Abstract
Previous studies have revealed that transforming growth factor-β-activated protein kinase 1 (TAB1) interacts with p38α and induces p38α autophosphorylation. Here, we examine the sequence requirements in TAB1 and p38α that drive their interaction. Deletion and point mutations in TAB1 reveal that a proline residue in the C terminus of TAB1 (Pro412) is necessary for its interaction with p38α. Furthermore, a cryptic D-domain-like docking site was identified adjacent to the N terminus of Pro412, putting Pro412 in the φB+3 position of the docking site. Through mutational analysis, we found that the previously identified hydrophobic docking groove in p38α is involved in this interaction, whereas the CD domain and ED domain are not. Furthermore, chimeric analysis with p38β (which does not bind to TAB1) revealed a previously unidentified locus of p38α comprising Thr218 and Ile275 that is essential for specific binding of p38α to TAB1. Converting either of these residues to the corresponding amino acid of p38β abolishes p38α interaction with TAB1. These p38α mutants still can be fully activated by p38α upstream activating kinase mitogen-activated protein kinase kinase 6, but their basal activity and activation in response to some extracellular stimuli are reduced. Adjacent to Thr218 and Ile275 is a site where large conformational changes occur in the presence of docking-site peptides derived from p38α substrates and activators. This suggests that TAB1-induced autophosphorylation of p38α results from conformational changes that are similar but unique to those seen in p38α interactions with its substrates and activating kinases.
Intracellular signal transduction pathways regulate cellular responses to physiological and nonphysiological stimuli. Specific protein-protein interactions determine the efficiency and fidelity of these signaling pathways. The family of mitogen-activated protein kinases (MAPKs) includes extracellular regulated protein kinase (ERK), c-Jun N-terminal kinase (JNK), stress-activated protein kinase, and p38 (7, 8, 24, 41, 48). MAPK activation results from phosphorylation in its activation lip at a single tyrosine-threonine residue by specific MAPK kinases (MKKs) (1). Inactivation of MAPK results from dephosphorylation of cognate residues by various tyrosine phosphatases and/or dual-specificity MAPK phosphatases (26, 47). MAPKs catalyze the phosphorylation of their targets, which include transcription factors and protein kinases (9, 15, 22, 25, 34, 37). Although a large number of molecules are involved in the phosphorylation cascades of MAPKs, each MAPK cascade is specifically regulated without cross talk inside cells. The molecular basis for the fidelity of these reactions is partly determined by the specificity of the interactions between kinases and substrates. Scaffolding proteins that organize pathways in specific modules through simultaneous binding of several components also play an important role in controlling the specificity of MAPK cascades (49). Although the kinase cascade is the primary activation mechanism of MAPKs, interaction with transforming growth factor-β-activated protein kinase 1 (TAK1)-binding protein 1 (TAB1) or phosphorylation on Tyr323 by tyrosine kinase Zap70 (12, 36) can also promote autoactivation of MAPK p38α, suggesting that autoactivation is a potential alternative mechanism of activation for other MAPKs as well.
MAPKs are known to interact with their substrates and processing enzymes (kinases and phosphatases) at sites outside the active site of the enzyme (5, 11, 50). The best-studied example of this is between D-domain peptides, which are found on substrates and processing enzymes, and hydrophobic docking sites found on MAPKs (2, 16, 21, 38, 45). D-domain peptides have a consensus motif (Arg/Lys)2-(X)2-6-φA-X-φB with a hydrophobic subregion that has been observed crystallographically bound to a hydrophobic groove in the C-terminal domain of the kinase (5). The site was later confirmed by mutagenic analysis, and a similar interaction was observed between JNK1 and JIP (20). Adjacent to the site is a region of negatively charged residues (Asp313, Asp315, and Asp316 in p38α) called the CD domain (43). In the crystallographic structure of p38α in the presence of substrate- and activator-derived D-domain peptides, allosteric conformational changes were observed that affected the structure of the activation loop (5). This suggests that docking-site interactions, in addition to determining the specificity of the kinase, also have a role in its activation.
TAB1 was originally identified as an interacting protein of TAK1 (39). We later found that it interacts with p38α but no other p38 family member (12), and that TAB1b, a splice variant of TAB1 in which the 69 C-terminal residues are replaced by a different 27-residue sequence, interacts only with p38α and not TAK1 (13). The interaction of TAB1 with p38α leads to p38α autophosphorylation on the dual phosphorylation sites in the activation lip both in vitro and in cells coexpressing TAB1 and p38α. TAB1-dependent p38α activation appears to play a role in some physiological and pathological processes such as injury during myocardial ischemia (42), maturation of monocyte-derived dendritic cells (30), maintenance of peripheral T-cell anergy (32), and intracellular infection of parasite-induced interleukin-12 production (27). On the other hand, phosphorylation of TAB1 by p38α has been observed and may play a role in negative feedback of TAK1 activation (6). Because of the unique nature of TAB1-p38α binding, we chose to use a mutagenic approach to investigate the physical basis for this interaction. We have mapped sequences in TAB1 required for p38α binding, and we have determined that Pro412 is especially important. With this in mind, a cryptic D-domain-like docking site was identified in TAB1 adjacent to the N terminus of Pro412, suggesting that TAB1 utilizes docking interactions with features similar to those of p38α substrates and activating enzymes. Important interaction sites in p38α were also identified, revealing that Ile116 and Gln120 of the hydrophobic docking groove and two previously unidentified residues, Thr218 and Ile275, are critical for TAB1 interactions. On the other hand, mutations in the CD (43) and ED (44) domains had no effect on p38α-TAB1 interaction. Therefore, the specific interaction between p38α and TAB1 involves both common and unique structural determinants.
MATERIALS AND METHODS
Construction of expression vectors.
The TAB1, Flag-p38α, Flag-p38β, Flag-p38α(CDmut), Flag-p38α(EDmut), and TAB1/Δ419-504 expression vectors were described in previous publications (12, 31). The TAB1 deletion mutants were generated by PCR. The p38α and p38β chimeras were created by PCR recombination. The TAB1, p38α, and p38β point mutations were generated with a QuickChange Site-Directed Mutagenesis kit (Stratagene).
Transfection of cells.
HEK293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM glutamine, and 100-μg/ml penicillin and streptomycin. Cells on six-well plates were transiently transfected with 1 μg (total) of plasmid DNA using Lipofectamine 2000 (Invitrogen).
Western blot and immunoprecipitation analysis.
Total-cell lysates were prepared using a lysis buffer composed of 20 mM Tris-HCl (pH 7.5), 120 mM NaCl, 10% glycerol, 1 mM Na3VO4, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1% Triton X-100. Equal loading of cell protein extracts in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was ensured using Bio-Rad's protein assay solution (Bio-Rad, Hercules, CA) and by staining the transferred nitrocellulose membranes with Ponceau's solution (Sigma, St. Louis, MO). Standard Western blot methods were then used (17). Rabbit polyclonal antibodies raised against bacterium-expressed recombinant His-TAB1β protein, anti-Flag M2 monoclonal antibody (Sigma, St. Louis, MO), and anti-phospho p38 (New England Biolabs, Beverly, MA) were used in immunoblotting. For coimmunoprecipitation, cell lysates prepared as described above were incubated with anti-Flag M2 beads (Sigma) and gently shaken for 4 h at 4°C. The beads were washed three times with the lysis buffer and one time with 50 mM Tris (pH 6.8). Then, 50 μl SDS sample buffer was added, and the samples were heated for 5 min at 100°C. The supernatant was applied to SDS-PAGE gels and was detected by immunoblotting.
