Abstract
CDO is a cell surface immunoglobulin superfamily member that positively regulates myogenic differentiation in vitro and in vivo and signals to posttranslationally activate myogenic basic helix-loop-helix (bHLH) transcription factors. The Cdo gene is also expressed in the dorsal aspect and midline structures of the developing central nervous system, and mice lacking CDO on the C57BL/6 background display holoprosencephaly with ∼80% penetrance, resulting in perinatal lethality. We report here that a fraction of Cdo−/− mice from this background have additional defects in brain development, including hydrocephalus and cortical thinning. Primary neural progenitor cultures from E14.5 Cdo−/− mutants display reduced proliferation, which may underlie the thinning. The cortical preplate and cortices of mutant animals also show reduced staining for β-tubulin III, indicating defective neuronal differentiation. CDO levels are strongly increased in cultured C17.2 neuronal precursor cells stimulated to differentiate; modulation of CDO levels in these cells by overexpression or interfering RNA approaches enhances or diminishes differentiation, respectively. Cotransfection of CDO enhances the activity of the neurogenic bHLH factor, neurogenin1, in reporter assays and enhances heterodimerization of neurogenin1 and E47. These results indicate that CDO promotes neuronal differentiation and support the hypothesis that CDO coordinates differentiation of multiple cell lineages by regulating the activity of tissue-specific bHLH factors.
The cerebral cortex originates from a sheet of neuroepithelial cells within the ventricular zone (VZ) of the dorsal telencephalon. In the mouse cerebral cortex, neurogenesis starts at embryonic day 11 (E11) and continues until E17. The balance between proliferation and differentiation of neural progenitor cells within the VZ is essential in determining the total number of neurons that comprise the neocortex (3, 8, 29, 38, 44, 45). Neurogenesis is mediated by neural basic helix-loop-helix (bHLH) transcription factors, including neurogenin1 (Ngn1) and Ngn2, Mash1, and NeuroD (4, 7, 27, 40). Neural bHLH factors are transcriptional activators that bind DNA as heterodimeric complexes with E proteins. Ngn1 and Ngn2 are selectively expressed in the VZ by dorsal telencephalic progenitors, where such cells begin differentiation, but not in the cortical plate, where fully differentiated neurons are situated (20, 28, 43). The transition from proliferation to differentiation during neurogenesis involves an increase in expression and activities of proneural bHLHs. However, the mechanisms that underlie this transition are not well understood.
CDO is a surface protein of the immunoglobulin (Ig) superfamily that is expressed at high levels in developing muscle, the central nervous system, and the midface (24, 32). Extensive studies on myogenesis suggest that CDO serves as a component of a receptor complex that includes cadherins and the related Ig proteins, BOC and neogenin (21, 23-25). CDO positively regulates differentiation of myoblast cell lines in vitro, and mice with a targeted Cdo mutation display delayed skeletal muscle development (10). CDO signals to posttranslationally activate myogenic bHLH transcription factors, which, like neural bHLH factors, function as heterodimers with E proteins (10). This appears to occur via enhanced heterodimer formation following CDO-mediated hyperphosphorylation of E proteins. A mechanism in which CDO signals to E proteins and shifts the equilibrium of their dimerization towards the heterodimeric state is appealing because it can be generalized to any cell type that expresses CDO and relies on tissue-specific bHLH factors that dimerize with E proteins, e.g., neural precursors. In addition to delayed myogenesis, mice lacking CDO display holoprosencephaly (HPE) with strain-specific severity (9; W. Zhang et al., unpublished data). HPE is the most common developmental defect of the forebrain and midface in humans (47, 48); some cases of HPE are accompanied by hydrocephalus (5, 26), a severe and often lethal birth defect in humans that results from the excess accumulation of cerebrospinal fluid in the developing brain.
The expression pattern of CDO and the proposed mechanism by which CDO activates myogenic bHLH factors predict that CDO may play an important role in neurogenesis. We report here that, in addition to HPE, Cdo−/− embryos display other defects in brain development, including hydrocephalus, thinning of the cortex, and reduced differentiation. Cultured primary neural progenitor cells from Cdo−/− embryos show reduced levels of proliferation, which may partly explain the cortical thinning. Additionally, CDO expression is induced during and positively regulates differentiation of a neural progenitor cell line in vitro. Finally, CDO enhances the transcriptional activity of Ngn1 in a reporter assay and heterodimerization of Ngn1 and E47. Our results suggest that CDO acts as a positive regulator of neurogenesis.
MATERIALS AND METHODS
Mice and embryos.
Cdo+/lacZ−1 (also known as Cdontm1Rsk) and Cdo+/lacZ−2 (also known as Cdontm2Rsk) mice of a mixed 129/Sv × C57BL/6 (B6) background have been previously described (9). Heterozygous males of the mixed background were backcrossed with C57BL/6 females for six generations; intercrosses of such Cdo+/− animals resulted in perinatal lethality in 80 to 85% of homozygous mutant offspring. The studies presented here are from these crosses. The phenotypes of mice homozygous for each mutation are similar, except that the CdolacZ−2 allele expresses lacZ in a pattern that mimics endogenous Cdo expression, while the CdolacZ−1 allele expresses lacZ extremely weakly. Mice of both lines were studied except in the case of β-galactosidase (β-Gal) staining, for which only CdolacZ−2 mice were used. Genotyping was performed by PCR analysis with a single forward primer (5′-GGAGGCTGAGTTAGGAGGATCACAAGTTCGAG-3′) and reverse primers specific for wild-type or mutant alleles (5′-ATAAGGCACTGGGAGATTATGGGGCGAG-3′ and 5′-GCGATG CCTGCTTGCCGAATATCATGGTG-3′, respectively). Noon of the plug date was designated E0.5.
β-Gal staining, histology, and immunohistochemistry.
For β-Gal staining, dissected embryos and brains were processed essentially as previously described (33) with the exception that fixation time was dependent on the age of the embryo and brains. The specimens were embedded in paraffin and sectioned at 8 to 10 μm. Histological examination of brain structures was performed by Nissl staining of paraffin sections; β-Gal-stained sections were counterstained with nuclear fast red. For immunohistochemistry, dissected embryos and brains were fixed in 4% paraformaldehyde and embedded in paraffin. Seven-micrometer-thick sections were stained with anti-TuJ1 (1:100; Sigma) and anti-phospho-histone H3 (1:200; Upstate Biotechnology) antibodies.
BrdU incorporation and TUNEL assay.
To monitor cell proliferation, pregnant mice were injected with 100 μg bromodeoxyuridine (BrdU; Sigma)/g of body weight. Two hours later, embryos were dissected and fixed with 4% paraformaldehyde, embedded in paraffin, and sectioned at 7 μm. Immunohistochemistry was performed with anti-BrdU antibody (1:200; Chemicon). Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) was performed according to the manufacturer's instructions (Roche).
Cell culture and transfections.
