Abstract
We observed that binding sites for the ubiquitously expressed transcription factor CP2 were present in regulatory regions of multiple erythroid genes. In these regions, the CP2 binding site was adjacent to a site for the erythroid factor GATA-1. Using three such regulatory regions (from genes encoding the transcription factors GATA-1, EKLF, and p45 NF-E2), we demonstrated the functional importance of the adjacent CP2/GATA-1 sites. In particular, CP2 binds to the GATA-1 HS2 enhancer, generating a ternary complex with GATA-1 and DNA. Mutations in the CP2 consensus greatly impaired HS2 activity in transient transfection assays with K562 cells. Similar results were obtained by transfection of EKLF and p45 NF-E2 mutant constructs. Chromatin immunoprecipitation with K562 cells showed that CP2 binds in vivo to all three regulatory elements and that both GATA-1 and CP2 were present on the same GATA-1 and EKLF regulatory elements. Adjacent CP2/GATA-1 sites may represent a novel module for erythroid expression of a number of genes. Additionally, coimmunoprecipitation and glutathione S-transferase pull-down experiments demonstrated a physical interaction between GATA-1 and CP2. This may contribute to the functional cooperation between these factors and provide an explanation for the important role of ubiquitous CP2 in the regulation of erythroid genes.
Transcriptional regulation is a key step in the commitment and differentiation of hemopoietic cells. This process is achieved by a network of interacting lineage-specific transcription factors acting in conjunction with more-general transcription factors. GATA-1 is a zinc finger transcription factor expressed within the hemopoietic system, in the erythroid and megakaryocytic lineages, and in mast cells and eosinophils. Knockout studies revealed that GATA-1 is required for the normal maturation of erythroid and megakaryocytic cells. Consistent with this, functional GATA-1 elements are known to regulate the expression of virtually all erythroid and megakaryocytic genes studied, including transcription factors such as EKLF and p45 NF-E2, which in turn regulate important aspects of the specification of these lineages. The transcriptional activity of GATA-1 is modulated by a complex network of directly interacting proteins, including FOG1, LMO-2, EKLF, p300, and Pu1; depending on the partner and on the cellular context, the outcome of these interactions can be synergism or cross-antagonism (5, 11, 32).
CP2 is a ubiquitously expressed transcription factor belonging to the Drosophila grainyhead-like gene family. CP2 consists of homo/heterodimers of several protein isoforms produced by alternative splicing, from two different genomic loci, LBP-1c and LBP-1a (the human homologs of mouse CP2c and CP2a). CP2 binds as a dimer to a CNRG (N5-6)CNRG DNA motif, present in diverse cellular and viral promoters (23, 48, 50). CP2c was originally identified by its ability to stimulate the transcription of the α-globin gene (19); it appears to be involved in the fetal erythroid expression of the γ-globin gene through the formation of a heterodimer with the erythroid transcription partner NF-E4 (the stage selector protein) (14, 51). Indeed, in transgenic experiments, the mutation of the stage selector protein binding site on the γ-globin promoter (the stage selector element [SSE]) shows that this region affects the γ- versus β-globin ratio of expression in early stages of fetal development (34). Moreover, overexpression of antisense CP2 mRNA in MEL cells undergoing erythroid differentiation in vitro not only suppresses α-globin expression but also impairs β-globin expression and hemoglobinization (8), suggesting that CP2 binding to the promoter is essential for optimal globin transcription in erythroid cells (7).
The transcriptional regulation of GATA-1 is coordinated by several regulatory elements located both 5′ to the transcriptional start site and in the first intron (25, 29, 30, 35, 39, 45). The region upstream of the erythroid promoter, also reported as hypersensitivity site 2 (HS2), contains a double GATA motif shown to be essential for the erythroid promoter activity (28, 42, 44). This sequence, foot printed in vivo in erythroid cells, represents a strong double GATA-1 binding site, which has been proposed to mediate autoregulation by GATA-1 itself. Mice harboring a 21-base-pair deletion in this motif in the endogenous GATA-1 locus (49) show a lack of eosinophil production, while platelets and mast cells appear normal. Erythropoiesis is also abnormal in these animals, which display a reduction in red cell number, hematocrit, and hemoglobin.
The common role of GATA-1 and CP2 in regulating erythroid genes suggested the possibility that this could be occurring cooperatively. Here, we show that this is true for GATA-1 gene transcription, in which the two factors act through adjacent binding sites present on the HS2 erythroid enhancer element. We show that CP2 binds to HS2, creating a ternary complex with GATA-1 and DNA, and that mutations in the CP2 consensus greatly impair HS2 activity in transient transfection assays with K562 cells. Adjacent GATA-1 and CP2 binding sites are also present in regulatory elements of several other genes expressed in the hematopoietic lineage. Among them, we show that a CP2 binding site adjacent to functionally relevant GATA-1 sites is important for the activities of the p45NF-E2 and EKLF promoters. Chromatin immunoprecipitation (ChIP) experiments reveal that CP2 is bound in vivo to the regulatory elements of the GATA-1, EKLF, and p45 NF-E2 genes and that at least for the first two genes, it is bound simultaneously to GATA-1. Finally, GATA-1 and CP2 can physically interact, even in the absence of DNA, as demonstrated by immunoprecipitation and glutathione S-transferase (GST) pull-down experiments. Taken together, these data suggest that adjacent GATA-1 and CP2 sites contribute to the regulation of the GATA-1, p45 NF-E2, and EKLF genes.
MATERIALS AND METHODS
Electrophoretic mobility shift assay (EMSA).
32P-labeled DNA probes were incubated for binding with 1 to 4 μg of nuclear extracts for 20 min at 15°C in a buffer containing 5% glycerol, 50 mM NaCl, 20 mM Tris, pH 7.9, 0.5 mM EDTA, 5 mM MgCl, 1 mM dithiothreitol (DTT), 100 ng/μl poly(dI-dC), and 50 ng/μl bovine serum albumin (BSA) in a 15-μl final reaction mixture. The reaction mixture was then loaded onto a 5% polyacrylamide gel (29:1, acrylamide/bisacrylamide ratio) and run at 4°C at 150 V for 3 h. Nuclear extracts were prepared according to standard protocols (21, 38).
Competition experiments were performed by incubating 50 to 100 ng of unlabeled oligonucleotides in the reaction mixture for 10 min, prior to the addition of 32P-labeled probes. The anti-CP2 and the anti-NF-E4 antibodies are described in references 14 and 51; the anti GATA-1 antibody N6 was obtained from Santa Cruz. The experiments with bacterial recombinant proteins (see below) were performed using 2 ng/μl of poly(dI-dC); EMSAs with proteins produced by in vitro coupled transcription-translation reactions (TNT kit; Promega) were carried out in the presence of 25 ng/μl of poly(dI-dC).
GATA-1 and EKLF probes were made by PCR using either the forward (FW) or the reverse (REV) 32P end-labeled primer (see below for sequences). All other probes used in this work were generated by labeling the “sense” strand oligonucleotide with 32P and T4 polynucleotide kinase and annealing it with the complementary oligonucleotide; in this case, only the sequence corresponding to the sense strand is reported below. Labeled probes were purified by polyacrylamide gel electrophoresis.
Primers used for probes generated by PCR (mutations are underlined) were as follows: GATA-1 WT FW, 5′-CGAGTCCATCTGATAAGACTTATC-3′; GATA-1 GATAmut5′ FW, 5′-CGAGTCCATCGTCTAAGACTTATC-3′; GATA-1 GATAmut3′ FW, 5′-CGAGTCCATCTGATAAGACTTACA-3′; GATA-1 REV, 5′-ATAAAGCCTGGATCCTGGGGCTTACGC-3′; EKLF FW, 5′-CCCCTACCTGATAGCGG-3′; and EKLF REV, 5′-CCTTTCAGGCATTATCAGACAC ACC-3′.
Oligonucleotide probes.
Short GATA-1 CP2 consensus oligonucleotides (nucleotides [nt] −695 to −660) were as follows: WT, 5′-CTGCTGCCCCAGAGCAGGCCAGAGCTGGCGTAAGC-3′; MUT 1/3, 5′-CTGCTGCCACAGAGCAGGTAAGAGCTGGCGTAAGC-3′; MUT 2/4, 5′-CTGCTGCCCCAGATTGAGCCAGATTGAGCGTAAGC-3′; and MUT 1-4, 5′-CTGCTGCCACAGATTGAGTAAGATTGAGCGTAAGC-3′.
p45 NF-E2 oligonucleotides (nt 450 to 493) were as follows: WT, 5′-CCTGCAGCTGATAAACCCCTTATCTGGCCCAGGCAGGGGAACCT-3′; MUT 1-3, 5′-CCTGCAGCTGATAAACCCCTTATCTGTCCTAGGTAGGGGAACCT-3′; and MUT 2/3, 5′-CCTGCAGCTGATAAACCCCTTATCTGGCCTAGGTAGGGGAACCT-3′.
EKLF oligonucleotides (nt −683 to −634) were as follows: WT, 5′-CCTACCTGATAGCGGCCTGAAACATCTGGTGTGTCTGATAATGCCTGAAA-3′; and MUT, 5′-CCTACCTGATAGCGGCCTGAAACATTTGGTGTGTTTGATAATGCCTGAAA-3′.
The following constructs were also used: α-globin CP2 site, 5′-TAGAGCAAGCACAAACCAGGCCAA-3′ (22); γ-globin SSE site, 5′-TCCAGTGAGGCCAGGGGCCGGCGGCTGGCTAGGGATGA-3′ (14); α-globin GATA site, 5′-GGCAACTGATAAGGATTCCCA-3′ (24); and γ-globin CCAAT box, 5′-GCCTTGCCTTGACCAATAGCCTTGACA-3′ (24).
Recombinant protein expression and purification.
