those of known mitotic kinesins CENPE and Rab6-KIFL than those of the nonmitotic kinesins KIF5A and KIF5B (Fig. 2B). These findings demonstrate that KIF14 expression is regulated in a manner consistent with it acting as a mitotic kinesin.
FIG. 2.
The mitotic accumulation of KIF14 and its cell cycle-regulated expression mirrors that of known mitotic kinesins. (A) Microarray profiling and cell cycle analysis were used to correlate changes in gene expression with distinct stages of the cell cycle in synchronized HCT116 cells sampled at 2-h intervals after release from a double-thymidine block. The percentage of cells resident within G2/M at each time point is defined by DNA content as determined by flow cytometry (primary y axis), and the relative amount of KIF14 mRNA at each time point is defined as the log10 ratio as determined by microarray analysis (secondary y axis). (B) The expression of various mitotic and nonmitotic kinesins was compared with that of KIF14.
Cellular localization of KIF14 during the cell cycle.
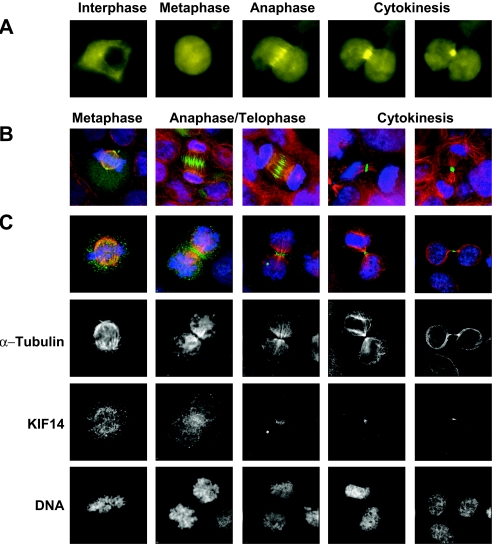
Characterization of the intracellular localization of specific mitotic kinesins to centrosomes (2), centromeres (30), and the midbody (5) have proven to be instrumental in deciphering where and how these kinesins function during mitosis. We therefore characterized the subcellular localization of both exogenously and endogenously expressed KIF14 throughout the cell cycle. Immunofluorescence microscopy was used to evaluate the localization of exogenously expressed KIF14-EGFP in 293T cells (Fig. 3A and B) and endogenous KIF14 in HeLa cells (Fig. 3C). KIF14-EGFP localization was restricted to the cytoplasm of interphase cells, while entry into mitosis triggered a redistribution of KIF14-EGFP to the nucleus (Fig. 3A). During prophase, KIF14 accumulated at the developing spindle poles and their associated microtubules; a similar distribution was observed in cells throughout metaphase (Fig. 3B and C). In contrast, during anaphase, KIF14 accumulated at the spindle midzone, and appeared increasingly concentrated at the midbody during telophase (Fig. 3A through C). In cells that were ready to undergo abscission, KIF14 was consolidated at the contractile ring (Fig. 3A through C) and colocalized with the midbody-associated proteins KNSL5, RhoA, and ECT2 (see Fig. S1 in the supplemental material).
FIG. 3.
Changes in KIF14 localization coincide with distinct cell cycle stages. Immunofluorescence microscopy was used to evaluate the localization of exogenously expressed KIF14-EGFP in HEK-293T cells and endogenously expressed KIF14 in HeLa cells. (A) Analysis of KIF14-EGFP in a single cell. (B) Localization of exogenously expressed KIF14-EGFP in separate cells at distinct stages of mitosis. (C) Localization of endogenously expressed KIF14 in separate cells at distinct stages of mitosis. DNA staining (Hoechst), blue channel; KIF14 localization, green channel; α-tubulin localization, red channel.
Distinct changes in ploidy are linked with KIF14 siRNA efficacy.
