Abstract
Nuclear migration and positioning within cells are critical for many developmental processes and are governed by the cytoskeletal network. Although mechanisms of nuclear-cytoskeletal attachment are unclear, growing evidence links a novel family of nuclear envelope (NE) proteins that share a conserved C-terminal SUN (Sad1/UNC-84 homology) domain. Analysis of Caenorhabditis elegans mutants has implicated UNC-84 in actin-mediated nuclear positioning by regulating NE anchoring of a giant actin-binding protein, ANC-1. Here, we report the identification of SUN1 as a lamin A-binding protein in a yeast two-hybrid screen. We demonstrate that SUN1 is an integral membrane protein located at the inner nuclear membrane. While the N-terminal domain of SUN1 is responsible for detergent-resistant association with the nuclear lamina and lamin A binding, lamin A/C expression is not required for SUN1 NE localization. Furthermore, SUN1 does not interact with type B lamins, suggesting that NE localization is ensured by binding to an additional nuclear component(s), most likely chromatin. Importantly, we find that the luminal C-terminal domain of SUN1 interacts with the mammalian ANC-1 homologs nesprins 1 and 2 via their conserved KASH domain. Our data provide evidence of a physical nuclear-cytoskeletal connection that is likely to be a key mechanism in nuclear-cytoplasmic communication and regulation of nuclear position.
The nuclear envelope (NE) is a double-membrane structure that separates chromatin from the cytoplasm, thereby allowing regulation of DNA replication and gene expression in eukaryotic cells. Nuclear pore complexes span the double membrane and regulate the passage of molecules between the cytoplasm and the nucleus (16). The outer nuclear membrane (ONM) is contiguous with, and biochemically similar to, the endoplasmic reticulum (ER). In contrast, the inner nuclear membrane (INM) contains a unique set of integral membrane proteins. Both nuclear pore complexes and INM proteins are anchored by association with the nuclear lamina, a network of lamin intermediate filaments that underlies the INM. The lamina, together with the associated INM proteins, provides structural support for the NE and sites for attachment of chromatin to the nuclear periphery (reviewed in reference 11).
Most mammalian cells express two classes of lamin protein, types A and B (reviewed in reference 26). A-type lamins, the major isoforms of which are lamins A and C, are alternative splice products of the LMNA gene (8, 23). B-type lamins are mainly composed of lamins B1 and B2, which are encoded by separate genes (LMNB1 and LMNB2, respectively). A- and B-type lamins differ in their patterns of expression. While type B lamins are found in all nucleated somatic cells, type A lamins are absent in early embryos, their expression correlating with terminal differentiation (33). This has led to the suggestion that A-type lamins, although not essential for individual cell survival, are involved in determining differentiation patterns, possibly through effects on chromatin organization. Support for such a role comes from the finding that a wide range of human genetic disorders, each predominantly affecting distinct groups of tissues, from muscle to neurons and adipocytes, are caused by mutations in the LMNA gene (reviewed in reference 27).
INM proteins are an expanding family of integral membrane proteins that includes the lamin B receptor (LBR), lamina-associated polypeptide 1 (LAP1), LAP2, and emerin (3). INM proteins have a structure comprising a nucleoplasmic N-terminal domain (NTD), which confers NE localization through lamin and/or chromatin binding; one or more transmembrane domains; and a C-terminal region. In all but two proteins so far examined (MAN1 and LBR), the C-terminal region is located in the NE lumen and is generally very short, suggesting that it simply serves as a membrane anchor. Most INM proteins are thus thought to have structural roles within the nucleus through their interactions with lamins and chromatin. Interestingly, a new family of NE proteins, containing a large, conserved C-terminal SUN (Sad1/UNC-84 homology) domain (13, 20), instead appears to play a role in nuclear positioning, potentially by connecting the NE to the cytoskeleton.
Nuclear migration and positioning within cells are mainly dependent upon the microtubule network and the associated microtubule motor protein dynein (reviewed in references 31, 32, and 36) but can also be influenced by the actin cytoskeleton (41). Caenorhabditis elegans possesses two SUN domain proteins, UNC-84 and Ce-SUN1, which have been implicated in actin- and microtubule-dependent processes, respectively. Preliminary studies indicate that Ce-SUN1 (also reported as matefin (10) is required for nuclear-centrosome attachment via dynein-mediated anchoring of a novel protein, ZYG-12, to the ONM (21). On the other hand, unc-84 mutants display defects in certain nuclear anchoring and migration events during development of the organism (20) and the UNC-84 protein is required for the localization of a giant actin-binding protein, ANC-1, to the ONM (41, 42). ANC-1 has an N-terminal calponin homology domain, responsible for binding actin, and a C-terminal KASH (Klarsicht/ANC-1/Syne-1 homology) domain, which contains a transmembrane domain and is responsible for NE localization of ANC-1. These two domains are separated by an extended unique repeat region that is predicted to act as a linker spanning the distance between the NE and the actin cytoskeleton (41). Nesprins (also reported as Syne, Myne, and NUANCE) are the mammalian homologs of ANC-1 and exist as multiple alternatively spliced isoforms of two genes, those for nesprins 1 and 2. As a result, the structures of nesprins are highly variable, in both length and the presence of the conserved N-terminal actin-binding (calponin homology) and C-terminal NE localization (KASH) domains (1, 25, 48-50). Shorter nesprin isoforms have been found to interact with lamins and emerin and are thus thought to contribute to a scaffold at the INM (24).
Two predicted mammalian SUN proteins, SUN1 and SUN2, have been identified in database searches (20), and SUN2 was recently reported to be a typical INM protein with a luminal C-terminal domain (CTD) (14). Conversely, the topology of C. elegans SUN proteins has not been determined. Indeed, some models propose that UNC-84 may reside in the ONM (17, 30). This speculation is in part due to the fact that no interacting partners of SUN proteins at the INM have yet been identified. In particular, although it is widely assumed that NE localization is conferred by binding to the nuclear lamina, interaction between SUN proteins and lamins has so far not been demonstrated.
Having identified mutations in the LMNA gene as causative of familial partial lipodystrophy (38), we sought to discover lamin A-interacting proteins that may be involved in the disease mechanism. We performed a yeast two-hybrid interaction screen with the CTD of lamin A as bait and identified the adipocyte differentiation factor SREBP1 (18). In the same screen, we identified SUN1 as another novel lamin A-binding protein. Here we report the characterization of this potentially important interaction and provide evidence that SUN1 is an integral INM protein. While the NTD of SUN1 is responsible for the anchoring at the INM, the CTD interacts with nesprin isoforms in the NE lumen, thereby forming a direct physical link between the cytoplasm and the nucleus.
MATERIALS AND METHODS
Yeast two-hybrid screen.
A yeast two-hybrid interaction screen was performed as described previously (18). Briefly, bait vector pGBDU-mLMNA379, encoding amino acids 379 to 664 of lamin A, and a pGAD-GH mouse 3T3-L1 adipocyte cDNA library (kind gift of A. Saltiel) were sequentially transformed into yeast strain PJ69-4a and interacting clones were selected by growth on synthetic dropout medium lacking uracil, leucine, histidine, and adenine and supplemented with 10 mM 3-aminotriazole.
Protein sequence comparisons and structure prediction.
cDNA sequences were compared with BLAST (National Center for Biotechnology Information), and protein sequences were compared and aligned with ClustalW. Motif predictions were performed with ExPaSy software (www.expasy.org), including TMpred and Coils. Genomic structure was predicted and mouse homologues identified with Ensembl (www.ensembl.org).
Plasmid constructs.
pGEX-hLaminA389, pCI-LMNA, and pCI-mycLMNA have been described previously (18). pCI-LMNC was generated by PCR with a 3′ primer containing the entire lamin C-specific sequence. The PCR product was then used to replace the lamin A-specific tail of pCI-LMNA. The lamin B1 cDNA was excised from IMAGE clone 3445475 with SalI and NotI and cloned into the XhoI and NotI sites of the pCIneo expression vector (Promega). pBS-LMNB2 was a kind gift of E. Schirmer. The pCDNA3-nesprin2β and pEGFP-nesprin constructs have been described previously (47, 48).
The murine SUN1 cDNA was cloned into pCIneo by first introducing an XbaI restriction site into pGADGH-mSUN1, 258 bp downstream of the stop codon, with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The insert was then subcloned into pCIneo via the EcoRI-XbaI restriction sites. 5′ and 3′ myc and hemagglutinin (HA) tags were added to the SUN1 cDNA by PCR amplification with primers containing the myc or HA tag sequence. Internal myc tags were introduced after codons 355 and 456 by first introducing a unique MluI site at these locations by site-directed mutagenesis. The myc tag sequence was then ligated into these MluI sites following annealing of oligonucleotides comprising the myc tag sequence flanked by MluI sticky ends.
