Abstract
The translation elongation factor 2 in eukaryotes (eEF-2) contains a unique posttranslationally modified histidine residue, termed diphthamide, which serves as the only target for diphtheria toxin and Pseudomonas aeruginosa exotoxin A. Diphthamide biosynthesis is carried out by five highly conserved proteins, Dph1 to Dph5, and an as-yet-unidentified amidating enzyme. The evolutionary conservation of the complex diphthamide biosynthesis pathway throughout eukaryotes implies a key role for diphthamide in normal cellular physiology. Of the proteins required for diphthamide synthesis, Dph3 is the smallest, containing only 82 residues. In addition to having a role in diphthamide biosynthesis, Dph3 is also involved in modulating the functions of the Elongator complex in yeast. To explore the physiological roles of Dph3 and to begin to investigate the function of diphthamide, we generated dph3 knockout mice and showed that dph3+/− mice are phenotypically normal, whereas dph3−/− mice, which lack the diphthamide modification on eEF-2, are embryonic lethal. Loss of both dph3 alleles causes a general delay in embryonic development accompanied by lack of allantois fusion to the chorion and increased degeneration and necrosis in neural tubes and is not compatible with life beyond embryonic day 11.5. The dph3−/− placentas also developed abnormally, showing a thinner labyrinth lacking embryonic erythrocytes and blood vessels. These results attest to the physiological importance of Dph3 in development. The biological roles of Dph3 are also discussed.
Eukaryotic translation elongation factor 2 (eEF-2) contains a unique posttranslationally modified histidine residue, which was termed diphthamide due to the fact that it serves as the only target for diphtheria toxin (DT) and the closely related Pseudomonas aeruginosa exotoxin A (ETA). Diphthamide is formed by stepwise additions to the side chain of the His715 (His699 in yeast) residue of eEF-2, beginning with the transfer of the 3-amino-3-carboxypropyl group of S-adenosylmethionine to the imidazole C-2 of the precursor histidine residue and followed by trimethylation of the resulting amino group, using S-adenosylmethionine as the methyl donor, to produce diphthine (refer to Fig. 1A in reference 11). The final step is an ATP-dependent amidation of the carboxyl group, yielding diphthamide. DT and ETA specifically catalyze the transfer of ADP-ribose from NAD to diphthamide, thereby inactivating eEF-2, shutting down cellular protein synthesis, and causing cell death. Although diphthamide has been found in all eukaryotic organisms and archaebacteria, it is not present in eubacteria. Thus, DT and ETA have evolved a specific mechanism for targeting eukaryotic protein synthesis machinery without inactivating the analogous elongation factor (EF-G) present in the bacterial pathogens that produce them.
FIG. 1.
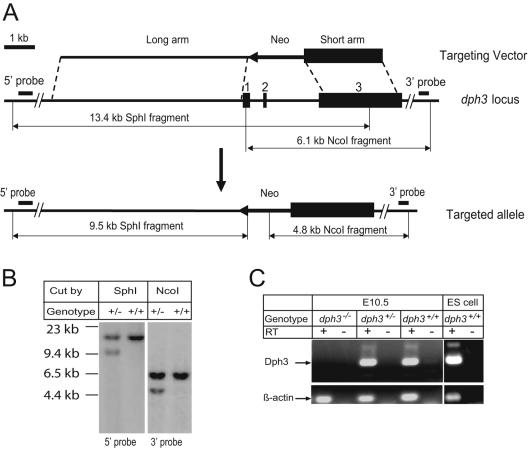
Targeted deletion of dph3 in mice. (A) Schematic representation of the targeting strategy of dph3. Top row represents the targeting vector, in which the Neo cassette (the arrowed bar indicating the direction of transcription), is flanked by the homologous arms. The middle row is the dph3 genomic locus, with the numbered filled boxes representing the three dph3 exons. The bottom row is the targeted locus, in which the entire coding sequence, including exons 1 and 2 and the 5′ end of exon 3, is replaced by the Neo cassette. (B) An example of Southern blot analysis of a correctly targeted ES cell clone using probes shown in panel A. (C) RT-PCR analyses showing that dph3 is expressed in ES cells as well as in wild-type and heterozygous, but not homozygous, E10.5 embryos.