CD spectroscopy.
Circular dichroism (CD) spectra were recorded at room temperature using an Aviv 202 Series Circular Dichroism spectrometer, model 62DS (Aviv Instruments, Lakewood, NJ). The CD spectra were obtained in 10 mM phosphate buffer (pH 7.4) using a cell with a 0.2-cm path length. p38α was expressed as described previously in the BL21(DE3) strain as an N-terminal His6 fusion protein and was purified by Ni-nitrilotriacetic acid agarose (QIAGEN) and Mono Q (Pharmacia) columns. The purified protein was dialyzed against an incubation buffer (10 mM phosphate; pH 7.4). The peptides SKGKSKRKKDLRISCNSK (MKK3) and SSAQSTSKTSVTLSLVMPSQ (TAB1) were custom synthesized and purified by Invitrogen (San Diego, CA). A threefold molar excess of the MKK3 or TAB1 peptides was added to p38α at a concentration of 20 μg/ml in the incubation buffer overnight at 4°C. The samples were incubated at room temperature for an additional hour and then had their CD spectra measured. The MKK3, TAB1, and p38α peptides were measured as controls.
Reporter gene assay.
Cells were grown on 35-mm-diameter multiwell plates and transiently transfected with GAL4-responsive luciferase plasmid or the NF-κB-dependent luciferase reporter plasmid. A β-galactosidase expression plasmid (pCMV-β-gal,]; Clontech, Palo Alto, CA) was used to control for transfection efficiency. The total amount of DNA for each transfection was kept constant by using the empty vector pcDNA3. Cell extracts were prepared 24 h later, and β-galactosidase and luciferase activities were measured.
In vitro pull down assay.
GST fusion protein of TAB1β expressed in Escherichia coli strain BL21 was bound to glutathione-Sepharose 4B beads (Amersham Biosciences, Uppsala, Sweden) by incubating the beads with bacteria lysates and subsequently washing them. The amount of bound protein was about 2 mg/ml, as estimated by SDS-PAGE. A total of 40 μl of the 50% slurry of beads was added to 200 μl of cell lysate generated from 5 × 105 cells, and they were incubated at 4°C for 3 h. The beads were washed three times with lysis buffer and then subjected to Western blot analysis.
Protein kinase assays.
In vitro kinase assays were carried out at 37°C for 30 min using immunoprecipitates as kinases, 5 μg of kinase substrate, 250 μM ATP, and 10 μCi of [γ-32P]ATP in 20 μl of kinase reaction buffer as previously described (18, 23). Reactions were terminated by the addition of Laemmli sample buffer. Reaction products were resolved by 12% SDS-PAGE, and the extent of protein phosphorylation was visualized by autoradiography.
RESULTS
Proline 412 in TAB1 is required for TAB1 to bind to p38α.
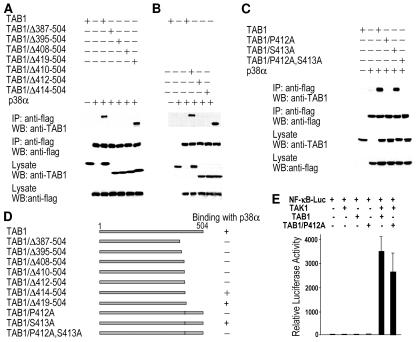
In previous studies, we determined that TAB1 residues between Ser373 and Val418 are required for interaction with p38α (12). To further analyze which sequences in TAB1 are required for binding to p38α, we generated additional deletion and point mutants in TAB1. We coexpressed wild-type TAB1 and TAB1 mutants with Flag-p38α and carried out coimmunoprecipitation assays. Consistent with our previous report (12), wild-type TAB1 and TAB1/Δ419-504 coprecipitated with p38α (Fig. 1A). The progressive deletion of TAB1 C-terminal sequences showed that the sequence between 408 and 418 was required (Fig. 1A). Further deletions showed that while TAB1/Δ414-504 interacts with p38α, deletions of two more C-terminal amino acids, Pro412 and Ser413, abolished TAB1's interaction with p38α (Fig. 1B). The point mutant TAB1/S413A had little effect on TAB1's interaction with p38α. In contrast, the mutant TAB1/P412A could not be pulled down by Flag-p38α (Fig. 1C). Figure 1D summarizes the length and mutation sites of the TAB1 mutants used in Fig. 1A to C and their ability to bind with p38α. To determine whether TAB1/P412A expressed in cells is a functional protein, we measured its activity toward activation of TAK1. It is known that coexpression of TAB1 with TAK1 in cells leads to TAK1 activation, which in turn activates the NF-κB reporter (39). We expressed the NF-κB promoter-driven luciferase reporter together with or without TAK1, TAB1, and TAB1/P412A in different combinations. As previously reported (39), coexpression of TAB1 with TAK1 led to high expression of the NF-κB reporter gene (Fig. 1E). TAB1/P412A appeared to function in a manner similar to that of wild-type TAB1 in the activation of TAK1 because similar induction of NF-κB reporter gene expression was observed when TAB1/P412A was coexpressed with TAK1 (Fig. 1E). Therefore, Pro412 is essential for TAB1's interaction with p38α but not TAK1.
FIG. 1.
Pro412 of TAB1 is required for TAB1-p38α interaction. (A) TAB1 C-terminal truncated mutants TAB1/Δ387-504, TAB1/Δ395-504, TAB1/Δ408-504, and TAB1/Δ419-504 were coexpressed with Flag-p38α in 293 cells. The cells were lysed 24 h after transfection. One-third of the cell lysates was analyzed by Western blotting (WB) with anti-TAB1 and anti-Flag antibodies. The rest of the cell lysates were subjected to immunoprecipitation (IP) with anti-Flag antibodies, and the immunoprecipitates were then analyzed by Western blotting with anti-TAB1 and anti-Flag. (B) The same experiments as shown in panel A except TAB1 mutants with deletions after amino acids 409 (TAB1/Δ410-504), 411 (TAB1/Δ412-504), and 413 (TAB1/Δ414-504) were used in the coexpression. (C) The interaction between TAB1 or TAB1 mutants and p38α was analyzed as described in the legend to panel A. The TAB1 mutants with P412 changed to alanine (TAB1/P412A), S413 changed to alanine (TAB1/S413A), and P412 and S413 changed to alanine (TAB1/P412A/S413A) are used. (D) The TAB1 mutants used in panels A to C are summarized with bar graphs and their abilities to bind with p38α are indicated by + and −. (E) 293 cells were transfected with the NF-κB reporter plasmid together with expression vectors of TAK1, TAB1, or TAB1/P412A in different combinations as indicated. Luciferase activity was measured 24 h after transfection.
A kinase interaction motif similar to the docking sites in MKK3 and myocyte enhancer factor 2A (MEF2A) appears to exist in TAB1.