All SUN1 deletion constructs were generated by PCR amplification of the relevant regions of the SUN1 cDNA. For maltose-binding protein (MBP) pull-down assays, products were subcloned into pMALc2G (New England Biolabs). For mammalian expression, the PCR primers incorporated a myc tag at the 5′ terminus and products were subcloned into pCIneo. A C-terminally myc-tagged emerin construct was generated with a cDNA kindly provided by J. Ellis. The cDNA was subcloned into pCIneo, and the 3′ end was replaced by PCR amplification with a primer containing the myc tag sequence.
In vitro translation and pull-down assays.
35S-labeled SUN1, lamin, and nesprin proteins were produced by in vitro translation of the relevant plasmids with the TNT T7 quick coupled transcription-translation system according to the manufacturer's instructions (Promega). Glutathione-S-transferase (GST) pull-down assays were performed as previously described (18). Briefly, GST and GST-laminA389 expression was induced in Escherichia coli BL21 cells by addition of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Pull-down assays were performed with modified NETN buffer (1.5% Nonidet P-40, 1 mM EDTA, 20 mM Tris-HCl [pH 8], 200 mM NaCl) by binding GST fusion proteins to glutathione-Sepharose beads (Amersham Biosciences) and then incubating them with in vitro-translated proteins. For MBP pull-down assays, expression of MBP-SUN1 fusion proteins was performed by transformation of E. coli BL21 cells with the appropriate pMAL-SUN1 constructs and induction with 0.2 mM IPTG in the presence of 0.2% (wt/vol) glucose. Pull-down assays were performed by binding to amylose resin (New England Biolabs) in modified NETN buffer.
Antibodies.
Anti-SUN1 antibodies were generated by inducing MBP-SUN1-C expression in E. coli BL21 cells and isolating the protein purified with amylose resin. The purified fusion protein was used to immunize rabbits (Cambridge Research Biochemicals), and the antibody was affinity purified from the resulting rabbit serum with an Affigel 15 affinity column (Bio-Rad).
Monoclonal anti-lamin A/C antibody XB10 was a kind gift from B. Burke, and rabbit anti-lamin A/C antibodies were generously provided by L. Gerace and E. Schirmer. Anti-myc 9E10 antibodies were obtained from Zymed. Rabbit anti-HA tag and mouse anti-CBP antibodies were from Santa Cruz Biotechnologies. Rabbit anti-green fluorescent protein (GFP) antibodies were purchased from AbCam. Anti-tubulin antibodies were obtained from Sigma. Monoclonal anti-LAP1 antibodies (RL13) were obtained from Affinity Bioreagents. Anti-nesprin 2 (N2) antibodies have been described previously (48).
Cell culture and transfections.
NIH 3T3, U2OS, and Lmna−/− mouse embryo fibroblast (MEF) cells (kind gift of B. Burke) were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum and antibiotics at 37°C and 5% CO2. Transfection of pCI-neo and pCDNA3 constructs was performed with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's recommendation.
Cell extracts, immunoprecipitation, and immunoblotting.
Cells were grown to confluence on 10-cm dishes. To prepare total cell extracts for immunoblotting, the cell pellets were resuspended in phosphate-buffered saline (PBS) and an equal volume of Laemmli buffer was then added. Nuclei were prepared by an adaptation of the method described by Collas et al. (4). Briefly, cells were removed from plates by scraping, washed in PBS, and incubated in approximately 20 volumes of cold hypotonic buffer (10 mM HEPES [pH 7.4], 2 mM MgCl2, 25 mM KCl, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride [PMSF]) containing Complete protease inhibitor cocktail (Roche) for 45 min at 4°C to swell the cells. Cells were lysed by 100 strokes in a Dounce homogenizer, and 0.5 volume of hypotonic buffer containing 24% sucrose was added. Nuclei were pelleted by centrifuging at 400 × g at 4°C for 10 min, and in some instances the supernatant (cytoplasmic fraction) was collected. Nuclei were washed once in hypotonic buffer containing 8% sucrose. For further fractionation, nuclei were resuspended in extraction buffer (10 mM HEPES [pH 7.4], 2 mM MgCl2, 1 mM PMSF) supplemented with either 7 M urea or 1% Triton X-100 and 50 to 500 mM NaCl, as indicated. Extracts were incubated on ice for 30 min and then centrifuged at 16,000 × g for 10 min to obtain soluble and insoluble fractions.
For immunoprecipitations, cells were scraped into lysis buffer (10 mM HEPES [pH 7.4], 5 mM EDTA, 50 mM NaCl, 1% Triton X-100) supplemented with 1 mM PMSF and protease inhibitor cocktail (Roche), followed by sonication and centrifugation at 16,000 × g and 4°C for 10 min. Lysates were precleared with protein A-Sepharose (Amersham Biosciences) for 1 h and then incubated with 2 μg of antibody for 2 h at 4°C. Antibodies were precipitated by addition of protein A-Sepharose for a further 1 h, pelleted at 1,000 × g, and then washed in lysis buffer minus protease inhibitors.
For immunoblotting, samples were boiled in an equal volume of 2× Laemmli buffer, resolved on 7.5% polyacrylamide gels, and transferred to nitrocellulose membrane. Following incubation with primary antibodies, these were detected with horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibodies (Sigma) and visualized with the ECL plus Western blotting detection system (Amersham Biosciences).
Indirect immunofluorescence microscopy.
Cells grown on glass coverslips were fixed in methanol at −20°C and processed for indirect immunofluorescence microscopy as previously described (7). Where indicated, cells were instead fixed in 4% paraformaldehyde and permeabilized either with 0.5% Triton X-100 at room temperature for 5 min or with 40 μg/ml digitonin on ice for 2 to 5 min. Cells were washed in PBS and incubated with antibodies diluted in PBS-3% bovine serum albumin. Secondary antibodies were goat anti-rabbit AlexaFluor 488 and donkey anti-mouse AlexaFluor 594 (Molecular Probes Inc.). DNA was stained with 0.2 μg/ml Hoechst 33258 (Sigma). Coverslips were mounted in 80% glycerol-3% n-propyl gallate (in PBS). Fluorescence microscopy was performed with a Nikon TE300 inverted microscope with an ORCA ER charge-couple device camera (Hamamatsu) and Openlab 4.02 software (Improvision). Images were processed with Adobe Photoshop (Adobe Systems).
RESULTS
Identification of SUN1 as a lamin A-interacting protein.
We previously described the use of a yeast two-hybrid interaction screen with the CTD of lamin A as bait to identify transcription factor SREBP1 as a novel lamin A interactor (18). Sequencing of 12 out of the 19 clones obtained in the screen identified a total of five interacting proteins. Two clones encoded the full-length 913-amino-acid murine SUN1 (mSUN1) protein (Fig. 1A).
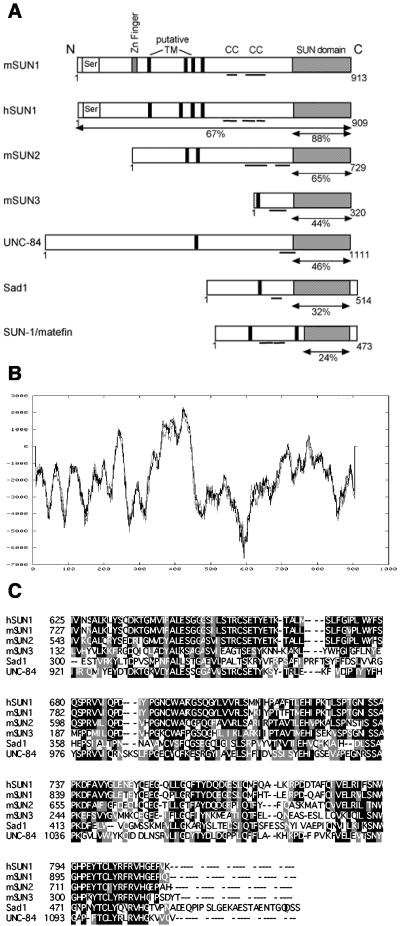
FIG. 1.
SUN protein family sequence alignments and predicted structures. (A) Schematic representation of SUN protein structures highlighting the serine-rich domain (Ser), zinc (Zn) finger motif, transmembrane (TM) domains predicted by TMpred, coiled coils (CC), and conserved SUN domain (shaded box). Percent homology with mSUN1 over the relevant region (arrows) is shown for each homolog. (B) mSUN1 hydropathy plot obtained with TMpred software, predicting four transmembrane domains at amino acids 231 to 254, 358 to 383, 386 to 407, and 413 to 431. (C) Alignment of the C-terminal SUN domains of SUN protein family members. Identical residues and conservative substitutions are shown in black and gray, respectively.