10T1/2 cells were cultured as previously described (10). C17.2 cells were cultured in Dulbecco's modified Eagle medium (DMEM) with 7% fetal bovine serum-5% horse serum and passaged at 50% confluence every 2 days. To induce differentiation, 80% confluent cultures were shifted to DMEM with 2% fetal bovine serum. Transfections were performed with FuGene6 (Roche). For reporter assays, C17.2 cells cultured in 12-well plates were cotransfected with the reporter construct EB7/Luc, which contains three copies of an E box from the NeuroD2 promoter (200 ng), and 5 ng (each) of pCS2+Ngn1 and pCS2+E12 (14). Luciferase assays were performed using a dual luciferase assay system (Promega).
For interfering RNA (RNAi) studies, five sequences against mouse Cdo were tested initially. Oligonucleotides corresponding to the Cdo sequences were cloned into the pSilencer 2.0-U6 vector (Ambion), and 9.0 μg of the individual vectors or parental vector was transfected with 1.0 μg of an enhanced green fluorescent protein (EGFP) expression vector (Clontech) into C17.2 cells with FuGene6. GFP-positive cells were isolated 24 h later with a FACDiVa flow cytometer (Becton Dickinson), and identical numbers of cells were replated. The cultures were harvested after 48 h and analyzed for CDO expression by Western blotting. The most consistently effective sequence was chosen for further use (AGCTTTTCCAAAAAACAGCGTTGGTGCCGTTGTGTCTCTTGAACAC AACGGCACCAACGCTGCGG; CAGCGTTGGTGCCGTTGTG).
Primary neural progenitor cultures and BrdU incorporation.
Neurosphere cultures were prepared from cortices of E14.5 embryos (15). Briefly, cells were resuspended in serum-free DMEM/F12 medium containing N2 supplement, B27 supplement, basic fibroblast growth factor (10 ng/ml), epidermal growth factor (10 ng/ml), and penicillin and streptomycin (all from Invitrogen). To analyze their proliferative capacity, cells were seeded onto plates coated with poly-l-ornithine (Sigma) and laminin (Invitrogen). The following day, cells were refed with medium containing basic fibroblast growth factor and epidermal growth factor and incubated with 20 μM BrdU for 2 h. Cultures were then fixed with 4% paraformaldehyde, followed by immunostaining with anti-BrdU antibody (1:200; Chemicon).
Western blot analysis, immunoprecipitation, and immunocytochemistry.
Western blot analyses were done as previously described (22). Primary antibodies used were anti-CDO (22), anti-Nestin (R401; Hybridoma Bank), anti-cadherin (Sigma), anti-β-tubulin III (TuJ1; Sigma), anti-NeuroD (Santa Cruz), and anti-GFAP (for glial fibrillary acidic protein; Sigma). For coimmunoprecipitation studies, six myc-epitope tags were introduced at the N terminus of Ngn1 within the pCS2 expression vector. 10T1/2 and C17.2 cells in 10-cm plates were cotransfected with 0.5 μg of either control pCS2 vector or pCS2/Ngn1(myc) and 9.5 μg of either the control pBabePuro vector or pBabePuro/CDO vector and analyzed as previously described (10). Cell lysates were coimmunoprecipitated with anti-E47 antibody (Santa Cruz) and Western blotted for expression of Ngn1 with anti-myc (9E10; Mount Sinai School of Medicine Hybridoma Core Facility), anti-E47 (Santa Cruz), and anti-CDO (23). For immunocytochemistry, C17.2 cells cultured in six-well plates were cotransfected with 25 ng EGFP expression vector and 450 ng of either the control pBabePuro vector or pBabePuro/CDO vector. Thirty-six hours to 48 h later, cells were fixed with 4% paraformaldehyde for 10 min, permeabilized with phosphate-buffered saline-0.5% Triton X-100, blocked with phosphate-buffered saline-5% goat serum, and probed with anti-TuJ1 (1:200; Sigma) and Alexa Fluor 568 goat anti-mouse antibody (Molecular Probes).
RESULTS
CDO deficiency causes a complex phenotype in brain development, including hydrocephalus and cortical thinning.
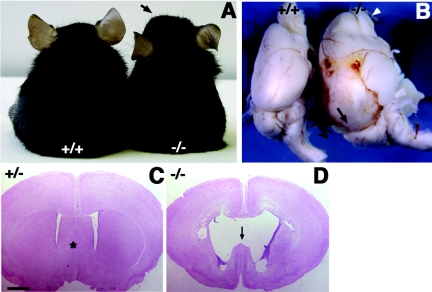
Mice homozygous for a targeted mutation of Cdo on a mixed 129/Sv × B6 background display facial microsigns of HPE and ∼35% perinatal lethality. After Cdo+/− mice of mixed background were backcrossed for six generations onto the B6 background, intercrosses of such animals revealed 80 to 85% perinatal lethality among Cdo−/− offspring, due to severe forms of HPE (51). The remaining 15 to 20% of Cdo−/− mice, whose phenotype is described here, did not display overt signs of HPE, survived beyond the perinatal period, but died within 4 to 12 weeks of birth. Prior to death, these mice exhibited a dome-shaped head (Fig. 1A), limb weakness, and immobility, suggesting they had developed hydrocephalus, which sometimes accompanies HPE (2). Brains from such Cdo−/− mice displayed surface bleeding, and the forebrains were highly enlarged with dilated blood vessels and olfactory bulbs (Fig. 1B). Coronal sections revealed enlarged lateral ventricles with a disruption of the septum, consistent with hydrocephalus (Fig. 1C and D). Taken together, CDO deficiency in the B6 background results in abnormal brain development, including HPE and hydrocephalus.
FIG. 1.
Hydrocephalus in Cdo−/− mice. (A) Ten-week-old littermates of the indicated genotype. The arrow indicates the dome-shaped head of the Cdo−/− mouse. (B) Brains from 8-week old wild-type and Cdo−/− littermates. The Cdo−/− brain and olfactory bulbs (arrowhead) exhibit dramatic swelling and hemorrhage, compared to the those of the wild-type littermate. (C and D) Coronal sections of brains from 3-week-old littermates. Brains were stained with nuclear fast red. Enlarged lateral ventricles and disruption of the septum (marked with an asterisk in the Cdo+/− section) are observed in the Cdo−/− brain. Size bar, 1 mm.