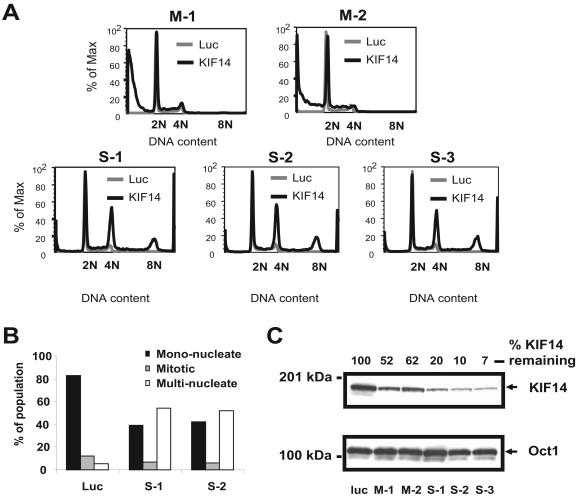
The concentration of KIF14 at the midbody raised the possibility that, in addition to its reported role in regulating chromosome congression and alignment (33), KIF14 might function during mammalian cell cytokinesis. Therefore, we expanded our examination of KIF14 function by using siRNAs to silence KIF14 in HeLa cells. Initial results suggested that different siRNAs elicited different phenotypes (unpublished data). To more firmly establish the cellular phenotypes resulting from KIF14 silencing, we evaluated several KIF14-specific siRNA duplexes. Total RNA isolated from cells 24 h posttransfection was analyzed using quantitative PCR, and only those siRNAs that reduced KIF14 mRNA levels by more than 50% (unpublished data) were selected for further study. Using these validated siRNAs, cell cycle profiles of HeLa cells at 72 h after transfection were generated and compared to those of HeLa controls transfected with a siRNA directed against luciferase (Fig. 4A).
FIG. 4.
Distinct changes in ploidy are linked with KIF14 siRNA efficacy. (A) HeLa cells transfected with KIF14-specific moderate and strong siRNAs, M-1, M2, S-1, S-2, and S-3, were analyzed by flow cytometry 72 h after transfection. The DNA content of KIF14-specific siRNA-transfected cells is compared to that of cells transfected with a luciferase-specific siRNA. (B) Cells transfected with either of two polyploid-inducing KIF14-specific siRNAs (S-1 or S-2) were analyzed by immunofluorescence microscopy at 72 h after transfection. Staining with α-tubulin and Hoechst allowed the evaluation of nuclear morphology of single cells. A total of 100 cells were evaluated for each siRNA duplex, and the number of mononuclear, multinuclear, and mitotic cells was compared to those of a luciferase (Luc) siRNA control. (C) HeLa cells were transfected with a panel of KIF14-specific siRNAs linked to different ploidy phenotypes as described in Fig. 4A. Cells were harvested 24 h after transfection, and equal amounts of protein were analyzed by immunoblotting with anti-KIF14 antibody. Cells transfected with an siRNA targeting luciferase, a gene not expressed in mammalian cells, served as the negative control. Those siRNAs that reduced KIF14 expression by less than 50% are labeled as moderate. Those siRNAs that reduced KIF14 expression more than 80% are labeled as strong. The percent of KIF14 remaining for each sample was calculated using Oct1 as a load control.
We observed two distinct ploidy phenotypes in HeLa cells in response to KIF14 silencing. Four of 11 validated siRNAs elicited an increase in hypoploid cells that was greatest by 3 days posttransfection (Fig. 4A) (unpublished data). An increase in annexin V staining preceded the increase in hypoploid cells in response to the silencing of KIF14 with these siRNAs, confirming death by apoptosis (unpublished data). The remaining seven siRNAs induced a phenotype that was associated with an increase in cells exhibiting tetraploid (4N) and polyploid (more than 4N) DNA content (Fig. 4A) (unpublished data) that was caused not by an increase in mitotic cells but by a marked accumulation of multinucleated cells (Fig. 4B and see Fig. S2 in the supplemental material). Importantly, a comparison of protein knockdown generated with a representative set of KIF14 siRNAs allowed for the segregation of these two phenotypes based upon siRNA efficacy (Fig. 4C). “Strong” (S) siRNAs that generated the robust depletion of KIF14 (80% or greater) were responsible for the induction of the hyperploid phenotype, while “moderate” (M) siRNAs that generated less severe KIF14 silencing (less than 50%) induced the hypoploid phenotype. These data clearly demonstrate that siRNA efficacy can be used to segregate distinct phenotypes associated with KIF14 depletion.
Time-lapse microscopy reveals multiple cell cycle defects associated with KIF14 silencing.