Based on software algorithms that predict protein structural motifs, mSUN1 comprises distinct N- and C-terminal regions separated by up to three clustered putative transmembrane domains, with a fourth weaker potential membrane-spanning region positioned 100 residues N terminal to them (Fig. 1B). The N terminus contains a serine-rich region and a Cys2/His2 zinc finger domain, whereas the C terminus contains two potential coiled-coil regions and the SUN domain, which is so named because of its homology with the Schizosaccharomyces pombe Sad1 and C. elegans UNC-84 proteins (13, 20) (Fig. 1C).
Searches of the mouse Ensembl database revealed that the mSUN1 gene is composed of 23 exons spanning 50 kb on chromosome 5G1. This region is syntenic with the region on human chromosome 7 containing hSUN1 (7p22); thus, the proteins are likely to be true orthologs. From the initial comparison, it appeared that hSUN1 lacked 87 amino acids encoded by exons 7 and 8 of mSUN1. However, analysis of the hSUN1 genomic sequence revealed that these two exons are in fact present within a predicted intron, resulting in a protein of 909 rather than the reported 822 amino acids. Database searches identified cDNA clones containing this additional sequence, suggesting that SUN1 may undergo alternative splicing. Two potential mSUN1 orthologs, mSUN2 and mSUN3, were also identified (Fig. 1). Comparison of SUN protein sequences revealed that mouse SUN1 and SUN2 both share significantly higher homology with UNC-84 than with Ce-SUN1 (Table 1). Indeed, UNC-84 itself has greater sequence conservation with the mouse SUN domain proteins than with Ce-SUN1, suggesting that both mammalian SUN proteins probably evolved from UNC-84 and may have similar functions.
TABLE 1.
Percent homology between C. elegans and mouse SUN domain proteins
| Protein | % Homology with:
|
||
|---|---|---|---|
| mSUN1 | mSUN2 | UNC-84 | |
| mSUN2 | 64 | ||
| UNC-84 | 44 | 37 | |
| Ce-SUN-1 | 21 | 24 | 24 |
Probing of a mouse multiple-tissue Northern blot with an mSUN1 cDNA fragment identified a 4.3-kb band in all of the tissues examined, while in brain tissue a 9-kb band was also detected (data not shown). Thus, SUN1 appears to be widely expressed and may be subject to alternative splicing in some tissues.
The NTD of mSUN1 is responsible for lamin A binding and oligomerization.
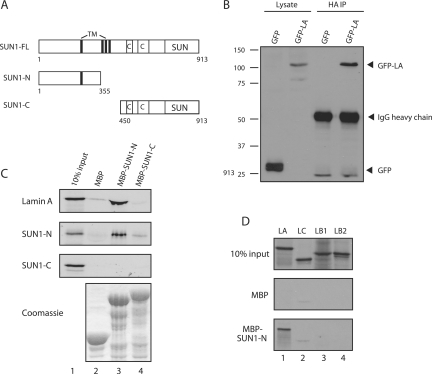
To confirm that SUN1 is a lamin A-binding protein, immunoprecipitation experiments were performed. Due to the high insolubility of both proteins when present at endogenous levels (see below), U2OS cells were cotransfected with plasmids encoding HA-tagged SUN1 together with either GFP or GFP-lamin A. Anti-HA antibodies efficiently coprecipitated GFP-lamin A, but not GFP alone (Fig. 2B). Pull-down experiments with MBP-fused SUN1 NTD or CTD constructs (encoding residues 1 to 355 and 450 to 913, respectively; Fig. 2A) revealed that the NTD of SUN1 is responsible for lamin A binding (Fig. 2C, top). In a reciprocal experiment with the GST-fused human lamin A CTD (residues 389 to 664), the SUN1 NTD showed only a weak interaction with the lamin A CTD (data not shown). This suggests that additional sequences, in the N-terminal head or coiled-coil rod domain of lamin A, are required for optimal SUN1 binding.
FIG. 2.
The NTD of SUN1 interacts with lamin A and with itself. (A) Schematic representation of the SUN1 constructs used. TM, transmembrane. (B) Cell lysates were made from U2OS cells transfected with HA-SUN1 together with either GFP (lanes 1 and 3) or GFP-lamin A (lanes 2 and 4). HA-SUN1 was immunoprecipitated with anti-HA antibodies and bound to protein A-Sepharose beads. Initial lysates (lanes 1 and 2) and immunoprecipitates (IP; lanes 3 and 4) were immunoblotted with anti-GFP antibodies to detect coprecipitating proteins. Positions of molecular size markers (kilodaltons) are shown on the left. IgG, immunoglobulin G. (C) 35S-labeled lamin A, SUN1-N, and SUN1-C proteins were produced by in vitro translation (lanes 1). The proteins were each incubated with MBP (lanes 2), MBP-SUN1-N (lanes 3), or MBP-SUN1-C (lanes 4) previously bound to amylose resin. Following resolution by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, bound proteins were detected by autoradiography. The Coomassie-stained gel shows equal expression of the MBP fusion proteins. (D) Lamin A (LA), lamin C (LC), lamin B1 (LB1), and lamin B2 (LB2) were produced by in vitro translation (top) and bound to MBP alone (middle) or to MBP-SUN1-N (bottom) previously immobilized on amylose beads. Bound proteins were detected by autoradiography.
Complementation studies of C. elegans unc-84 mutants previously indicated that UNC-84 may dimerize (20). We investigated the possibility that SUN1 dimerizes by performing pull-down assays with MBP fusions of the SUN1 NTD and CTD. In vitro-translated SUN1-N was capable of binding to MBP-SUN1-N but not to MBP-SUN1-C (Fig. 2C), demonstrating that the NTD of SUN1, as well as binding lamin A, is responsible for oligomerization of the protein. In contrast, SUN1-C did not bind to either itself or the NTD.
SUN1 is an NE protein.
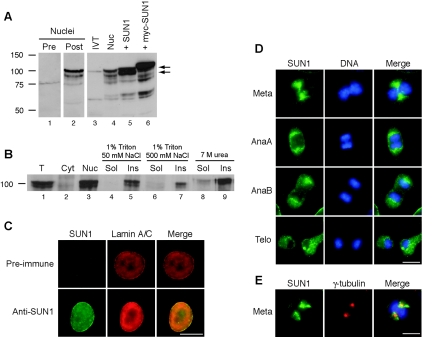
Polyclonal rabbit antisera were raised against the CTD of murine SUN1 (residues 450 to 913) and affinity purified. The predicted molecular mass of mSUN1 is 101 kDa. In protein extracts of NIH 3T3 cells, this antibody (SSHR1) recognized a doublet of approximately 100 and 90 kDa which was present in a nuclear fraction but was absent when the extract was probed with preimmune serum (Fig. 3A, lanes 1 and 2). To confirm that this doublet represents SUN1 protein, NIH 3T3 cells were transfected with untagged and myc-tagged SUN1 constructs. The doublet increased in intensity in SUN1-transfected cells, and a slightly higher-molecular-weight band was seen in myc-SUN1-transfected cells (Fig. 3A, lanes 4 to 6).
FIG. 3.
SUN1 is an NE protein. (A) Nuclei were produced from untransfected NIH 3T3 cells, and total extracts were produced from NIH 3T3 cells transiently transfected with untagged or myc-tagged SUN1 constructs, as indicated. In vitro-translated (IVT) SUN1 was generated from pCI-mSUN1. Protein samples were immunoblotted and probed with either preimmune rabbit serum (lane 1) or affinity-purified anti-SUN1 antibodies (lanes 2 to 6). Arrows indicate the mSUN1 doublet. Positions of molecular size markers (kilodaltons) are shown on the left. (B) Total (T), cytoplasmic (Cyt), and nuclear (Nuc) fractions were produced from NIH 3T3 cells. The nuclear fraction was then subjected to further extraction with buffers containing Triton X-100, salt, and urea, as indicated. Following centrifugation, soluble (Sol) and insoluble (Ins) fractions were produced. All fractions were analyzed by immunoblotting with anti-SUN1 antibodies. (C to E) Immunofluorescence localization of SUN1 in interphase (C) and mitotic (D and E) cells. In panel C, cells were stained with anti-lamin A/C antibody XB10 (red) and either preimmune serum or affinity-purified SUN1 antibodies (green). In panel D, cells stained for SUN1 (green) are shown at different stages of mitosis: metaphase (Meta), anaphase A (AnaA), anaphase B (AnaB), and telophase (Telo). In panel E, a metaphase cell is shown costained with SUN1 and mouse anti-γ-tubulin antibodies. DNA is shown in blue. Scale bars in panels C to E, 10 μm.