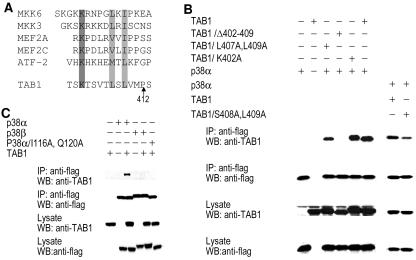
Because Pro412 is essential for TAB1-p38α interaction and the sequence C-terminal of Pro412 is not required for this interaction, we closely examined the sequence N-terminal to Pro412 in TAB1. A D-domain like motif adjacent to the N terminus of Pro412 was found. Figure 2A shows a sequence alignment of TAB1 with docking motifs in p38α substrates and activators. The motif in TAB1 varied from the consensus motif found in p38α substrates and modifying enzymes in that it has only a single positively charged amino acid (K-X4-φA-X-φB). To determine whether this D-domain plays a role in TAB1-p38α interaction, we generated a few D-domain mutations in TAB1. Deletion of the D-domain-like sequence in TAB1 (TAB1/Δ402-409) abolished TAB1-p38α interaction (Fig. 2B), confirming that this D-domain like motif is a docking site for p38α. However, as its sequence already indicated, the D-domain in TAB1 was atypical. Mutations of both φA and φB (TAB1/L407A; L409A) or single φB (TAB1/S408A/L409A) in TAB1 only reduced TAB1's affinity to p38α (Fig. 2B). Mutating K402 (TAB1/K402A) did not affect TAB1's interaction with p38α (Fig. 2B). Therefore, the D-domain in TAB1 has both similarities and differences in comparison to previously described D-domains.
FIG. 2.
K-X4-φA-X-φB motif in TAB1 is involved in TAB1-p38α interaction. (A) Sequence alignments of the K-X4-φA-X-φB motif near P412 in TAB1 compared to the docking sites of some known p38α activators and substrates. (B) TAB1 or TAB1 mutants with a deletion of amino acids 402 to 409 (TAB1/Δ402-409), L407 and L409 changed to alanine (TAB1/L407A,L409A), K402 changed to alanine (TAB1/K402A), or S408 and L409 changed to alanine (TAB1/S409A,L410A) were coexpressed with p38α in 293 cells. The interaction between p38α and TAB1 or the TAB1 mutant was analyzed as in Fig. 1A. (C) p38α, p38α mutated in the docking groove (p38α/I116A/Q120A), or p38β was coexpressed with TAB1. The interaction between TAB1 and p38α, the p38α mutant, or p38β was analyzed as in described in the legend to Fig. 1A.
Crystallographic structures of p38α bound to peptides from MEF2A or MKK3 have revealed that φA-X-φB residues of the docking site bind to p38α in a groove composed in part of Ile116 and Gln120 (5). To determine whether these two residues are also required for p38α to interact with TAB1, we performed coimmunoprecipitation assays, which revealed that Ile116 and Gln120 mutations abolished the interaction between p38α and TAB1 (Fig. 2C). Thus, the hydrophobic docking groove apparently played a role in TAB1-p38α interaction. However, it should be noted that p38β had a sequence identical to that of p38α in the docking groove (see Fig. S1 in the supplemental material) but could be coimmunoprecipitated with TAB1 (Fig. 2C). Therefore, another domain(s) in p38α must be required for the specific interaction between p38α and TAB1.
CD and ED domains are not required for p38α-TAB1 interaction.
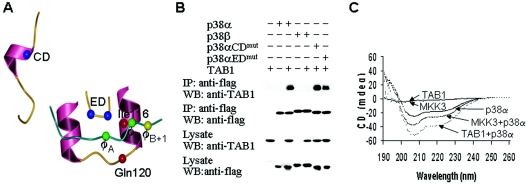
The CD domain in p38α (Asp313, Asp315, and Asp316) was shown to be required for its interaction with its activators and substrates (43), and the ED domain (Glu160 and Asp161) was shown to confer the specificity of p38α in binding to its substrates (44). The positions of the CD domain, ED domain, and the hydrophobic docking groove in p38α are shown in Fig. 3A. We examined whether CD and ED domains of p38α are required for TAB1 binding and found that mutations in the CD domain (p38αCDmut) or the ED domain (p38αEDmut) did not have any effect on p38α-TAB1 interaction (Fig. 3B). Therefore, p38α binding to TAB1 is dissimilar to that of p38α binding to other activators and substrates.
FIG. 3.
p38α's interaction with TAB1 is different from its interaction with upstream kinases and substrates. (A) Part of the p38α structure shows the CD domain, the ED domain, and the hydrophobic docking groove bound to the MEF2A peptide (p38α/pepMEF2A; Protein Data Bank [PDB] file 1LEW). (B) p38α, p38α mutated in the CD domain (p38αCDmut) or ED site (p38αEDmut), or p38β was coexpressed with TAB1. The interaction between TAB1 and p38α, p38α mutants, or p38β was analyzed as in Fig. 1A. (C) MKK3 peptide SKGKSKRKKDLRISCNSK or TAB1 peptide SSAQSTSKTSVTLSLVMPSQ and p38α were incubated alone or together, and the CD spectra were analyzed.
It is known that the R/K-X4-φA-X-φB peptide from MKK3 and MEF2A causes conformational changes of p38α upon binding (5). We therefore synthesized a K-X4-φA-X-φB peptide of TAB1 and used CD spectrum analysis to compare conformation changes of p38α caused by R/K-X4-φA-X-φB peptides from TAB1 and MKK3. As shown in Fig. 3C, TAB1 and MKK3 peptides caused changes in the CD spectra of p38α and the changes were different for each peptide. The change in the CD absorption was large, which may come both from the induced structure of the peptide and from rigidification of the structure local to the TAB1-binding site (35, 40). Support for this possibility comes from the observed rigidification (B-factor changes) of p38α induced by peptides derived from MEF2A or MKK3 (5). The CD spectra suggest that there are similarities and differences between p38α interactions with peptides from TAB1 and MKK3.
Two regions in the C-terminal domain of p38α are required for its binding to TAB1.
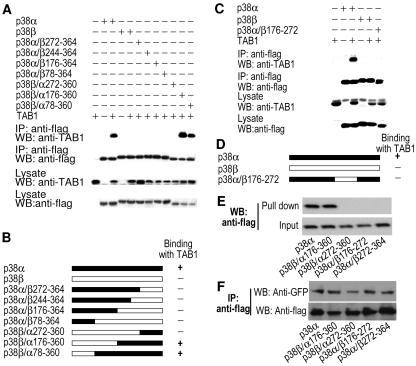
We then sought to determine which residues in p38α confer the specificity of TAB1-p38α interaction. p38β is a very close homologue of p38α (33, 51); the two homologues interact similarly with activating kinases and substrates, but p38β cannot bind to TAB1 (12). We constructed a series of p38α/β chimeras by swapping the corresponding regions of these two proteins. A coimmunoprecipitation assays showed that the chimera p38α/β272-364 (in which the 88 C-terminal amino acids of p38α were replaced by a p38β sequence) did not bind TAB1 (Fig. 4A). Therefore, residues at the C terminus of p38α are required for TAB1 binding. Also, the region spanning amino acids 176 to 272 in p38α appeared to be required for interaction with TAB1, since p38β/α176-360 interacted with TAB1 while p38β/α272-360 did not (Fig. 4A). Diagrams of these mutants and their ability to binding with TAB1 are shown in Fig. 4B.
FIG. 4.