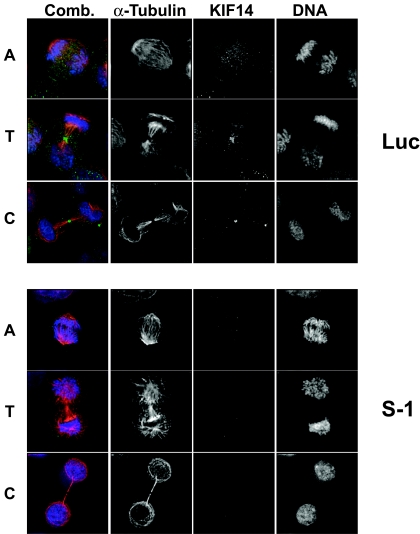
Time-lapse microscopy was utilized to characterize the fate of HeLa cells transfected with either a strong or a moderate KIF14 siRNA. The single prominent phenotype associated with strong silencing of KIF14 was failure to complete cytokinesis. Cells transfected with strong siRNAs were able to segregate chromosomes, proceed through anaphase, initiate furrow formation, ingress the furrow and elongate the midbody in manners similar to those of luciferase siRNA controls (Fig. 5 and 6; see Videos S1 and S2 in the supplemental material). However, midbody cleavage did not occur in KIF14-depleted cells, resulting in midbody collapse and binucleated cells (see Video S2 in the supplemental material). The only marked difference we observed between control and KIF14-depleted cells when staining for KIF14 was a significant reduction in the concentration of KIF14 at the midbody of late-stage mitotic cells (Fig. 5). The inability of KIF14-depleted cells to complete cytokinesis is consistent with the increase in ploidy observed by both flow cytometry and microscopy (Fig. 4B; see Fig. S2 in the supplemental material). In addition, extensive endoreduplication was toxic to HeLa cells in long-term colony-forming assays as multinucleate cells were observed to undergo apoptosis after subsequent failed attempts at mitosis (unpublished data).
FIG. 5.
Strong silencing of KIF14 eliminates midbody concentration of KIF14 but does not prevent chromosome segregation, progression to telophase, or furrow ingression. Immunofluorescence microscopy was used to evaluate chromosome segregation and furrow ingression in KIF14 siRNA-transfected HeLa cells. DNA staining (Hoechst), blue channel; KIF14 localization, green channel; α-tubulin localization, red channel. A, anaphase, T, telophase, C, cytokinesis.
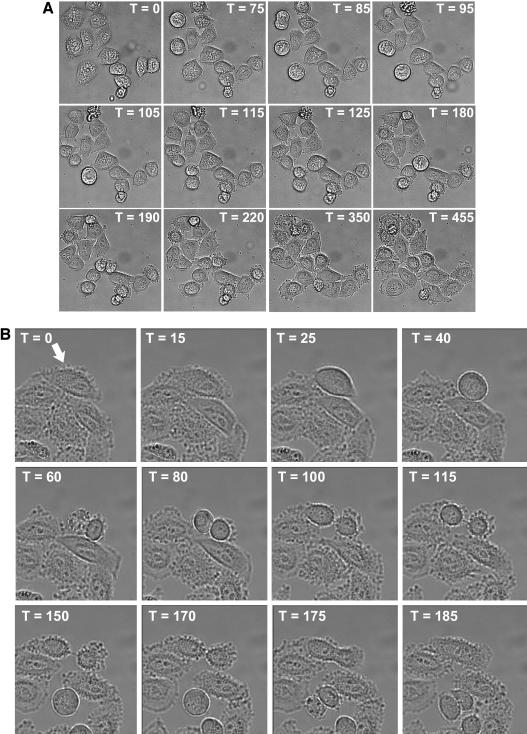
FIG. 6.
Strong silencing of KIF14 prevents midbody cleavage and induces cytokinesis failure. HeLa cells were transfected with either a luciferase-specific siRNA (A) or the S-1 KIF14-specific siRNA (B). Twenty hours posttransfection, time-lapse imaging was initiated and images were generated at 5 min intervals for 20 h. The elapsed time for each sequence of images is denoted in minutes with the initial image set at time (T) zero.