For the expression of the GST-GATA-1 fusion protein, the murine GATA-1 cDNA was cloned in the polylinker region of the pMGSTev expression vector (10), in frame with the GST moiety. GST-CP2 fusion protein was produced from the pGEX vector as described previously (14). The Escherichia coli BL21 strain cells were transformed with the above plasmids, cultures were grown at mid-logarithmic phase (0.6 A600), and protein expression was induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C. GST-CP2 and GST-GATA-1 proteins present in the soluble fraction were bound to GST-Sepharose 4B (Amersham Bioscience) and purified according to the manufacturer's instructions.
For EMSA experiments, the GST moiety of CP2 was cleaved with thrombin (Amersham Biosciences). Briefly, the eluted GST-CP2 protein was dialyzed against a buffer containing 20 mM HEPES, pH 7.9, 100 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.5 mM DTT, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and then incubated overnight with thrombin at room temperature. The cleaved GST moiety was removed by binding to GST-Sepharose 4B resin at room temperature. The unbound fraction containing the CP2 cleaved protein was recovered and stored in aliquots at −80°C.
Plasmids. (i) GATA-1 reporter plasmids.
The GATA-1 mouse promoter region from nucleotide −856 to nucleotide −655 flanked by BamHI sites was obtained from the previously described pSVo GATA-1 plasmid (28) and cloned into the compatible BglII site present in the polylinker of the pGL2 Basic luciferase reporter vector (Promega). The mouse GATA-1 minimal promoter from the same pSVo GATA plasmid (HindIII fragment from nucleotides −330 to −31) was transferred to the HindIII site of the pGL2 polylinker, immediately downstream to BglII.
To generate mutations in the CP2 boxes, a two-step PCR approach was used: in the first series of PCRs, mutations were introduced by amplifying the wild-type template sequence with REV oligonucleotides, carrying the desired mutations, corresponding to the 3′ end of the enhancer sequences, in conjunction with a common FW primer carrying an external BamHI site for subsequent cloning. The amplified fragments were gel purified and used as a template for the second round of PCRs with the same FW primer and a common REV primer, external to mutations, to introduce a 3′ BamHI site. The resulting fragments were then BamHI digested and cloned in the compatible BglII site of pGL2 Basic vector upstream to the GATA-1 minimal promoter.
Primers for GATA-1 constructs were as follows: FW-856 BamHI, 5′-CATAAAGCTTGGATCCACTCTGGGTGTCACCTC-3′; REV-655mut 1/3, 5′-CTGGGGCTTACGCCAGCTCTTACCTGCTCTGTGGCA-3′; REV-655mut 2/4, 5′-CTGGGGCTTACGCTCAATCTGGCTCAATCTGGGGCA-3′; REV-655mut 1-4, 5′-CTGGGGCTTACGCCAACTCTTACCTACTCTGTGGCAGCAGATAAGTCTTATGACATGGACTCG-3′; and REV-655BamHI, 5′-ATAAAGCCTGGATCCTGGGGCTTACGC-3′.
(ii) p45 NF-E2 reporter plasmids.
The p45 NF-E2 pGL2 plasmids (wild type and mutated in GATA-1 sites) have been described previously (27). To generate the two mutations in CP2 boxes, we used a two-step PCR approach, using a pGEM-T (Promega) plasmid containing the p45 NF-E2 mouse fetal promoter (nucleotides 233 to 605) as a template (provided by B. Giglioni). In the first series of PCRs, partially overlapping primers carrying the desired mutations were used in conjunction with external primers on the pGEM-T flanking sequences to obtain the 5′ and 3′ portions of the fragment of interest. The amplified fragments were then gel purified and used as a template for a second round of PCRs with the same external pGEM-T primers. The resulting fragments were then cut with MboI and cloned in the compatible BglII site of pGL2 Basic vector.
Primers used for p45 mutants were as follows: pGEM-T FW, 5′-TAATACGACTCACTATA-3′; pGEM-T REV, 5′-ATTGGTGACACTATAGAA-3′; mut 2/3 sense, 5′-GATAAACCCCTTATCTGGCCTAGGTAGGGG-3′; and mut 1-3 sense, 5′-GATAAACCCCTTATCTGTCCTAGGTAGGGG-3′.
(iii) EKLF reporter plasmids.
The EKLF HS1 fragment was obtained by digestion with ApaI and NcoI of a PCR fragment from MEL cell DNA (nucleotides −938 to −24), amplified with oligonucleotides FW (5′-ACGTCTCGAGAACGGCATACTAGCTGCAGCTC-3′) and REV (5′-ACGTGGATCCGGCTCCTGTCTGCCCACATC-3′).
The fragment was blunted and cloned into the SmaI site of the pGL2 Basic vector containing the GATA-1 minimal promoter. Again, the mutations in CP2 boxes 2 to 3 were obtained by subsequent PCRs: overlapping sense and antisense oligonucleotides, together with the above external primers, were used to generate templates for the second round of PCR, carried out with the same external primers. The EKLFmut sense primer was 5′-GCGGCCTGAAACATTTGGTGTGTTTGATAATG-3′. The resulting mutated fragment was ApaI/NcoI digested, blunted, and cloned into the SmaI site of the pGL2 Basic vector containing the GATA-1 minimal promoter. All constructs obtained by PCR were sequenced to exclude the presence of undesired mutations.
Transfection experiments.
K562 human erythroleukemic cells were grown in RPMI 1640 medium supplemented with l-glutamine and 5% fetal bovine serum. Exponentially growing K562 cells (1 × 107 to 2 × 107) were electroporated at 400 V and 960 μF with a Bio-Rad apparatus in 800 μl of phosphate-buffered saline with 20 μg of plasmid. To normalize experiments for transfection efficiency, 2 μg of pRL-TK plasmid carrying the Renilla luciferase gene under the control of the ubiquitous TK promoter was cotransfected in each sample. After 48 h, total cellular extracts were prepared and the double luciferase activity was measured according to the Promega Dual-Luciferase reporter system protocol. All experiments were repeated in triplicate with at least three independent plasmid preparations.
ChIP assay.
ChIP assays were performed as described previously (15, 16). Isolated DNA fragments were purified with a QIAquick spin kit (QIAGEN), and 2 μl from a 40-μl DNA extraction was amplified quantitatively by real-time PCR with the GATA-1, NF-E2, or EKLF promoter-specific primer or MyoD primer as a negative control (37). For ChIP/chromatin reimmunoprecipitation (Re-ChIP) experiments, we immunoprecipitated the soluble chromatin fraction with antibody to CP2, washed it, and released the bound immune DNA complexes in 20 mM DTT solution for 30 min at 37°C. We resuspended the precipitates in 1 volume of the immunoprecipitated dilution buffer and performed a Re-ChIP with antibody to GATA-1. We also carried out control ChIP to test antibody specificity with either a control (no antibody) or normal rabbit serum.
The primers used were as follows: GATA-1 (forward), 5′-CTTTCCTACCCTATCCCACTCCTCG-3′; GATA-1 (reverse), 5′-TGCTGGATTTGAACTAGAGCCTGTG-3′; P45 NF-E2 (forward), 5′-GTTAAGGTATGGCCCAAATGACCCT-3′; P45 NF-E2 (reverse), 5′-AAGTTGTGGAAAGAGGCAAGCAGAC-3′; EKLF (forward), 5′-ATTGAACGCCAGGCTAATTTGAAGA-3′; EKLF (reverse), 5′-GAAAAGGCAGAAAGGGTATTCTGGG-3′; MyoD (forward), 5′-TGCAAGGCGTGCAAGCGCAAGAC-3′; and MyoD (reverse), 5′-CTCGATATAGCGGATGGCGTTG-3′.
In vitro protein-protein interaction assay.
The plasmid used to produce GATA-1 in vitro in the reticulocyte cell-free coupled transcription-translation system (TNT kit, Promega) was derived from the above-mentioned pMGSTev plasmid by removing the GST coding sequence by a double SpeI-NcoI digestion. The resulting ends were then blunted with Vent polymerase and religated. The CP2 cDNA was inserted into a pBluescript plasmid under the T7 promoter. 35S-labeled proteins were synthesized in the presence of [35S]methionine (Amersham). We incubated 1 μg of purified GST or GST fusion proteins with the above 35S-labeled proteins for 2 h at 4°C, adjusting the reaction mixture to a final volume of 40 μl with a buffer containing 50 mM Tris HCl, pH 7.5, 100 mM NaCl, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, and protein inhibitor cocktail (Roche). After incubation, 60 μl of 50% glutathione-Sepharose beads was added together with 500 μl of binding buffer (150 mM NaCl, 10 mM Tris HCl, pH 8, 0.3% NP-40, 1 mM DTT, PMSF, protein inhibitor cocktail, 0.25% BSA). Incubation was continued for 90 min at 4°C. Beads were collected by centrifugation and washed three times with 1 ml of IPP250 buffer (20 mM Tris HCl, pH 7.9, 250 mM NaCl, 0.05% NP-40, 1 mM DTT, PMSF, protein inhibitor cocktail). Proteins bound to the resin were eluted by boiling in 20 μl of Laemmli buffer, loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, and visualized by autoradiography after drying of the gel.
In vivo protein-protein interaction assay.
K562 nuclear extract (50 μg) was preincubated with 7.5 μl of protein G agarose beads (Kirkegaard & Perry Laboratories) and 100 μl of NDB buffer (100 mM KCl, 20 mM Tris HCl, pH 7.8, 0.5 mM EDTA, 1 μg/μl BSA, 0.1% NP-40, 5 mM β-mercaptoethanol, 20% glycerol) for 1 h at 4°C. Precleared nuclear extracts were then incubated with 2 μg of anti-GATA-1 antibody (N6; Santa Cruz) or with 2 μg of anti-T antigen antibody (Santa Cruz) as a negative control for 4 h at 4°C. One hundred microliters of 50% protein G agarose beads was added to the reaction for 1 h at 4°C and then washed three times with 150 μl of NDB buffer. Bound proteins were eluted by boiling in 20 μl of Laemmli buffer, loaded onto a 10% SDS-PAGE gel, blotted, and probed for the presence of CP2.