The two regions in p38α that distinguish p38α from p38β in TAB1 binding. (A) p38α, the p38α-p38β chimeras, or p38β was coexpressed with TAB1. The interaction between TAB1 and p38α, the chimeras, or p38β was analyzed as in Fig. 1A. (B) Diagrams of the chimeras of p38α and p38β and a summary of their binding with TAB1. (C) p38α, p38α/β176-272, or p38β was coexpressed with TAB1. The interaction between TAB1 and p38α, p38α/β176-272, or p38β was analyzed as described in the legend to Fig. 1A. (D) Diagram of the chimera p38α/β176-272. (E) A total of 40 ml GST-TAB1β bound to glutathione-agarose was added to cell lysates from the 293 cells transfected with the expression vector of Flag-p38α, Flag-p38β/α176-360, Flag-p38β/α272-360, Flag-p38α/β176-272, or Flag-p38α/β272-364. The agarose beads were washed and subjected to Western blot analysis with anti-Flag antibodies. The levels of these Flag-tagged proteins in the cell lysates were also determined by Western blot analysis with anti-Flag-antibodies. (F) The GFP-MK2 expression vector was cotransfected with Flag-p38α, Flag-p38β/α176-360, Flag-p38β/α272-360, Flag-p38α/β176-272, or Flag-p38α/β272-364 expression plasmids. Immunoprecipitations were performed with anti-Flag antibodies 24 h after transfection. Immunoprecipitates were analyzed by Western blot with anti-GFP and anti-Flag antibodies.
To confirm that the region from amino acids 176 to 272 in p38α is required for p38α-TAB1 interaction, we constructed p38α/β176-272, a chimera of p38α and p38β. p38α/β176-272 did not interact with TAB1 (Fig. 4C and D). We also used in vitro pull-down to detect the interaction between TAB1 and p38α mutants. Because recombinant TAB1β was better than TAB1 in protein stability when expressed in Escherichia coli, we used it in in vitro pull-down experiments. Equal amounts of agarose beads that bound with GST-TAB1β were added into cell lysates from the 293 cells that had been transfected with expression plasmids of Flag-p38α, Flag-p38β/α176-360, Flag-p38β/α272-360, Flag-p38α/β176-272, or Flag-p38α/β272-364. p38α and p38β/α176-360, but not the others, were pulled down by TAB1β (Fig. 4E), which was consistent with our coimmunoprecipitation results (Fig. 4A and C). To determine whether the p38α mutants that cannot bind with TAB1 still interacted with other p38α-binding partners, we coexpressed green fluorescent protein (GFP)-tagged MAPK-activated protein kinase 2 (MK2) with Flag-p38α, Flag-p38β/α176-360, Flag-p38β/α272-360, Flag-p38α/β176-272, or Flag-p38α/β272-364 in 293 cells. Immunoprecipitations were performed with anti-Flag antibodies, and the presence of GFP-MK2 in the immunoprecipitates was determined by Western blot analysis using anti-GFP antibodies. All p38α mutants interacted with MK2 (Fig. 4F). In conclusion, both the region spanning amino acids 176 to 272 and the region spanning amino acids 273 to 360 in p38α are required for p38α-TAB1 interaction.
Thr218 is critical for p38α-TAB1 interaction.
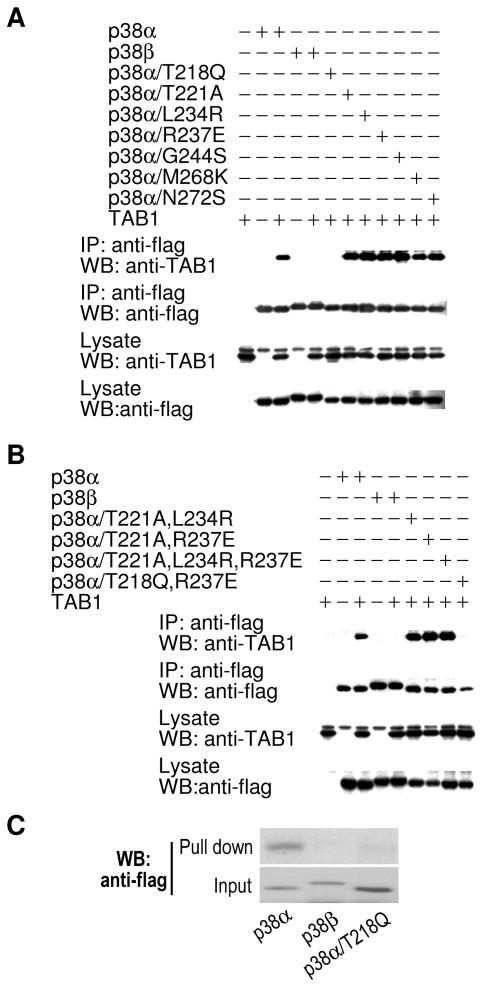
Next, we introduced point mutations to determine which amino acid(s) in the region from Asp176 to Asn272 of p38α is required for p38α-TAB1 interaction. We mutated p38α at seven amino acid sites between Asp176 and Asn272 that differed from the corresponding residues in p38β. As shown in Fig. 5A, mutation of Thr218 (p38α/T218Q) abolished p38α-TAB1 interaction, while the other six point mutations had no effect. To determine whether double- or triple-amino-acid point mutations had an additive effect on p38α-TAB1 interaction, we generated three double mutations and one triple mutation. Coimmunoprecipitation assays showed that multiple amino acid mutations did not effect p38α-TAB1 interaction unless T218Q was included in the mutation (Fig. 5B). We also performed in vitro pull-down experiment, which confirmed that p38α/T218Q did not bind to TAB1 (Fig. 5C). Therefore, we concluded that Thr218 is a critical residue for p38α-TAB1 interaction.
FIG. 5.
Thr218 in p38α is required for its interaction with TAB1. (A) p38α, p38β, or p38α with point mutations at T218, T221, L234, R237, G244, M268, or N272 (to the corresponding amino acids from p38β at the same positions) was coexpressed with TAB1. Their interaction with TAB1 was analyzed as described in the legend to Fig. 1A. (B) p38α, p38β, or p38α with two or three point mutations at T218, T221, L234, or R237 (to the corresponding amino acids from p38β at the same positions) was coexpressed with TAB1. Their interaction with TAB1 was analyzed as described in the legend to Fig. 1A. (C) In vitro pull-down of Flag-p38α, Flag-p38β, or Flag-p38α/T218Q by TAB1β was performed as described in the legend to Fig. 4E.
Ile 275 is critical for p38α-TAB1 interaction.
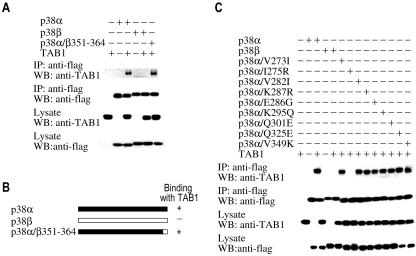
We next determined which amino acids from Val273 to Ser360 in p38α are required for p38α-TAB1 interaction. The C-terminals of p38α and p38β differ after Pro351 (see Fig. S1 in the supplemental material). We made a p38α/β351-364 chimera and examined its interaction with TAB1 (Fig. 6A and B). TAB1 coimmunoprecipitated with wild-type p38α and p38α/β351-364. Therefore, these 10 C-terminal amino acids in p38α do not play a role in the selective interaction between p38α and TAB1.
FIG. 6.
Ile275 in p38α is required for its interaction with TAB1. (A) p38α, p38β, or p38α/β351-364 was coexpressed with TAB1. Their interaction with TAB1 was analyzed as described in the legend to Fig. 1A. (B) Diagram of p38α/β351-364. (C) p38α, p38β, or p38α with single point mutations at V273, I275, V282, K287, E286, K295, E301, Q325, or V349 (to the corresponding amino acids from p38β at the same positions) was coexpressed with TAB1. Their interaction with TAB1 was analyzed as described in the legend to Fig. 1A.
We then focused on the region from Val273 to Pro351 in p38α and generated point mutations in p38α using the corresponding amino acids from p38β. Nine p38α mutants were synthesized, and their interactions with TAB1 were examined by coimmunoprecipitation. One of these mutants, p38α/I275R, cannot coimmunoprecipitate with TAB1. Therefore, we concluded that Ile275 is another critical residue for the specific interaction between p38α and TAB1.