Recent work has identified all the proteins required for diphthamide biosynthesis (10, 11, 13, 14), with the exception of an unidentified amidating enzyme required at the last step. The proteins are designated Dph1 to Dph5. Although the physiological functions of diphthamide remain elusive, the conservation of Dph1 to Dph5 in all eukaryotes implies that this unique residue has a significant biological role in eukaryotes. Supporting this notion is recent work identifying dph1 as OVCA1 (for ovarian cancer gene 1) (11), a newly identified tumor suppressor gene (1) that plays important roles in the regulation of cell proliferation, embryonic development, and tumorigenesis (1).
Of the five known proteins required for diphthamide biosynthesis, Dph3 is the only one that has a documented role in other processes. Unlike the other four Saccharomyces cerevisiae dph mutants that have at most a subtle phenotype, yeast dph3 mutants have growth defects and are temperature sensitive and drug sensitive (3, 11). In yeast, dph3 was independently identified as Kti11 (dairy yeast Kluyveromyces lactis toxin insensitive gene 11), a gene required for the toxic action of zymocin to yeast (2, 3). Dph3 has been shown to physically associate with Elongator complex, the well-recognized target of zymocin (2, 8). Elongator complex, which consists of the six subunits Elp1 to Elp6 (15), is evolutionarily conserved from yeast to humans (6) and was originally identified as a novel component of RNA polymerase II holoenzyme. Deletion of dph3 in yeast causes the truncation of approximately 200 residues at the amino terminus of Elp1 (2), apparently through a controlled proteolysis, thus linking the role of Dph3 in zymocin action to the function of Elongator complex. Elongator complex can specifically bind to the hyperphosphorylated form of RNA polymerase II, thereby modulating its activity during transcription elongation (5, 15, 19). Very recently, both the Elongator complex and Kti11 were shown to be required for posttranscriptional modification of tRNAs at the wobble position of the anticodon (7). Furthermore, this tRNA modification makes the tRNAs susceptible to cleavage by zymocin (12).
To explore the physiological roles of Dph3 and to begin to investigate the function of diphthamide, in this work we have generated a dph3 null mutation in mice using a gene-targeting approach. We find that loss of both dph3 alleles causes a general delay in embryonic development, which is not compatible with life beyond embryonic day 11.5 (E11.5), attesting to the biological importance of Dph3.
MATERIALS AND METHODS
Gene targeting and genotyping.
A clone with a 20-kb insert containing the dph3 gene was isolated from a 129SVJ mouse genomic DNA library (Stratagene, La Jolla, CA). A 5.8-kb NcoI fragment upstream of dph3 exon 1 was subcloned into the HpaI site downstream of the neomycin resistance gene cassette (Neo) within pKO Scrambler NTKV-1904 (Stratagene). Next, a 2.8-kb BamHI fragment containing the majority of dph3 exon 3 was inserted into a BamHI site upstream of the Neo cassette, and the targeting construct was isolated after NotI digestion (Fig. 1A). W9.5 mouse embryonic stem (ES) cells with a 129SVJ background were transfected with the targeting construct and selected with G418. The resistant ES colonies were analyzed by Southern blot analysis for homologous recombination (Fig. 1B). The ES cells heterozygous for the targeted mutation were microinjected into C57BL/6 blastocysts, and resulting male chimeras were mated with C57BL/6 females to obtain germ line transmission. Thus, the mice generated and used in this study are on a mixed B6;129 genetic background. Mice were maintained on standard chow diet (National Institutes of Health, Bethesda, MD) with a light/dark cycle consisting of 14 h of light and 10 h of darkness. These studies were approved by the Animal Care and Use Committee of the National Institute of Allergy and Infectious Diseases. For genotyping, genomic DNA was isolated from ES cells or mouse tails by using a DNeasy tissue kit (QIAGEN, Valencia, CA). Samples were digested with SphI and hybridized with a labeled 0.5-kb 5′ probe (amplified using primers 5′-AAGCCAAGAATGCCACAGAGCC-3′ and 5′-TTACAGCAGAGAACAAAGGATGGG-3′) or were digested with NcoI and hybridized with a 400-bp 3′ probe (amplified using primers 5′-GCAAAAGTAGCAAGGAAGCCATTG-3′ and 5′-GGTCAAAGGTGGGGTAGAAACC-3′) (Fig. 1B). Subsequent genotyping was performed by PCR of mouse tail DNA or embryo DNA isolated from the yolk sacs using a REDExtract-N-Amp tissue PCR kit (Sigma, St. Louis, MO). Genotyping-PCR using GT1 (5′-AGTGGACGGAGAACTGCGCAACGCT-3′) and GT2 (5′-TGTCTCCGAGTCCTCGTCATATTGA-3′) generates a 290-bp band from the wild-type allele, whereas using GT1 and GT3 (5′-GAGGATTGGGAAGACAATAGCAGGCA-3′) generates a 202-bp band from the mutant allele. The ES cell injections, screening, and initial mouse breeding work were performed by Ozgene Pty. Ltd., Western Australia, Australia, under contract from the National Institute of Allergy and Infectious Diseases, NIH.
