Abstract
Several receptor protein tyrosine phosphatases (RPTPs) are cell adhesion molecules involved in homophilic interactions, suggesting that RPTP outside-in signaling is coupled to cell contact formation. However, little is known about the mechanisms by which cell density regulates RPTP function. We show that the MAM family prototype RPTPκ is cleaved by three proteases: furin, ADAM 10, and γ-secretase. Cell density promotes ADAM 10-mediated cleavage and shedding of RPTPκ. This is followed by γ-secretase-dependent intramembrane proteolysis of the remaining transmembrane part to release the phosphatase intracellular portion (PIC) from the membrane, thereby allowing its translocation to the nucleus. When cells were treated with leptomycin B, a nuclear export inhibitor, PIC accumulated in nuclear bodies. PIC is an active protein tyrosine phosphatase that binds to and dephosphorylates β-catenin, an RPTPκ substrate. The expression of RPTPκ suppresses β-catenin's transcriptional activity, whereas the expression of PIC increases it. Notably, this increase required the phosphatase activity of PIC. Thus, both isoforms have acquired opposing roles in the regulation of β-catenin signaling. We also found that RPTPμ, another MAM family member, undergoes γ-secretase-dependent processing. Our results identify intramembrane proteolysis as a regulatory switch in RPTPκ signaling and implicate PIC in the activation of β-catenin-mediated transcription.
The phosphorylation of cellular proteins on tyrosine residues is reversible and regulated by the coordinated and competing actions of two enzyme families: protein tyrosine kinases and protein tyrosine phosphatases (PTPs). The PTP family is structurally diverse and includes both receptor-like and cytoplasmic enzymes. The majority of the receptor PTPs (RPTPs) contain two catalytic domains: a membrane-proximal domain (D1), which is responsible for mainly catalysis, and a membrane-distal domain (D2), which contains little or no phosphatase activity (45). The extracellular (E) portion exhibits broad structural variation. The MAM (meprin/A5 glycoprotein/PTPmu) family of RPTPs, including RPTPκ, -μ, -ρ, -λ, and PCP-2, are characterized by the presence of the MAM domain at their N termini (3, 6, 18, 29, 46). Additional structural features of the extracellular portions involve one immunoglobulin-like domain and four fibronectin type III domains which are commonly found in cell adhesion molecules.
Functionally, MAM family RPTPs have been recognized as homophilic cell adhesion receptors. In fact, when expressed at the cell surface, the extracellular fragments of the enzymes can mediate cell aggregation (3, 39). These trans-interactions are highly specific, i.e., RPTPκ proteins interact with only each other and there is no heterophilic binding between, for instance, RPTPκ and RPTPμ. Moreover, the expression of MAM family RPTPs is increased in high-cell density cultures (11, 12). These observations have led to the proposal that these enzymes could directly sense cell-cell contact (4, 45) and thereby mediate contact inhibition of cell growth, a process that is disturbed in many tumors. Indeed, RPTPκ is localized at sites of cell-cell contact where it associates with β-catenin (11), which indirectly (via α-catenin) links E-cadherin to the actin cytoskeleton.
Transmembrane proteases of the ADAM family (containing disintegrin and metalloprotease domains) have been implicated in the cleavage and shedding of cytokines, cytokine receptors, growth factors and their receptors, amyloid precursor protein (APP), cell adhesion molecules, and Notch and its ligand Delta (41). The exposure of cells to phorbol esters, calcium ionophores, G protein-coupled receptor ligands, and high cell densities can induce specific ADAM-mediated shedding events. Receptor proteins with type I membrane orientation cleaved by ADAMs can further undergo so-called intramembrane proteolysis that has now been identified as a key signal transduction pathway in which receptors become processed within their transmembrane regions, resulting in the release of bioactive intracellular signaling fragments. Intramembrane cleavage is mediated by the γ-secretase complex, which is composed of presenilin (PS), nicastrin, PEN-2, and APH-1 (13). Presenilins have been demonstrated to mediate the cleavage of Notch, APP, low-density lipoprotein receptor-related protein, cadherins, CD44, the receptor tyrosine kinase ErbB-4, and other receptors (7, 20). Some released domains, like the Notch intracellular domain (ICD) or CD44 ICD, can enter the cell nucleus, where they function as transcriptional coactivators to initiate the expression of target genes. RPTPs are thought to initiate tyrosine phosphorylation-dependent signaling at the cell surface. We have found that RPTPκ, after becoming cleaved by furin, undergoes proteolytic processing via ADAM 10. Notably, cleavage by the sheddase is promoted at a high cell density. Following this processing step, the transmembrane product is further cleaved by γ-secretase, resulting in the release of the phosphatase intracellular portion (PIC) to the cytoplasm. By taking advantage of recombinant PIC, we observed the localization of this fragment in the nucleus and demonstrated that it binds to β-catenin. Whereas the great majority of β-catenin associates with the E-cadherin C terminus and is thus membrane bound, nonjunctional β-catenin can translocate to the nucleus to activate LEF/TCF transcription factors (26). Alternatively, when phosphorylated by GSK3β, cytoplasmic β-catenin is rapidly degraded by the ubiquitin-proteasome system. Although β-catenin tyrosine phosphorylation is critically involved in tumor progression (25, 36), its regulation is poorly characterized. Also, while the molecular consequences of this phosphorylation are not well understood, an involvement in the regulation of β-catenin-mediated transcription has been suggested (37). We identify RPTPκ as a negative regulator of β-catenin tyrosine phosphorylation in cells and show that PIC retains the ability to dephosphorylate the protein. We further show that the RPTPκ and PIC isoforms serve opposing roles in regulating β-catenin-mediated transcription, with RPTPκ as a suppressor and PIC as an activator.