We determined which band of the doublet corresponds to full-length SUN1 by comparison with in vitro-translated SUN1 protein. Western blotting detected a protein of approximately 100 kDa that comigrated with the upper band of the NIH 3T3 SUN1 doublet (Fig. 3A, lanes 3 and 4). Together, these results indicate that SUN1 is produced as a protein of 100 kDa and that the lower band of 90 kDa most likely represents an alternative splice form or a truncated product generated in cells.
Biochemical fractionation experiments revealed that SUN1 is exclusively located in the nuclear fraction and is resistant to both Triton X-100 and high-salt extraction (Fig. 3B), consistent with SUN1 being a nuclear lamina-associated protein. Furthermore, SUN1 was resistant to extraction with 7 M urea, indicating that it is membrane associated.
To determine the subcellular localization of endogenous SUN1 in NIH 3T3 cells, indirect immunofluorescence microscopy was performed. We found that SUN1 localizes exclusively to the nucleus and colocalizes with lamin A/C at the NE (Fig. 3C). The distribution of SUN1 was also observed in mitotic cells (Fig. 3D). In prometaphase and metaphase, following breakdown of the NE, SUN1 became concentrated around two foci on either side of the condensed chromosomes. These foci partially colocalized with the spindle poles, as judged by costaining with antibodies against the centrosomal protein γ-tubulin (Fig. 3E). By anaphase, SUN1 was mainly dispersed throughout the cytoplasm, although enrichment at the polar face of the chromosomes was still evident in some cells (Fig. 3D). These results are in agreement with recent observations of NE breakdown in live cells (2, 35), which suggest that the NE is mechanically torn apart by forces generated by microtubule-associated dynein attached to the NE. In these studies, membrane-associated NE proteins were found to be enriched around the spindle poles until early metaphase and moved toward the poles in a microtubule-dependent manner. Colocalization experiments with γ-tubulin indicated that SUN1 did not localize to the centrosome or pericentrosomal region at any other stage of the cell cycle (data not shown).
The NTD of SUN1 is responsible for anchoring to the nuclear lamina.
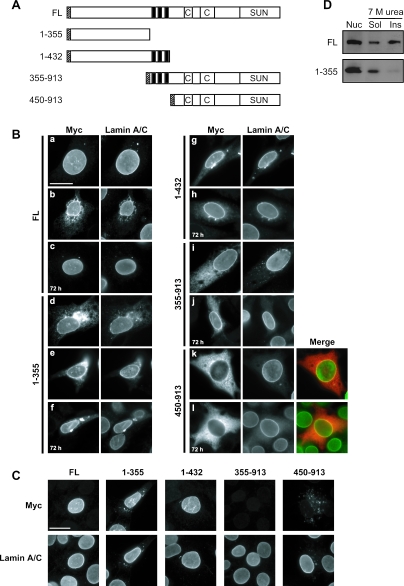
To investigate the requirements for SUN1 localization at the NE, various myc-tagged SUN1 constructs with N- or C-terminal deletions were generated, as shown in Fig. 4A. In addition to the SUN1(1-355) construct mentioned previously, a further NTD construct, SUN1(1-432), was generated that also included the three clustered membrane-spanning sequences. Likewise, an additional CTD construct that included the three transmembrane domains, SUN1(355-913), was produced. These constructs and full-length myc-SUN1 were transiently expressed in NIH 3T3 cells and localized with antibodies against the myc tag. Twenty-four hours after transfection, full-length myc-SUN1 colocalized with lamin A/C at the NE but, in highly expressing cells, was also found in cytoplasmic aggregates or had a cytoplasmic distribution reminiscent of ER accumulation (Fig. 4B, a and b). The latter finding suggests that binding sites for SUN1 at the NE are easily saturated. Consistent with this, at 72 h posttransfection, myc-SUN1 was present at lower levels and was found exclusively at the NE (Fig. 4B, c). Similar properties have recently been reported for the INM protein nurim (15).
FIG. 4.
The NTD of SUN1 is responsible for anchoring to the nuclear lamina. (A) Schematic representation of the myc-tagged SUN1 constructs. Labeling is as in Fig. 2A, except that hatched boxes indicate locations of myc tags. (B) NIH 3T3 cells were transiently transfected with myc-SUN1 constructs as indicated and fixed in methanol 24 or 72 h (c, f, h, j, and l) posttransfection. Cells were then costained with anti-myc 9E10 (left side) and 3262 anti-lamin A/C (right side) antibodies. In parts k and l, apparent colocalization of myc-SUN1(450-913) with lamin A/C at the NE is indicated by yellow areas in the merged color images [red, myc-SUN1(450-913); green, lamin A/C]. (C) Transfected NIH 3T3 cells were subjected to pre-extraction with 0.5% Triton X-100 in PBS for 5 min on ice, immediately fixed in methanol, and then subjected to immunofluorescence staining as for panel B. Scale bars, 10 μm. (D) NIH 3T3 cells were transfected with myc-tagged SUN1 constructs encoding the full-length (FL) protein or the NTD (amino acids 1 to 355). Nuclei (Nuc) were isolated and incubated in extraction buffer containing 7 M urea, separated into soluble (Sol) and insoluble (Ins) fractions, and immunoblotted with anti-myc 9E10 antibodies.
Like the full-length protein, the NTD construct SUN1(1-432) localized mainly to the NE, indicating that the NTD of the protein contains all of the necessary sequences for NE targeting (Fig. 4B, g and h). Although the shorter SUN1(1-355) protein showed a greater degree of cytoplasmic staining at 24 h, by 72 h the protein was mainly concentrated at the NE, suggesting that it may require more time to fully incorporate (Fig. 4B, d to f). Surprisingly, the CTD construct SUN1(355-913) also concentrated at the NE 24 h after transfection, with additional cytoplasmic accumulation that was largely lost by 72 h (Fig. 4B, i and j). In contrast, SUN1(450-913) appeared evenly distributed throughout the cytoplasm at all times (Fig. 4b, k and l). Costaining with lamin A/C indicated some overlap with SUN1(450-913); however, at the resolution of light microscopy, it was not possible to determine whether this represented SUN1(450-913) on the outer or inner face of the NE or indeed within the NE lumen (Fig. 4B, k and l, merge).
INM proteins associated with the insoluble nuclear lamina and matrix have been shown to be resistant to detergent extraction (6, 9, 15) (Fig. 3B). To determine which of the SUN1 domains is responsible for interaction with the nuclear lamina, transfected cells were therefore subjected to pre-extraction with 0.5% Triton X-100 72 h after transfection and the myc-SUN1 proteins were then detected by immunofluorescence microscopy. Full-length SUN1 and the NTD constructs SUN1(1-432) and SUN1(1-355) all remained highly expressed at the NE following Triton X-100 pre-extraction, and any cytoplasmic staining was lost. In contrast, both SUN1(355-913) and SUN1(450-913) were entirely absent from both the NE and the cytoplasm (Fig. 4C). These results correlate with our in vitro pull-down assay data, indicating that the NTD of SUN1 is responsible for NE localization through interaction with lamin A at the INM.
Previous studies on the requirements for NE localization have indicated that, in addition to a nucleoplasmic lamina/chromatin-interacting domain, a membrane-spanning domain is also required (28, 39, 45). The NE localization of SUN1(1-355) therefore suggested that it too contains a functional transmembrane domain, presumably the weaker domain predicted to lie at residues 231 to 254 (Fig. 1). We tested the membrane association of SUN1(1-355) by extraction of transfected NIH 3T3 cells with 7 M urea and found that while most of the full-length SUN1 protein was resistant to extraction, SUN1(1-355) was found almost exclusively in the soluble fraction (Fig. 4D). This result indicates that residues 1 to 355 do not contain a functional transmembrane domain and that, like lamins themselves, the nucleoplasmic domain of SUN1 is able to associate with the NE in the absence of a membrane-spanning region.
SUN1 does not require lamin A/C for NE localization.