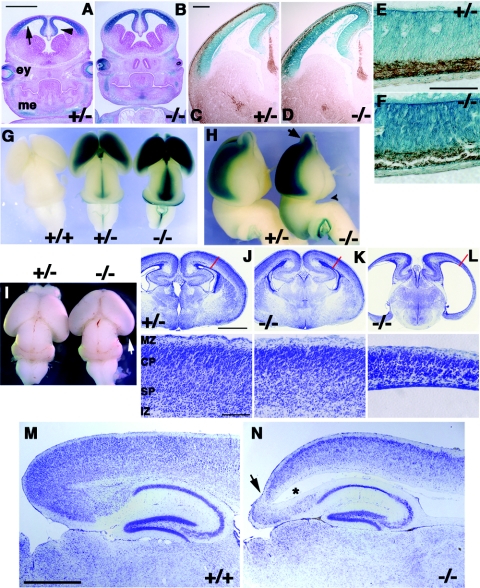
We have previously shown that Cdo is expressed in the developing central nervous system. During embryogenesis and early postnatal development, expression is primarily observed in the dorsal ventricular and subventricular zones of the telencephalon, diencephalon, and midbrain (32). We further studied Cdo expression in Cdo+/− and Cdo−/− animals by taking advantage of mice carrying the CdolacZ−2 allele, a functional lacZ knockin that is also likely a null allele. These mice produce β-Gal activity in a pattern that mimics endogenous Cdo expression as observed by RNA in situ hybridization (9, 32). In coronal sections of the forebrain at E13.5, β-Gal activity was detected throughout dorsal regions of the developing cortex of Cdo+/− and Cdo−/− embryos (Fig. 2A and B). Immunostaining with an antibody to β-tubulin III (TuJ1), a pan-neuronal marker that is expressed early during neuronal differentiation, suggested that most Cdo-expressing cells reside in the proliferating, TuJ1-negative neuroepithelium (Fig. 2C to F). A dorsally restricted pattern of Cdo expression was maintained in later stages of brain development, as seen in whole-mount β-Gal staining of brains from E16.5 embryos, in which expression expanded along with developing dorsal structures of the telencephalon (Fig. 2G and H). High levels of β-Gal activity was also seen in the midline of the midbrain and hindbrain and the olfactory bulbs (Fig. 2G and H). Anatomically, the forebrain of Cdo−/− mutant embryos exhibited an enlarged gap between the hypothalamus and pons, indicating a slight truncation of the forebrain (Fig. 2H). Moreover, the posterior telencephalon of E18.5 Cdo−/− brains often showed a translucent appearance, indicating thinning of the cortex (Fig. 2I). This transparent appearance was specific to the posterior telencephalon and detected only at later developmental stages (e.g., E18.5), suggesting the defect may reflect alterations in corticogenesis rather than defects in early telencephalon patterning associated with HPE.
FIG. 2.
Expression of Cdo in proliferating cortical neural progenitors and defects in cortical thickness in Cdo−/− mice. (A and B) Coronal sections from E13.5 littermates of the indicated genotype. Sectioned embryos stained for β-Gal activity (blue) reveal Cdo expression in the neuroepithelium of the developing cortex (arrow) and hippocampus (arrowhead). Cdo is also expressed in the developing eye (ey) and the condensing mesenchyme of the craniofacial region (me). Size bar, 1 mm. (C and D) Sections of E13.5 embryos were doubly stained for Cdo expression (β-Gal activity; blue) and β-tubulin III to label the differentiated layer of the cortex (Tuj1 antibody; brown). Note that the Tuj1+ differentiated layer (the preplate) is devoid of β-Gal staining, indicating that Cdo is expressed in proliferating neural progenitors. Size bar, 0.2 mm. (E and F) Higher magnification micrographs of sections from (C, D). Size bar, 0.1 mm. (G and H) Dissected E16.5 brains stained for Cdo expression (β-Gal activity; blue). Dorsal (G) and lateral (H) views of brains of the indicated genotype document Cdo expression in dorsal regions and olfactory bulbs (H, arrow). The arrowhead in panel H marks an expanded gap between the hypothalamus and pons, indicating a truncation of the forebrain. (I) Brains from newborn littermate pups. Extreme thinning of the cortex in the Cdo−/− brain is visible as a translucent area in the posterior region (arrow). (J to L, top) Nissl-stained sections from E18.5 littermates of the indicated genotype. The two Cdo−/− mutants shown reveal the range of severity in cortical thinning observed (indicated by the red lines), with the mutant in panel J showing a severe phenotype. Note that this mutant also displayed HPE. (Bottom) Abnormal cortical organization is observed in both Cdo−/− cortices. IZ, intermediate zone; SP, subplate; CP, cortical plate; MZ, marginal zone. Size bar, 1 mm (top); 0.2 mm (bottom). (M and N) Sagittal sections of brains from 3-week-old Cdo+/+ and Cdo−/− littermates were Nissl stained to reveal thinning and disorganization of the cortex in the Cdo−/− brain. In addition, the Cdo−/− brain shows an ectopic ventricle (marked with an asterisk) and extreme thinning in the posterior cortex (arrow), indicating hydrocephalus. Size bar, 0.5 mm.
To analyze CDO's role in cortical neurogenesis, we histologically examined cortices from E18.5 and postnatal day 21 (P21) mutant and control littermates. Thinning of the cortex was observed with high penetrance in Cdo−/− animals but with variable severity. In general, mice with HPE displayed the most severe thinning, but even mice without evidence of HPE had abnormally thin cortices (Fig. 2J and L). Sagittal sections of brains from P21 mice also showed thinner cortices (particularly in the posterior region) and ectopic ventricles in the Cdo mutants, the latter phenotype indicative of hydrocephalus (Fig. 2M and N).
Reduced proliferation of primary neural progenitor cells from Cdo mutant cortices.
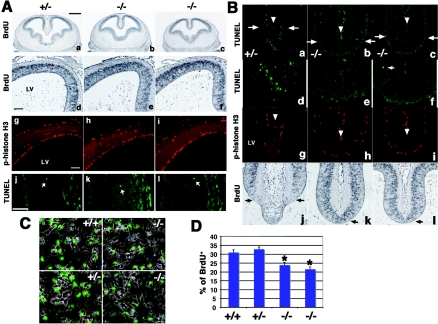
To gain insight into potential mechanisms of the defective corticogenesis seen in Cdo−/− mice, we examined cell death and proliferation in the dorsal telencephalon of embryos at several stages. To assess proliferation, BrdU incorporation was measured 2 h following a pulse of BrdU. As expected, incorporation was observed at E13.5 in the VZ of the telencephalon (Fig. 3Aa to f). However, no obvious difference between Cdo−/− and control littermates was detected (Table 1). Moreover, immunostaining for phospho-histone H3 also failed to reveal a difference between E13.5 control and mutant cortices (Fig. 3Ag to i). Phospho-histone H3 immunostaining of sections of E15.5 and E18.5 cortices also did not show any differences between control and Cdo−/− embryos (Table 1).
FIG. 3.