MATERIALS AND METHODS
Antibodies.
Anti-RPTPκJM is a polyclonal antibody raised against the cytoplasmic juxtamembrane region of RPTPκ (residues 783 to 904). Anti-RPTPκEC antibody was directed against the whole extracellular portion. Antisera were processed for affinity purification by using the antigen coupled to CNBr Sepharose. The extracellular fragment of the phosphatase was produced in HEK293 cells and secreted into the cell supernatant. Polyclonal anti-RPTPμJM antiserum was directed against the juxtamembrane part (residues 773 to 917) of RPTPμ. Polyclonal anti-β-catenin antibody was directed against a glutathione S-transferase fusion protein containing residues 374 to 781 of human β-catenin. The polyclonal anti-ERK2 antibody was from Santa Cruz Biotechnology, and the monoclonal antibody anti-hemagglutinin (HA) was obtained from Babco. Polyclonal antibody to ADAM 15 was raised against the peptide H-CDHSEAQKYRDF-OH coupled to keyhole limpet hemocyanin. Antibodies directed towards ADAM 10 and ADAM 17 were purchased from Chemicon.
Plasmids.
The RPTPκ LNTR cleavage-site mutant has been previously described (18). The PΔE-HA construct was obtained by using the QuikChange site-directed mutagenesis kit (Stratagene). To this end, nucleotide sequences 5′-AGAGTGGTCAAAATAGCAGG-3′ and 5′-TGCTATTTTGACCACTCTGGATCCCAGGAGAGGCCAAGGAGAGAGGAGCAAG-3′ were used to delete most of the extracellular sequence (RPTPκ nucleotide 82 to 2244) and to fuse the leader peptide sequence to the four-amino-acid extracellular stalk. PIC-HA (starting at nucleotide 2317) was generated by PCR amplification with nucleotides 5′-GGAATTCGCCACCATGATTGTAAAAAAGAGCAAACTTGCTAA-3′ and 5′-CTGAATTCAGAGTCTGAAATTCA-3′. The resulting EcoRI-EcoRI fragment lacking the extracellular and transmembrane regions was used to replace the wild-type sequence. PICΔJM was generated by PCR using the primer sequences 5′-GGAATTCGCCACCATGGATCAAAATAGAGC-3′ and 5′-CTGAATTCAGAGTCTGAAATTCATCCTTG-3′. ATG was introduced in front of nucleotide 2722 (amino acid 908 in the RPTPκ precursor sequence) and the resulting EcoRI/EcoRI fragment was used to replace the corresponding fragment of the RPTPκ-HA sequence. PIC-CS1-HA harbors a C-to-S transition at position C1082 and was generated in the same way as PIC-HA (see above), except that the RPTPκ-CS1 sequence was used as template. Enhanced green fluorescent protein (GFP)-PIC, starting with amino acid 774 within the RPTPκ sequence, was obtained by PCR using the primer sequences 5′-CCGCTCGAGGTATTGTAAAAAAGAGCAAACTTGCT-3′ and 5′-CCCAAGCTTCTAAGATGATTCCAGGTACTCC-3′. The fragment was subcloned into vector pEGFP-C1 (BD Biosciences Clontech) via XhoI/HindIII. Constitutive active Src (SrcYF) harbors a transition of tyrosine residue 529 to phenylalanine.
Biotinylation of cell surface proteins.
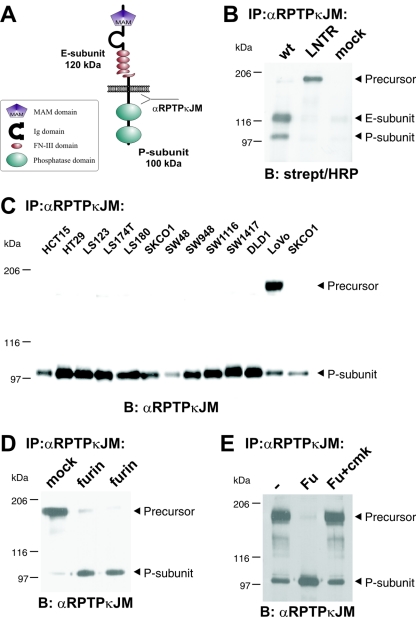
Cell surface biotinylation was carried out on transfected cells by using 0.5 mg/ml sulfo-NHS-LC-biotin (Pierce) according to the manufacturer. RPTPκ was immunoprecipitated by an antibody to the intracellular juxtamembrane part (anti-RPTPκJM), followed by Western blot analysis with horseradish peroxidase-conjugated streptavidin (Fig. 1B).
FIG. 1.