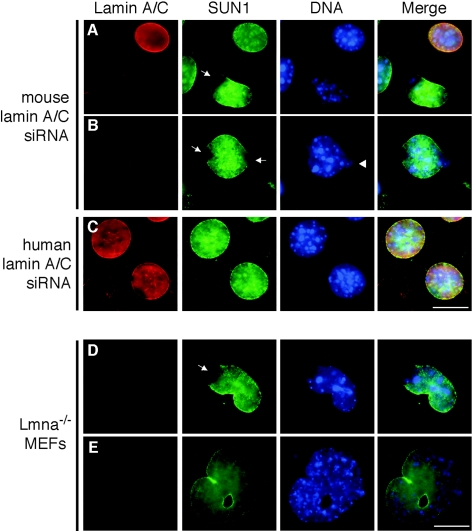
Our results indicate that the NTD of SUN1 is necessary for association with the nuclear lamina and is responsible for interaction with lamin A. This raised the question of whether NE localization of SUN1 is dependent upon lamin A binding. To test this, SUN1 distribution was assessed in NIH 3T3 cells in which lamin A/C expression was depleted by RNA interference. In cells transfected with mouse lamin A/C-specific small interfering RNA oligonucleotides (siRNAs), SUN1 still localized to the NE but, in some cells, was absent from one or both poles of the nucleus (Fig. 5A and B). These nuclei tended to exhibit an abnormal morphology, with chromatin apparently extruding from the confines of the nucleus. In contrast, no reduction in lamin A/C and no abnormalities in SUN1 distribution were apparent in cells treated with a control human lamin A/C siRNA (Fig. 5C), which has a single-nucleotide mismatch with the mouse sequence. SUN1 was also still present at the NE in lamin A/C knockout (Lmna−/−) MEFs, with a similar absence of SUN1 from the poles of the nucleus (Fig. 5D and E). The same phenomenon has been noted for other NE proteins, including LAP2, Nup153, and lamin B, and is thought to represent sites of NE herniation (43).
FIG. 5.
Lamin A/C is not required for localization of SUN1 to the NE. Subcellular localization of SUN1 in RNA interference-treated NIH 3T3 (A to C) or Lmna knockout MEF cells (D and E). NIH 3T3 cells were transfected with a mouse (A and B) or a control human (C) lamin A/C siRNA and processed for immunofluorescence microscopy 48 h after transfection. Cells were stained with anti-lamin A/C XB10 (red) and anti-SUN1 (green) antibodies. Hoechst staining of DNA is in blue. Arrows indicate disrupted SUN1 localization at the poles of nuclei. The arrowhead indicates apparent leakage of DNA from the nucleus. Scale bars, 10 μm.
These results suggest that lamin A/C is not essential for localization of SUN1 to the NE and that additional nuclear proteins are likely to bind SUN1. Since many NE proteins are capable of interaction with both type A and B lamins, we compared the interaction of MBP-fused SUN1(1-355) with all four major lamin isoforms (A, C, B1, and B2) by in vitro pull-down assay. While lamin A was efficiently pulled down by the SUN1 NTD, neither lamin B1 nor B2 showed a significant interaction (Fig. 2D). Surprisingly, lamin C also did not interact with SUN1, indicating that the interaction is lamin A specific. This is in keeping with the notion that the main site of interaction is the CTD of lamin A. Taken together, our results suggest that localization of SUN1 to the NE is not entirely dependent on either type A or B lamins and that an additional binding partner must be responsible for nuclear anchoring of SUN1 in the absence of lamin A.
The CTD of SUN1 is located in the NE lumen.
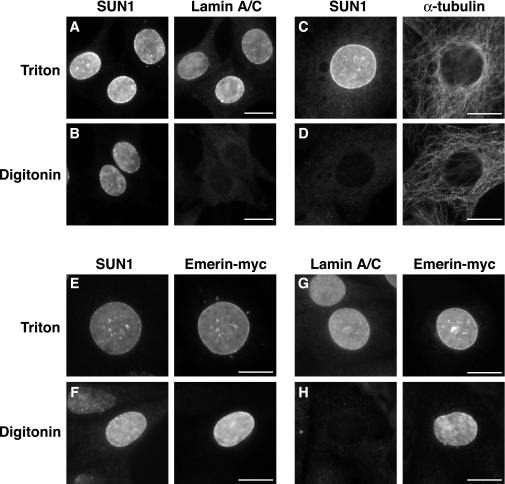
Having demonstrated that the NTD of SUN1 is nucleoplasmic, we next sought to determine the location of the CTD. Since the number of transmembrane domains present in the protein could not be accurately predicted from the hydropathy plot (Fig. 1B), the CTD could be either luminal or nucleoplasmic, depending on whether there is an odd or even number of membrane-spanning regions, respectively. To address this, paraformaldehyde-fixed NIH 3T3 cells were permeabilized either with digitonin for 5 min, leaving the INM intact and the nucleoplasm inaccessible to antibodies, or with Triton X-100, which permeabilizes the entire NE. In Triton X-100-permeabilized cells, as expected, the CTD of SUN1 and nuclear lamin A/C were recognized by their respective antibodies (Fig. 6A). When cells were instead permeabilized with digitonin, neither lamin A/C nor CBP was detectable, demonstrating that the NE was impermeable to antibodies. In contrast, the CTD of SUN1 remained accessible to SUN1 antibodies in the same digitonin-treated cells, suggesting that this domain is located in the NE lumen (Fig. 6B and data not shown). To confirm that the ONM was permeabilized in the presence of digitonin, thereby allowing access to the NE lumen, NIH 3T3 cells were also transfected with an emerin construct engineered with a myc tag at its C terminus to act as a marker for the NE lumen. Following digitonin permeabilization, both the myc tag and the SUN1 CTD were visible, whereas nucleoplasmic lamins A and C were not detected (Fig. 6E to H). By reducing exposure to digitonin to 2 min, we were able to selectively permeabilize only the plasma membrane, as judged by cytoplasmic α-tubulin staining (Fig. 6C to D). In these cells, SUN1 was not detected, demonstrating that the SUN1 CTD does not reside on the outer face of the NE. Together, these results indicate that the CTD of SUN1 is located in the NE lumen.
FIG. 6.
The CTD of SUN1 is located in the NE lumen. NIH 3T3 cells left untransfected (A to D) or transfected with pCI-emerin-myc (E to H) were fixed with 4% paraformaldehyde and permeabilized with either 0.5% Triton X-100 (A, C, E, and G) or 40 μg/ml digitonin on ice for 5 min (B, D, and F) or 2 min (H). Immunofluorescence staining was performed with antibodies against SUN1, α-tubulin, lamin A/C (3262), and the myc epitope, as indicated. Scale bars, 10 μm.
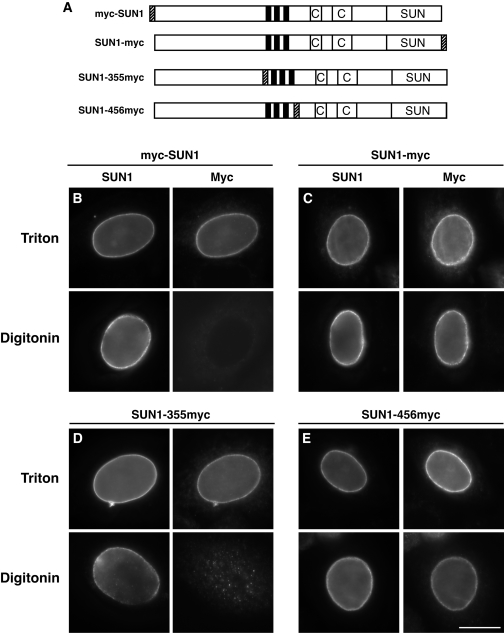
We further probed the topology of SUN1 with a series of SUN1 constructs with myc tags engineered at different sites: at the N terminus (myc-SUN1), at the C terminus (SUN1-myc), and internally, after either residue 355 (SUN1-355myc) or 456 (SUN1-456myc). The latter two constructs introduced the myc tag just upstream and downstream of the three predicted transmembrane domains, respectively (Fig. 7A). U2OS cells were transiently transfected with all four myc-tagged SUN1 constructs and permeabilized for 5 min with either Triton X-100 or digitonin. Like anti-SUN1 antibodies, anti-myc antibodies were able to detect both SUN1-myc and SUN1-456myc in digitonin-treated cells (Fig. 7C and E), indicating that the entire CTD is luminal. In contrast, 9E10 antibodies did not detect either myc-SUN1 or SUN1-355myc at the NE following digitonin permeabilization (Fig. 7B and D).
FIG. 7.
Probing the topology of SUN1 using myc-tagged SUN1 constructs. (A) Schematic representation of the myc-tagged SUN1 constructs used. Positions of myc tags are shown as hatched boxes. (B to E) U2OS cells were transiently transfected with SUN1 constructs with a myc tag engineered at either the N terminus (B) or the C terminus (C) or internally following residue 355 (D) or 456 (E). The cells were fixed with 4% paraformaldehyde and permeabilized with either Triton X-100 at room temperature (top) or with digitonin at 4°C (bottom) and then costained with anti-SUN1 (left side) and anti-myc 9E10 (right side) antibodies. Scale bar, 10 μm.
Taken together, our results convincingly demonstrate that SUN1 has the topology of a typical INM protein, with a nucleoplasmic NTD and a luminal CTD, and that it possesses either one or three centrally located transmembrane domains.
SUN1 interacts with the luminal KASH domain of nesprins 1 and 2.