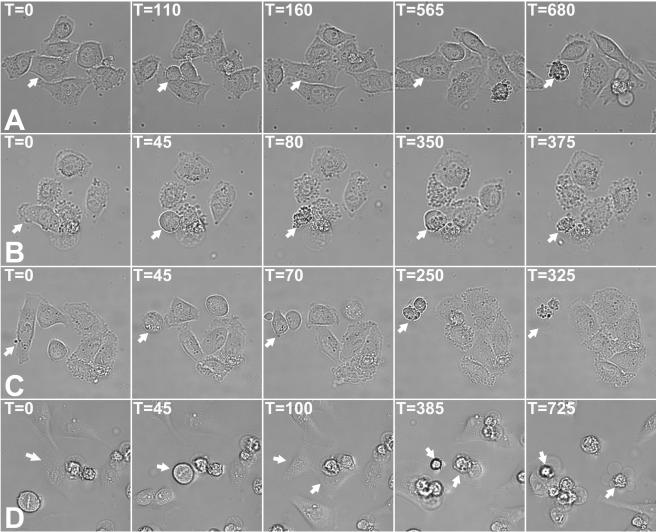
Moderate KIF14 siRNAs could be differentiated from stronger siRNAs by their inductions of multiple phenotypes. Whereas strong KIF14 siRNAs could induce several rounds of endoreduplication (see Video S3 in the supplemental material) prior to the induction of apoptosis, the most common phenotype associated with moderate KIF14 siRNAs was cytokinesis failure and binucleation followed by apoptosis that occurred after mitotic exit (Table 2 and Fig. 7A; see Video S4 in the supplemental material). In addition to this binucleation followed by apoptosis phenotype, moderate KIF14 silencing was also observed to induce apoptosis of cells upon mitotic entry (Table 2 and Fig. 7B; see Video S5 in the supplemental material) while attempting cytokinesis (Table 2 and Fig. 7C; see Video S6 in the supplemental material) or during interphase after completion of cytokinesis (Table 2 and Fig. 7D; see Video S7 in the supplemental material). Furthermore, the progression from prophase to telophase in cells transfected with either moderate or strong KIF14 siRNAs was similar to that of control cells and did not exceed 90 min (Fig. 6 and 7; unpublished data). This is in contrast to the several-hours-long delay at the metaphase-to-anaphase transition reported to occur in response to esiRNA-mediated silencing of KIF14 (33). Taken together, these data reveal a concentration-dependent requirement for KIF14 at distinct stages during the cell cycle.
TABLE 2.
Penetrance of apoptotic phenotypes associated with moderate KIF14 silencinga
| Cell cycle stageb | % Penetrance of:
|
|
|---|---|---|
| Luciferase | M-1 | |
| Interphase | 3 | 21 |
| Metaphase | 0 | 28 |
| Cytokinesis | 0 | 8 |
| Postbinucleation | 0 | 42 |
Time-lapse microscopy was utilized to determine the percentage of penetrance. HeLa cells were transfected with either a luciferase control siRNA or the moderate KIF14 siRNA M-1. Time-lapse was initiated 30 h posttransfection and was terminated 24 h later. The baseline level of apoptosis was determined by counting the number of apoptotic events that occurred within a population of 100 luciferase siRNA-transfected cells. For M-1 transfected cells, 75 apoptotic events were scored.
All apoptotic events were segregated based upon the stage of the cell cycle where apoptosis occurred.
FIG. 7.
Moderate silencing of KIF14 induces apoptosis at multiple points in the cell cycle. HeLa cells were transfected with M-1 KIF14-specific siRNA and time-lapse imaging was initiated at 20 h posttransfection (A, B, and C) or 40 h posttransfection (D). Apoptosis can be seen to occur after failed cytokinesis and binucleation (A), prior to progression beyond metaphase (B), during an attempt at cytokinesis (C), and after completion of cytokinesis (D). Images were generated at 5 min intervals for 20 h, and for each sequence of images, the initial image is denoted as time (T) zero.