Analysis of proliferation and apoptosis in the lateral cortex of control and Cdo−/− embryos and primary neural progenitor cultures. (A) Coronal sections of cortices from E13.5 Cdo+/− (a and d) and two Cdo−/− (b, c, e, and f) littermates immunostained for BrdU incorporation. Similar sections were analyzed by immunostaining with phospho-histone H3 antibodies for mitotic cells (g to i) and TUNEL assay for apoptotic cells (j to l). Mitotic cells were observed adjacent to the lateral ventricle (LV) of the telencephalon; cells that appear positive for phospho-histone H3 staining that are distal to this region are red blood cells that display autofluorescence. TUNEL-positive cells (arrows) were infrequent in the lateral cortices of these embryos. No changes in proliferation or apoptosis were observed in Cdo−/− embryos; see Table 1 for quantification. (B) Alterations in cell death and proliferation in the dorsomedial telencephalon of Cdo−/− embryos. Coronal sections of E13.5 embryos analyzed by TUNEL assay (a to f) and immunostaining for phospho-histone H3 (g to i). The altered appearance of the dorsomedial telencephalon of Cdo−/− embryos is visible at low magnification (a to c); the areas below the arrows in panels a to c are shown in higher magnification in (d to f), which reveals substantial cell death in the dorsomedial telencephalon of the control embryo, whereas cell death is rarely observed in this region of Cdo−/− embryos. The choroidal cleft is indicated by the arrowhead in (a to c and g to i). (g to i) As in panel A, mitotic cells are located adjacent to the LV, and the presence of staining in distal regions and in the choroidal cleft is due to autofluorescence of red blood cells. Note also that the Cdo+/− telencephalon contains fewer mitotic cells than the Cdo−/− telencephalons (g to i). Sections immunostained for BrdU incorporation in the dorsomedial telencephalon (j to l) show that the Cdo+/− telencephalon contains fewer BrdU-positive cells than the Cdo−/− telencephalons. (C) Immunostaining for BrdU incorporation (green) in primary neural progenitor cells isolated from cortices of two Cdo−/− and two control littermate E14.5 embryos. (D) Quantification of experiment shown in panel C. These experiments were repeated with similar results with neural progenitors isolated from two mutant and two control embryos each from three independent litters. Asterisks indicate that BrdU incorporation in Cdo−/− cultures is significantly different from BrdU incorporation in Cdo+/+ cultures; P < 0.01 by analysis of variance.
TABLE 1.
Quantification of cell proliferation and apoptosis in lateral cortices of Cdo+/− and Cdo−/− micea
| Embryonic age | Assay | Genotype
|
||
|---|---|---|---|---|
| Cdo+/− | Cdo−/− | Cdo−/− | ||
| E13.5 | BrdU incorporation | 591 ± 50 | 606 ± 57 | 583 ± 44 |
| Phospho-histone H3 | 40 ± 8.7 | 40 ± 4.6 | 41 ± 5.8 | |
| TUNEL | 3.3 ± 0.6 | 2.7 ± 0.6 | 3.0 ± 0.6 | |
| E15.5 | Phospho-histone H3 | 21.3 ± 5.6 | 21 ± 6.1 | |
| TUNEL | 2.8 ± 1.2 | 2.7 ± 1.0 | ||
| E18.5 | Phospho-histone H3 | 12.4 ± 2.9 | 12.0 ± 6.6 | |
| TUNEL | 16.3 ± 1.3 | 15.8 ± 2.2 | ||
Multiple sections from the embryos shown in Fig. 3 are quantified in this table. Analyses of sections from multiple animals produced similar results.
Since these assays may have failed to reveal more subtle differences in cell proliferation within the lateral cortex, cultured neural progenitor cells were also examined. Primary neural progenitors from E14.5 cortices were derived from neurosphere cultures of several different Cdo−/− and control littermate embryos, and the cultures were analyzed for their proliferative capacity by BrdU incorporation. Cdo−/− neural progenitor cell cultures displayed a reduced proliferation rate (24% and 22% BrdU incorporation), compared to the control cultures (31% and 33% BrdU incorporation) (Fig. 3C and D). These data suggest that Cdo−/− neural progenitor cells do indeed proliferate more slowly than control cells and that this may underlie, at least in part, the cortical thinning seen in Cdo−/− embryos.
To determine whether abnormalities of cortical thickness reflected increased levels of apoptosis, TUNEL assays of sections of E13.5, E15.5, and E18.5 embryos were performed. Apoptotic cells were detected infrequently in the lateral cortices of control or homozygous mutant E13.5 embryos (Fig. 3Aj to Al), and the numbers of TUNEL-positive cells between control and mutant cortices were similar (Table 1). While these results suggest that alterations in apoptosis are not responsible for the cortical thinning phenotype of Cdo−/− mice, we cannot rule out the involvement of other forms of cell death. However, levels of cell death were not obviously different between Cdo+/+ and Cdo−/− neural progenitor cultures (data not shown).
In contrast to the lateral cortices, the dorsal midline region of the telencephalon exhibited a significant difference between mutant and control embryos in cell proliferation and death. Regionally selective cell proliferation and death in the dorsal midline of the telencephalon is believed to be involved in creating the midline recess (19, 31). Consistent with this notion, the ventrally extending tip of the dorsal midline (the lamina terminalis) of control telencephalons showed reduced proliferation relative to the rest of the midline region, as observed by a lower rate of BrdU incorporation and scarcity of phospho-histone H3-positive cells (Fig. 3Bg and j). Furthermore, this tip region contained many TUNEL-positive cells (Fig. 3Ba and d). Interestingly, the dorsal midline of Cdo−/− cortices exhibited an abnormal structure, lacking the thinned lamina terminalis. In contrast to control animals, BrdU incorporation and anti-phospho-histone H3 staining were seen throughout the dorsal telencephalic midline structures of Cdo−/− mice (Fig. 3Bh, i, k, and l). Moreover, no obvious cell death was observed in this region of the mutants (Fig. 3Bb, c, e, and f). This abnormal cell proliferation and apoptosis in the dorsal midline region of the Cdo−/− telencephalon may be related to the HPE phenotypes seen with these mice.
Regulation of neuronal differentiation by CDO.
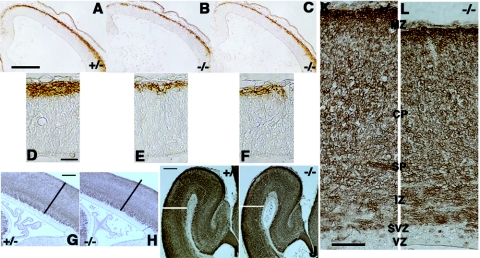
To examine whether lack of Cdo alters cell differentiation in the lateral cortex, we examined expression of the neural marker, β-tubulin III, at various stages of cortical development in control and Cdo−/− embryos. TuJ1+ cell clusters were similar in Cdo+/− and Cdo−/− sections at E10.5, indicating that neurogenesis initiated normally in the absence of CDO (data not shown). However, analysis of sections from E13.5 cortices showed that the staining intensity and thickness of the TuJ1+ preplate was reduced in Cdo−/− embryos, compared to control littermates (Fig. 4A to F), indicating a decrease in neuronal differentiation. Consistent with this observation, the TuJ1+ cortical layers of E15.5 and E18.5 Cdo−/− embryos were reduced in size relative to those of control embryos (Fig. 4G to L), particularly the cortical plate at E18.5, suggesting impaired neuronal differentiation of progenitor cells in cortices lacking CDO.
FIG. 4.