RESULTS
CP2 binds within the HS2 enhancer region of the GATA-1 gene to sites adjacent to the GATA-1 palindromic site.
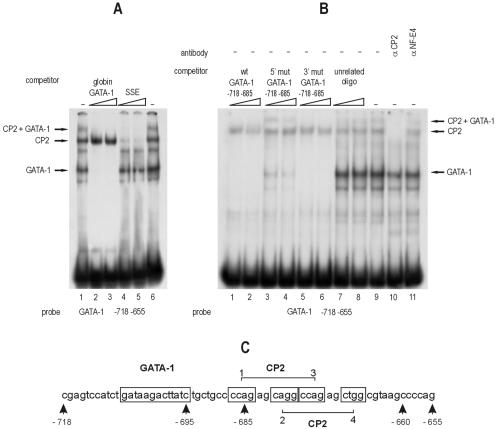
In order to better characterize the erythroid GATA-1 HS2 enhancer, we analyzed by EMSA the region from nt −718 to nt −655 relative to the transcription initiation site, containing the previously described double GATA-1 binding site (28) and its 3′ flanking region, which extends to the end of the 3′ border of the mouse/human homology region. The incubation of a 32P-labeled fragment corresponding to this region with K562 nuclear extracts generated a complex pattern in the fast-migrating GATA-1 bands (28) and other previously undescribed slower bands (Fig. 1A, lane 1). Surprisingly, an unlabeled oligonucleotide corresponding to the GATA-1 binding site from the α-globin promoter not only competed for the two faster bands but also affected the formation of the upper slower band, suggesting that the latter might also contain GATA-1.
FIG. 1.
Binding of CP2 to the GATA-1 HS2 enhancer element. (A) EMSA using an oligonucleotide (positions −718 to −655) encompassing the GATA-1 CP2 binding motifs present in the HS2 enhancer with K562 cell nuclear extract. The effects of excess unlabeled GATA-1 binding (globin GATA-1) and CP2 binding (SSE) competitors are shown. The positions of the GATA-1, CP2, and GATA-1-plus-CP2 complexes are indicated. Note that both competitors abolish the upper band, demonstrating that it contains both CP2 and GATA-1. (B) Unlabeled oligonucleotides comprising the wild-type (wt) or mutated GATA-1 palindrome do not affect the binding of CP2 to the GATA-1 HS2 enhancer oligonucleotide in competition experiments. Note that the weak GATA-1 binding mutant (5′ mut GATA-1, lanes 3 to 4) competes almost completely for the GATA-1 band, while it leaves the upper CP2-plus-GATA-1 band essentially intact. A specific anti-CP2 antibody, but not an anti-NF-E4 antibody, abolishes the CP2 band (lanes 10 to 11). The positions of the GATA-1, CP2, and GATA-1-plus-CP2 complexes are indicated. (C) Schematic representation of the GATA-1 and CP2 binding sites on the mouse GATA-1 promoter.
Immediately downstream to the GATA-1 double site, we noticed a putative double CP2 consensus site (CnRG5/6CnRG/C), extending from position −687 to position −667 (Fig. 1C). To test whether CP2 alone, or its heterodimeric complex with the erythroid subunit NF-E4, could bind to this putative consensus, we set up a competition experiment with an oligonucleotide containing the SSE from the γ-globin promoter (14). As shown in lanes 4 and 5 in Fig. 1A, the unlabeled SSE oligonucleotide efficiently abolished both upper bands, confirming that they contain CP2 and/or NF-E4 and suggesting that the slower band (also competed for by the GATA consensus element) might contain CP2 and/or NF-E4 and GATA-1 molecules (Fig. 1A, compare lanes 2 to 3 with lanes 4 to 5).
To confirm the abilities of CP2 and/or NF-E4 to bind the HS2 GATA core sequence, we carried out experiments with specific antibodies against CP2 and NF-E4: the former, but not the latter, antibody was able to compete for the formation of the upper doublet (Fig. 1B, lanes 10 and 11), suggesting the absence of NF-E4 in these CP2 complexes. Consistent with this, an oligonucleotide carrying the CP2 consensus from the α-globin promoter and known to bind CP2 in its homodimeric form was also able to compete for the upper complexes (data not shown).
To determine whether CP2 binding to this region was affected by the integrity of the double GATA-1 site, we tested probes carrying mutations in either the 3′ low-affinity or the 5′ high-affinity GATA-1 site (28). In this experiment, a labeled oligonucleotide (nt −718 to −655) comprising both the GATA-1 and CP2 sites was competed for by a “short” unlabeled oligonucleotide carrying either the wild-type or the mutated GATA-1 sites but not the CP2 binding sites. Neither the wild-type nor the mutated competitor oligonucleotide significantly affected the CP2 band (Fig. 1B, lanes 1 to 6), indicating that the binding of CP2 is not dependent on the binding of GATA-1. Interestingly, while the oligonucleotide with the 3′ GATA-1 mutation fully competed for both GATA-1 and GATA-1-plus-CP2 bands (as the wild-type oligonucleotide does), the 5′ GATA-1 mutant oligonucleotide (a weak GATA-1 competitor [28]) competed for the GATA-1 band more efficiently than the GATA-1-plus-CP2 band (Fig. 1B, lanes 3 to 4). This suggested that the GATA-1-plus-CP2 complex may be more stable on target DNA than the simple GATA-1 complex. These data were confirmed by direct binding experiments with labeled normal or mutated oligonucleotides (data not shown).
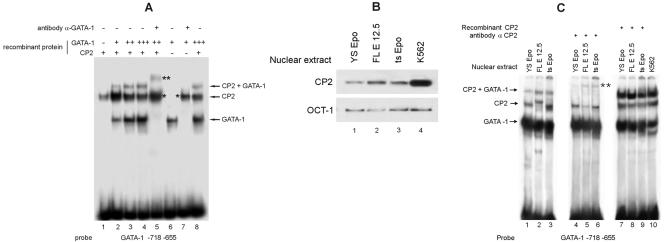
To better characterize the formation of the upper CP2 bands on the HS2 GATA sequence, we repeated our analysis using recombinant proteins. GATA-1 was produced by a coupled in vitro transcription/translation reaction, while CP2 was expressed in bacteria as a GST fusion protein and then cleaved to remove the GST moiety. As shown in Fig. 2A, lanes 1 and 6, respectively, CP2 and GATA-1 bound the HS2 probe, as expected. The addition of increasing amounts of GATA-1 to a constant amount of CP2 (Fig. 2A, lanes 2 to 4) progressively led to the formation of a new band migrating more slowly than the band generated by CP2 alone, suggesting the simultaneous binding of the two proteins on the same DNA molecule. This new band contained GATA-1, as confirmed by the use of an anti-GATA-1 antibody (Fig. 2A, lane 5). In fact, when the anti-GATA-1 antibody was added in the presence of the GATA-1 protein alone (Fig. 2A, compare lane 7 with lane 6), the GATA-1 band was supershifted to a position close to that corresponding to the band generated by the CP2 protein alone (lane 1). In the presence of both CP2 and GATA-1, the addition of the anti-GATA-1 antibody clearly supershifted not only the prominent GATA-1 lower band but also the upper band, resulting from the presence of both GATA-1 and CP2 in the reaction mixture (see double asterisk in Fig. 2A, lane 5). Similar results were obtained with K562 extracts (data not shown).
FIG. 2.
Binding of GATA-1 and CP2 to the GATA-1 HS2 enhancer element. (A) Studies of recombinant GATA-1 and CP2 confirm that both proteins bind to the HS2 enhancer element. Note that antibody against GATA-1 supershifts both the GATA-1 and the GATA-1-plus-CP2 bands (lanes 5 and 7, asterisks). The positions of the GATA-1, CP2, and GATA-1-plus-CP2 complexes are indicated. (B) Western analysis of extracts from primary fetal liver cells from E12.5 day embryos (FL E 12.5) and immortalized mouse yolk sac (YS Epo) and bone marrow (ts Epo) growth factor-dependent cell lines with anti-CP2 antibody. Anti-OCT-1 antibody served as the loading control. (C) The GATA-1-plus-CP2 band was obtained using extracts from various hematopoietic cell types (lanes 1 to 3); compare lanes 1 to 3 with lanes 4 to 6, where the anti-CP2 antibody was added. Recombinant CP2 added to nuclear extracts from the same hematopoietic cell extracts generates a strong GATA-1-plus-CP2 band (lanes 7 to 9); compare lanes 7 to 9 with lane 10, K562 nuclear extracts.
Among hematopoietic cell lines expressing GATA-1, there is variability of CP2 levels. For example, K562 cells have high levels of CP2 in comparison to GATA-1 levels (Fig. 2B, lane 4), whereas a number of immature mouse hematopoietic cells, such as primary fetal liver cells (E12.5) and immortalized bone marrow and yolk sac-derived growth factor-dependent progenitor cell lines (4), show low CP2 levels (Fig. 2B, lanes 1 to 3). Extracts from all these cells contain abundant GATA-1, as evidenced on the EMSA results shown in Fig. 2C using the GATA-1 promoter probe (lanes 1 to 3). CP2 binding to the probe with these extracts mirrored the protein levels observed on the Western blot, and a weak GATA-1-plus-CP2 complex was also observed which was ablated with the addition of anti-CP2 antiserum (Fig. 2C, lanes 4 to 6). The addition of recombinant CP2 to nuclear extracts of the immature mouse cells augmented the GATA-1/CP2 band, which became clearly evident (Fig. 2C, lanes 7 to 10). Overall, these results indicate that when high levels of both GATA-1 and CP2 are present, the GATA-1-plus-CP2 complex forms readily.