RPTPκ is a two-subunit receptor at the cell surface after furin-mediated processing. (A) Scheme depicting the two-subunit structure of RPTPκ. The fragment sizes are indicated. Ig, immunoglobulin. (B) Cell surface-presented RPTPκ is a two-subunit enzyme, composed of the E subunit and the PTP domain-containing P subunit. 293 cells were transfected with wild-type RPTPκ and the convertase cleavage site mutant RPTPκ-LNTR, in which the dibasic sequence motif RTKR located in the membrane-proximal fibronectin type III domain was replaced by LNTR. Cells were surface biotinylated prior to lysis under standard conditions as described in Materials and Methods. RPTPκ was immunoprecipitated by an antibody to the intracellular juxtamembrane part (anti-RPTPκJM), followed by blotting with horseradish peroxidase (HRP)-conjugated streptavidin. (C) Accumulation of the precursor in LoVo cells that are devoid of functional furin. RPTPκ protein was analyzed in a panel of colon carcinoma cell lines by immunoprecipitation and blotting with anti-RPTPκJM antibody. (D) Stable expression of furin in LoVo cells restores processing of RPTPκ. The phosphatase was immunoprecipitated from two LoVo cell clones stably expressing human furin and from a vector-transfected clone (mock) for comparison. (E) Purified furin cleaves RPTPκ within the membrane-proximal fibronectin type III domain at the sequence RTKR in vitro. Furin-null, LoVo cell-derived RPTPκ was immunoprecipitated and incubated for 1 h at 37°C with phosphate-buffered saline (PBS) (−), purified recombinant mouse furin (Fu) or purified recombinant furin previously treated with the inhibitor decRVKR-cmk (Fu+cmk). Abbreviations: α, anti; B, blotting; IP, immunoprecipitation.
RNA interference.
The transfection of 21-nucleotide, small-interfering-RNA (siRNA) duplexes (Dharmacon) for targeting endogenous genes in Caki-1 cells was carried out using Lipofectamine 2000 (Invitrogen) and siRNA duplex according to the manufacturer's protocol. Cells were assayed 2 days after transfection. Specific silencing of targeted genes was confirmed by Western blotting. Sequences of the siRNAs used have been described earlier (10). The pSUPER.retro plasmid (OligoEngine) expressing RPTPκ siRNA was directed against nucleotides 392 to 412 of the RPTPκ coding sequence.
Cell culture, transfections, and retroviral infections.
All of the cell lines (American Type Culture Collection) were routinely grown according to the supplier's instructions. To analyze cell density-dependent cleavage, MDA-MB-453 cells and MDA-MB-468 cells were plated at different cell densities for 24 h in RPMI medium supplemented with Ultroser G serum substitute to prevent the formation of cell aggregates during the seeding procedure. HEK293 and LoVo cells were transfected using the calcium phosphate-DNA coprecipitation method. Transiently transfected HEK293 cells were assayed and lysed 24 to 36 h after the removal of precipitates. Monoclonal LoVo cell clones stably expressing furin were obtained after selection in the presence of G418 (500 μg/ml). COS-7 cells were transiently transfected with Polyfect (QIAGEN) according to the manufacturer's instructions. Mouse embryonic fibroblasts from ADAM 10−/− mice and respective wild-type animals were generated and characterized as described previously (38). For retroviral infections, the amphotropic packaging cell line Phoenix A was transfected with respective pSUPER retro (Oligoengine) plasmids using calcium phosphate-chloroquine. Thirty-six hours after transfection, the viral supernatant was collected and used to infect subconfluent target cells (1 × 105 cells/6-cm dish). Monoclonal ACHN kidney cancer cell lines that were stably expressing scrambled or RPTPκ-directed siRNA were generated by selecting retrovirally infected cells in medium containing puromycin (1.5 μg/ml) for 5 days and subsequent clonal separation.
Membrane fractionation.
Cells were washed and scraped in phosphate-buffered saline (PBS), pelleted by centrifugation (5 min at 3,500 rpm), and incubated in hypotonic buffer (20 mM HEPES [pH 7.2], 10 mM KCl, 1.5 mM MgCl2, 0.1 mM EGTA, 5 mM EDTA, 10 mM Na4P2O7, 1 mM phenylmethylsulfonyl fluoride, 0.1 μg/ml aprotinin, 10 mM NaF, 1 mM Na3VO4) for 10 min on ice. Cells were broken using a Dounce homogenizer (30 strokes), and nuclei were pelleted by centrifugation (10 min at 3,500 rpm). Nucleus-free supernatant was subjected to ultracentrifugation at 100,000 × g for 1 h to separate the membrane (pellet) from the cytosolic fraction (supernatant). The pellet was washed with hypotonic buffer and solubilized with membrane solubilization buffer, followed by centrifugation at 100,000 × g for 1 h. The resulting supernatant was termed membrane fraction.
Cell treatment.
The furin inhibitor decanoyl-Arg-Val-Lys-Arg-chloromethylketone (Calbiochem) was included in the incubation mixture at 100 μmol/liter. If not otherwise indicated, cells were treated with the phenothiazine derivatives trifluoperazine (TFP), chlorpromazine, promazine, and fluphenazine at a concentration of 100 μM. A phorbol ester, phorbol myristate acetate (PMA), and ionomycin were used at concentrations of 1 and 10 μM. To analyze S3 cleavage, confluent cells were treated with the γ-secretase inhibitors DAPT {N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester} (2 μM) and L-658458 (5 μM) for 16 h. The inhibition of the proteasome in cells was achieved by treatment with 5 μM MG132, 5 μM lactacystin, or 5 μM epoxomycin for 16 h. Cells were treated with BB-94 (Batimastat) at a concentration of 5 μM. Following 20 min of BB-94 incubation, TFP was added to cells for 15 min. Cells were treated with epidermal growth factor (EGF) for 5 min at a final concentration of 200 ng/ml. Anti-RPTPκEC antibody was added to the cell supernatant for 5, 15, or 30 min at concentrations of 10 μg/ml.