Embryo dissection and histology.
Entire conceptuses and placentas were isolated between E8.5 and E12.5 from pregnant mice and photographed. A part of a yolk sac or embryo was used for genotyping. For histological analyses, the embryos and placentas were fixed for 24 h in 2% paraformaldehyde in phosphate-buffered saline, embedded in paraffin, sectioned, and stained with hematoxylin-eosin.
Reverse transcription-PCR (RT-PCR).
Total RNA was isolated from feeder-independent mouse ES cells and E10.5 embryos by using TRIzol reagent and treated with DNase I (Invitrogen, Carlsbad, CA) to remove contaminating genomic DNA. Identical amounts of total RNA (0.5 μg) from dph3+/+, dph3+/−, and dph3−/− E10.5 embryos were hybridized to oligo(dT)12-18. The first-strand cDNAs were synthesized by adding 1 μl (200 units) SuperScript reverse transcriptase (RT) (Invitrogen), 1 μl dNTP mix (10 mM each), 2 μl dithiothreitol (0.1 M) in 20-μl reaction mixtures in accordance with the product manuals. PCRs were performed to amplify dph3 cDNA fragment by using primers 5′-CGGTGTTTCACGACGAGGTGGAGAT-3′ and 5′-AACTAACTCCTTGTTGGTTGAAGGTGC-3′ or to amplify β-actin cDNA fragment by using primers 5′-GACCTCTATGCCAACACAGT-3′ and 5′-TAGGAGCCAGAGCAGTAATC-3′.
ADP ribosylation assay.
Mouse ES cells and E9.5 and E10.5 embryos with different dph3 genotypes were directly lysed in modified radioimmunoprecipitation assay buffer containing protease inhibitors (9). The protein concentrations were determined using bicinchoninic acid protein assay reagent (Pierce, Rockford, IL). ADP ribosylation reactions were performed at room temperature for 30 min in a final volume of 20 μl containing 5 μg of tissue lysate, 2 ng fully nicked DT, 100 nM [32P-adenylate]NAD (Amersham Biosciences Corp., Piscataway, NJ), 20 mM Tris-HCl (pH 7.5), 50 mM dithiothreitol, 1 mM EDTA. The reactions were terminated by boiling in sodium dodecyl sulfate (SDS) sample buffer, and the samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) followed by autoradiography. The ADP ribosylation status of eEF-2 was also examined by native PAGE followed by Western blot analysis using an anti-eEF-2 antibody as described previously (10, 11). Briefly, portions (20 μg) of the tissue lysates were incubated with 60 ng fully nicked DT at room temperature for 30 min and then were subjected to native PAGE using 4% Tris-glycine gradient gels (Novex). Prior to loading, the cell lysates were incubated for 10 min at room temperature in 1× native sample buffer (Novex). The proteins were then transferred to nitrocellulose membranes overnight, a procedure followed by Western blotting with a goat antibody (catalog no. sc-13004; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) directed to a linear peptide at the carboxyl terminus of eEF-2. The ADP-ribosylated eEF-2, which has two added negatively charged phosphate groups, migrates faster than unmodified eEF-2 (10, 11).
RESULTS
Generation of dph3+/− ES cells and mice.