DISCUSSION
The human genome contains 45 kinesins (16), and the functional characterization of each member of this motor protein superfamily remains a challenge. In this report, we have demonstrated that the putative KIF14 motor domain exhibits microtubule-dependent ATPase activity in vitro, confirming that KIF14 is in fact a kinesin. Our data also establish that, like other mitotic kinesins (31), KIF14 gene expression is under cell cycle regulation as an elevation of KIF14 transcripts coincides with mitotic progression. In addition, we have demonstrated that KIF14 localizes to the cytoplasm of interphase cells and redistributes to the nucleus, where it associates with the developing spindle in early mitotic cells. Coincident with the progression of cells beyond anaphase, KIF14 accumulates at the midbody, becoming concentrated at the contractile ring prior to the completion of cytokinesis in a manner that mimics the subcellular localization of kinesins known to regulate cytokinesis (1, 5, 9, 15, 32). We have determined that strong RNAi-mediated depletion of KIF14 can induce cytokinesis failure, resulting in binucleated cells. Furthermore, less efficacious KIF14 silencing expands the phenotypes associated with KIF14 depletion to include apoptosis that occurs upon entry into mitosis, while attempting cytokinesis, or after mitotic exit. However, it is important to note that the 90% reduction of KIF14 protein levels reported here, although significant, is not a substitute for gene ablation. Therefore, it is possible that complete elimination of KIF14 could cause additional phenotypes not detected in this study. Taken together, these data (i) indicate that KIF14 associates with the mitotic spindle and functions at multiple points during mitosis and (ii) clearly define KIF14 as a mitotic kinesin whose function is essential for cytokinesis. In addition, these data demonstrate that with RNAi, it is possible to elicit distinct phenotypes over a range of gene expression levels. This is reminiscent of the use of short hairpin RNA to induce epiallelic hypomorphs of TP53 in mice (8).
The effects of KIF14 silencing in HeLa cells that we observed differ from what was reported to occur in response to esiRNA-mediated silencing of KIF14 (33). Specifically, an esiRNA pool targeting KIF14 was shown to disrupt chromosome congression and alignment, resulting in a prolonged delay at the metaphase-to-anaphase transition (33). Moreover, this study did not report an increase in apoptosis or a defect in cytokinesis nor did a study that reported the absence of any mitotic phenotype in response to siRNA-mediated silencing of the KIF14 ortholog KLP38B in Drosophila S2 cells (7). However, mutants in KLP38B have been linked with cytokinesis failure (21) and defects in chromosome segregation (17). It is important to point out that, in the Drosophila S2 cell experiments, siRNA efficacy was not determined as experiments evaluating mRNA or protein knockdown were not reported (7). Additionally, the mRNA silencing reported for the KIF14 esiRNA pool used by Zhu et al. was approximately 60%, moderate by our definition, but how this reduction in mRNA affected protein levels was not evaluated (33). Furthermore, given the limitation that each siRNA duplex exhibits a unique “off-target” gene expression profile when introduced into a cell (11), the validation of phenotypes obtained by using siRNA pools is best confirmed by reproducing the phenotype with multiple single siRNAs.
Our experiments do not confirm a role for KIF14 in chromosome congression and alignment as we did not observe the metaphase-to-anaphase delay described by Zhu et al. despite testing several KIF14 siRNAs with different efficacies. Importantly, we show by using multiple siRNAs that, in response to strong KIF14 silencing, HeLa cells progress unimpeded through mitosis and are able to initiate furrow formation and ingression. However, these cells were unable to cleave the intracellular bridge connecting the dividing cells, resulting in midbody collapse, cell fusion, and binucleation. Together, these data support a role for KIF14 during cell abscission.
In addition to cytokinesis failure, we observed an increase in cell death in response to moderate KIF14 silencing that is preceded by an increase in annexin V staining confirming death by apoptosis (unpublished data). Time-lapse microscopy revealed that cell death in response to moderate KIF14 silencing occurred at three distinct points: prior to anaphase, during cytokinesis, or after mitotic exit. It is difficult to speculate as to why moderate KIF14 silencing is able to trigger this acute apoptotic phenotype, while the predominant, immediate effect caused by strong KIF14 silencing is cytokinesis failure. One possibility is that KIF14 may indirectly, through some yet-to-be-defined cargo, serve to enable the function of a mitotic sensor able to elicit an apoptotic signal in response to mitotic spindle abnormalities. Such a sensor function could operate throughout mitosis and would be diminished or eliminated as a consequence of strong KIF14 silencing. Importantly, the link we observed between siRNA efficacy and phenotypic outcome indicates that different stages of the cell cycle can be disrupted by changing the level of KIF14 expression and this could explain the diversity of phenotypes reported for both KIF14 (33) (this report) and its Drosophila ortholog (21) (17).