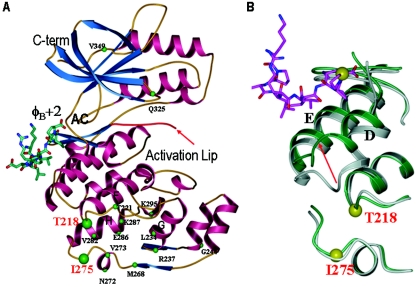
Figure 7A summarizes the location of the point mutations we made in p38α. Thr218 and Ile275 are near (about 20 Å) the hydrophobic docking groove observed crystallographically in p38α-pepMEF2A and p38α-pepMKK3 complexes, in the linker region between helices F and G, and at L14, right before helix H. This was one locus of conformational change in p38α-pepMEF2A (Fig. 7B) and p38α-pepMKK3 (5). The locations of Thr218, Ile275, and other domains in the three-dimensional structure of p38α and the corresponding amino acids in a modeled structure of p38β can be found in Figure S2 in the supplemental material.
FIG. 7.
(A) A summary of point mutations on the three-dimensional structure of p38α complexed with pepMEF2A (PDB file 1LEW). Mutations affecting TAB1 binding are shown as large green spheres. Mutations that do not affect TAB1 binding are shown as small spheres. Placement of the φB+2 residue in pepMEF2A is labeled. φB+3 could bind in the active site. Note that the activation loop is missing in 1LEW. (B) The location of residues Thr218 and Ile275 in relation to the p38α docking groove and the MEF2A docking-site peptide (PDB file 1LEW; green) overlaid with uncomplexed p38α (1P38; gray). Note that the peptide-induced conformational changes in p38α helices D and E and the linker between them.
Converting residues 218 and 275 in p38β to corresponding amino acids of p38α leads to weak p38β-TAB1 interaction.
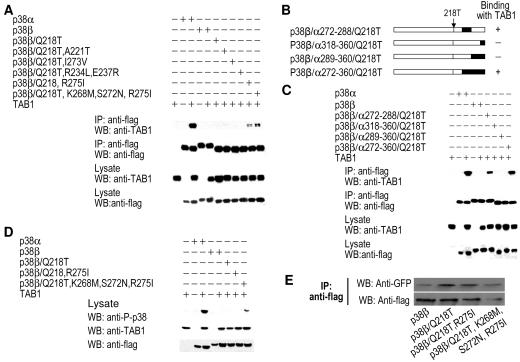
We next evaluated whether converting key amino acids in p38β to corresponding amino acids in p38α can result in an interaction between p38β and TAB1. We made a series of point mutations in p38β by converting various amino acids (Thr218, Ile275, and a few others) to their corresponding amino acids in p38α, and we then examined their interaction with TAB1 by coimmunoprecipitation assays. As shown in Fig. 8A, the mutant p38β/Q218T,R275I bound to TAB1, although the interaction was relatively weak. Therefore, both Thr218 and Ile275 were required for p38β-TAB1 interaction (Fig. 8A). Additionally, mutations made to make p38β more like p38α-enhanced TAB1 coimmunoprecipitation.
FIG. 8.
The interaction between p38β mutants and TAB1. (A) p38α, p38β, or p38β with point mutations at Q218; Q218 and A221; Q218 and I273; Q218, R243 and E237; Q218 and R275; or Q218, K268, S272, and R275 (to the corresponding amino acids from p38α at the same positions) was coexpressed with TAB1. The interaction with TAB1 was analyzed as described in the legend to Fig. 1A. (B) Diagrams of p38β mutants with Q218 mutated to threonine and C-terminal fragments replaced by sequences from p38α. (C) p38α, p38β, or the p38β mutants described in the legend to panel B were coexpressed with TAB1. Their interaction with TAB1 was analyzed as described in the legend to Fig. 1A. (D) p38α, p38β, or p38β with point mutations at Q218, Q218, and R275 or at Q218, K268, S272, and R275 (to the corresponding amino acids from p38α at the same positions) was coexpressed with TAB1. The phosphorylation of p38α, p38β, or the p38β mutants was determined by Western blotting analysis with anti-phospho-p38 antibodies. (E) GFP-MK2 was coexpressed with Flag-p38β, Flag-p38β/Q218T, Flag-p38β/Q218T,R275I, or Flag-p38β/Q218T,K268M,S272N,R275I. Immunoprecipitations were performed with anti-Flag antibodies 24 h after transfection. Immunoprecipitates were analyzed by Western blotting with anti-GFP and anti-Flag antibodies.
Since none of the aforementioned p38β mutants could strongly interact with TAB1 compared with p38α, it seemed likely that more C-terminal sequences of p38α are needed for optimal binding to TAB1. Because Thr218 is required for p38α-TAB1 interaction, we generated p38β/Q218T and then made p38β/p38α chimeras (Fig. 8B). The chimera p38β/Q218T/p38α272-360 coimmunoprecipitated fairly well with TAB1 (Fig. 8C). Additional mapping showed that the region from Asn272 to Met288 in p38α was important, while the region from Leu289 to Ser360 was not (Fig. 8C). We also studied whether the forced p38β-TAB1 interaction induces p38β phosphorylation. As shown in Fig. 8D, p38β could be phosphorylated when it interacted with TAB1. To determine whether these p38β mutants retain their ability to interact with MK2, we coexpressed GFP-MK2 with Flag-p38β, Flag-p38β/Q218T, Flag-p38β/Q218T/R275I, or p38β/Q218,K268M,S272N,R275I and determined their interaction by coimmunoprecipitation assays. All of these p38β mutants interacted with MK2 (Fig. 8E). Our data indicated that while the double mutant p38β/Q218T/R275I binds to TAB1, optimal interaction required the longer C-terminal sequence of p38α.
Mutations in p38α that selectively abolish its interaction with TAB1 affect some aspects of its activation.
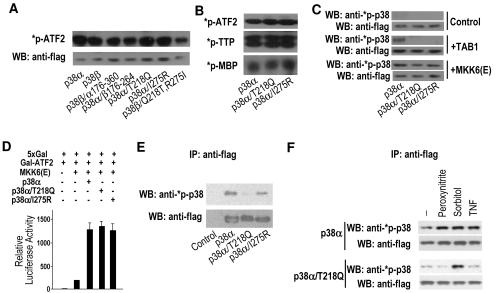
To determine whether the p38α and p38β mutants that either lose or gain a specific interaction with TAB1 have the same or different enzymatic activity, we expressed Flag-p38α, Flag-p38β, Flag-p38β/α176-360, Flag-p38α/β176-264, Flag-p38α/T218Q, Flag-p38α/I275R, or Flag-p38β/Q218T,R275I in 293 cells and stimulated the cells with hyperosmolarity (0.4 M sorbitol) to activate these kinases. Flag-tagged p38α, p38β, and the various mutants were immunoprecipitated by anti-Flag antibodies, and the immunoprecipitates were used as enzymes in an in vitro kinase assay with GST-ATF2 (1 to 109) as a substrate. p38α, p38β, and the mutants phosphorylated ATF2 in vitro and their activities were comparable (Fig. 9A). We also tested other p38 substrates, including tristetraprolin and myelin basic protein, and did not find any difference in substrate specificity among p38α and its mutants (Fig. 9B). This suggests that the mutations affecting the interaction between TAB1 and either p38α or p38β do not affect the kinase activity of p38α or p38β.
FIG. 9.