SUN domain proteins differ from most other INM proteins in possessing a large CTD. This suggests that they have a significant functional role within the NE lumen, presumably through protein-protein interactions. UNC-84 has been shown to be required for ONM localization of the giant actin-binding protein ANC-1 in C. elegans, yet a direct interaction between the two proteins has not been identified. We therefore sought to determine whether SUN1 interacts with the mammalian ANC-1 homologs, nesprins, by coimmunoprecipitation.
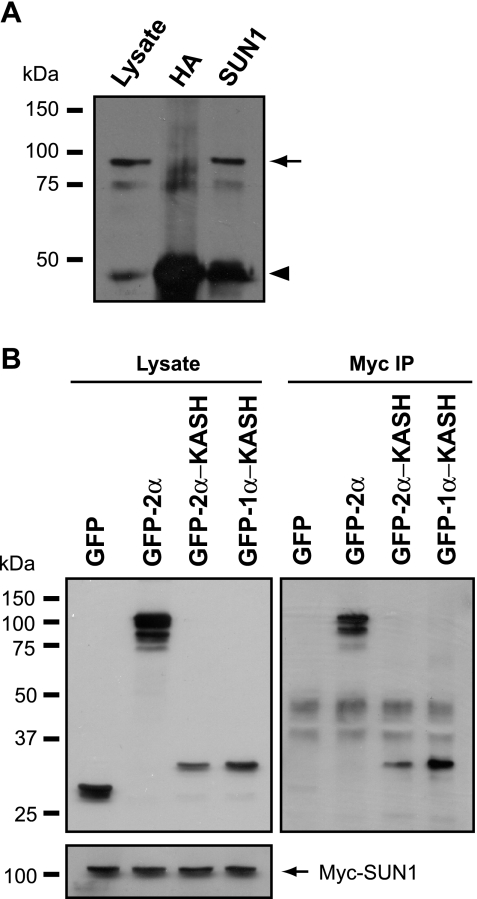
U2OS cells were cotransfected with myc-SUN1 and nesprin 2β, and the myc-SUN1 was immunoprecipitated with SUN1 antibodies. Nesprin 2β was efficiently coprecipitated by the anti-SUN1 antibodies but not by nonspecific anti-HA antibodies (Fig. 8A). In complementary experiments, GFP-nesprin 2α, but not GFP alone, was found to coprecipitate myc-SUN1 (Fig. 8B). Furthermore, the luminal KASH domains of both nesprin 1α and nesprin 2α were sufficient for this interaction.
FIG. 8.
The SUN1 CTD interacts with the KASH domain of nesprins 1 and 2. (A) Lysates of U2OS cells transiently transfected with myc-SUN1 and nesprin 2β were subjected to immunoprecipitation with either anti-SUN1 or nonspecific anti-HA tag antibodies. The initial lysate and immunoprecipitates were then immunoblotted with anti-nesprin N2 antibodies. The arrow and arrowhead indicate coprecipitated nesprin 2β and the antibody heavy chain, respectively. (B) U2OS cells were cotransfected with myc-SUN1 and GFP or GFP-nesprin fusions, as indicated. Expression of GFP- and myc-tagged proteins was confirmed by immunoblotting (upper and lower left parts, respectively). Anti-myc immunoprecipitates (IP) were probed with GFP antibodies to detect coprecipitating GFP fusion proteins (right side).
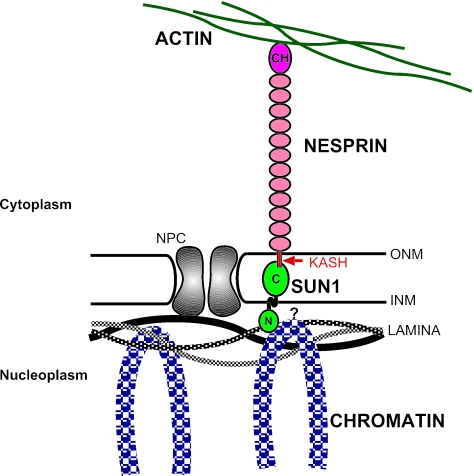
Together, our data strongly support a model whereby SUN1 and nesprins, located in the INM and ONM, respectively, form a bridge across the NE and provide a direct physical connection between the nuclear lamina (and potentially chromatin) and the cytoskeleton (Fig. 9).
FIG. 9.
Model of SUN1 topology and interactions at the NE. SUN1 is anchored at the INM through interaction of the nucleoplasmic NTD with lamin A and other nuclear factors, most likely chromatin (indicated by the question mark). The CTD of SUN1 lies in the NE lumen, where it interacts with nesprins that contain a C-terminal membrane-spanning KASH domain, resulting in their anchoring at the ONM. The N terminus of long nesprin isoforms binds to actin. As a result, a direct physical connection between the nuclear interior and the actin cytoskeleton is created. NPC, nuclear pore complex; CH, calponin homology domain.
DISCUSSION
In a yeast two-hybrid interaction screen, we have identified murine SUN1 as a lamin A-binding protein, thus demonstrating interaction of SUN domain proteins with the nuclear lamina. With antibodies raised against the CTD, we have confirmed SUN1 as an integral membrane protein of the NE: SUN1 is associated with the urea-insoluble nuclear fraction of NIH 3T3 cells and, in immunofluorescence studies, localizes to the nuclear periphery. In support of our findings, SUN1 was previously isolated in two subcellular proteomics approaches involving NE fractionation and sequencing of isolated proteins by mass spectrometry (5, 37).
SUN1 interacts with lamin A but not with other lamin isoforms.
The interaction between lamin A and SUN1 was confirmed both by in vitro pull-down assay and by coimmunoprecipitation, thus providing strong evidence that the interaction occurs in vivo. The interaction appeared much stronger with full-length lamin A than with the CTD fragment used for the initial yeast two-hybrid screen, suggesting that sequences in the coiled-coil domain of lamin A may also be required for optimal SUN1 binding. However, it cannot be ruled out that this effect was produced simply by the fact that the tagging of the proteins differed in the two experiments (GST-lamin A versus MBP-SUN1), and we are currently investigating the binding requirements in greater detail. Surprisingly, we could not detect an interaction with the alternative splice product lamin C, which differs from lamin A only in lacking the C-terminal 98 residues. This suggests that the binding site for SUN1 includes the extreme C terminus of lamin A.
Less surprisingly, we found that SUN1 does not interact with B-type lamins. Studies of other INM proteins indicate that they exhibit different lamin isoform binding specificities; for example, LBR and emerin preferentially bind to lamin B and lamin C, respectively (44, 46).
SUN1 may bind to chromatin.
While SUN1 interacts with lamin A, we found that the presence of lamin A/C is not required for SUN1 NE localization since SUN1 remains exclusively located at the NE in cells lacking lamin A/C. In contrast, emerin has been shown to redistribute to the ER in cells lacking lamin A/C, indicating that it relies solely upon lamin A/C for anchoring at the INM (43). Our findings suggest that SUN1 also binds to at least one other nuclear factor to ensure INM targeting. Many INM proteins interact with chromatin-associated proteins, providing a membrane anchor important for chromatin organization (12). A similar interaction between SUN1 and chromatin may therefore confer NE localization in the absence of type A lamins.
It has been suggested that C. elegans UNC-84 dimerizes (20), and here we provide evidence from GST pull-down experiments that the NTD of SUN1 is also capable of self-association. The functional significance of oligomerization is not known. One possibility is that, similar to other NE proteins, including lamins (19, 34) and LBR (46), SUN1 binds directly to DNA, a function which often requires protein dimerization. The presence of a zinc finger motif within the NTD of SUN1 is also worth noting in this regard. Direct DNA binding may thus provide the additional anchor for SUN1 at the NE.
Both the NTD and CTD of SUN1 are capable of NE localization.
In an attempt to define the sequences required for NE targeting, we used a combination of biochemical and immunofluorescence localization techniques to study the behavior of a range of SUN1 deletion mutants. Firstly, we used in vitro pull-down assays to demonstrate that the NTD of SUN1, encompassing residues 1 to 355, is responsible for lamin A binding. This suggested that, like other INM proteins, the NTD of SUN1 anchors the protein at the INM through interaction with the nuclear lamina. Surprisingly, however, we found that both the NTD and CTD of SUN1 are capable of NE localization when expressed in conjunction with the three central transmembrane domains, indicating that both termini contain NE-targeting sequences. In the absence of transmembrane domains, the CTD of SUN1 was highly expressed in the cytoplasm. Interestingly, there was also significant overlap with lamin A/C staining at the NE, suggesting that the CTD may interact with components of the NE. Given our finding that the CTD of SUN1 interacts with nesprins and previous studies that have shown that the KASH domain of nesprins is responsible for their NE localization, it is likely to be an interaction between these two domains that is responsible for NE localization of the SUN1 CTD.