The CP2 binding sites present on the GATA-1 HS2 enhancer contribute functionally to its activity.
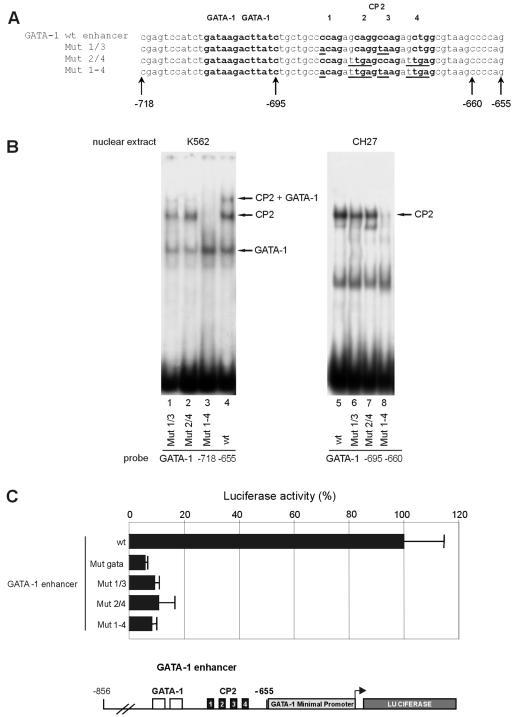
To assess the functional relevance of CP2 binding on the GATA-1 promoter, we analyzed in transient transfection assays with K562 cells a series of mutants in the CP2 double consensus site in the HS2 region. In these experiments, the HS2 element from nt −856 to nt −655 was cloned into a pGL2 plasmid immediately upstream of the 330-nt GATA-1 minimal promoter to drive the activity of a luciferase reporter gene. Since there is a double CP2 consensus in the GATA-1 HS2 region, with four CP2 half consensus motifs (CNRG) comprising two partially overlapping CNRG-6-CNRG complete motifs, we prepared three different mutants, abolishing the first CP2 consensus (Mut 1/3), the second one (Mut 2/4), or both of them (Mut 1-4), as shown in Fig. 3A. To test the efficiency of these mutations in disrupting CP2 binding, we carried out a series of EMSAs (Fig. 3B) on DNA sequences, which contain either the GATA-1 double site (nt −718 to −655) or the double CP2 region alone (nt −695 to −655) (Fig. 3B and data not shown). None of the mutations affected GATA-1 binding (Fig. 3B, left panel, compare lanes 1 to 4). Mutations abolishing the single CP2 consensus (Mut 1/3 and Mut 1/4) (Fig. 3B, left panel) only slightly reduced the CP2 band, as residual binding occurred on the remainder of the CP2 site (lanes 1 and 2). A more substantial reduction was observed in the higher-order GATA-1-plus-CP2 complex with these mutants. Mutation of both sites completely abolished binding of both CP2 and the higher order complex containing GATA-1 plus CP2 (Fig. 3B, lane 3). Reduction in CP2 binding to the single site mutants and loss of CP2 binding to the Mut 1-4 probe were confirmed with lymphoid CH27 nuclear extracts, which contain CP2 but not GATA-1 (Fig. 3B, right panel).
FIG. 3.
CP2 sites on the GATA-1 HS2 enhancer substantially contribute to the transcriptional activity of the GATA-1 gene. (A) Schematic of the wild-type (wt) and mutated GATA-1 HS2 enhancer constructs. The GATA-1 and CP2 consensus sites are boldface, and the mutated bases in the CP2 consensus sites are underlined. (B) The effect of mutations on protein binding. EMSAs with the wild-type and mutated oligonucleotides were performed with extracts from K562 (erythroid) or CH27 (nonerythroid) cells. The positions of the GATA-1, CP2, and GATA-1-plus-CP2 complexes are indicated. The most extensive mutation (Mut 1-4) totally abolished CP2 binding (lanes 3 and 8), but mutations in the single CP2 boxes (Mut 1/3 and Mut 2/4) still allowed significant CP2 binding on the intact CP2 site (lanes 1, 2, 6, and 7). The CH27 nuclear extracts contain CP2 but not GATA-1. (C) Functional luciferase reporter assays with K562 cells of mutants shown in panel A and an additional construct carrying a mutation of the GATA-1 binding site. All three CP2 mutations greatly reduced the ability of the HS2 enhancer linked to the GATA-1 minimal promoter to drive the luciferase reporter, shown schematically in the lower panel.
The above mutations were then introduced into a pGL2-based construct containing the HS2 element from nt −856 to nt −655 immediately upstream of the GATA-1 330-nt minimal promoter element (28) to drive the expression of a firefly luciferase reporter gene. An additional mutation in the double GATA-1 motif (28) was tested in parallel. The resulting plasmids were transiently transfected into K562 cells, and the results of these experiments are summarized in Fig. 3C. All results are normalized for the efficiency of transfection, determined by cotransfection of a Renilla luciferase reporter under the control of the ubiquitous thymidine kinase promoter, and apply to a series of at least three experiments in duplicate. All three tested CP2 mutations (either in the single CP2 boxes [Mut 1/3 and Mut 2/4] or in both sites [Mut 1-4]) drastically reduced the transcriptional activity of the GATA-1 HS2 element (less than 10% of the activity of the intact HS2 element). The effect of the mutations in the CP2 boxes was similar to that of the mutation in the GATA boxes, indicating a substantial role for the CP2 consensus in the regulation of this element in K562 cells. Of note, Mut 1/3 and Mut 2/4, which did not greatly reduce CP2 binding in the EMSA (but do affect the GATA-1-plus-CP2 complex; see above) had a clear functional effect, suggesting the need for an intact CP2 double consensus element in vivo (and thus the formation of the CP2-plus-GATA-1 complex).
Adjacent GATA-1 and CP2 binding sites are present in several genes expressed in hematopoietic cells.
To explore the possibility that adjacent sites for GATA-1 and CP2 could represent a common “regulatory module” shared by several genes expressed in the hematopoietic system, we searched for regulatory regions containing GATA-1 binding sites flanked by putative CP2 sites (Table 1). Among the identified hits, we focused our attention on sequences known to be relevant for the regulation of p45 NF-E2 and EKLF, which both code for transcription factors expressed in, and important for, the erythroid and (in the case of NF-E2) megakaryocytic differentiation programs. The EKLF region studied lies within erythroid hypersensitive site 1 mapped from positions −700 to −650 and known to contain a functionally relevant GATA-1 site flanked by other poorly characterized elements (1, 2, 9). The p45 NF-E2 sequence containing adjacent GATA-1 and CP2 sites constitutes the core of the “fetal” proximal p45 NF-E2 1b promoter (27).
TABLE 1.
Adjacent GATA-1 and CP2 sites in erythroid regulatory elements
| Binding site (reference)a | Sequenceb |
|---|---|
| mGATA-1 | CGAGTCCATCTgataaGACttatcTGCTGCCCCAGAGCAGGCCAGAGCTGGCGTAAGCCCCA |
| mEKLF | CTACCTgatagCGGCCTGAAACATCTGGTGTGTCTgataatGCCTGAA |
| mp45/NF-E2 | CCTGCAGCTgataaACCCCttatcTGGCCCAGGCAGGGGAACCTAAAGTAAGGATGGG |
| mGfi-1b (36) | AGCTGTCTCctatctGTGGCCACCAGTCTGGACCTGTGTCCCCTTGGGGAGTTGTT |
| hRBTN-2* | GCTCCGCCctatcagatagACAACCAGGCCACCAAGAGGCCCAGCCCTCCAAACCCTGG |
| hUroIIIsynth. (41) | TGTCTTTCCAAGTgatatCAACTGCTAACATGCTCTTTCTTGGCcttatcAGTGACAGGGGTCT |
| hPBGD* | CACTGGGGAACCTGTGCTGAGTCACTGAAGgatagATTCAttatctACAACAGACAAGGCACTTGA |
| hALAS2 (3) | TCAGCCTGGCACCctatcTCTGGTCTGCCAGCTGGTCTCTCAGGCTGTA |
| mEpoR* | GGCTGTAGcttatcTGTCCCCAGCCTGCAAAGCTGGCCCCGCCCCCTGGAAGGAGCTGCC |
| hAnkyrin (12) | CGACAGCAAGCGCCTCTGGGGCCgataagGCCCTCGGGGGCCTGGCCCGCACGTCACA |
| hPlat.Glyc.IIB* | AGTCTTTTcttatcAAACTGGAACCCCAAGTAACTTGCTGAGCAACGGGCAGAGCAAAG |
| mTransferrinR (18) | GGTTCAGGCTGCCTGTCAGGGCtgataaGGGGCCCTC |
| hGlobinLCR HS2* | AGAGTGATGACTCctatctGGGTCCCCAGCAGGATGCTTACAGGGCAG |
*, information based on the Transcription Regulatory Regions Database (20) and other published data (see references).
GATA-1 binding sites are in lowercase letters; CP2 binding sites are underlined.