Extract preparation and immunoprecipitation.
Cells were lysed, and proteins were solubilized on ice for 20 min in ice-cold lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 2 mM EDTA, 10% glycerin, 1% Triton X-100, 10 mM Na4P2O7, 1 mM phenylmethylsulfonyl fluoride, 0.1 μg/ml aprotinin, 10 mM NaF, 1 mM Na3VO4). Lysates were precleared by centrifugation at 12,500 × g for 20 min at 4°C. Prior to immunoprecipitation, lysates were adjusted for equal protein concentrations and the appropriate antibody and protein A-Sepharose were added to the lysate and incubated for 4 h at 4°C. Precipitates were washed four times with HNTG buffer (20 mM HEPES [pH 7.5], 150 mM NaCl, 10% glycerol, 0.1% Triton X-100), and sodium dodecyl sulfate sample buffer was added.
TCA precipitation.
Cells were washed twice with serum-free medium. Following cell stimulation or basal shedding, cell medium was centrifuged at 2,000 × g for 5 min to pellet-detached cells. Afterwards, supernatants were centrifuged at 20,000 × g to remove cell debris and subjected to trichloroacetic acid (TCA) precipitation. Briefly, samples were mixed with an equal volume of 20% TCA, incubated on ice for 30 min, and centrifuged for 15 min and supernatants were carefully removed. Pellets were washed in ice-cold acetone and centrifuged for 10 min, dried, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer, and heated at 65°C for 3 min.
Immunofluorescence analysis.
COS-7 and NIH 3T3 cells were fixed with 4% paraformaldehyde for 10 min, exposed to 0.2% Triton X-100 in PBS for 10 min, blocked with PBG (0.5% bovine serum albumin and 0.045% gelatin in PBS) plus 5% goat serum, and then incubated for 2 h with anti-HA or anti-RPTPκJM antibody. After being washed three times with blocking buffer, the cells were incubated for 2 h with fluorescein isothiocyanate-conjugated secondary antibodies (Molecular Probes). Cells were subsequently washed with PBS and mounted with Fluoromount G (Biozol) for observation. Immunofluorescence slides were viewed and analyzed using a Leica confocal microscope.
Reporter assays.
HCT116 cells were plated on 24-well tissue culture plates 24 h before transfection. Lipofectamine was used to cotransfect cells with 2 ng of an internal control (pRL-CMV), 100 ng of a reporter construct (pGL3-OT or pGL3-OF), and the indicated RPTPκ expression vectors. pGL3-OT is an improved pTOPFLASH vector and contains an optimized TCF-binding site upstream of a luciferase reporter gene, whereas pGL3-OF contains a mutated site that does not bind TCF. Thirty-six hours after transfection, luciferase activities were measured and normalized for background Renilla luciferase activities by using the dual luciferase reporter assay system (Promega). Normalized values were corrected for nonspecific transcription by subtracting pGL3-OF values.
RESULTS
Furin is required for S1 processing of RPTPκ in the secretory pathway.
The RPTPκ protein consists of two subunits that are noncovalently attached to each other: the transmembrane P subunit (100 kDa) that harbors two PTP domains and the extracellular E subunit (120 kDa) that covers most of the extracellular sequence (Fig. 1A). Both subunits are generated from one precursor protein by proteolytic processing at the dibasic cleavage site (RTKR) located within the membrane-proximal fibronectin type III domain (11, 18). Surface biotinylation of HEK293 cells transfected with wild-type RPTPκ revealed the presence of the two-subunit structure at the cell surface (Fig. 1B). In contrast, a cleavage-site mutant, with the motif RTKR replaced by LNTR, is transported to the cell surface in the full-length form.
Mammalian subtilisin-like protein convertases (PCs) have been shown to target dibasic sequence motifs and mediate constitutive processing of precursor proteins in the trans-Golgi network. Four of them, namely, furin, PACE4, PC5/6, and LPC, are ubiquitously expressed, and PC5 has been shown to mediate the cleavage of RPTPμ (5), a closely related phosphatase of the MAM family. An analysis of RPTPκ protein expression in colon carcinoma cells revealed that furin-deficient LoVo cells accumulate the precursor (Fig. 1C). Importantly, LoVo cells do not express functional furin due to mutations in both furin alleles (42, 43) but the expression of active furin in these cells by stable transfection restored cleavage (Fig. 1D). Likewise, the precursor was specifically cleaved at the RTKR site to generate the E and P subunits when recombinant furin was added to LoVo cell-derived RPTPκ in vitro (Fig. 1E). These results demonstrate the involvement of furin in RPTPκ processing in the secretory pathway to generate the two-subunit receptor. In analogy to the processing of the Notch protein by furin at the dibasic motif RQRR (27), RPTPκ cleavage at RTKR is hereafter referred to as S1 processing and RTKR is referred to as the S1 site.
Proteolytic processing of RPTPκ at a second site (S2) induced by cell density and trifluoperazine treatment.