The NTD of SUN1 targets the protein to the NE through association with the insoluble nuclear lamina.
To convincingly establish which domain of SUN1 is responsible for anchoring at the INM, we utilized the fact that resistance to detergent extraction can be used as a criterion for determining protein association with the insoluble nuclear lamina and matrix (6, 9, 15). We found that only SUN1 fragments including the NTD (residues 1 to 355) were retained at the NE following Triton X-100 pre-extraction, whereas those containing the CTD alone were efficiently solubilized. These results indicate that it is the NTD of SUN1, encompassing residues 1 to 355, that is responsible for the tight association of the protein with the nuclear lamina. Consistent with our data, the NTD of SUN2, including its two predicted transmembrane domains, is responsible for its targeting to the NE (14). In contrast, the association of the SUN1 CTD with the NE must occur via a different type of interaction, most likely with nesprins, that is sensitive to detergent extraction.
SUN1 does not require transmembrane sequences for NE targeting.
Targeting of proteins to the INM has previously been shown to require both a transmembrane domain and lamin/chromatin-interacting sequences within the nucleoplasmic domain of the protein (28, 40, 45). The N-terminal nucleoplasmic domains of LBR, emerin, and MAN1 are sufficient to target these proteins to the nucleus, presumably because of the presence of a bipartite nuclear localization signal in each of these domains. However, NE targeting was achieved only when the domains were attached to a transmembrane sequence, whether this was the native sequence or a heterologous sequence from an unrelated protein. Since the SUN1 NTD construct used in our studies was able to localize to the NE, this suggested that it contains both a nucleoplasmic interaction domain and a functional transmembrane domain. The first transmembrane domain of SUN1 predicted from hydropathy plots lies within the NTD, at amino acids 231 to 254 (Fig. 2A). Intriguingly, however, we found that, unlike full-length SUN1, SUN1(1-355) was soluble in 7 M urea, indicating that it is not membrane associated and does not contain a bona fide transmembrane domain. It is not clear why SUN1 should behave differently from other INM proteins that have been studied in this respect; however, more extensive interactions at the INM remain one possibility.
SUN1 is an INM protein with a luminal CTD.
We have used both endogenous SUN1 and a series of SUN1 constructs with myc tags engineered at various sites in digitonin permeabilization experiments that probe the topology of SUN1 at the NE. Our results demonstrate that the NTD, including residues 1 to 355, is located at the inner face of the NE, along with lamin A/C. Conversely, the CTD, including residues 450 to 913, resides in the NE lumen, together with the C terminus of emerin. This topology is typical of many NE proteins, including emerin, LAP1, LAP2, and nesprins (12). Indeed, the CTD of SUN2 has also been reported to be located in the NE lumen (14). On the other hand, the topologies of the C. elegans SUN domain proteins, UNC-84 and Ce-SUN1, are unknown and have been the focus of some debate (30, 42).
SUN1 interacts with cytoplasmic nesprins to form a bridge across the NE.
While the NTD of SUN1 interacts with the nuclear lamina, we have also demonstrated an interaction between SUN1 and nesprins. In addition to the ONM localization demonstrated for the large nesprin isoforms, many of the short isoforms are thought to localize to the INM through interactions with the nuclear lamina (24). This raises the possibility that nesprins may interact either with the nucleoplasmic NTD of SUN1 or with the luminal CTD. Here we have demonstrated that the conserved luminal KASH domain of both nesprins 1 and 2 is responsible for interaction with SUN1 and thus that the luminal CTD of SUN1 is capable of interaction with nesprins in the ONM.
Through its direct interaction with both the nuclear lamina and cytoplasmic nesprins, SUN1 therefore forms part of a physical bridge across the NE, linking the nuclear lamina to the cytoskeleton. An analogous mechanism involving Ce-lamin, UNC-84, and ANC-1 has been proposed in C. elegans. Although no physical interactions among these proteins have been detected, ANC-1 is dependent upon UNC-84 for its NE localization and, in turn, UNC-84 requires Ce-lamin for correct NE anchoring (17, 42).
The roles of SUN proteins in nuclear-cytoplasmic connection.
Nuclear-cytoskeletal connections are crucial for a wide variety of developmental processes, ranging from pronuclear migration following fertilization, through to establishing cell polarity in differentiated cells. The phenotypes displayed by unc-84 and Ce-sun1 mutants indicate the importance of SUN domain proteins in developmental processes and that they may play a role in attachment to both the actin and microtubule networks (10, 20, 21, 42).
The major function of SUN proteins appears to be in controlling nuclear position within the cell. Indeed, the existence of a cytoplasmically exposed NE “receptor” anchored by attachment to the nuclear lamina has previously been proposed as a mechanism for attachment of the cytoskeleton to the nucleus in the control of nuclear migration (31). Interestingly, physical connections between the plasma membrane, cytoskeleton, and nucleoskeleton have been demonstrated in experiments involving micromanipulation of the cell surface (22). Here, tugging of the cell surface via beads bound to integrins led to changes in cytoskeletal and nuclear organization, including reorientation of nucleoli. A further function of the SUN-nesprin NE bridge may therefore be to act as a mechanical signal transduction pathway in response to external stimuli.
ADDENDUM
During the revision of the manuscript and in agreement with our data, Padmakumar et al. reported SUN1 as an NE protein required for nesprin 2 localization to the NE (29).
Acknowledgments
We thank A. Saltiel (University of Michigan) for providing the 3T3-L1 yeast two-hybrid library and M. Pfuhl (University of Leicester, Leicester, United Kingdom) for help with protein sequence alignments. We are also grateful to J. Ellis (King's College, London, United Kingdom) for the emerin cDNA, B. Burke (University of Florida, Gainesville) for anti-lamin A/C XB10 antibodies and Lmna−/− MEFs, and E. Schirmer (University of Edinburgh, Edinburgh, United Kingdom) for the lamin B2 construct and 3262 lamin A/C antibodies.
This work was funded by grants to R.C.T. and S.S. from the British Heart Foundation and to S.S. and A.M.F. from the Wellcome Trust. S.S. is a Research Councils United Kingdom research fellow.
REFERENCES
- 1.Apel, E. D., R. M. Lewis, R. M. Grady, and J. R. Sanes. 2000. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 275:31986-31995. [DOI] [PubMed] [Google Scholar]
- 2.Beaudouin, J., D. Gerlich, N. Daigle, R. Eils, and J. Ellenberg. 2002. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108:83-96. [DOI] [PubMed] [Google Scholar]
- 3.Burke, B., and C. Stewart. 2002. Life at the edge: the nuclear envelope and human disease. Nat. Rev. Mol. Cell Biol. 3:575-585. [DOI] [PubMed] [Google Scholar]
- 4.Collas, P., K. Le Guellec, and K. Tasken. 1999. The A-kinase-anchoring protein AKAP95 is a multivalent protein with a key role in chromatin condensation at mitosis. J. Cell Biol. 147:1167-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dreger, M., L. Bengtsson, T. Schoneberg, H. Otto, and F. Hucho. 2001. Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc. Natl. Acad. Sci. USA 98:11943-11948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis, J. A., M. Craxton, J. R. Yates, and J. Kendrick-Jones. 1998. Aberrant intracellular targeting and cell cycle-dependent phosphorylation of emerin contribute to the Emery-Dreifuss muscular dystrophy phenotype. J. Cell Sci. 111(Pt. 6):781-792. [DOI] [PubMed] [Google Scholar]
- 7.Faragher, A. J., and A. M. Fry. 2003. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol. Biol. Cell 14:2876-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher, D. Z., N. Chaudhary, and G. Blobel. 1986. cDNA sequencing of nuclear lamin-A and lamin-C reveals primary and secondary structural homology to intermediate filament proteins. Proc. Natl. Acad. Sci. USA 83:6450-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foisner, R., and L. Gerace. 1993. Integral membrane-proteins of the nuclear-envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell 73:1267-1279. [DOI] [PubMed] [Google Scholar]