Defective neuronal differentiation in Cdo−/− cortices from various developmental stages. (A to C) Coronal sections of the telencephalon from E13.5 littermates of the indicated genotypes immunostained with TuJ1 antibody (brown). Note that the Cdo−/− cortices show weaker staining and that the differentiated preplate is thinner than the Cdo+/− cortex. Size bar, 0.2 mm. (D to F) Higher-magnification micrographs of sections from panels A to C. Size bar, 0.02 mm. (G and H) Sagittal sections of E15.5 Cdo+/− and Cdo−/− littermates immunostained with TuJ1 antibody (brown). Size bar, 0.15 mm. (I and J) Coronal sections of E18.5 Cdo+/− and Cdo−/− littermates immunostained withTuJ1 antibody (brown). Size bar, 0.2 mm. Note that the Cdo−/− cortices show thinning of the differentiated, Tuj1-positive layers relative to the control cortices (black and white lines in panels G and H and I and J, respectively). (K and L) Higher-magnification micrograph of E18.5 cortices. MZ, marginal zone, CP; cortical plate, SP; subplate, IZ; intermediate zone, SVZ; subventricular zone, VZ; ventricular zone. Size bar, 0.05 mm.
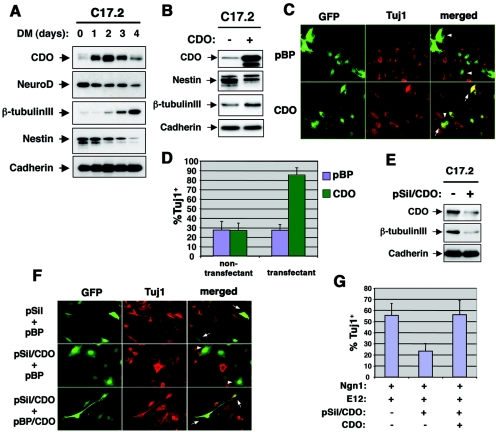
To further explore the possibility that CDO regulates neuronal differentiation, we utilized C17.2 cells, a clonal, multipotent neuronal precursor line originally derived from the external germinal layer of the neonatal mouse cerebellum (41, 42). C17.2 cells proliferate in serum-rich medium (GM) but differentiate when cultured in serum-poor differentiation medium (DM), as monitored by expression of β-tubulin III (Fig. 5A). β-Tubulin III expression was induced 2 days after transfer of C17.2 cells to DM and increased dramatically 4 days after transfer. Conversely, expression of nestin, a marker of proliferating progenitors, was strongly decreased when these cells differentiated. NeuroD was detected throughout the time course of differentiation, but its levels diminished steadily. Interestingly, exponentially growing C17.2 cells expressed low levels of CDO, which were strongly but transiently increased when these cells were challenged to differentiate (Fig. 5A). Furthermore, this increase in CDO expression preceded neuronal differentiation, as detected by expression of β-tubulin III.
FIG. 5.
CDO promotes neuronal differentiation of C17.2 neural progenitor cells. (A) Western blot analysis of CDO, NeuroD, β-tubulin III, nestin, and cadherin expression in C17.2 cells during a time course of neuronal differentiation. Subconfluent cells were shifted to DM, and cells were harvested at the indicated time points. (B) Western blot analysis of CDO, nestin, β-tubulin III, and cadherin expression by C17.2 cells transiently transfected with either control (−) or CDO (+) expression vectors. Transfection efficiency was 40 to 50%, with similar results in three independent experiments. (C) Immunostaining for β-tubulin III expression by C17.2 cells transiently transfected with EGFP expression vector and either control (pBP) or CDO expression vectors. Arrowheads indicate Tuj1-negative transfectants, and arrows indicate Tuj1-positive transfectants. (D) Quantification of four independent experiments shown in panel C. The y axis represents the percentage of TuJ1-positive cells. (E) Western blot analysis of CDO, β-tubulin III, and cadherin expression by C17.2 cells transiently transfected with control siRNA vector (−) or pSil/CDO RNAi vector (+). Similar results were obtained with three independent experiments. (F) Immunostaining for β-tubulin III expression by C17.2 cells transiently transfected with expression vectors for GFP, Ngn1, and E12 and as indicated with combinations of pSil, pSil/CDO, pBP, and pBP/CDO. Arrowheads indicate Tuj1-negative transfectants, and arrows indicate Tuj1-positive transfectants. (G) Quantification of four independent experiments shown in panel F. The y axis represents the percentage of TuJ1-positive transfectants.
To determine if CDO plays a functional role in C17.2 cell differentiation, we assessed whether increasing or decreasing CDO levels in these cells would result in a corresponding augmentation or inhibition of differentiation. Initial attempts at stable overexpression of CDO in C17.2 cells failed, suggesting that constitutively high levels of CDO might drive these cells to differentiate precociously, exiting the cell cycle such that they were lost to the pool of stable transfectants (data not shown). Consistent with this possibility, C17.2 cultures that overexpressed CDO via transient transfection in 40 to 50% of the cells displayed decreased levels of nestin and increased levels of β-tubulin III relative to control vector transfectants, even when cultured in GM (Fig. 5B). To quantify this more accurately, C17.2 cells were transiently cotransfected with a GFP vector and either a control or CDO expression vector. Forty-eight hours later, the cultures were fixed and immunostained with TuJ1 antibody. As shown in Fig. 5C and D, ∼30% of nontransfectants from both cultures were Tuj1+. Approximately 30% of the control vector transfectants were also TuJ1+, whereas ∼85% of the CDO transfectants were TuJ1+. Overexpression of CDO therefore enhances neuronal differentiation of C17.2 cells.
To assess the effect of reducing CDO levels on differentiation of C17.2 cells, an RNAi approach was taken. A target sequence from the mouse CDO coding region was inserted into the pSilencer (pSil) vector, and C17.2 cells were transiently transfected with either control pSil or pSil/CDO. To avoid the difficulty of knocking down CDO levels at the same time point that they were being dramatically increased by transfer of the cells to DM, the transfected populations were cultured for 3 days in GM, a condition that resembles ∼24 h in DM (J.-S. Kang, unpublished data). Western blot analysis revealed that CDO protein levels were reduced by pSil/CDO under these conditions, as was induction of β-tubulin III (Fig. 5E). To further assess the role of endogenous CDO in C17.2 cell differentiation, we stimulated differentiation of cells maintained in GM by cotransfection of expression vectors for Ngn1 and its dimeric partner, E12. A GFP vector and either pSil or pSil/CDO were cotransfected; 36 h later, the cells were fixed and immunostained with TuJ1 antibody. As shown in Fig. 5F and G, ∼55% of the pSil-transfected cells were TuJ1+, whereas only 25% of the pSil/CDO-transfected cells were Tuj1+. Furthermore, cotransfection of a rat CDO expression vector, which is not fully conserved at the sequence used to knock down endogenous mouse CDO and is impervious to the effects of the RNAi vector (data not shown), increased the percentage of TuJ1+ cells to control levels, indicating that the effects of the CDO RNAi were specific. Taken together, the overexpression and RNAi studies suggest that CDO levels are rate limiting for neuronal differentiation of C17.2 cells.