Activity and activation of p38α mutants that cannot interact with TAB1. (A) Expression vector of Flag-p38α, Flag-p38β, Flag-p38β/α176-360, Flag-p38α/β176-264, Flag-p38α/T218Q, Flag-p38α/I275R, or Flag-p38β/Q218T,R275I was transfected into in 293 cells. At 24 h after transfection, the cells were treated with 0.4 M sorbitol for 30 min. Flag-tagged proteins were immunoprecipitated with anti-Flag antibodies. An in vitro kinase assay was performed using the immunoprecipitates as kinases and GST-ATF2 as a substrate. The levels of immunoprecipitated Flag-proteins were determined by Western blot analysis with anti-Flag antibodies. (B) Flag-p38α, Flag-p38α/T218Q, and Flag-p38α/I275R were prepared by immunoprecipitations as described in the legend to panel A and used as kinases. GST-ATF2, tristetraprolin, or myelin basic protein was used as a substrate in the kinase assays. (C) Flag-p38α, Flag-p38α/T218Q, or Flag-p38α/I275R was coexpressed with control (empty vector), TAB1, or MKK6(E) in 293 cells as indicated. Total cell lysates were analyzed 24 h after transfection by Western blotting with anti-phospho-p38 and anti-Flag antibodies. (D) Expression vectors of a luciferase reporter gene under the control of 5× GAL4-binding site (5xGal), GAL4-binding domain fused with ATF2 activation domain (Gal-ATF2), MKK6(E), Flag-p38α, Flag-p38α/T218Q, or Flag-p38α/I275R were transfected into 293 cells in different combinations as indicated. Luciferase activity was measured 24 h later. (E) 293 cells were transfected with empty (control), Flag-p38α, Flag-p38α/T218Q, or Flag-p38α/I275R expression vectors. Flag-tagged proteins were immunoprecipitated with anti-Flag antibodies 48 h after transfection. The immunoprecipitates were analyzed by Western blotting using anti-phospho-p38 and anti-Flag antibodies. (F) Flag-p38α or Flag-p38α/T218Q was expressed in 293 cells. At 48 h after transfection, the cells were treated with peroxynitrite (500 μM), tumor necrosis factor (100 ng/ml), or sorbitol (0.4 M) for 5, 30, and 30 min, respectively. Flag-p38α or Flag-p38α/T218Q was immunoprecipitated with anti-Flag antibodies. The phosphorylation and the levels of Flag-p38α or Flag-p38α/T218Q were analyzed by Western blotting with anti-phospho-p38 and anti-Flag antibodies.
To determine if the p38α mutants that cannot bind to TAB1 can still be activated by its upstream kinases, we coexpressed p38α, p38α/T218Q, or p38α/I275R expression vectors with control (empty), TAB1, or dominant active MKK6 [MKK6(E)] mutant expression vectors. p38α phosphorylation was determined by Western blot analysis with anti-phospho-p38 antibodies. Coexpression of TAB1 resulted in phosphorylation of p38α but not p38α/T218Q or p38α/I275R (Fig. 9C). Coexpression of MKK6(E) led to phosphorylation of p38α and the two mutants (Fig. 9C). To further confirm the activation of p38α/T218Q and p38α/I275R by MKK6, we used an ATF2-dependent reporter. As shown in Fig. 9D, p38α, p38α/T218Q, and p38α/I275R all enhanced MKK6(E)-induced ATF2 reporter gene expression. Therefore, T218 and I275 mutations in p38α do not influence MKK6-mediated p38α activation.
Basal activity of p38α can be detected, but a relatively large amount of protein is needed. We overexpressed Flag-p38α, Flag-p38α/T218Q, and Flag-p38α/I275R in 293 cells and isolated them by immunoprecipitation with anti-Flag antibodies. The basal activities of p38α and its mutants were analyzed by Western blot analysis using anti-phospho-p38 antibodies, and the amounts of these proteins were analyzed by Western blotting with anti-Flag antibodies. p38α/T218Q had a much lower level of basal activity than p38α (Fig. 9E). The basal level phosphorylation of p38α/I275R was between those of p38α and p38α/T218Q. To determine whether abolishing p38α-TAB1 interaction affects p38α activation by extracellular stimuli, we transiently expressed Flag-p38α and Flag-p38α/T218Q in 293 cells and stimulated the cells with peroxynitrite, sorbitol, or tumor necrosis factor. Flag-p38α and Flag-p38α/T218Q were immunoprecipitated, and their protein levels and enzymatic activities in the immunoprecipitates were determined by Western blot analysis with anti-Flag and anti-phospho-p38 antibodies, respectively. p38α was activated by all three stimuli, while p38α/T218Q was activated by hyperosmolarity (sorbitol) but not others. Therefore, interaction with TAB1 plays some role in stimulus-induced p38α activation.
DISCUSSION
The interaction between TAB1 and p38α is unique among MAP kinases in that p38α is the only MAP kinase known to autophosphorylate naturally (29). There are no known homologues of TAB1 acting on different MAP kinases. In this study, we made several observations that bring us closer to understanding how TAB1 induces p38α autophosphorylation. First, we found that Pro412 of TAB1 is essential for TAB1 binding to p38α. This residue is in the φB+3 position of a putative D-domain. Although it is atypical, we were able to confirm that the sequence K402TSVTLSLVMP412 in TAB1 is a docking site for p38α. Furthermore, we found that p38α/I116A and p38α/Q120A, which are mutated in the previously identified docking groove for the φAXφB motif, are unable to interact with TAB1. We also found that the CD and ED domains do not contribute to this interaction. In the structure of p38α/pepMEF2A (5), a proline residue is present in φB+2 but is not tightly bound to the surface of p38α. Model building suggests that Pro412 in TAB1 could make closer contacts near the active site of p38α. Lastly, through a series of mutagenic analyses, we found that Thr218 and Ile275 of p38α are essential for TAB1 binding. These residues are apparently specific for TAB1 interaction, since they have yet to appear in screens of p38α interaction sites in other studies, and we show that their mutations have no effect on p38α binding with MK2. In addition, the p38 family members that cannot interact with TAB1 share the same upstream kinases (MKK3 and MKK6), and downstream substrates have amino acids replacements in one or both of these positions. Therefore, both common and unique sites in p38α contribute to its specific interactions with TAB1.
Thr218 and Ile275 are near the hydrophobic docking groove (Fig. 7), but model building suggests that a continuous polypeptide in TAB1 would not be able to bind to both the docking groove and the cleft flanked by Thr218 and Ile275. Therefore, it seems likely that some other part of TAB1 may interact with this region. Since the C-terminal portion of TAB1 (Arg333 to Pro504) is sufficient to bind p38α but insufficient to cause p38α phosphorylation (data not shown) and since the C-terminal up to Ser413 is not required for TAB1 to interact with p38α (Fig. 1), the sites that are involved in the interaction with the cleft flanked by Thr218 and Ile275 may be outside the region from Arg333 to Ser413. Thr218 and Ile275 are adjacent to helix D, which undergoes conformational changes upon p38α binding to D-domain peptides derived from MEF2A and MKK3 (Fig. 7). The fact that TAB1 binds in the docking groove also suggests that similar conformational changes are occurring in the p38α-TAB1 complex. While the MAP kinases JNK1 (20) and ERK2 (H. Zhou and E. J. Goldsmith, unpublished data) do not change shape in this locus in the presence of D-domain docking sites, both p38α and JNK1 are known to undergo long-range conformational changes that affect the conformation and order of the activation loop. Therefore, a plausible model is that TAB1 binding utilizes and enhances the conformational change induced by the docking site and Pro412, forming a unique conformation of p38α that is capable of autophosphorylation.