Our characterization of KIF14 as a mitotic kinesin may be functionally significant to its identification as a poor-prognosis breast cancer gene (28). Our data are consistent with the hypothesis that elevated expression of KIF14 serves as a reporter for highly proliferative cancers as increased proliferation has been shown to strongly correlate with poor-prognosis breast cancer (27). Alternatively, the recent identification of KIF14 as a candidate oncogene (4) raises the interesting possibility that mitotic spindle or cytokinesis defects that might result from KIF14 overexpression could lead to increased genetic instability and progression towards aneuploidy in certain cell types or genetic backgrounds (6, 20). It will be important to determine whether the elevated expression of KIF14 serves to promote oncogenesis, as this would warrant evaluation of KIF14 as a target for new antineoplastic compounds.
During the review of the manuscript, work by Gruneberg et al. was published demonstrating that KIF14 and citron kinase act together to promote cytokinesis (7a). Notably, this study demonstrated that the localization of KIF14 and citron kinase to the midbody is interdependent and that depletion of citron kinase induced cytokinesis failure. Given the findings of Gruneberg et al., it will be important to determine whether moderate silencing of citron kinase expression is able to phenocopy the effects of moderate KIF14 silencing we report here.
Supplementary Material
Acknowledgments
We thank David Wiest, Nelson Chau, and Marcia Gordon for critical review of the manuscript.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abaza, A., J. M. Soleilhac, J. Westendorf, M. Piel, I. Crevel, A. Roux, and F. Pirollet. 2003. M phase phosphoprotein 1 is a human plus-end-directed kinesin-related protein required for cytokinesis. J. Biol. Chem. 278:27844-27852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blangy, A., H. A. Lane, P. d'Herin, M. Harper, M. Kress, and E. A. Nigg. 1995. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83:1159-1169. [DOI] [PubMed] [Google Scholar]
- 3.Case, R. B., D. W. Pierce, N. Hom-Booher, C. L. Hart, and R. D. Vale. 1997. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell 90:959-966. [DOI] [PubMed] [Google Scholar]
- 4.Corson, T. W., A. Huang, M. S. Tsao, and B. L. Gallie. 2005. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene 24:4741-4753. [DOI] [PubMed] [Google Scholar]
- 5.Fontijn, R. D., B. Goud, A. Echard, F. Jollivet, J. van Marle, H. Pannekoek, and A. J. Horrevoets. 2001. The human kinesin-like protein RB6K is under tight cell cycle control and is essential for cytokinesis. Mol. Cell. Biol. 21:2944-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujiwara, T., M. Bandi, M. Nitta, E. V. Ivanova, R. T. Bronson, and D. Pellman. 2005. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437:1043-1047. [DOI] [PubMed] [Google Scholar]
- 7.Goshima, G., and R. D. Vale. 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Gruneberg, U., R. Neef, X. Li, E. H. Chan, R. B. Chalamalasetty, E. A. Nigg, and F. A. Barr. 2006. KIF14 and citron kinase act together to promote efficient cytokinesis. J. Cell Biol. 172:363-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemann, M. T., J. S. Fridman, J. T. Zilfou, E. Hernando, P. J. Paddison, C. Cordon-Cardo, G. J. Hannon, and S. W. Lowe. 2003. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat. Genet. 33:396-400. [DOI] [PubMed] [Google Scholar]
- 9.Hill, E., M. Clarke, and F. A. Barr. 2000. The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J. 19:5711-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes, T. R., M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Shoemaker, R. Stoughton, A. P. Blanchard, S. H. Friend, and P. S. Linsley. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19:342-347. [DOI] [PubMed] [Google Scholar]
- 11.Jackson, A. L., S. R. Bartz, J. Schelter, S. V. Kobayashi, J. Burchard, M. Mao, B. Li, G. Cavet, and P. S. Linsley. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21:635-637. [DOI] [PubMed] [Google Scholar]