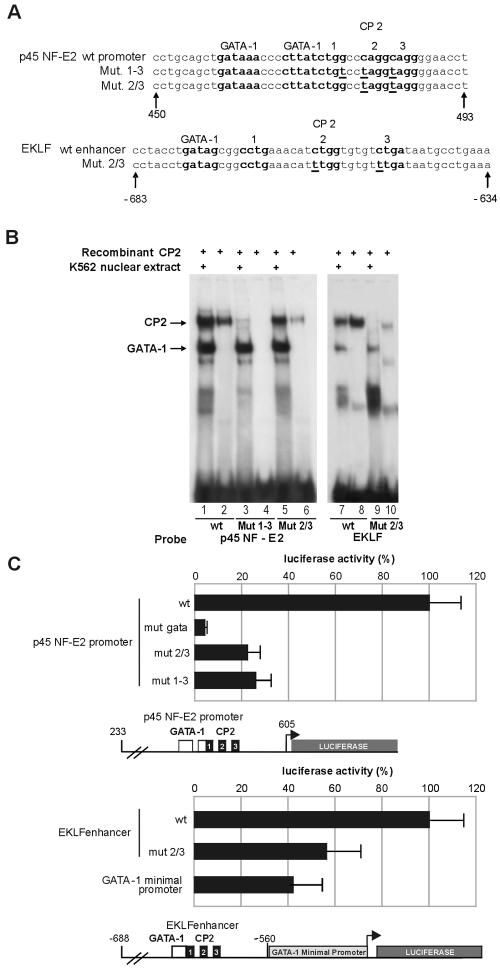
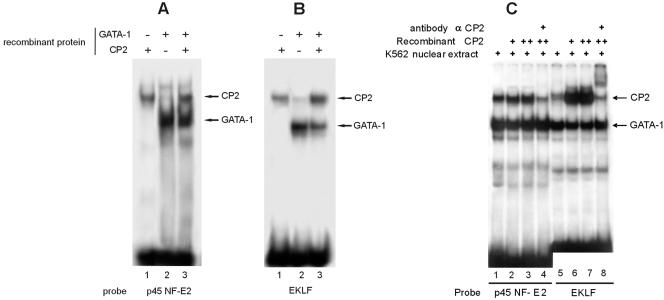
To test the effective ability of CP2 to bind to these consensus sequences on the EKLF and p45 NF-E2 regulatory elements, we performed an EMSA using 32P-labeled fragments encompassing the regions of interest (see Table 1 and Fig. 5 for sequences). Both EKLF and p45 NF-E2 sequences bound GATA-1 and CP2 when recombinant proteins (Fig. 4A and B) and K562 extracts (Fig. 4C, lanes 1 and 5) were used. In these experiments, the CP2 band was weaker than that formed using the GATA-1 oligonucleotide (compare Fig. 4A and B with Fig. 1 and 2), and little, if any, ternary complex with GATA-1 and CP2 was formed when additional recombinant CP2 was added to the K562 nuclear extract (Fig. 4C, lanes 2 to 4 and 6 to 8). This indicates that the ternary GATA-1-plus-CP2 complex may be unstable under the experimental conditions adopted. Alternatively, the simultaneous binding of GATA-1 and CP2 may be constrained by the relative arrangement of the binding sites. However, changing the spacing between GATA-1 and CP2 binding sites did not affect the results (data not shown).
FIG. 5.
Mutations in the CP2 binding sites in the p45-NF-E2 and EKLF promoters inhibit their transcriptional activity in K562 cells. (A) Schematic of the wild-type (wt) and mutated p45 NF-E2 and EKLF promoters. The GATA-1 and CP2 binding sites are boldface and labeled, and the mutated bases are underlined. (B) EMSA with K562 nuclear extract or recombinant CP2 and the wild-type and mutant oligonucleotides shown in panel A. The mutations abolished CP2 binding but did not affect GATA-1 binding, as demonstrated by both direct binding (lanes 1 to 10) and competition experiments (not shown). (C) Functional luciferase reporter assays with K562 cells of mutants shown in panel A. In experiments with the p45 NF-E2 promoter, a construct carrying a mutation of the GATA-1 binding site was also tested. In the experiments with the EKLF promoter, the GATA-1 minimal promoter was tested as a control. Mutations in the CP2 binding sites significantly reduced the transcriptional activity of the transfected constructs, which are shown schematically below the respective bar graphs.
FIG. 4.
Sequences from the fetal 1b proximal p45-NF-E2 promoter and EKLF erythroid hypersensitive site 1 bind CP2. Binding of recombinant CP2 and GATA-1 to the p45 NF-E2 promoter (A) and the EKLF promoter (B) was assessed by EMSA. The positions of the GATA-1 and CP2 complexes are indicated. (C) The effects of the addition of recombinant CP2 and anti-CP2 antibody on GATA-1 and CP2 complex formation on the p45 NF-E2 and EKLF promoters were assessed by EMSA. The positions of the GATA-1 and CP2 complexes are indicated.
The CP2 binding sites present on p45 NF-E2 and EKLF promoters functionally contribute to their activity.
To test the functional relevance of the CP2 binding sites identified in the p45 and EKLF regulatory elements, we again prepared a series of CP2 binding mutants and assayed them in transient transfections in K562 cells. In the p45 NF-E2 promoter, we made two different mutations in the CP2 consensus: p45 NF-E2 Mut 1-3 altered all three half CP2 boxes, whereas p45 NF-E2 Mut 2/3 left intact the first half box that partially overlaps with the GATA-1 consensus. This second mutant was designed to avoid any possible perturbation of the adjacent GATA-1 site (Fig. 5A). Similarly, we mutated two of the potential CP2 half sites in EKLF erythroid hypersensitive site 1 (Fig. 5A). Both p45 NF-E2- and EKLF-mutated oligonucleotides failed to bind CP2 in EMSA experiments when used as labeled probes in direct binding experiments (Fig. 5B) or as unlabeled competitors versus the GATA-1 HS2 probe (data not shown). The CP2 mutations in both p45 NF-E2 and EKLF competitor oligonucleotides did not affect their ability to compete for GATA-1 binding, showing that the mutation(s) of the CP2 site(s) does not affect the binding of GATA-1 to the adjacent GATA-1 motifs (Fig. 5B and data not shown).
The above mutations in the p45 NF-E2 element were introduced into the 373-nt fragment corresponding to the fetal/1b intronic promoter (from nt 233 to 605) to drive transcription of the luciferase reporter gene in the pGL2 basic plasmid (27). To test the contribution of the CP2 sites in the EKLF element, we cloned an EKLF promoter fragment from nt −688 to nt −560 immediately upstream to the GATA-1 minimal promoter into the same pGL2vector. The resulting plasmids were then transiently transfected into K562 cells. Both CP2 mutations in the p45 NF-E2 promoter reduced the transcriptional activity by over 70% (Fig. 5C). With this construct, the residual activity was significantly higher than that of the mutant in the GATA-1 HS2 site (∼5% of the wild-type activity). EKLF erythroid hypersensitive site 1, when placed upstream to the GATA-1 minimal promoter, led to a 2.5-fold increase in the GATA-1 minimal promoter activity, which was almost completely abrogated by loss of CP2 binding (Fig. 5C). Taken together, these data demonstrate the relevant role of CP2 in activating transcription from GATA-1, p45 NF-E2, and EKLF promoters.
CP2 binds to the GATA-1, EKLF, and p45 NF-E2 regulatory elements in vivo.
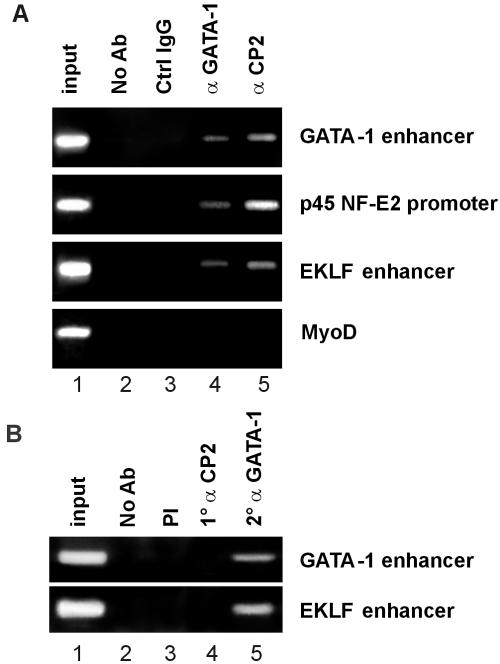
To further assess the potential role of GATA-1 and CP2 in the regulation of the GATA-1, EKLF, and p45 NF-E2 genes, we performed ChIP assays with K562 cells. As shown in Fig. 6A, all three erythroid regulatory elements were occupied by GATA-1 and CP2 in vivo. No amplification of these elements was observed in the absence of specific antiserum or with normal rabbit serum. To determine whether GATA-1 and CP2 occupied the erythroid regulatory elements simultaneously, we performed a ChIP/Re-ChIP experiment in which chromatin from K562 cells was initially immunoprecipitated with anti-CP2 antibody and the selected chromatin was secondarily immunoprecipitated with anti-GATA-1 antibody (Fig. 6B). Both the GATA-1 and EKLF regulatory elements were specifically immunoprecipitated by anti-GATA-1 antibody from the washed precipitate from the CP2 antibody, indicating that CP2 and GATA-1 cooccupy these elements. In contrast, although GATA-1 and CP2 appear to participate in the regulation of p45 NF-E2, we were unable to demonstrate simultaneous occupancy of the factors on this target element under these experimental conditions.
FIG. 6.
Binding of CP2 and GATA-1 to erythroid gene regulatory elements. (A) ChIP with anti-GATA-1 or anti-CP2 antibody. Chromatin from K562 cells was immunoprecipitated using antiserum to CP2 or GATA-1. Normal rabbit serum (Ctrl IgG) samples and samples without antibody (No Ab) served as the controls. Quantitative PCR was performed with primer pairs to amplify the erythroid gene regulatory elements (GATA-1 enhancer, p45 NF-E2 promoter, and EKLF enhancer) or the MyoD gene as a control. The input chromatin is shown. (B) ChIP/Re-ChIP with anti-GATA-1 and anti-CP2 antibodies. Chromatin from K562 cells was sequentially immunoprecipitated with anti-CP2 and then anti-GATA-1 antibodies prior to quantitative PCR with the erythroid gene regulatory elements shown. Controls were as described for panel A.
CP2 and GATA-1 proteins physically interact in the absence of DNA.