Sequence alignments of mammalian MAPKs reveals that several, including p38γ and p38δ, have a threonine at the position corresponding to Thr218 in p38α, but none contains an isoleucine corresponding to Ile275. This sequence information is consistent with our conclusion that both Thr218 and Ile275 are required for selective interaction between p38α and TAB1. The requirement of the common docking groove in p38α for its interaction with TAB1 is consistent with our data that an R/K-X4-φA-X-φB motif in TAB1 is involved in the interaction. We found that Pro412 at the +3 position to φB in TAB1 is required for p38α binding (Fig. 1). Proline residues can be found adjacent to the C terminus of the R/K-X4-φA-X-φB motif of several p38α-interacting proteins (Fig. 2A), although none is exactly at the +3 position of φB. To date, there is no report indicating that proline residues adjacent to the C terminus of the R/K-X4-φA-X-φB motif of MAPK-interacting proteins are required for their binding to MAPK. Perhaps Pro412 in TAB1 sits in a relatively hydrophobic pocket of p38α and this hydrophobic interaction is essential for TAB1-p38α interaction. The involvement of the common docking groove in p38α-TAB1 interactions may also contribute to the phosphorylation of TAB1 by p38α (6). However, p38α phosphorylation site mutations in TAB1 to either alanine or aspartic acid residues did not affect p38α-TAB1 interactions by coimmunoprecipitation assays (data not shown), suggesting that p38α-TAB1 interactions and TAB1 phosphorylation are independent events. Furthermore, TAB1 can be efficiently phosphorylated by other MAPKs, and no interaction between these MAPKs and TAB1 can be detected by coimmunoprecipitation assays (6, 12).
Although autophosphorylation is a common mechanism for the activation of other kinases, such as receptor tyrosine kinases (14, 19, 28, 46), its role in MAPK activation was not realized until the discovery of a number of gain-of-function mutants in ERK and p38α by genetic screens and in vitro mutagenesis (3, 4, 10). Although the mutation sites of these gain-of-function mutants affect different regions of the kinases, all of the mutations seem to enhance autophosphorylation and lead to constitutive activation of MAPKs. Very recently, Salvador et al. showed that phosphorylation on Tyr323 in p38α by tyrosine kinase Zap70 also led to p38α auto-activation (36). It is highly possible that a common conformational change in MAPKs underlies autophosphorylation initiated through different mechanisms. Crystallographic studies of docking-site interactions in MAPKs have already suggested that MAPKs can adopt different conformations within the activation loop in response to binding by other proteins (5), and the action of TAB1 binding may be related to the conformational changes induced by docking-site interactions.
Supplementary Material
Acknowledgments
We thank Luke Leman for CD spectrum analysis.
This work was supported in part by NIH grants GM37696 and AI41637, and funds from Xiamen University, and DK46993 and I1128 from the Welch Foundation to E.J.G.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Anderson, N. G., J. L. Maller, N. K. Tonks, and T. W. Sturgill. 1990. Requirement for integration of signals from two distinct phosphorylation pathways for activation of MAP kinase. Nature 343:651-653. [DOI] [PubMed] [Google Scholar]
- 2.Barsyte-Lovejoy, D., A. Galanis, and A. D. Sharrocks. 2002. Specificity determinants in MAPK signaling to transcription factors. J. Biol. Chem. 277:9896-9903. [DOI] [PubMed] [Google Scholar]
- 3.Bell, M., R. Capone, I. Pashtan, A. Levitzki, and D. Engelberg. 2001. Isolation of hyperactive mutants of the MAPK p38/Hog1 that are independent of MAPK kinase activation. J. Biol. Chem. 276:25351-25358. [DOI] [PubMed] [Google Scholar]
- 4.Brunner, D., N. Oellers, J. Szabad, W. H. Biggs III, S. L. Zipursky, and E. Hafen. 1994. A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell 76:875-888. [DOI] [PubMed] [Google Scholar]
- 5.Chang, C. I., B. E. Xu, R. Akella, M. H. Cobb, and E. J. Goldsmith. 2002. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol. Cell 9:1241-1249. [DOI] [PubMed] [Google Scholar]
- 6.Cheung, P. C., D. G. Campbell, A. R. Nebreda, and P. Cohen. 2003. Feedback control of the protein kinase TAK1 by SAPK2a/p38α. EMBO J. 22:5793-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cobb, M. H., and E. J. Goldsmith. 1995. How MAP kinases are regulated. J. Biol. Chem. 270:14843-14846. [DOI] [PubMed] [Google Scholar]
- 8.Davis, R. J. 1994. MAPKs: new JNK expands the group. Trends Biochem. Sci. 19:470-473. [DOI] [PubMed] [Google Scholar]
- 9.Dong, C., R. J. Davis, and R. A. Flavell. 2002. Map kinases in the immune response. Annu. Rev. Immunol. 20:55-72. [DOI] [PubMed] [Google Scholar]
- 10.Emrick, M. A., A. N. Hoofnagle, A. S. Miller, L. F. Ten Eyck, and N. G. Ahn. 2001. Constitutive activation of extracellular signal-regulated kinase 2 by synergistic point mutations. J. Biol. Chem. 276:46469-46479. [DOI] [PubMed] [Google Scholar]
- 11.Enslen, H., J. Raingeaud, and R. J. Davis. 1998. Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J. Biol. Chem. 273:1741-1748. [DOI] [PubMed] [Google Scholar]
- 12.Ge, B., H. Gram, F. di Padova, B. Huang, L. New, R. J. Ulevitch, Y. Luo, and J. Han. 2002. MAPKK-independent activation of p38α mediated by TAB1-dependent autophosphorylation of p38α. Science 295:1291-1294. [DOI] [PubMed] [Google Scholar]
- 13.Ge, B., X. Xiong, Q. Jing, J. L. Mosley, A. Filose, D. Bian, S. Huang, and J. Han. 2003. TAB1β (transforming growth factor-β-activated protein kinase 1-binding protein 1β), a novel splicing variant of TAB1 that interacts with p38α but not TAK1. J. Biol. Chem. 278:2286-2293. [DOI] [PubMed] [Google Scholar]
- 14.Gotoh, Y., and J. A. Cooper. 1998. Reactive oxygen species- and dimerization-induced activation of apoptosis signal-regulating kinase 1 in tumor necrosis factor-alpha signal transduction. J. Biol. Chem. 273:17477-17482. [DOI] [PubMed] [Google Scholar]
- 15.Guan, K. L. 1994. The mitogen activated protein kinase signal transduction pathway: from the cell surface to the nucleus. Cell. Signal. 6:581-589. [DOI] [PubMed] [Google Scholar]
- 16.Gum, R. J., and P. R. Young. 1999. Identification of two distinct regions of p38 MAPK required for substrate binding and phosphorylation. Biochem. Biophys. Res. Commun. 266:284-289. [DOI] [PubMed] [Google Scholar]
- 17.Han, J., J. D. Lee, P. S. Tobias, and R. J. Ulevitch. 1993. Endotoxin induces rapid protein tyrosine phosphorylation in 70Z/3 cells expressing CD14. J. Biol. Chem. 268:25009-25014. [PubMed] [Google Scholar]
- 18.Han, J., J.-D. Lee, L. Bibbs, and R. J. Ulevitch. 1994. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 265:808-811. [DOI] [PubMed] [Google Scholar]