- 10.Fridkin, A., E. Mills, A. Margalit, E. Neufeld, K. K. Lee, N. Feinstein, M. Cohen, K. L. Wilson, and Y. Gruenbaum. 2004. Matefin, a Caenorhabditis elegans germ line-specific SUN-domain nuclear membrane protein, is essential for early embryonic and germ cell development. Proc. Natl. Acad. Sci. USA 101:6987-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldberg, M., A. Harel, and Y. Gruenbaum. 1999. The nuclear lamina: molecular organization and interaction with chromatin. Crit. Rev. Eukaryot. Gene Expr. 9:285-293. [DOI] [PubMed] [Google Scholar]
- 12.Gotzmann, J., and R. Foisner. 1999. Lamins and lamin-binding proteins in functional chromatin organization. Crit. Rev. Eukaryot. Gene Expr. 9:257-265. [DOI] [PubMed] [Google Scholar]
- 13.Hagan, I., and M. Yanagida. 1995. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129:1033-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodzic, D. M., D. B. Yeater, L. Bengtsson, H. Otto, and P. D. Stahl. 2004. Sun2 is a novel mammalian inner nuclear membrane protein. J. Biol. Chem. 279:25805-25812. [DOI] [PubMed] [Google Scholar]
- 15.Hofemeister, H., and P. O'Hare. 2005. Analysis of the localization and topology of nurim, a polytopic protein tightly associated with the inner nuclear membrane. J. Biol. Chem. 280:2512-2521. [DOI] [PubMed] [Google Scholar]
- 16.Kiseleva, E., M. W. Goldberg, J. Cronshaw, and T. D. Allen. 2000. The nuclear pore complex: structure, function, and dynamics. Crit. Rev. Eukaryot. Gene Expr. 10:101-112. [PubMed] [Google Scholar]
- 17.Lee, K. K., D. Starr, M. Cohen, J. Liu, M. Han, K. L. Wilson, and Y. Gruenbaum. 2002. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol. Biol. Cell 13:892-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lloyd, D., R. C. Trembath, and S. Shackleton. 2002. A novel interaction between lamin A and SREBP1: implications for partial lipodystrophy and other laminopathies. Hum. Mol. Genet. 11:769-777. [DOI] [PubMed] [Google Scholar]
- 19.Luderus, M. E. E., J. L. Denblaauwen, O. J. B. Desmit, D. A. Compton, and R. Vandriel. 1994. Binding of matrix attachment regions to lamin polymers involves single-stranded regions and the minor groove. Mol. Cell. Biol. 14:6297-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malone, C. J., W. D. Fixsen, H. R. Horvitz, and M. Han. 1999. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development 126:3171-3181. [DOI] [PubMed] [Google Scholar]
- 21.Malone, C. J., L. Misner, N. Le Bot, M. C. Tsai, J. M. Campbell, J. Ahringer, and J. G. White. 2003. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115:825-836. [DOI] [PubMed] [Google Scholar]
- 22.Maniotis, A. J., C. S. Chen, and D. E. Ingber. 1997. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA 94:849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKeon, F. D., M. W. Kirschner, and D. Caput. 1986. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature 319:463-468. [DOI] [PubMed] [Google Scholar]
- 24.Mislow, J. M., J. M. Holaska, M. S. Kim, K. K. Lee, M. Segura-Totten, K. L. Wilson, and E. M. McNally. 2002. Nesprin-1α self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett. 525:135-140. [DOI] [PubMed] [Google Scholar]
- 25.Mislow, J. M., M. S. Kim, D. B. Davis, and E. M. McNally. 2002. Myne-1, a spectrin repeat transmembrane protein of the myocyte inner nuclear membrane, interacts with lamin A/C. J. Cell Sci. 115:61-70. [DOI] [PubMed] [Google Scholar]
- 26.Moir, R. D., T. P. Spann, and R. D. Goldman. 1995. The dynamic properties and possible functions of nuclear lamins. Int. Rev. Cytol. 162B:141-182. [DOI] [PubMed] [Google Scholar]
- 27.Mounkes, L., S. Kozlov, B. Burke, and C. L. Stewart. 2003. The laminopathies: nuclear structure meets disease. Curr. Opin. Genet. Dev. 13:223-230. [DOI] [PubMed] [Google Scholar]
- 28.Ostlund, C., J. Ellenberg, E. Hallberg, J. Lippincott-Schwartz, and H. J. Worman. 1999. Intracellular trafficking of emerin, the Emery-Dreifuss muscular dystrophy protein. J. Cell Sci. 112:1709-1719. [DOI] [PubMed] [Google Scholar]
- 29.Padmakumar, V. C., T. Libotte, W. Lu, H. Zaim, S. Abraham, A. A. Noegel, J. Gotzmann, R. Foisner, and I. Karakesisoglou. 2005. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 118:3419-3430. [DOI] [PubMed] [Google Scholar]
- 30.Raff, J. W. 1999. The missing (L)UNC? Curr. Biol. 9:R708-R710. [DOI] [PubMed] [Google Scholar]
- 31.Reinsch, S., and P. Gonczy. 1998. Mechanisms of nuclear positioning. J. Cell Sci. 111:2283-2295. [DOI] [PubMed] [Google Scholar]
- 32.Reinsch, S., and E. Karsenti. 1997. Movement of nuclei along microtubules in Xenopus egg extracts. Curr. Biol. 7:211-214. [DOI] [PubMed] [Google Scholar]
- 33.Rober, R. A., K. Weber, and M. Osborn. 1989. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development 105:365-378. [DOI] [PubMed] [Google Scholar]
- 34.Rzepecki, R., S. S. Bogachev, E. Kokoza, N. Stuurman, and P. A. Fisher. 1998. In vivo association of lamins with nucleic acids in Drosophila melanogaster. J. Cell Sci. 111:121-129. [DOI] [PubMed] [Google Scholar]
- 35.Salina, D., K. Bodoor, D. M. Eckley, T. A. Schroer, J. B. Rattner, and B. Burke. 2002. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108:97-107. [DOI] [PubMed] [Google Scholar]
- 36.Schatten, G. 1982. Motility during fertilization. Int. Rev. Cytol. 79:59-163. [DOI] [PubMed] [Google Scholar]
- 37.Schirmer, E. C., L. Florens, T. Guan, J. R. Yates, 3rd, and L. Gerace. 2003. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301:1380-1382. [DOI] [PubMed] [Google Scholar]
- 38.Shackleton, S., D. J. Lloyd, S. N. J. Jackson, R. Evans, M. F. Niermeijer, B. M. Singh, H. Schmidt, G. Brabant, S. Kumar, P. N. Durrington, S. Gregory, S. O'Rahilly, and R. C. Trembath. 2000. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat. Genet. 24:153-156. [DOI] [PubMed] [Google Scholar]
- 39.Soullam, B., and H. J. Worman. 1993. The amino-terminal domain of the lamin-B receptor is a nuclear-envelope targeting signal. J. Cell Biol. 120:1093-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soullam, B., and H. J. Worman. 1995. Signals and structural features involved in integral membrane-protein targeting to the inner nuclear-membrane. J. Cell Biol. 130:15-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Starr, D. A., and M. Han. 2003. ANChors away: an actin based mechanism of nuclear positioning. J. Cell Sci. 116:211-216. [DOI] [PubMed] [Google Scholar]
- 42.Starr, D. A., and M. Han. 2002. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science 298:406-409. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan, T., D. Escalante-Alcalde, H. Bhatt, M. Anver, N. Bhat, K. Nagashima, C. L. Stewart, and B. Burke. 1999. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147:913-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaughan, O. A., M. Alvarez-Reyes, J. M. Bridger, J. L. V. Broers, F. C. S. Ramaekers, M. Wehnert, G. E. Morris, W. G. F. Whitfield, and C. J. Hutchison. 2001. Both emerin and lamin C depend on lamin A for localization at the nuclear envelope. J. Cell Sci. 114:2577-2590. [DOI] [PubMed] [Google Scholar]
- 45.Wu, W., F. Lin, and H. J. Worman. 2002. Intracellular trafficking of MAN1, an integral protein of the nuclear envelope inner membrane. J. Cell Sci. 115:1361-1371. [DOI] [PubMed] [Google Scholar]
- 46.Ye, Q., and H. J. Worman. 1994. Primary structure-analysis and lamin-B and DNA-binding of human LBR, an integral protein of the nuclear-envelope inner membrane. J. Biol. Chem. 269:11306-11311. [PubMed] [Google Scholar]
- 47.Zhang, Q., C. Ragnauth, M. J. Greener, C. M. Shanahan, and R. G. Roberts. 2002. The nesprins are giant actin-binding proteins, orthologous to Drosophila melanogaster muscle protein MSP-300. Genomics 80:473-481. [PubMed] [Google Scholar]
- 48.Zhang, Q., C. D. Ragnauth, J. N. Skepper, N. F. Worth, D. T. Warren, R. G. Roberts, P. L. Weissberg, J. A. Ellis, and C. M. Shanahan. 2005. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J. Cell Sci. 118:673-687. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, Q., J. N. Skepper, F. Yang, J. D. Davies, L. Hegyi, R. G. Roberts, P. L. Weissberg, J. A. Ellis, and C. M. Shanahan. 2001. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J. Cell Sci. 114:4485-4498. [DOI] [PubMed] [Google Scholar]
- 50.Zhen, Y. Y., T. Libotte, M. Munck, A. A. Noegel, and E. Korenbaum. 2002. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell Sci. 115:3207-3222. [DOI] [PubMed] [Google Scholar]