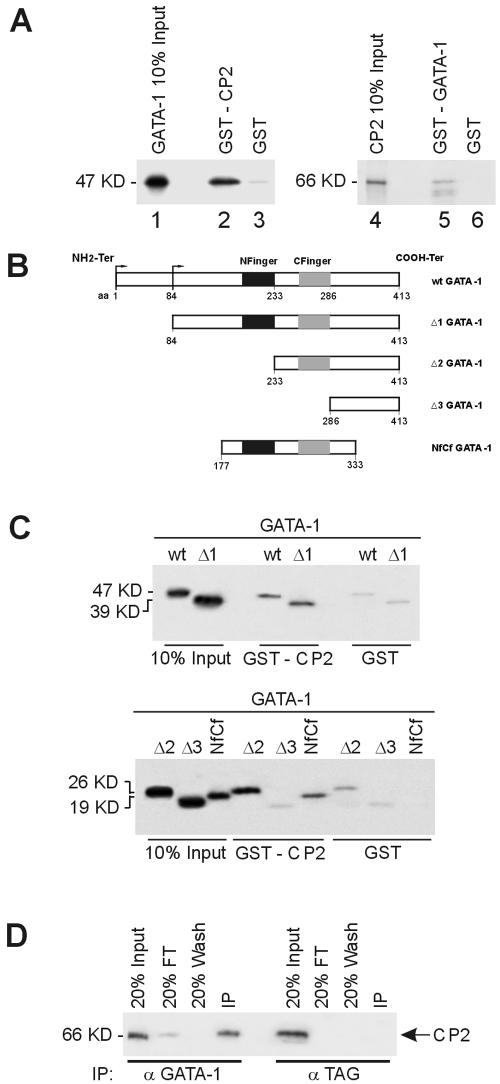
As CP2 and GATA-1 were shown to bind to adjacent sites on several genes, we wondered whether these two transcription factors might directly interact. To test this hypothesis, we performed a series of GST pull-down experiments in which either GST-GATA-1 or GST-CP2 immobilized on glutathione-Sepharose beads was assayed for its ability to capture in vitro-transcribed/translated 35S-labeled CP2 or GATA-1, respectively. As shown in Fig. 7A (left panel), [35S]GATA-1 was retained on a GST-CP2 resin (lane 2) but not on GST alone (lane 3). In a reciprocal experiment (Fig. 7A, right panel), [35S]CP2 was retained on the GST-GATA-1 resin (lane 5) but not on GST alone (lane 6). Under the same conditions, GATA-1 was able to bind to another known partner (EKLF) but failed to interact with a protein not known to interact (Cf1, RNA cleavage factor 1), confirming the specificity of the CP2-GATA-1 interaction (data not shown).
FIG. 7.
Direct physical interaction between GATA-1 and CP2. (A) Purified GST or GST fusion proteins containing full-length CP2 (GST-CP2) preadsorbed to glutathione-Sepharose beads were incubated with 35S-labeled in vitro-transcribed/translated GATA-1 (lanes 1 to 3). Specifically bound protein was eluted from washed beads and visualized by autoradiography after SDS-PAGE. Input represents 10% of the in vitro-translated GATA-1 used in the assay. Lanes 4 to 6 utilized glutathione-Sepharose-bound GST-GATA-1 and 35S-labeled CP2. (B) Schematic of truncation mutants used to map the GATA-1 regions responsible for the interaction with CP2. (C) GST or GST-CP2 protein coupled to glutathione-Sepharose was incubated with 35S-labeled wild type or truncated GATA-1 mutants shown in panel A. Input represents 10% of the in vitro-translated GATA-1 or mutant protein used in the assay. The molecular masses of the proteins are shown. (D) K562 cell extract was immunoprecipitated with either anti-GATA-1 or an unrelated antibody (anti-TAG) as the control. Immunoprecipitates were fractionated by SDS-PAGE and immunoblotted with anti-CP2 antibody. CP2 protein was detected in precipitates from anti-GATA-1 antibody but not from the control.
As different GATA-1 domains are known to mediate its interaction with several other transcription factors and to be differentially required for proper GATA-1 activity during development (5, 40), we mapped the GATA-1 regions responsible for its interaction with CP2. To this end, we produced a series of progressive N-terminal deletions of GATA-1 (Fig. 7B) and tested them for their ability to be retained on GST-CP2 resin. GATA-1 mutant protein lacking the 83 N-terminal amino acids specifically bound GST-CP2 but not GST alone (Fig. 7C, upper panel). A mutant protein lacking this region and the N-terminal zinc finger (amino acids 1 to 233) also continued to bind to GST-CP2 (Fig. 7C, lower panel). However, the further deletion of the residual COOH-terminal finger (amino acids 1 to 286) abolished the interaction with GST-CP2, with comparable levels of binding observed with GST alone (Fig. 7C, lower panel). A mutant GATA-1 protein containing both zinc fingers (amino acids 177 to 333) was able to interact specifically with CP2 (Fig. 7C, lower panel).
To confirm the ability of CP2 to interact with GATA-1 in the cellular environment, we immunoprecipitated K562 nuclear extracts with either anti-GATA-1 antibody (α-GATA-1) or an unrelated antibody (α-TAG) and probed the precipitates with anti-CP2 antibody (Fig. 7D). CP2 was efficiently coimmunoprecipitated with GATA-1, but no nonspecific immunoprecipitation was observed when we used the unrelated antibody. Taken together, these data demonstrate that GATA-1 directly interacts with CP2.
DISCUSSION
The transcription factor GATA-1 is a critical regulator of the activity of a large number of hematopoietic cell-restricted genes (5, 11, 47). Here, we show that a strong hematopoietic enhancer (the HS2 element of the mouse GATA-1 gene), known to be completely dependent on GATA-1 for its activity, additionally requires the binding of CP2 to a sequence adjacent to the GATA-1 binding site. Juxtaposed GATA-1-CP2 sites are present in regulatory elements of several hematopoietic-restricted genes, and CP2 binding is necessary for optimal activity of at least two additional genes encoding hematopoietic transcription factors p45 NF-E2 and EKLF. The further observation that the CP2 protein physically interacts with the GATA-1 protein in the region of the COOH-terminal zinc finger, where other transcription factors also bind, suggests that CP2 binding to GATA-1 might modulate its interaction with other transcription factors as well as its transcriptional activity.
Adjacent GATA-1 and CP2 binding motifs are essential for GATA-1 HS2 activity in transfection experiments.
GATA-1 controls essential stages of the differentiation and proliferation of hematopoietic progenitor cells, acting on a large number of target genes (6, 11). The levels of expression of GATA-1 and its physical interactions with other transcription factors (FOG1, Tal-1/SCL, Lmo-2, ETS-1, Pu-1, etc.) play critical roles in determining the differentiation of multipotent progenitors into specific hematopoietic lineages. The expression of GATA-1 is itself determined by several regulatory elements spread along the gene and characterized by chromatin DNase I hypersensitivity in hematopoietic progenitors (25, 29-31, 35, 45). These elements are partially redundant, as loss of single elements only moderately affects overall GATA-1 expression but rather diminishes its activity in a subset of cell lineages (13, 26, 42, 45). The HS2 enhancer is one of the best-studied elements. A high-affinity palindromic GATA-1 binding sequence in HS2 is the site of positive autoregulation by GATA-1 itself. In transfection studies, the activity of HS2 is largely dependent on the GATA-1 binding sequence (28). In vivo, mutation of the GATA-1 motif, within the context of the endogenous gene, decreases GATA-1 expression and cell differentiation in the eosinophilic lineage, indicating an important role of high GATA-1 levels in this lineage (49). Red cell production is also affected in this mutant strain, with mice exhibiting diminished red cell, hematocrit, and hemoglobin levels (49). However, as the other GATA-1 HS elements are intact, it is still unclear to what extent HS2 contributes to GATA-1 expression in other lineages. Aside from the GATA-1 binding elements, no other functionally important transcription factor binding sites in HS2 have been identified, leaving open the question as to whether HS2 requires only the GATA-1 binding sites.
Our experiments show that CP2 binds efficiently to a double CP2 binding element located immediately 3′ to the GATA-1 binding motif in HS2 (Fig. 1). Each of the two CP2 binding sites (the 1/3 and 2/4 boxes) is able to bind CP2, as indicated by experiments in which one of the two boxes was individually mutated (Fig. 3B). However, when both boxes were intact, we did not detect any band with slower mobility than that observed with the single-box CP2 mutants, even at a high protein concentration (Fig. 3 and 4 and data not shown). This suggests that although each of the two CP2 motifs may bind a CP2 dimer, only one CP2 dimer at a time is able to interact with the DNA or to form a stable complex under the electrophoretic conditions. Functionally, the binding of CP2 is essential, as mutation of even a single CP2 element in HS2 decreases the activity in transfection experiments with K562 cells to levels as low as those obtained in the absence of the GATA-1 binding site (Fig. 3C). Additionally, in vitro experiments (Fig. 3B, right panel) show that single CP2 mutants, Mut 1/3 and Mut 2/4 oligonucleotides, bind less CP2 than the wild-type oligonucleotide; this difference is further amplified, particularly for Mut 1/3, when the binding is carried out at low protein and oligonucleotide concentrations (data not shown).
One possible explanation for the functional effect of single CP2 mutants is that the existence of two adjacent functional sites might increase the affinity for CP2 or the stability of the bound CP2, limiting the off rate of the CP2-DNA complex. According to this hypothesis, we would suggest that in transfected cells, the probability of CP2 occupancy of the HS2 region is low when one of the CP2 sites is mutated, and thus, the formation of the functional GATA-1 complex is relatively unlikely. Note that the formation of the GATA-1-plus-CP2 complex on DNA is impaired when one of the two CP2 sites (Mut 1/3 and Mut 2/4) is mutated (Fig. 3B, lanes 1 and 2). Additionally, it is worth mentioning that the Mut 1-4 construct (in which all four CP2 half sites are mutated), when transfected in nonerythroid NIH 3T3 cells, is expressed at 80% ± 0.35% compared to the wild-type construct and that the simultaneous cotransfection of both CP2 and GATA-1 stimulates both the wild-type and CP2-mutated sequence at similar levels (3.5 ± 0.75% and 3.1 ± 0.8%, respectively). Thus, the functional interaction of CP2 and GATA-1 may require the involvement of additional hematopoietic-specific transcription factors and/or chromatin modifications not detectable in an EMSA. Overall, these results suggest that within this enhancer, the activity of GATA-1 bound to its cognate site may be largely, if not completely, dependent on collaboration with CP2. In vivo experiments with mice will be needed to confirm this hypothesis.