To investigate the role of Dph3 in development, we disrupted the dph3 gene by homologous recombination in mouse ES cells using a vector that replaced exon 1, exon 2, and a small portion of exon 3 with a Neo cassette (Fig. 1A), in which the targeting region covered the entire coding sequence for Dph3 (82 residues). ES cell clones in which one allele was targeted were identified by Southern blot analyses with probes flanking the targeted region (Fig. 1A and B). One of these ES clones was used to generate chimeric mice. Germ line transmission was achieved with chimeras from this clone, as was evident from Southern blot analysis of the offspring. RT-PCR analyses showed that dph3, while expressing well in wild-type ES cells and in dph3+/+ and dph3+/− embryos, did not express in dph3−/− embryos (Fig. 1C). Mice heterozygous for the dph3 mutant allele were viable and fertile. In breeding of both sexes of heterozygous mice to wild-type mice, the mutant allele was detected in approximately 50% of progeny. These results demonstrated that the heterozygosity of dph3 is phenotypically unapparent and that the dph3 gene is not an imprinting gene.
Loss of dph3 results in general delay in mouse development and embryonic lethality at E11.5.
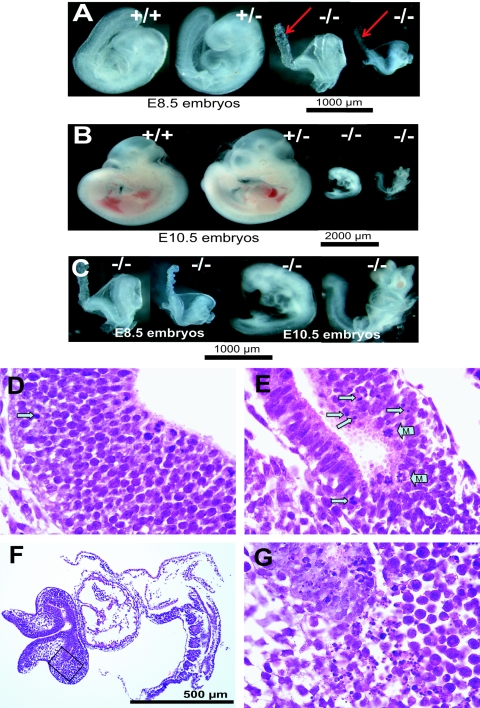
While dph3 heterozygous mice are phenotypically normal, genotyping of offspring from the heterozygous intercrosses did not yield viable homozygous null mice, indicating that loss of both dph3 alleles results in embryonic lethality (Table 1). Subsequently, staged embryos from the heterozygous crosses were analyzed. At E8.5 to E10.5, dph3−/− embryos were detectable (Table 1). However, all these dph3−/− embryos were phenotypically abnormal, being much smaller than their dph3+/+ and dph3+/− littermates (Fig. 2A through C). The allantoides of these null embryos did not fuse with the chorions (Fig. 2A), apparently because of the general delay of development in these embryos. The dph3−/− embryos were viable, growing and developing to E10.5 stage, although with a low growth rate compared to their dph3+/+ and dph3+/− littermates (Fig. 2A through C). The dph3−/− embryos did not turn until E10.5, whereas the well-developed dph3+/+ and dph3+/− embryos had turned at E8.5 (Fig. 2A). The small size of the E10.5 dph3−/− embryos in comparison to the E8.5 wild-type embryos (Fig. 2A and C) reflected a severe delay in development of at least 2 days. Histological analyses revealed that these dph3−/− embryos were undergoing increased apoptosis and cell necrosis in the neural tubes (Fig. 2D and E), which was associated with smaller and thinner neural tube structure, indicating these abnormal embryos were in the process of degeneration. Increased mitotic figures were also observed in the neuroepithelial cells in dph3−/− embryos (Fig. 2E). At E11.5 and E12.5, the homozygous embryos were reabsorbed and thus could not be genotyped. These results demonstrate that the loss of dph3 causes a general delay in mouse embryo development and progressive embryo cell degeneration that is incompatible with life beyond embryonic day 11.5.
TABLE 1.
Genotyping analyses reveal that dph3−/− mice are embryonic lethal
| Developmental stage | No. of progeny
|
||||
|---|---|---|---|---|---|
| Genotyped as:
|
Reabsorbedb | Total | |||
| +/+ | +/− | −/−a | |||
| Adult | 54 | 89 | 0 | 143 | |
| E12.5 | 11 | 27 | 0 | 18 | 56 |
| E11.5 | 7 | 7 | 0 | 7 | 21 |
| E10.5 | 11 | 20 | 7* | 38 | |
| E9.5 | 20 | 38 | 17* | 75 | |
| E8.5 | 8 | 24 | 8* | 40 | |
* indicates abnormal, small embryos.