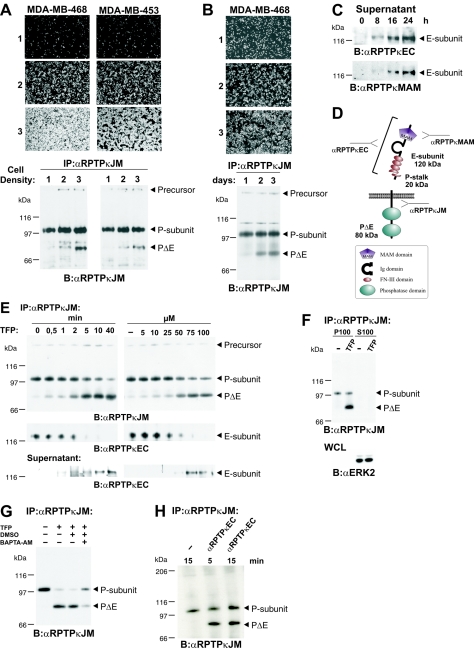
RPTPs are thought to be involved in the density-dependent inhibition of cell growth. For instance, cell treatment with the PTP inhibitor vanadate decreased density-dependent growth inhibition (19). Moreover, membrane fractions derived from dense cells showed increased PTP activities compared to the activities of those derived from sparse cells (35). It is therefore not surprising that the expression of several RPTPs, like DEP-1 (34) and the MAM family members RPTPκ (11) and RPTPμ (12), was found to be upregulated in cell monolayers. We observed a previously undescribed RPTPκ-specific product of 75 kDa (hereafter designated PΔE to specify the lack of most of the extracellular portion; see below) that accumulated in high-density cultures in a panel of breast, renal, melanoma, and colon carcinoma cell lines (data not shown). For instance, when MDA-MB-468 and MDA-MB-453 breast carcinoma cells were incubated at sparse, medium, and high densities for 24 h, the accumulation of PΔE was found to be increased in high-density culture compared to that in low-density culture (Fig. 2A). In contrast, the expression levels of the precursor and the P subunit were not markedly affected in these cell lines. In another approach, cells were seeded at equal densities and those cells grown for longer times reached higher densities and, thus, expressed increased levels of PΔE as well (Fig. 2B). Moreover, the E subunit of RPTPκ was detected in cell supernatants derived from confluent cell layers (Fig. 2C), suggesting that it had been shed from the cell surfaces. We therefore hypothesized that the generation of PΔE and shedding of the RPTPκ extracellular fragment into the cell medium is achieved by cleavage at an extracellular site close to the membrane that we named S2 (since it is located downstream of S1). A scheme depicting the expected products derived from cleavage at S1 and S2 is shown in Fig. 2D.
FIG. 2.
Proteolytic processing at a second site (S2 cleavage) results in generation of PΔE and shedding of RPTPκ. (A) Mammary carcinoma cell lines MDA-MB-468 and MDA-MB-453 were seeded at increasing cell densities (10, 50, and 100% [panels and lanes 1, 2, and 3, respectively]) and incubated for 24 h in serum-free medium containing 2% (vol/vol) Ultroser G. RPTPκ was immunoprecipitated with anti-RPTPκJM antibody and analyzed as indicated. (B) MDA-MB-468 cells were seeded at equal cell densities in medium containing 10% fetal calf serum and incubated for 1, 2, or 3 days (panels 1, 2, and 3, respectively). (C) Time-dependent accumulation of the E subunit in cell supernatants. Conditioned media were collected from confluent MDA-MB-468 cells at indicated time points and concentrated by TCA precipitation. The E subunit was detected with antibodies to the extracellular part (anti-RPTPκEC) and the MAM domain of RPTPκ (anti-RPTPκMAM). (D) Scheme depicting the expected fragments generated by cleavage at S1 and S2. The fragment sizes are indicated. (E) Time and concentration dependence of TFP-induced S2 cleavage and shedding. MDA-MB-468 cells were seeded at confluence, washed, and incubated in serum-free medium. The cells were treated with 100 μM TFP for different incubation times (left panel) or for 20 min with various concentrations of TFP as indicated (right panel). RPTPκ was immunoprecipitated from cell lysate with anti-RPTPκJM antibody (upper panel). The blot was reprobed with an antibody to the extracellular part of RPTPκ (anti-RPTPκEC) (middle panel). Conditioned media were collected, proteins concentrated by TCA precipitation and probed with anti-RPTPκEC antibody (lower panel). (F) PΔE is bound to the plasma membrane. 786-O cells were incubated with 100 μM TFP for 30 min and resuspended in hypotonic buffer. Soluble (S100) and membrane-bound (P100) proteins were separated by membrane fractionation. RPTPκ was immunoprecipitated from both fractions (upper panel). As a control, fractions were analyzed for the soluble protein ERK2 (lower panel). WCL, whole-cell lysate. (G) Inhibitory effect of the membrane-permeable calcium chelator BAPTA-AM on TFP-induced S2 processing. Cells were incubated for 2 h with (+) or without (−) 10 μM BAPTA-AM. DMSO, dimethyl sulfoxide. (H) Anti-RPTPκEC antibody treatment of MDA-MB-468 cells leads to rapid cleavage of endogenously expressed RPTPκ. Cells were starved and incubated with or without (−) anti-RPTPκEC antibody at 10 μg/ml as indicated. Abbreviations: α, anti; B, blotting; IP, immunoprecipitation.