- 19.Heldin, C. H. 1995. Dimerization of cell surface receptors in signal transduction. Cell 80:213-223. [DOI] [PubMed] [Google Scholar]
- 20.Heo, Y. S., S. K. Kim, C. I. Seo, Y. K. Kim, B. J. Sung, H. S. Lee, J. I. Lee, S. Y. Park, J. H. Kim, K. Y. Hwang, Y. L. Hyun, Y. H. Jeon, S. Ro, J. M. Cho, T. G. Lee, and C. H. Yang. 2004. Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125. EMBO J. 23:2185-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holland, P. M., and J. A. Cooper. 1999. Protein modification: docking sites for kinases. Curr. Biol. 9:R329-R331. [DOI] [PubMed] [Google Scholar]
- 22.Ip, Y. T., and R. J. Davis. 1998. Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Curr. Opin. Cell Biol. 10:205-219. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, Y., C. Chen, Z. Li, W. Guo, J. A. Gegner, S. Lin, and J. Han. 1996. Characterization of the structure and function of a new mitogen-activated protein kinase (p38β). J. Biol. Chem. 271:17920-17926. \sbauto1\saauto1 [DOI] [PubMed]
- 24.Johnson, G. L., and R. R. Vaillancourt. 1994. Sequential protein kinase reactions controlling cell growth and differentiation. Curr. Opin. Cell Biol. 6:230-238. [DOI] [PubMed] [Google Scholar]
- 25.Karin, M., and T. Hunter. 1995. Transcriptional control by protein phosphorylation: signal transmission from the cell surface to the nucleus. Curr. Biol. 5:747-757. [DOI] [PubMed] [Google Scholar]
- 26.Keyse, S. M. 1995. An emerging family of dual specificity MAP kinase phosphatases. Biochim. Biophys. Acta 1265:152-160. [DOI] [PubMed] [Google Scholar]
- 27.Kim, L., R. L. Del, B. A. Butcher, T. H. Mogensen, S. R. Paludan, R. A. Flavell, and E. Y. Denkers. 2005. p38 MAPK autophosphorylation drives macrophage IL-12 production during intracellular infection. J. Immunol. 174:4178-4184. [DOI] [PubMed] [Google Scholar]
- 28.Leung, I. W., and N. Lassam. 1998. Dimerization via tandem leucine zippers is essential for the activation of the mitogen-activated protein kinase kinase kinase, MLK-3. J. Biol. Chem. 273:32408-32415. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, T. S., P. S. Shapiro, and N. G. Ahn. 1998. Signal transduction through MAP kinase cascades. Adv. Cancer Res. 74:49-139. [DOI] [PubMed] [Google Scholar]
- 30.Matsuyama, W., M. Faure, and T. Yoshimura. 2003. Activation of discoidin domain receptor 1 facilitates the maturation of human monocyte-derived dendritic cells through the TNF receptor associated factor 6/TGF-beta-activated protein kinase 1 binding protein 1 beta/p38 alpha mitogen-activated protein kinase signaling cascade. J. Immunol. 171:3520-3532. [DOI] [PubMed] [Google Scholar]
- 31.New, L., Y. Jiang, and J. Han. 2003. Regulation of PRAK subcellular location by p38 MAP kinases. Mol. Biol. Cell 14:2603-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohkusu-Tsukada, K., N. Tominaga, H. Udono, and K. Yui. 2004. Regulation of the maintenance of peripheral T-cell anergy by TAB1-mediated p38 alpha activation. Mol. Cell. Biol. 24:6957-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono, K., and J. Han. 2000. The p38 signal transduction pathway: activation and function. Cell. Signal. 12:1-13. [DOI] [PubMed] [Google Scholar]
- 34.Pearson, G., F. Robinson, T. Beers Gibson, B. E. Xu, M. Karandikar, K. Berman, and M. H. Cobb. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22:153-183. [DOI] [PubMed] [Google Scholar]
- 35.Receveur-Brechot, V., J. M. Bourhis, V. N. Uversky, B. Canard, and S. Longhi. 2006. Assessing protein disorder and induced folding. Proteins 62:24-45. [DOI] [PubMed] [Google Scholar]
- 36.Salvador, J. M., P. R. Mittelstadt, T. Guszczynski, T. D. Copeland, H. Yamaguchi, E. Appella, A. J. Fornace, Jr., and J. D. Ashwell. 2005. Alternative p38 activation pathway mediated by T cell receptor-proximal tyrosine kinases. Nat. Immunol. 6:390-395. [DOI] [PubMed] [Google Scholar]
- 37.Schaeffer, H. J., and M. J. Weber. 1999. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol. Cell. Biol. 19:2435-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharrocks, A. D., S. H. Yang, and A. Galanis. 2000. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci. 25:448-453. [DOI] [PubMed] [Google Scholar]
- 39.Shibuya, H., K. Yamaguchi, K. Shirakabe, A. Tonegawa, Y. Gotoh, N. Ueno, K. Irie, E. Nishida, and K. Matsumoto. 1996. TAB1: an activator of the TAK1 MAPKKK in TGF-beta signal transduction. Science 272:1179-1182. [DOI] [PubMed] [Google Scholar]
- 40.Sreerama, N., and R. W. Woody. 2004. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 383:318-351. [DOI] [PubMed] [Google Scholar]
- 41.Su, B., and M. Karin. 1996. Mitogen-activated protein kinase cascades and regulation of gene expression. Curr. Opin. Immunol. 8:402-411. [DOI] [PubMed] [Google Scholar]
- 42.Tanno, M., R. Bassi, D. A. Gorog, A. T. Saurin, J. Jiang, R. J. Heads, J. L. Martin, R. J. Davis, R. A. Flavell, and M. S. Marber. 2003. Diverse mechanisms of myocardial p38 mitogen-activated protein kinase activation: evidence for MKK-independent activation by a TAB1-associated mechanism contributing to injury during myocardial ischemia. Circ. Res. 93:254-261. [DOI] [PubMed] [Google Scholar]
- 43.Tanoue, T., M. Adachi, T. Moriguchi, and E. Nishida. 2000. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2:110-116. [DOI] [PubMed] [Google Scholar]
- 44.Tanoue, T., R. Maeda, M. Adachi, and E. Nishida. 2001. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 20:466-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanoue, T., and E. Nishida. 2003. Molecular recognitions in the MAP kinase cascades. Cell. Signal. 15:455-462. [DOI] [PubMed] [Google Scholar]
- 46.van der Geer, P., T. Hunter, and R. A. Lindberg. 1994. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu. Rev. Cell Biol. 10:251-337. [DOI] [PubMed] [Google Scholar]
- 47.Ward, Y., S. Gupta, P. Jensen, M. Wartmann, R. J. Davis, and K. Kelly. 1994. Control of MAP kinase activation by the mitogen-induced threonine/tyrosine phosphatase PAC1. Nature 367:651-654. [DOI] [PubMed] [Google Scholar]
- 48.Waskiewicz, A. J., and J. A. Cooper. 1995. Mitogen and stress response pathways: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr. Opin. Cell Biol. 7:798-805. [DOI] [PubMed] [Google Scholar]
- 49.Whitmarsh, A. J., and R. J. Davis. 1998. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem. Sci. 23:481-485. [DOI] [PubMed] [Google Scholar]
- 50.Yang, S. H., A. Galanis, and A. D. Sharrocks. 1999. Targeting of p38 mitogen-activated protein kinases to MEF2 transcription factors. Mol. Cell. Biol. 19:4028-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zarubin, T., and J. Han. 2005. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 15:11-18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.