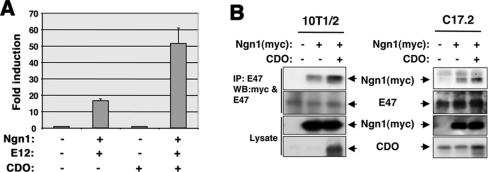
One way that CDO positively regulates myogenesis is via posttranslational activation of myogenic bHLH factors (e.g., MyoD, myogenin, and Myf5). CDO-mediated signaling results in enhanced heterodimerization of MyoD with E47 and E12 (10), the ubiquitously expressed E protein binding partners of tissue-specific bHLH factors, including neural bHLH factors. As enhanced heterodimerization appears to be a consequence of CDO-stimulated phosphorylation of the E proteins (10) and as the neural bHLH factors, Ngn1 and Ngn2, are temporally and spatially coexpressed with CDO in the dorsal telencephalon during neurogenesis (20, 28, 43), it is tempting to speculate that one way that CDO regulates neurogenesis is by modulation of neural bHLH activity. To test this possibility, the ability of CDO to stimulate Ngn1-mediated transcription was examined. An Ngn1-responsive luciferase reporter construct that contains three copies of an E box from the neuroD2 promoter was largely inactive in transiently transfected C17.2 cells in GM but could be stimulated by cotransfection of expression vectors for Ngn1 and E12 (Fig. 6A). CDO enhanced Ngn1/E12-dependent luciferase activity ∼3.5 fold, but did not activate the reporter in the absence of Ngn1/E12, suggesting that, as seen with myogenic bHLH factors, CDO stimulates Ngn1 activity. To test whether CDO enhances heterodimerization of Ngn1 with E proteins, myc-tagged Ngn1 [Ngn1(myc)] was transfected into 10T1/2 and C17.2 cells with or without CDO; 48 h later, cell lysates were subjected to immunoprecipitation with anti-E47 antibody. As shown in Fig. 6B, more Ngn1(myc) coimmunoprecipitated with E47 in the presence of cotransfected CDO than in its absence. These data suggest that CDO can enhance heterodimerization of Ngn1 and E proteins and that this may be one way in which the activity of neural bHLH transcription factors are regulated during neurogenesis.
FIG. 6.
CDO enhances the activity of Ngn1 and its heterodimerization with E proteins. (A) Ngn1-specific reporter gene activity. C17.2 cells were cotransfected in growth medium with an Ngn-responsive luciferase reporter construct and the indicated combinations of expression vectors for Ngn1, E12, and CDO. The luciferase activity was assessed 48 h after transfection and normalized to an internal control reporter (Renilla luciferase). Activity is reported as fold stimulation above that obtained with cells transfected with the reporter plasmid but only control vectors lacking cDNA. Values are means of triplicate determinations performed four independent times. (B) Coimmunoprecipitation of Ngn1(myc) and E47 in C17.2 neural progenitors and 10T1/2 cells. Cells were cotransfected with the indicated combinations of expression vectors for Ngn1 and CDO. Twenty-four hours later, cells were shifted to DM and cultured further for 24 h. Cells were lysed and lysates were subjected to coimmunoprecipitation with antibody to the endogenous E47 protein. Immunoprecipitations were Western blotted for Ngn1(myc), E47, and CDO expression.
DISCUSSION
The results provided here and elsewhere (51) indicate that mice lacking CDO display complex defects in brain development in a strain-dependent fashion. Cdo−/− mice of a mixed genetic background show microsigns of holoprosencephaly, generally restricted to the midface region, while brain development is grossly normal (9). In contrast, mice backcrossed for at least six generations onto the B6 background show multiple defects. Approximately 80% of Cdo−/− mice from this background have severe forms of HPE due to reduced Sonic hedgehog (Shh) signaling specifically in the ventral forebrain; these mice die or are culled by their mothers shortly after birth (51). However, the remaining ∼20% of homozygous mutants do not display overt holoprosencephalic phenotypes and survive the perinatal period. These mice provided the opportunity to visualize additional phenotypes associated with loss of CDO.
In addition to HPE, loss of CDO in the B6 background resulted in hydrocephalus and a thinned neocortex. Hydrocephalus is a potentially lethal birth defect in humans that results from excess accumulation of cerebrospinal fluid in the developing brain. A common cause of congenital hydrocephalus is aqueduct stenosis (1, 37). Cdo is expressed in the subcommissural organ and the roof of the aqueduct, defects in which can result in hydrocephalus; histologically, however, these structures appeared to be normal in Cdo−/− animals (data not shown). While hydrocephalus was also only observed in the B6 background, mice of the mixed background showed slightly expanded lateral ventricles (data not shown), suggesting that distinct genetic backgrounds may modify a predisposition of Cdo−/− animals to ventricular dilation. Interestingly, a similar observation has been made with mice carrying a targeted mutation of the gene encoding L1, which like CDO is a member of the Ig/FNIII family. L1 mutant mice develop severe hydrocephalus only in the B6 background, whereas in a mixed background these mice exhibit mild expansion of the lateral ventricles (12, 13, 17, 39). Mutations in the human L1 gene are associated with X-linked hydrocephalus as part of a syndrome called CRASH (corpus callosum hypoplasia, retardation, adducted thumbs, spastic paraplegia, and hydrocephalus) (18, 49). Hydrocephalus is sometimes observed in human HPE patients and, while CDO is clearly a candidate human HPE gene, it is also possible that mutations in CDO contribute to human hydrocephalus. The human CDO gene (also known as CDON) resides at chromosome 11q24.2, a region implicated in hydrolethalus, a rare recessively inherited lethal malformation syndrome characterized by hydrocephalus with absent midline structures of the brain (46). Two likely hydrolethalus cases with features of HPE have been reported (2). A hydrolethalus-associated mutation was recently identified in a gene of unknown function, designated HYLS1 (30). Although the human CDON gene falls just outside a critical region mapped by recombination events in affected families, HYLS1 and CDON are separated by only ∼70 kb, and it is possible that loss of CDO function might play a role in hydrolethalus alongside HYLS1 mutations.
Cdo−/− mice of the B6 background also showed abnormally thin cerebral cortices. This is consistent with the observation of reduced proliferation of cultured primary neural progenitor cells from Cdo−/− embryos. Cdo expression in the developing brain is most prevalent in dorsal structures, specifically in regions that contain proliferating neural progenitors, such as the cortical VZ. CDO has been implicated in two activities that might account for thinning of the cortex. First, CDO functions to positively regulate Shh signaling in specific regions of the developing embryo. Cdo is expressed transiently in the prechordal plate mesendoderm and notochord, and defects in Shh signaling from the former structure appear to underlie the HPE observed in Cdo−/− mice (51). Shh is also expressed in a layer-specific manner by differentiated cells in the neocortex and is suggested to regulate proliferation of Gli-positive neural progenitor cells during growth of the late embryonic and perinatal dorsal brain (11, 35). As CDO is expressed in proliferating VZ precursor cells, it is possible that a reduction in Shh signaling in these cells may contribute to the cortical thinning seen in Cdo−/− animals.