The accumulation of PΔE was observed when cells were grown for at least 12 to 24 h at high cell densities (Fig. 2A and B). We next sought to recapitulate S2 cleavage in a short-term assay. We therefore tested several compounds that are known to induce the shedding of cell surface proteins. Surprisingly, the treatment of renal carcinoma cells with TFP led to rapid accumulation of PΔE (Fig. 2E, upper panel). In MDA-MB-468 cells, the level of this product increased to a lesser extent in the presence of ionomycin but this effect was not observed in 786-O cells (see Fig. S1 in the supplemental material). Remarkably, PMA did not promote cleavage, even though it was reported to induce the shedding of members of another RPTP family, RPTP-LAR (leukocyte antigen related), and RPTPσ (1). TFP is one derivative of phenothiazines (see Fig. S2A in the supplemental material), a class of drugs that are widely used to control mental disorders like schizophrenia, depressions, and related conditions. Although phenothiazine-derived compounds are similar in structure, they do not induce S2 processing in general (see Fig. S2B in the supplemental material). It thus seems that the ability to induce shedding critically depends on CF3 and Cl substitutions at position 2 on the hydrophobic phenothiazine ring structure and/or the presence of piperazine-substituted side chains. TFP-induced S2 processing was found to be time and dose dependent (Fig. 2E). As expected, cleavage at S2 provoked the release of the E subunit from cells (Fig. 2E, middle panel), concomitant with the accumulation of this subunit in cell supernatants (lower panel). Concordantly, the C-terminal S2 cleavage product PΔE remains attached to the plasma membrane (Fig. 2F). These observations suggest that S2 processing induced by both cell density and TFP treatment targets the ectodomain of RPTPκ, leading to generation of transmembrane PΔE and shedding of the E subunit.
TFP has been reported as a shedding inducer of several cell surface receptors (8). One suggested mechanism of TFP action may involve the rapid increase in the cytoplasmic calcium concentration that is thought to be the result of interference with calmodulin function in cells (47). However, TFP does also block the dopamine receptor. We therefore asked whether calcium signaling is involved in the induction of cleavage. Indeed, the treatment of cells with the membrane-permeable calcium chelator 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetrakis(acetoxymethyl ester) (BAPTA-AM) partially blocked S2 cleavage (Fig. 2G), whereas BAPTA, a membrane-impermeable derivative, was not inhibitory (data not shown). This is in accordance with the observation that ionomycin at 10 μM induced cleavage at S2 in MDA-MB-468 cells (see Fig. S1 in the supplemental material) but to a much lesser extent. A simple explanation could be that the increase in the concentration of cytoplasmic calcium is higher in the case of treatment with less toxic TFP at 100 μM.
After having shown induction of RPTPκ S2 cleavage at a high cell density, we hypothesized that cell contact formation triggers cleavage. An interesting mechanistic explanation could be that, at points of cell contacts, there is an increased aggregation of RPTPκ proteins at the cell surface, which in turn could result in an induction of S2 processing. To test this, we took advantage of the affinity-purified antibody anti-RPTPκEC, which was raised against the extracellular portion of the phosphatase (Fig. 2D). This activating antibody was found to be an appropriate tool for studying the function of RPTPκ in the cellular context (see below) (Fig. 6B and C). We hypothesized that, when added to the cell medium, anti-RPTPκEC antibody could potentially aggregate the enzyme at the cell surface. Indeed, short-term treatment (5 min) with anti-RPTPκEC antibody caused the accumulation of PΔE in MDA-MB-468 cells (Fig. 2H), suggesting that RPTP aggregation results in the activation and cleavage of the phosphatase.
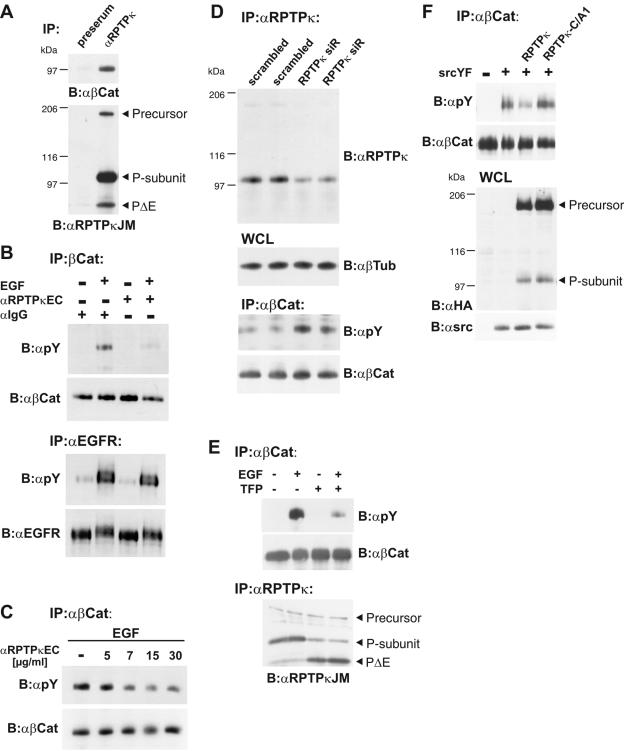
FIG. 6.