Reabsorbed embryos cannot be genotyped because only abnormal placentas fusing with dead embryos were found in these sites. These sites are likely to be the missing homozygous embryos.
FIG. 2.
Loss of Dph3 in mouse causes a general delay in embryonic development and lethality at midgestation. (A) Photographs of E8.5 embryos showing that dph3+/+ and dph3+/− embryos developed normally, while dph3−/− embryos were abnormally small and did not turn. The arrows indicate the allantoides that did not fuse with the chorions in the homozygous embryos. (B) Photographs of E10.5 embryos showing the delay in development of dph3−/− embryos. (C) The E8.5 and E10.5 dph3−/− embryos shown in panels A and B are presented at the same magnification to facilitate size comparison. This showed that both E8.5 and E10.5 dph3−/− embryos were still alive, but little growth occurred from E8.5 to E10.5, and the embryos turned at E10.5. Histological analyses of E9.5 dph3+/+ (D) and dph3−/− (E) embryos show that the dph3−/− embryos were undergoing increased apoptosis and cell necrosis in the neural tubes. Arrows in panels D and E identify apoptotic or necrotic cells; arrows with M identify cells in mitotic phase. (F) An E10.5 dph3−/− embryo shows developmental arrest (much smaller than dph3+/− and dph3+/+ embryos) and focal areas of necrosis and apoptosis. (G) High magnification of rectangle shown in panel F.
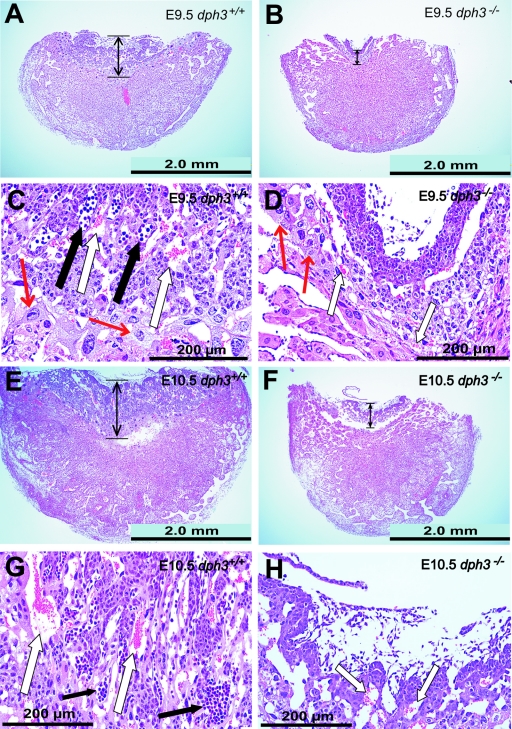
Abnormalities in placenta development were also recorded for dph3−/− embryos. At stage E9.5, although the dph3−/− and dph3+/+ placentas appeared to have the same size (Fig. 3A and B), histological analyses reveled that the labyrinth in dph3−/− placentas was much thinner and lacked immature nucleated erythrocytes or embryo-derived endothelium-lined blood vessels (Fig. 3A through D). However, other cell types, such as giant trophoblasts, which derive from the trophectoderm of the embryos (18), could be seen (Fig. 3C and D). These abnormalities in placental development are likely due to the lack of the fusions of the allantoides with the chorions (Fig. 2A), thus preventing the migration of the embryo-derived cells during placenta development. At stage E10.5, the defects observed as described above in dph3−/− placentas became more severe and were accompanied by a placenta size considerably smaller than that for dph3+/+ (Fig. 3E through H). At E12.5, the homozygous embryos were completely reabsorbed, and extensive hemorrhages were also observed in dph3−/− placentas at the junction with deciduae (data not shown). These results demonstrated that Dph3 is crucial for the development of mouse placenta.
FIG. 3.