Adjacent GATA-1 and CP2 sites are important for regulatory elements of EKLF and p45 NF-E2 genes.
A CP2 binding site adjacent to a GATA-1 binding site is present on the regulatory elements of EKLF and p45 NF-E2, two additional transcription factors. As for the GATA-1 HS2 element, an intact CP2 binding site is necessary for optimal activity of the EKLF and p45 NF-E2 regulatory elements. However, the reduction of activity observed upon mutation of the CP2 binding motif on these regulatory elements is not as strong as that observed with the GATA-1 HS2 element. The reasons for this difference are not known. This may be due to the HS2 GATA-1/CP2 motif being a much stronger functional element than the others. In fact, the first GATA-1 palindromic site has an unusually high affinity for GATA-1 (43). In addition, the CP2 motif is likely to confer efficient binding due to its configuration of four half-CP2 motifs. Indeed, a ternary complex between GATA-1, CP2, and the HS2 oligonucleotide is stable enough to be visualized in an EMSA, whereas a similar complex is not visible with the EKLF and p45 NF-E2 oligonucleotides (Fig. 5).
ChIP experiments indicate an in vivo participation of CP2 in the regulation of GATA-1, EKLF, and p45 NF-E2 genes.
The detection of CP2 occupying the regulatory elements of GATA-1, EKLF, and p45NF-E2 genes in vivo (Fig. 6) is in agreement with the functional role of CP2 ascribed on the basis of transfection experiments. Sequential ChIP with anti-CP2 and anti-GATA-1 antibodies demonstrates that both proteins are present on the GATA-1 and EKLF regulatory elements simultaneously. While we cannot precisely define what proportion of sites are cooccupied by both transcription factors, this result indicates that at least some sites may be regulated by the actual interaction of GATA-1 and CP2. In the context of the p45 NF-E2 promoter, we were able to perform ChIP on the element with both GATA-1 and CP2 antibodies, but the sequential ChIP was unsuccessful. This finding is compatible with a model in which one factor, for example, CP2, initially binds to its target site, establishing chromatin changes that favor the binding of the partner factor (GATA-1 in this example), which binds only after the release of the first factor. This, in turn, induces additional changes that increase transcriptional activity.
GATA-1-CP2 complex binding sites are common to several hematopoietic genes.
A search of 100 hematopoietic GATA-1-dependent regulatory elements identified GATA-1-CP2 composite binding sites in at least 13 of them (Table 1). One of these elements was studied previously: a mutation in a CP2 site adjacent to a GATA-1 site in the human uroporphyrinogen III synthase erythroid promoter is responsible for a type of inherited erythropoietic porphyria, with decreased gene expression (41). Additionally, two CP2 motifs lying at some distance (∼50 to 60 bp) from a GATA-1 site are essential for optimal expression of the α-globin gene (8, 22). In addition to the GATA-1 HS2 enhancer, we have shown that two other regulatory elements of genes encoding transcription factors (p45 NF-E2 and EKLF) require CP2 for optimal activity. Overall, the relatively high frequency of the GATA-1-CP2 composite motif among hematopoietic genes and the validation of the requirement of CP2 binding for activity (this paper and reference 41) suggest that this motif may represent a novel module for common regulation of a subset of genes. A precedent for a similar module exists for GATA-1 sites adjacent to Fli-1 binding sites, which regulate gene expression mainly in megakaryocytic cells (46).
The previously published gene targeting of CP2 (LBP-1c) in mice failed to show any significant alteration in the expression of erythroid genes. However, this was demonstrated to be due to compensation by the highly related factor LBP-1a/NF2d9 (33). Elucidation of the role of these factors in erythropoiesis will come with the analysis of mice lacking both these genes. Most of the CP2 isoforms are ubiquitously expressed, with the only partial exception being the CP2c/CP2b heteromer. Although this complex is more abundant in erythroid than in nonerythroid cells, it is not sufficient to confer erythroid-specific transcriptional activation (17). GATA-1, by its ability to interact with CP2, may be responsible for conferring erythroid-specific transcriptional activation properties to the ubiquitously expressed CP2.
CP2 interaction with the GATA-1 COOH-terminal zinc finger region suggests possible additional roles of CP2 in modulating GATA-1 activity.
The ability of the GATA-1 COOH-terminal zinc finger to physically interact with CP2 was unexpected on the basis of the functional experiments described in the legend to Fig. 3C. In fact, these experiments show a clear requirement for CP2 binding to DNA, in order to activate transcription. Thus, the simple binding of GATA-1 to its cognate element is not sufficient to recruit CP2 into a functionally active complex. Similarly, CP2 binding to the DNA is not able to functionally recruit GATA-1 into an active transcription complex in the absence of the GATA-1 binding site (Fig. 3C). What then is the significance of the CP2-GATA-1 protein-protein interaction? One possibility is that the binding of either CP2 or GATA-1 to its respective site helps to bring the partner onto its adjacent DNA binding site, with the formation of a stable complex. An additional, and not mutually exclusive, possibility is supported by the observation that CP2 binds to the COOH-terminal zinc finger region of GATA-1. Several other transcription factors are known to bind GATA-1 at the COOH-terminal zinc finger, positively or negatively modulating its activity (6, 11). Binding of CP2 to the zinc finger may positively or negatively influence the interaction of GATA-1 with other potential partners. These studies form the basis of our ongoing work.
Acknowledgments
This work was supported by Cofin MIUR 2002 grants (S.O. and A.R.), by Fondazione Cariplo, 2003 and 2005 (S.O.), by the NHMRC of Australia, by NIH grants PO1 HL53749-03 and RO1 HL69232-01 (S.M.J.), by Cancer Centre Support CORE grant P30 CA 21765, by the American Lebanese Syrian Associated Charities, and by the Assisi Foundation of Memphis (J.M.C.).
REFERENCES
- 1.Anderson, K. P., S. C. Crable, and J. B. Lingrel. 1998. Multiple proteins binding to a GATA-E box-GATA motif regulate the erythroid Kruppel-like factor (EKLF) gene. J. Biol. Chem. 273:14347-14354. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, K. P., S. C. Crable, and J. B. Lingrel. 2000. The GATA-E box-GATA motif in the EKLF promoter is required for in vivo expression. Blood 95:1652-1655. [PubMed] [Google Scholar]
- 3.Bekri, S., A. May, P. D. Cotter, A. I. Al Sabah, X. Guo, G. S. Masters, and D. F. Bishop. 2003. A promoter mutation in the erythroid-specific 5-aminolevulinate synthase (ALAS2) gene causes X-linked sideroblastic anemia. Blood 102:698-704. [DOI] [PubMed] [Google Scholar]
- 4.Cairns, L., M. Ciro, M. Minuzzo, F. Morle, J. Starck, S. Ottolenghi, and A. Ronchi. 2003. Induction of globin mRNA expression by interleukin-3 in a stem cell factor-dependent SV-40 T-antigen-immortalized multipotent hematopoietic cell line. J. Cell. Physiol. 195:38-49. [DOI] [PubMed] [Google Scholar]
- 5.Cantor, A. B., and S. H. Orkin. 2001. Hematopoietic development: a balancing act. Curr. Opin. Genet. Dev. 11:513-519. [DOI] [PubMed] [Google Scholar]
- 6.Cantor, A. B., and S. H. Orkin. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21:3368-3376. [DOI] [PubMed] [Google Scholar]
- 7.Chae, J. H., and C. G. Kim. 2003. CP2 binding to the promoter is essential for the enhanced transcription of globin genes in erythroid cells. Mol. Cells 15:40-47. [PubMed] [Google Scholar]
- 8.Chae, J. H., Y. H. Lee, and C. G. Kim. 1999. Transcription factor CP2 is crucial in hemoglobin synthesis during erythroid terminal differentiation in vitro. Biochem. Biophys. Res. Commun. 263:580-583. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X., M. Reitman, and J. J. Bieker. 1998. Chromatin structure and transcriptional control elements of the erythroid Kruppel-like factor (EKLF) gene. J. Biol. Chem. 273:25031-25040. [DOI] [PubMed] [Google Scholar]
- 10.Dettwiler, S., C. Aringhieri, S. Cardinale, W. Keller, and S. M. Barabino. 2004. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J. Biol. Chem. 279:35788-35797. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira, R., K. Ohneda, M. Yamamoto, and S. Philipsen. 2005. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol. Cell. Biol. 25:1215-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallagher, P. G., M. Romana, W. T. Tse, S. E. Lux, and B. G. Forget. 2000. The human ankyrin-1 gene is selectively transcribed in erythroid cell lines despite the presence of a housekeeping-like promoter. Blood 96:1136-1143. [PubMed] [Google Scholar]
- 13.Guyot, B., V. Valverde-Garduno, C. Porcher, and P. Vyas. 2004. Deletion of the major GATA1 enhancer HS 1 does not affect eosinophil GATA1 expression and eosinophil differentiation. Blood 104:89-91. [DOI] [PubMed] [Google Scholar]