β-Catenin is a cellular substrate of RPTPκ. (A) Coprecipitation of β-catenin with RPTPκ in 786-O renal carcinoma cells. Upper panel, detection of β-catenin in anti-RPTPκJM antibody immunoprecipitates. Preserum was used as a negative control. Lower panel, the blot was reprobed with anti-RPTPκJM antibody. (B) Stimulation of cells with anti-RPTPκEC antibody leads to decreased tyrosine phosphorylation of β-catenin but not of EGFR. MDA-MB-468 cells were treated with anti-RPTPκEC antibody (10 μg/ml) for 30 min prior to stimulation with EGF (200 ng/ml) for 5 min as indicated. −, no treatment. Immunoglobulin G-Fc antibody was used as a control. Upper panel, β-catenin was immunoprecipitated and probed for tyrosine phosphorylation with antibody 4G10 (anti-pY). Lower panel, EGFR was immunoprecipitated and probed for tyrosine phosphorylation. (C) Reduction in β-catenin tyrosine phosphorylation is antibody concentration dependent. Cell stimulation was performed with 200 ng/ml EGF for 5 min and various concentrations of anti-RPTPκEC antibody for 30 min as indicated. (D) Short interfering RNA (siR)-mediated knockdown of RPTPκ increases β-catenin tyrosine phosphorylation. Stably transfected ACHN cells were analyzed for RPTPκ expression andtyrosine phosphorylation of β-catenin. All cells were stimulated with EGF (200 ng/ml) for 5 min to induce the tyrosine phosphorylation of β-catenin. RPTPκ (top panel) and β-catenin (bottom panel) were immunoprecipitated and probed with indicated antibodies. Tubulin was used as loading control (middle panel). WCL, whole-cell lysates. (E) Effect of TFP treatment on β-Catenin tyrosine phosphorylation. Cells were incubated with (+) or without (−) TFP (100 μM) for 10 min prior to stimulation with EGF (200 ng/ml) for 5 min as indicated. β-Catenin (upper panel) and RPTPκ (lower panel) were immunoprecipitated and analyzed. (F) β-Catenin dephosphorylation depends on catalytic activity of the membrane-proximal PTP domain of RPTPκ. HEK293 cells were cotransfected with constitutive active Src (SrcYF) (+) or empty vector (−) and either RPTPκ-HA or catalytically inactive RPTPκ-C/A1-HA. To obtain maximal tyrosine phosphorylation of β-catenin, cells were starved for 12 h and the protein was immunoprecipitated and probed with the indicated antibody (upper panels). Whole-cell lysates (WCL) were analyzed for the expression of RPTPκ and SrcYF. Abbreviations: α, anti; B, blotting; IP, immunoprecipitation.
ADAM 10 accounts for S2 activity.
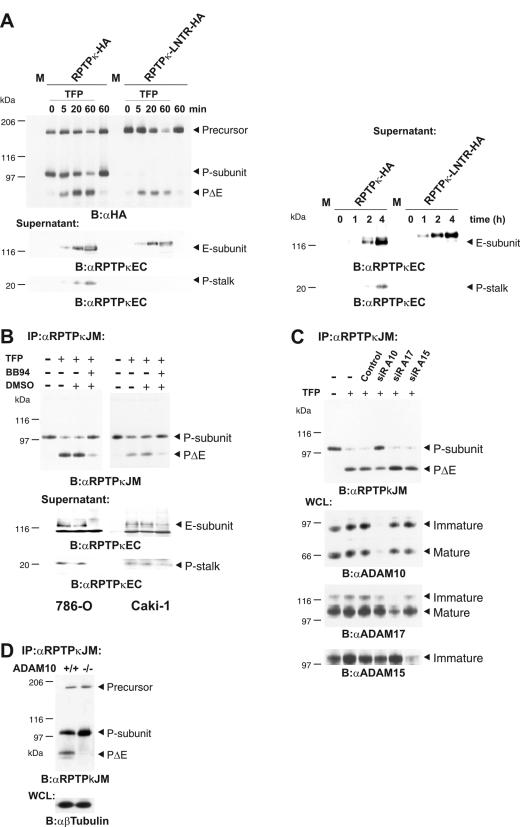
Furin-mediated processing of RPTPκ yields two subunit proteins (Fig. 1). Importantly, the extracellular subunit (E subunit) accumulated in cell supernatants in response to TFP treatment in a time- and concentration-dependent manner (Fig. 2E). To demonstrate that this accumulation is due to cleavage at S2, but not to subunit dissociation, we transfected wild-type phosphatase and the furin cleavage-site mutant (LNTR) into COS-7 cells and analyzed cell supernatants for RPTPκ-specific products. According to the scheme shown in Fig. 2D, we anticipated detection of both the E subunit and the short P subunit-derived extracellular stalk (P stalk). Indeed, both fragments were detected in response to treatment with TFP for at least 5 min (Fig. 3A, left lower panel). Basal shedding of the P stalk was also observed, but only after longer incubation periods at high cell densities (Fig. 3, right lower panel). In contrast, neither TFP-induced nor basal shedding caused P-stalk accumulation in medium derived from cells expressing the furin cleavage site mutant (LNTR). Concordantly, in both cases, the shed LNTR extracellular fragment is larger than the E subunit. These observations identify RPTPκ shedding as a consequence of cleavage at S2.
FIG. 3.