Developmental defects of placentas caused by the Dph3 null mutation. Histological analyses (low-power views) of E9.5 (A and B) and E10.5 (E and F) placentas show that the dph3−/− placentas had much thinner labyrinth layers (double-headed arrows). Panels C, D, G, and H (high-power views of labyrinth layers in panels A, B, E, and F, respectively) show that the labyrinth layers in dph−/− embryos lacked immature embryonic nucleated erythrocytes and embryonic blood vessels that derived from mesoderm. White arrows in panels C, D, G, and H, maternal mature erythrocytes; black arrows in panels C and G, embryonic blood vessels filled with embryonic nucleated erythroid precursors; red arrows in panels C and D, trophoblast giant cells.
Dph3 is required for diphthamide biosynthesis in mouse.
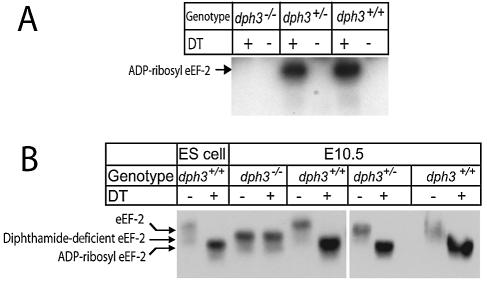
To test whether Dph3 is essential for diphthamide modification on eEF-2 in mice, as it is in yeast and CHO (Chinese hamster ovary) cells, whole-tissue lysates prepared from E9.5 and E10.5 dph3+/+, dph3+/−, and dph3−/− embryos were subjected to DT-mediated ADP ribosylation. The results showed that the eEF-2 from dph3−/− embryos could not be ADP ribosylated by DT, whereas that from dph3+/+ and dph3+/− embryos could (Fig. 4A and B), demonstrating that the role of Dph3 in diphthamide biosynthesis is conserved in the mouse. Analysis by native gels showed that the eEF-2 in dph3+/− embryos, like that from dph3+/+ embryos, could be completely ADP ribosylated by DT (Fig. 4B), indicating that one normal allele of dph3 is sufficient for diphthamide modification for all the eEF-2 in mouse embryos. The eEF-2 from dph3−/− embryos, which could not be ADP ribosylated, migrated on a native gel between the ADP-ribosylated and non-ADP-ribosylated forms of eEF-2 from dph3+/− and dph3+/+ embryos, as expected for diphthamide-deficient eEF-2 (10, 11). The eEF-2 from ES cells could also be ADP ribosylated by DT (Fig. 4B), consistent with the universal presence of diphthamide and the ubiquitous expression of the genes for diphthamide biosynthesis in eukaryotes.
FIG. 4.
Dph3 is required for diphthamide modification on eEF-2. (A) eEF-2 of dph3−/− embryos is resistant to ADP ribosylation by DT. Lysates from different dph3 genotype E10.5 embryos were incubated with [32P-adenylate]NAD in the absence or presence of DT. Reactions were terminated by boiling in SDS sample buffer, and the samples were analyzed by SDS-PAGE followed by autoradiography. (B) One allele of dph3 is sufficient for diphthamide modification of all eEF-2 in mouse. Lysates from ES cells and dph3−/−, dph3+/−, and dph3+/+ embryos were incubated with DT at room temperature for 30 min, and the lysates were then analyzed by native PAGE followed by Western blotting using an antibody against a linear peptide of the carboxyl terminus of eEF-2.
DISCUSSION
Unlike the other four proteins in the diphthamide pathway, Dph3 is unique in that its null mutations in yeast cause a reduction in growth rate, heat sensitivity (cannot grow at or above 38°C), and supersensitivity to transcription inhibitors. These observations imply that Dph3 has a role in other important biological processes beyond its role in diphthamide biosynthesis. In this study, we found that Dph3 plays an essential role in development; targeted deletion of both dph3 alleles in mice results in a severe developmental delay (2 to 3 days) and embryonic lethality at E11.5.
Understanding of the role(s) of Dph3 may be enhanced by considering the effects of the inactivation of other dph genes. Recently, Dph1/OVCA1 null mice were made and characterized (1). Similar to what we found for the Dph3 mutant mice, Dph1 was found to be important for normal mouse development. Dph1 null embryos on a mixed B6;129 background showed an approximately 1-day delay in mouse development, along with defects in multiple organ systems, and died immediately after birth due to malfunction of the lung (1). The observation of a phenotype in dph3−/− embryos more severe than that in dph1−/− embryos of the same genetic background is consistent with the evidence from yeast mutants that Dph3 is also required for other processes besides diphthamide biosynthesis. Although it is unlikely, we cannot rule out the possibility that Dph1, like Dph3, has functions besides that in diphthamide biosynthesis. Further investigation including the generation of Dph2, Dph4, or Dph5 null mice may help to answer this question and address the intriguing and unidentified role(s) of diphthamide in physiology.