An alternative, but nonmutually exclusive, possibility is that CDO regulates the activity of neurogenic bHLH factors required for neurogenesis. The ability of CDO to posttranslationally activate neurogenic bHLH factors was predicted from the mechanism by which it activates MyoD and other myogenic bHLH factors (10). CDO signaling results in enhanced heterodimer formation between MyoD and E proteins, most likely via hyperphosphorylation of the E proteins; this mechanism would be expected to be operative for any tissue-specific bHLH factor that relies on E protein heterodimerization for function. Consistent with this possibility is that (i) differentiation of such cells is reduced or delayed in the developing cortices of Cdo−/− mice, (ii) CDO levels are rate limiting for neural differentiation of C17.2 cells in vitro, and (iii) CDO enhances the activity of Ngn1 in transient reporter assays and heterodimerization of Ngn1 and E47. Taken together, these data further support the hypothesis that CDO may regulate differentiation of several different cell lineages via its effects on tissue-specific bHLH factors.
Although it is clear that proneural bHLH factors are required for the specification and differentiation of neural progenitors, their independent role in corticogenesis has been somewhat difficult to discern because of a compensatory mechanism between Ngns and Mash1 (6, 16, 36, 50). Analysis of early cortical development in Ngn1, Ngn2, and Mash1 single-mutant mice did not reveal overt defects in neurogenesis. However, double mutants for Ngn2 and Mash1 exhibited severe defects in cortical development, including a reduction of neurogenesis (34). The defect in development of Cdo−/− cortices appeared to be more severe than that of cortices lacking any single neural bHLH factor. This is consistent with the likelihood that, similar to its actions on members of the myogenic bHLH family, CDO activates multiple members of the neurogenic bHLH family and also that it exerts its effects via multiple signaling mechanisms.
Acknowledgments
We thank Stephen Tapscott for reagents and Ruth Simon and Anne Ozog for critical reading of the manuscript.
This work was supported by a grant from the NIH (DE15162 and AR46207) to R.S.K. J.-S.K. was supported by the T. J. Martell Foundation, and F.C. was supported by a predoctoral grant from the HHMI and a training grant from the NCI.
REFERENCES
- 1.Allan, R., R. Chaseling, N. Graf, and M. Dexter. 2005. Aqueduct stenosis—?benign. J. Clin. Neurosci. 12:178-182. [DOI] [PubMed] [Google Scholar]
- 2.Bachman, H., R. D. Clark, and W. Salahi. 1990. Holoprosencephaly and polydactyly: a possible expression of the hydrolethalus syndrome. J. Med. Genet. 27:50-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer, S., and J. Altman. 1991. Development of the endopiriform nucleus and the claustrum in the rat brain. Neuroscience 45:391-412. [DOI] [PubMed] [Google Scholar]
- 4.Bertrand, N., D. S. Castro, and F. Guillemot. 2002. Proneural genes and specification of neural cell types. Nat. Rev. Neurosci. 3:517-530. [DOI] [PubMed] [Google Scholar]
- 5.Brown, S. A., D. Warburton, L. Y. Brown, C. Y. Yu, E. R. Roeder, S. Stengel-Rutkowski, R. C. Hennekam, and M. Muenke. 1998. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat. Genet. 20:180-183. [DOI] [PubMed] [Google Scholar]
- 6.Casarosa, S., C. Fode, and F. Guillemot. 1999. Mash1 regulates neurogenesis in the ventral telencephalon. Development 126:525-534. [DOI] [PubMed] [Google Scholar]
- 7.Cau, E., G. Gradwohl, C. Fode, and F. Guillemot. 1997. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development 124:1611-1621. [DOI] [PubMed] [Google Scholar]
- 8.Caviness, V. J., T. Takahashi, and R. Nowakowski. 1995. Numbers, time and neocortical neurogenesis: a general developmental and evolutionary model. Trends Neurosci. 18:379-383. [DOI] [PubMed] [Google Scholar]
- 9.Cole, F., and R. S. Krauss. 2003. Microform of holoprosencephaly in mice that lack the Ig superfamily member Cdon. Curr. Biol. 13:411-415. [DOI] [PubMed] [Google Scholar]
- 10.Cole, F., W. Zhang, A. Geyra, J. S. Kang, and R. S. Krauss. 2004. Positive regulation of myogenic bHLH factors and skeletal muscle development by the cell surface receptor CDO. Dev. Cell 7:843-854. [DOI] [PubMed] [Google Scholar]
- 11.Dahmane, N., P. Sanchez, Y. Gitton, V. Palma, T. Sun, M. Beyna, H. Weiner, and A. Ruiz i Altaba. 2001. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development 128:5201-5212. [DOI] [PubMed] [Google Scholar]
- 12.Dahme, M., U. Bartsch, R. Martini, B. Anliker, M. Schachner, and N. Mantei. 1997. Disruption of the mouse L1 gene leads to malformation of the nervous system. Nat. Genet. 17:346-349. [DOI] [PubMed] [Google Scholar]
- 13.Demyanenko, G. P., A. Y. Tsai, and P. F. Maness. 1999. Abnormalities in neuronal process extension, hippocampal development, and the ventricular system of L1 knockout mice. J. Neurosci. 19:4907-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farah, M. H., J. M. Olson, H. B. Sucic, R. I. Hume, S. J. Tapscott, and D. L. Turner. 2000. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development 127:693-702. [DOI] [PubMed] [Google Scholar]
- 15.Federoff, S., and A. Richardson (ed.). 2001. Protocols for neural cell culture, 3rd ed. Humana Press, Totowa, NJ.