- 14.Jane, S. M., A. W. Nienhuis, and J. M. Cunningham. 1995. Hemoglobin switching in man and chicken is mediated by a heteromeric complex between the ubiquitous transcription factor CP2 and a developmentally specific protein. EMBO J. 14:97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, K. D., H. M. Christensen, B. Zhao, and E. H. Bresnick. 2001. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell 8:465-471. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, K. D., J. A. Grass, M. E. Boyer, C. M. Kiekhaefer, G. A. Blobel, M. J. Weiss, and E. H. Bresnick. 2002. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 99:11760-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang, H. C., J. H. Chae, Y. H. Lee, M.-A. Park, J. H. Shin, S.-H. Kim, S.-K. Ye, Y. S. Cho, S. Fiering, and C. G. Kim. 2005. Erythroid cell-specific α-globin gene regulation by the CP2 transcription factor family. Mol. Cell. Biol. 25:6005-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawabata, H., R. S. Germain, T. Ikezoe, X. Tong, E. M. Green, A. F. Gombart, and H. P. Koeffler. 2001. Regulation of expression of murine transferrin receptor 2. Blood 98:1949-1954. [DOI] [PubMed] [Google Scholar]
- 19.Kim, C. G., S. L. Swendeman, K. M. Barnhart, and M. Sheffery. 1990. Promoter elements and erythroid cell nuclear factors that regulate α-globin gene transcription in vitro. Mol. Cell. Biol. 10:5958-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolchanov, N. A., E. V. Ignatieva, E. A. Ananko, O. A. Podkolodnaya, I. L. Stepanenko, T. I. Merkulova, M. A. Pozdnyakov, N. L. Podkolodny, A. N. Naumochkin, and A. G. Romashchenko. 2002. Transcription Regulatory Regions Database (TRRD): its status in 2002. Nucleic Acids Res. 30:312-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liberati, C., M. R. Cera, P. Secco, C. Santoro, R. Mantovani, S. Ottolenghi, and A. Ronchi. 2001. Cooperation and competition between the binding of COUP-TFII and NF-Y on human epsilon- and gamma-globin gene promoters. J. Biol. Chem. 276:41700-41709. [DOI] [PubMed] [Google Scholar]
- 22.Lim, L. C., L. Fang, S. L. Swendeman, and M. Sheffery. 1993. Characterization of the molecularly cloned murine alpha-globin transcription factor CP2. J. Biol. Chem. 268:18008-18017. [PubMed] [Google Scholar]
- 23.Lim, L. C., S. L. Swendeman, and M. Sheffery. 1992. Molecular cloning of the α-globin transcription factor CP2. Mol. Cell. Biol. 12:828-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mantovani, R., N. Malgaretti, S. Nicolis, A. Ronchi, B. Giglioni, and S. Ottolenghi. 1988. The effects of HPFH mutations in the human gamma-globin promoter on binding of ubiquitous and erythroid specific nuclear factors. Nucleic Acids Res. 16:7783-7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDevitt, M. A., Y. Fujiwara, R. A. Shivdasani, and S. H. Orkin. 1997. An upstream, DNase I hypersensitive region of the hematopoietic-expressed transcription factor GATA-1 gene confers developmental specificity in transgenic mice. Proc. Natl. Acad. Sci. USA 94:7976-7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDevitt, M. A., R. A. Shivdasani, Y. Fujiwara, H. Yang, and S. H. Orkin. 1997. A “knockdown” mutation created by cis-element gene targeting reveals the dependence of erythroid cell maturation on the level of transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 94:6781-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moroni, E., T. Mastrangelo, R. Razzini, L. Cairns, P. Moi, S. Ottolenghi, and B. Giglioni. 2000. Regulation of mouse p45 NF-E2 transcription by an erythroid-specific GATA-dependent intronic alternative promoter. J. Biol. Chem. 275:10567-10576. [DOI] [PubMed] [Google Scholar]
- 28.Nicolis, S., C. Bertini, A. Ronchi, S. Crotta, L. Lanfranco, E. Moroni, B. Giglioni, and S. Ottolenghi. 1991. An erythroid specific enhancer upstream to the gene encoding the cell-type specific transcription factor GATA-1. Nucleic Acids Res. 19:5285-5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishimura, S., S. Takahashi, T. Kuroha, N. Suwabe, T. Nagasawa, C. Trainor, and M. Yamamoto. 2000. A GATA box in the GATA-1 gene hematopoietic enhancer is a critical element in the network of GATA factors and sites that regulate this gene. Mol. Cell. Biol. 20:713-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onodera, K., S. Takahashi, S. Nishimura, J. Ohta, H. Motohashi, K. Yomogida, N. Hayashi, J. D. Engel, and M. Yamamoto. 1997. GATA-1 transcription is controlled by distinct regulatory mechanisms during primitive and definitive erythropoiesis. Proc. Natl. Acad. Sci. USA 94:4487-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onodera, K., K. Yomogida, N. Suwabe, S. Takahashi, Y. Muraosa, N. Hayashi, E. Ito, L. Gu, M. Rassoulzadegan, J. D. Engel, and M. Yamamoto. 1997. Conserved structure, regulatory elements, and transcriptional regulation from the GATA-1 gene testis promoter. J. Biochem. (Tokyo) 121:251-263. [DOI] [PubMed] [Google Scholar]
- 32.Perry, C., and H. Soreq. 2002. Transcriptional regulation of erythropoiesis. Fine tuning of combinatorial multi-domain elements. Eur. J. Biochem. 269:3607-3618. [DOI] [PubMed] [Google Scholar]
- 33.Ramamurthy, L., V. Barbour, A. Tuckfield, D. R. Clouston, D. Topham, J. M. Cunningham, and S. M. Jane. 2001. Targeted disruption of the CP2 gene, a member of the NTF family of transcription factors. J. Biol. Chem. 276:7836-7842. [DOI] [PubMed] [Google Scholar]
- 34.Ristaldi, M. S., D. Drabek, J. Gribnau, D. Poddie, N. Yannoutsous, A. Cao, F. Grosveld, and A. M. Imam. 2001. The role of the −50 region of the human gamma-globin gene in switching. EMBO J. 20:5242-5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronchi, A., M. Ciro, L. Cairns, L. Basilico, P. Corbella, P. Ricciardi-Castagnoli, M. Cross, J. Ghysdael, and S. Ottolenghi. 1997. Molecular heterogeneity of regulatory elements of the mouse GATA-1 gene. Genes Funct. 1:245-258. [DOI] [PubMed] [Google Scholar]
- 36.Saleque, S., S. Cameron, and S. H. Orkin. 2002. The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 16:301-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawado, T., K. Igarashi, and M. Groudine. 2001. Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proc. Natl. Acad. Sci. USA 98:10226-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seshasayee, D., J. N. Geiger, P. Gaines, and D. M. Wojchowski. 2000. Intron 1 elements promote erythroid-specific GATA-1 gene expression. J. Biol. Chem. 275:22969-22977. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu, R., S. Takahashi, K. Ohneda, J. D. Engel, and M. Yamamoto. 2001. In vivo requirements for GATA-1 functional domains during primitive and definitive erythropoiesis. EMBO J. 20:5250-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solis, C., G. I. Aizencang, K. H. Astrin, D. F. Bishop, and R. J. Desnick. 2001. Uroporphyrinogen III synthase erythroid promoter mutations in adjacent GATA1 and CP2 elements cause congenital erythropoietic porphyria. J. Clin. Investig. 107:753-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi, S., K. Onodera, H. Motohashi, N. Suwabe, N. Hayashi, N. Yanai, Y. Nabesima, and M. Yamamoto. 1997. Arrest in primitive erythroid cell development caused by promoter-specific disruption of the GATA-1 gene. J. Biol. Chem. 272:12611-12615. [DOI] [PubMed] [Google Scholar]
- 43.Trainor, C. D., J. G. Omichinski, T. L. Vandergon, A. M. Gronenborn, G. M. Clore, and G. Felsenfeld. 1996. A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high-affinity interaction. Mol. Cell. Biol. 16:2238-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai, S. F., E. Strauss, and S. H. Orkin. 1991. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev. 5:919-931. [DOI] [PubMed] [Google Scholar]
- 45.Vyas, P., M. A. McDevitt, A. B. Cantor, S. G. Katz, Y. Fujiwara, and S. H. Orkin. 1999. Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the GATA-1 gene. Development 126:2799-2811. [DOI] [PubMed] [Google Scholar]
- 46.Wang, X., J. D. Crispino, D. L. Letting, M. Nakazawa, M. Poncz, and G. A. Blobel. 2002. Control of megakaryocyte-specific gene expression by GATA-1 and FOG-1: role of Ets transcription factors. EMBO J. 21:5225-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Welch, J. J., J. A. Watts, C. R. Vakoc, Y. Yao, H. Wang, R. C. Hardison, G. A. Blobel, L. A. Chodosh, and M. J. Weiss. 2004. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104:3136-3147. [DOI] [PubMed] [Google Scholar]
- 48.Yoon, J. B., G. Li, and R. G. Roeder. 1994. Characterization of a family of related cellular transcription factors which can modulate human immunodeficiency virus type 1 transcription in vitro. Mol. Cell. Biol. 14:1776-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, C., A. B. Cantor, H. Yang, C. Browne, R. A. Wells, Y. Fujiwara, and S. H. Orkin. 2002. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J. Exp. Med. 195:1387-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong, F., S. L. Swendeman, W. Popik, P. M. Pitha, and M. Sheffery. 1994. Evidence that levels of the dimeric cellular transcription factor CP2 play little role in the activation of the HIV-1 long terminal repeat in vivo or following superinfection with herpes simplex virus type 1. J. Biol. Chem. 269:21269-21276. [PubMed] [Google Scholar]
- 51.Zhou, W., D. R. Clouston, X. Wang, L. Cerruti, J. M. Cunningham, and S. M. Jane. 2000. Induction of human fetal globin gene expression by a novel erythroid factor, NF-E4. Mol. Cell. Biol. 20:7662-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]