- 12.Kapoor, T. M., T. U. Mayer, M. L. Coughlin, and T. J. Mitchison. 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150:975-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuznetsov, S. A., and V. I. Gelfand. 1986. Bovine brain kinesin is a microtubule-activated ATPase. Proc. Natl. Acad. Sci. USA 83:8530-8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence, C. J., R. K. Dawe, K. R. Christie, D. W. Cleveland, S. C. Dawson, S. A. Endow, L. S. Goldstein, H. V. Goodson, N. Hirokawa, J. Howard, R. L. Malmberg, J. R. McIntosh, H. Miki, T. J. Mitchison, Y. Okada, A. S. Reddy, W. M. Saxton, M. Schliwa, J. M. Scholey, R. D. Vale, C. E. Walczak, and L. Wordeman. 2004. A standardized kinesin nomenclature. J. Cell Biol. 167:19-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matuliene, J., and R. Kuriyama. 2002. Kinesin-like protein CHO1 is required for the formation of midbody matrix and the completion of cytokinesis in mammalian cells. Mol. Biol. Cell 13:1832-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miki, H., M. Setou, K. Kaneshiro, and N. Hirokawa. 2001. All kinesin superfamily protein, KIF, genes in mouse and human. Proc. Natl. Acad. Sci. USA 98:7004-7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molina, I., S. Baars, J. A. Brill, K. G. Hales, M. T. Fuller, and P. Ripoll. 1997. A chromatin-associated kinesin-related protein required for normal mitotic chromosome segregation in Drosophila. J. Cell Biol. 139:1361-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morii, H., T. Takenawa, F. Arisaka, and T. Shimizu. 1997. Identification of kinesin neck region as a stable alpha-helical coiled coil and its thermodynamic characterization. Biochemistry 36:1933-1942. [DOI] [PubMed] [Google Scholar]
- 19.Nakagawa, T., Y. Tanaka, E. Matsuoka, S. Kondo, Y. Okada, Y. Noda, Y. Kanai, and N. Hirokawa. 1997. Identification and classification of 16 new kinesin superfamily (KIF) proteins in mouse genome. Proc. Natl. Acad. Sci. USA 94:9654-9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nigg, E. A. 2001. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2:21-32. [DOI] [PubMed] [Google Scholar]
- 21.Ohkura, H., T. Torok, G. Tick, J. Hoheisel, I. Kiss, and D. M. Glover. 1997. Mutation of a gene for a Drosophila kinesin-like protein, Klp38B, leads to failure of cytokinesis. J. Cell Sci. 110:945-954. [DOI] [PubMed] [Google Scholar]
- 22.Phelps, M. A., A. B. Foraker, and P. W. Swaan. 2003. Cytoskeletal motors and cargo in membrane trafficking: opportunities for high specificity in drug intervention. Drug Discov. Today 8:494-502. [DOI] [PubMed] [Google Scholar]
- 23.Sablin, E. P. 2000. Kinesins and microtubules: their structures and motor mechanisms. Curr. Opin. Cell Biol. 12:35-41. [DOI] [PubMed] [Google Scholar]
- 24.Taylor, S. S., and F. McKeon. 1997. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell 89:727-735. [DOI] [PubMed] [Google Scholar]
- 25.Tripet, B., R. D. Vale, and R. S. Hodges. 1997. Demonstration of coiled-coil interactions within the kinesin neck region using synthetic peptides. Implications for motor activity. J. Biol. Chem. 272:8946-8956. [DOI] [PubMed] [Google Scholar]
- 26.Vale, R. D., and R. J. Fletterick. 1997. The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 13:745-777. [DOI] [PubMed] [Google Scholar]
- 27.van Diest, P. J., E. van der Wall, and J. P. Baak. 2004. Prognostic value of proliferation in invasive breast cancer: a review. J. Clin. Pathol. 57:675-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van 't Veer, L. J., H. Dai, M. J. van de Vijver, Y. D. He, A. A. Hart, M. Mao, H. L. Peterse, K. van der Kooy, M. J. Marton, A. T. Witteveen, G. J. Schreiber, R. M. Kerkhoven, C. Roberts, P. S. Linsley, R. Bernards, and S. H. Friend. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415:530-536. [DOI] [PubMed] [Google Scholar]
- 29.Whitfield, M. L., L. X. Zheng, A. Baldwin, T. Ohta, M. M. Hurt, and W. F. Marzluff. 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20:4188-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yen, T. J., D. A. Compton, D. Wise, R. P. Zinkowski, B. R. Brinkley, W. C. Earnshaw, and D. W. Cleveland. 1991. CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J. 10:1245-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yen, T. J., G. Li, B. T. Schaar, I. Szilak, and D. W. Cleveland. 1992. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature 359:536-539. [DOI] [PubMed] [Google Scholar]
- 32.Zhu, C., and W. Jiang. 2005. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc. Natl. Acad. Sci. USA 102:343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu, C., J. Zhao, M. Bibikova, J. D. Leverson, E. Bossy-Wetzel, J. B. Fan, R. T. Abraham, and W. Jiang. 2005. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell 16:3187-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.