- 16.Fode, C., Q. Ma, S. Casarosa, S.-L. Ang, D. J. Anderson, and F. Guillemot. 2000. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 14:67-80. [PMC free article] [PubMed] [Google Scholar]
- 17.Fransen, E., R. D'Hooge, G. Van Camp, M. Verhoye, J. Sijbers, E. Reyniers, P. Soriano, H. Kamiguchi, R. Willemsen, S. K. Koekkoek, C. I. De Zeeuw, P. P. De Deyn, A. Van der Linden, V. Lemmon, R. F. Kooy, and P. J. Willems. 1998. L1 knockout mice show dialated ventricles, vermis hypoplasia and impaired exploration patterns. Hum. Mol. Genet. 7:999-1009. [DOI] [PubMed] [Google Scholar]
- 18.Fransen, E., V. Lemmon, G. Van Camp, L. Vits, P. Coucke, and P. J. Willems. 1995. CRASH syndrome: clinical spectrum of corpus callosum hypoplasia, retardation, adducted thumbs, spastic paraparesis and hydrocephalus due to mutations in one single gene, L1. Eur. J. Hum. Genet. 3:273-284. [DOI] [PubMed] [Google Scholar]
- 19.Furuta, Y., D. W. Piston, and B. L. Hogan. 1997. Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development 124:2203-2212. [DOI] [PubMed] [Google Scholar]
- 20.Gradwohl, G., C. Fode, and F. Guillemot. 1996. Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev. Biol. 180:227-241. [DOI] [PubMed] [Google Scholar]
- 21.Kang, J.-S., J. L. Feinleib, S. Knox, M. A. Ketteringham, and R. S. Krauss. 2003. Pro-myogenic members of the Ig and cadherin families associate to positively regulate differentiation. Proc. Natl. Acad. Sci. USA 100:3989-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang, J.-S., M. Gao, J. L. Feinleib, P. D. Cotter, S. N. Guadagno, and R. S. Krauss. 1997. CDO: an oncogene-, serum-, and anchorage-regulated member of the Ig/fibronectin type III repeat family. J. Cell Biol. 138:203-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang, J.-S., P. J. Mulieri, Y. Hu, L. Taliana, and R. S. Krauss. 2002. BOC, an Ig superfamily member, associates with CDO to positively regulate myogenic differentiation. EMBO J. 21:114-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang, J.-S., P. J. Mulieri, C. Miller, D. A. Sassoon, and R. S. Krauss. 1998. CDO, a Robo-related cell surface protein that mediates myogenic differentiation. J. Cell Biol. 143:403-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang, J. S., M.-J. Yi, W. Zhang, J. L. Feinleib, F. Cole, and R. S. Krauss. 2004. Netrins and neogenin promote myotube formation. J. Cell Biol. 167:493-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinsman, S. L., L. L. Plawner, and J. S. Hahn. 2000. Holoprosencephaly: recent advances and new insights. Curr. Opin. Neurobiol. 13:127-132. [DOI] [PubMed] [Google Scholar]
- 27.Lee, J. E. 1997. Basic helix-loop-helix genes in neural development. Curr. Opin. Neurobiol. 7:13-20. [DOI] [PubMed] [Google Scholar]
- 28.Ma, Q., L. Sommer, P. Cserjesi, and D. J. Anderson. 1997. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing Notch ligands. J. Neurosci. 17:3644-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McConnell, S. K. 1995. Constructing the cerebral cortex: neurogenesis and fate determination. Neuron 15:761-768. [DOI] [PubMed] [Google Scholar]
- 30.Mee, L., H. Honkala, O. Kopra, J. Vesa, S. Finnila, I. Visapaa, T. Sang, G. Jackson, R. Salonen, M. Kestila, and L. Peltonen. 2005. Hydrolethalus syndrome is caused by a missense mutation in a novel gene HYLS1. Hum. Mol. Genet. 14:1475-1488. [DOI] [PubMed] [Google Scholar]
- 31.Monuki, E. S., and C. A. Walsh. 2001. Mechanisms of cerebral cortical patterning in mice and humans. Nat Neurosci. 4(Suppl.):1199-1206. [DOI] [PubMed] [Google Scholar]
- 32.Mulieri, P. J., A. Okada, D. A. Sassoon, S. K. McConnell, and R. S. Krauss. 2000. Developmental expression pattern of the cdo gene. Dev. Dyn. 219:40-49. [DOI] [PubMed] [Google Scholar]
- 33.Nait-Oumesmar, B., B. Stecca, G. Fatterpekar, T. Naidich, J. Corbin, and R. A. Lazzarini. 2002. Ectopic expression of Gcm1 induces congenital spinal cord abnormalities. Development 129:3957-3964. [DOI] [PubMed] [Google Scholar]
- 34.Nieto, M., C. Schuurmans, O. Britz, and F. Guillemot. 2001. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron 29:401-413. [DOI] [PubMed] [Google Scholar]
- 35.Palma, V., and A. Ruiz i Altaba. 2003. Hedgehog-Gli signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development 131:337-345. [DOI] [PubMed] [Google Scholar]
- 36.Parras, C. M., C. Schuurmans, R. Scardigli, J. Kim, D. J. Anderson, and F. Guillemot. 2002. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 16:324-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Figares, J. M., A. J. Jimenez, and E. M. Rodriguez. 2001. Subcommissural organ, cerebrospinal fluid circulation, and hydrocephalus. Microsc. Res. Tech. 52:591-607. [DOI] [PubMed] [Google Scholar]
- 38.Price, D., S. Aslam, L. Tasker, and K. Gillies. 1997. Fates of the earliest generated cells in the developing mouse neocortex. J. Comp. Neurol. 377:414-422. [PubMed] [Google Scholar]
- 39.Rolf, B., M. Kutsche, and U. Bartsch. 2001. Severe hydrocephalus in L1-deficient mice. Brain Res. 891:247-252. [DOI] [PubMed] [Google Scholar]
- 40.Ross, S. E., M. E. Greenberg, and C. D. Stiles. 2003. Basic helix-loop-helix factors in cortical development. Neuron 39:13-25. [DOI] [PubMed] [Google Scholar]
- 41.Ryder, E., E. Snyder, and C. Cepko. 1990. Establishment and characterization of multipotent neural cell lines using retrovirus vector-mediated oncogene transfer. J. Neurobiol. 21:356-375. [DOI] [PubMed] [Google Scholar]
- 42.Snyder, E., D. Deitcher, C. A. Walsh, S. Arnold-Aldea, E. Hartwieg, and C. Cepko. 1992. Multipotent neural cell lines can engraft and participate in development of mouse cerebellum. Cell 68:33-51. [DOI] [PubMed] [Google Scholar]
- 43.Sommer, L., Q. Ma, and D. J. Anderson. 1996. Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol. Cell. Neurosci. 8:221-241. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi, T., R. Nowakowski, and V. J. Caviness. 1995. The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J. Neurosci. 15:6046-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi, T., R. Nowakowski, and V. J. Caviness. 1994. Mode of cell proliferation in the developing mouse neocortex. Proc. Natl. Acad. Sci. USA 91:375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Visapaa, I., R. Salonen, T. Varilo, P. Paavola, and L. Peltonen. 1999. Assignment of the locus for hydrolethalus syndrome to a highly restricted region on 11q23-25. Am. J. Hum. Genet. 65:1086-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallis, D., and M. Muenke. 2000. Mutations in holoprosencephaly. Hum. Mutat. 16:99-108. [DOI] [PubMed] [Google Scholar]
- 48.Wallis, D. E., and M. Muenke. 1999. Molecular mechanisms of holoprosencephly. Mol. Genet. Metab. 68:126-138. [DOI] [PubMed] [Google Scholar]
- 49.Yamasaki, M., J. Thompson, and V. Lemmon. 1997. CRASH syndrome: mutations in L1CAM correlate with severity of the disease. Neuropediatrics 28:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yun, K., S. Fischman, J. E. Johnson, M. H. De Angelis, G. Weinmaster, and J. L. R. Rubenstein. 2002. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development 129:5029-5040. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, W., J.-S. Kang, F. Cole, M.-J. Yi, and R. S. Krauss. Cdo functions at multiple points in the Sonic Hedgehog pathway and Cdo-deficient mice accurately model human holoprosencephaly. Dev. Cell, in press. [DOI] [PubMed]