In addition to the general delay in development, embryonic fibroblast cells isolated from dph1−/− embryos display growth defects, with significantly increased doubling times associated with an increase in the G1-phase population and a reduction in the S-phase population, indicating arrest of the cell cycle in G1 phase (1). Interestingly, these growth defects can be rescued by p53 deficiency, suggesting a genetic interaction between dph1 and p53 (1). If the defects in Dph1 null mice are due solely to the deficiency in diphthamide biosynthesis on eEF-2, we might infer that diphthamide modification of eEF-2 is needed to maintain a high rate of protein synthesis. Even a small decrease in protein synthesis rates may be deleterious during embryogenesis, where cell growth and division are operating at or near maximum rates.
Dph3 has been extensively studied since its first description in 2002. At least two functions in yeast for this small protein have now been documented. In addition to its essential role in diphthamide biosynthesis, Dph3 is also involved in regulating the functions of Elongator complex, thereby regulating the sensitivity of yeast to zymocin (2, 3). Therefore, yeast mutants mutated in any of the elp1 to elp6 genes display phenotypic changes the same as those observed in dph3 mutants (4, 8). Although the link that was established between Dph3 and Elongator was in yeast, it is likely that their relationship also stands in mammalian cells because of the high conservation of these proteins in evolution. Thus, the defects of Dph3 null embryos likely are caused by the losses of function of both diphthamide biosynthesis and Elongator. As noted earlier, recent work shows that both Dph3 and Elongator are required for modification of tRNAs, and an absence of these modifications could explain a general loss of accuracy and efficiency in protein synthesis that could account for the phenotypes seen in yeast and in our dph3 null mice. Although Dph3 acts along with Elongator in several ways that control translational efficiency (7, 12), Elongator is not involved in the diphthamide biosynthetic pathway, because the elp1, elp2, and elp3 yeast mutants are able to synthesize diphthamide and thus are sensitive to DT (data not shown).
Dph3 is a small acidic protein with only 82 residues, containing 20 and 19 negatively charged residues in yeast and humans, respectively. Recently, the structure of yeast Dph3 was defined using nuclear magnetic resonance, by which it was shown that it belongs to the CSL-class zinc-binding family of proteins, which contain a single Zn2+ bound to four conserved cysteine residues (17). The large number of conserved negatively charged residues form two negatively charged surfaces that cover a majority of the molecule, suggesting that Dph3 can interact with positively charged molecules (17). Dph3 can interact with Elongator, a complex of Dph1 and Dph2, eEF-2, and ribosomal proteins Rps7A and Rps19A (2), suggesting that it can regulate multiple biological processes. In fact, human Dph3 was also identified using a two-hybrid screen as DelGEF (deafness locus-associated putative guanine nucleotide exchange factor protein)-interacting protein 1 (DelGIP1) and was reported to negatively regulate the interaction between DelGEF and Sec5, thereby negatively regulating the extracellular secretion of proteoglycans by HeLa cells (16).
In this work, we demonstrate a crucial role for Dph3 in mouse development. Further investigations are needed to reveal the detailed molecular mechanisms by which this small protein acts in the regulation of the functions of Elongator and the other diphthamide biosynthesis proteins.
Acknowledgments
We thank Mary Jo Danton and Jie Liu for assistance with genomic library screening, Mahtab Moayeri and Thomas Bugge for assistance with design and administration of mouse experiments, and Chuxia Deng and Cuiling Li for mouse ES cell handling.
This research was supported by the intramural research program of the National Institute of Allergy and Infectious Diseases (NIAID), NIH, and a NIAID contract to SoBran, Inc.
REFERENCES
- 1.Chen, C. M., and R. R. Behringer. 2004. Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis. Genes Dev. 18:320-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fichtner, L., D. Jablonowski, A. Schierhorn, H. K. Kitamoto, M. J. Stark, and R. Schaffrath. 2003. Elongator's toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol. Microbiol. 49:1297-1307. [DOI] [PubMed] [Google Scholar]
- 3.Fichtner, L., and R. Schaffrath. 2002. KTI11 and KTI13, Saccharomyces cerevisiae genes controlling sensitivity to G1 arrest induced by Kluyveromyces lactis zymocin. Mol. Microbiol. 44:865-875. [DOI] [PubMed] [Google Scholar]
- 4.Frohloff, F., L. Fichtner, D. Jablonowski, K. D. Breunig, and R. Schaffrath. 2001. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 20:1993-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert, C., A. Kristjuhan, G. S. Winkler, and J. Q. Svejstrup. 2004. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell 14:457-464. [DOI] [PubMed] [Google Scholar]
- 6.Hawkes, N. A., G. Otero, G. S. Winkler, N. Marshall, M. E. Dahmus, D. Krappmann, C. Scheidereit, C. L. Thomas, G. Schiavo, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 2002. Purification and characterization of the human elongator complex. J. Biol. Chem. 277:3047-3052. [DOI] [PubMed] [Google Scholar]
- 7.Huang, B., M. J. Johansson, and A. S. Bystrom. 2005. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11:424-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jablonowski, D., F. Frohloff, L. Fichtner, M. J. Stark, and R. Schaffrath. 2001. Kluyveromyces lactis zymocin mode of action is linked to RNA polymerase II function via Elongator. Mol. Microbiol. 42:1095-1105. [DOI] [PubMed] [Google Scholar]
- 9.Liu, S., and S. H. Leppla. 2002. Cell surface tumor endothelium marker 8 cytoplasmic tail-independent anthrax toxin binding, proteolytic processing, oligomer formation, and internalization. J. Biol. Chem. 278:5227-5234. [DOI] [PubMed] [Google Scholar]
- 10.Liu, S., and S. H. Leppla. 2003. Retroviral insertional mutagenesis identifies a small protein required for synthesis of diphthamide, the target of bacterial ADP-ribosylating toxins. Mol. Cell 12:603-613. [DOI] [PubMed] [Google Scholar]
- 11.Liu, S., G. T. Milne, J. G. Kuremsky, G. R. Fink, and S. H. Leppla. 2004. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol. Cell. Biol. 24:9487-9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu, J., B. Huang, A. Esberg, M. J. Johansson, and A. S. Bystrom. 2005. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA 11:1648-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattheakis, L. C., W. H. Shen, and R. J. Collier. 1992. DPH5, a methyltransferase gene required for diphthamide biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:4026-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattheakis, L. C., F. Sor, and R. J. Collier. 1993. Diphthamide synthesis in Saccharomyces cerevisiae: structure of the DPH2 gene. Gene 132:149-154. [DOI] [PubMed] [Google Scholar]
- 15.Otero, G., J. Fellows, Y. Li, T. de Bizemont, A. M. Dirac, C. M. Gustafsson, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3:109-118. [DOI] [PubMed] [Google Scholar]
- 16.Sjolinder, M., J. Uhlmann, and H. Ponstingl. 2004. Characterisation of an evolutionary conserved protein interacting with the putative guanine nucleotide exchange factor DelGEF and modulating secretion. Exp. Cell Res. 294:68-76. [DOI] [PubMed] [Google Scholar]
- 17.Sun, J., J. Zhang, F. Wu, C. Xu, S. Li, W. Zhao, Z. Wu, J. Wu, C. Z. Zhou, and Y. Shi. 2005. Solution structure of Kti11p from Saccharomyces cerevisiae reveals a novel zinc-binding module. Biochemistry 44:8801-8809. [DOI] [PubMed] [Google Scholar]
- 18.Watson, E. D., and J. C. Cross. 2005. Development of structures and transport functions in the mouse placenta. Physiology 20:180-193. [DOI] [PubMed] [Google Scholar]
- 19.Wittschieben, B. O., G. Otero, T. de Bizemont, J. Fellows, H. Erdjument-Bromage, R. Ohba, Y. Li, C. D. Allis, P. Tempst, and J. Q. Svejstrup. 1999. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4:123-128. [DOI] [PubMed] [Google Scholar]