ADAM 10 is involved in S2 processing. (A) RPTPκ subunit dissociation does not contribute to TFP-induced or basal shedding. RPTPκ constructs used here are HA tagged at the C terminus. COS-7 cells were transfected with cDNAs of RPTPκ, RPTPκ-LNTR, or pRK5 vector (M). Left panel, cells were washed, incubated in serum-free medium, and stimulated with 100 μM TFP for the times indicated. Upper panel, cell lysates were blotted and probed with anti-HA antibody. Lower panels, detection of shed RPTPκ fragments. Conditioned media were collected, and proteins were concentrated by TCA precipitation and probed with anti-RPTPκEC antibody. Right panel, time course of RPTPκ basal shedding at a high cell density. Transfected COS-7 cells were incubated in serum-free medium for the times indicated in the text. Conditioned media were processed as described above. (B) Metalloprotease inhibitor BB-94 diminishes S2 cleavage and shedding of RPTPκ in 786-O and Caki-1 cells. Prior to stimulation, cells were washed and serum-free medium was added. Cells were pretreated either with (+) or without (−) the metalloprotease inhibitor BB-94 (5 μM) or dimethyl sulfoxide (DMSO) and were then stimulated with 100 μM TFP for 15 min. Upper panel, RPTPκ was immunoprecipitated with anti-RPTPκJM antibody. Lower panel, conditioned media were collected, and proteins were concentrated by TCA precipitation, blotted, and probed with anti-RPTPκEC antibody. (C) ADAM 10 is involved in TFP-induced S2 cleavage. Caki-1 cells were transfected with siRNA (siR) directed against ADAM 10 (A10), ADAM 15 (A15), and ADAM 17 (A17). After 48 h, cells were treated or not treated (−) with 100 μM TFP for 15 min. Upper panel, RPTPκ was immunoprecipitated with anti-RPTPκJM antibody. Lower panels, specific silencing of ADAM expression was confirmed by immunoblot analyses of whole-cell lysate with antibodies to ADAM 10, ADAM 15, and ADAM 17. While immature and mature isoforms were detected for ADAM 10 and ADAM 17, no mature isoform was observed for ADAM 15. (D) ADAM 10-mediated cleavage at a high cell density. RPTPκ was immunoprecipitated from confluent ADAM 10+/+ and ADAM 10−/− fibroblasts. Abbreviations: α, anti; B, blotting; IP, immunoprecipitation; WCL, whole-cell lysate.
Membrane-integrated, Zn-dependent proteases of the ADAM family have been implicated in the shedding of numerous cell surface proteins (41). In order to identify the enzymatic activity that is responsible for the processing of RPTPκ at S2, we treated a panel of cell lines with the metalloprotease inhibitor BB-94. Figure 3B demonstrates that the inhibition of metalloprotease activity diminished the accumulation of PΔE in cells and concomitantly reduced the amount of shed fragments in cell supernatants. Importantly, BB-94 treatment had the same effects on basal shedding at a high cell density and led to accumulation of the P subunit (data not shown). We then asked for the involvement of individual ADAMs by specifically reducing their endogenous expressions using the siRNA approach. Here we chose to target ADAM 10, 15, and 17 since ADAM 10 has been implicated in the regulation of cell adhesion, ADAM 15 was found to be localized at sites of cell contacts, and ADAM 17 (TACE) is involved in the majority of observed shedding events (41). As Fig. 3C shows, S2 processing was diminished in the case of ADAM 10 knockdown but not by interfering with the expressions of ADAM 15 and ADAM 17. Likewise, cleavage was diminished in confluent embryonic fibroblasts derived from ADAM 10 knockout mice (38). This was accompanied by the accumulation of the P subunit in ADAM 10−/− fibroblasts (Fig. 3D). By contrast, processing was not affected in embryonic ADAM 17−/− fibroblasts (data not shown). These results demonstrate that ADAM 10 is an important mediator of RPTPκ S2 cleavage in untreated confluent cells and upon stimulation with calmodulin inhibitors.
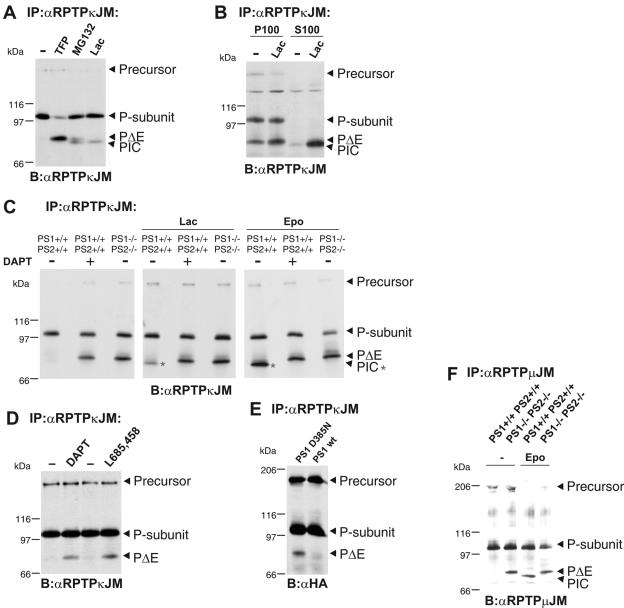
Presenilin 1-dependent S3 processing of membrane-bound PΔE leads to generation of PIC.
In the course of our study, we tested the effects of a panel of protease inhibitors on RPTPκ S2 cleavage (data not shown). These studies revealed, however, that the inhibition of the proteasome with inhibitors MG132 and lactacystin led to the accumulation of an additional isoform that we named PIC (phosphatase intracellular portion) (Fig. 4A). Thus, endogenous PIC levels in cells are low but get stabilized by the inhibition of the proteasome. PIC is approximately 5 kDa smaller than PΔE and is found exclusively in the soluble fraction (Fig. 4B). In contrast, PΔE was detected in only the membrane preparation. We observed an increased rate of PΔE generation during the fractionation procedure, which was blocked when BB94 was included in the cell lysate (data not shown). This suggests that transmembrane proteases, such as ADAM 10, may get increased access to the receptor phosphatase in the membrane fraction and, in this way, may lead to an increased rate of S2 cleavage (Fig. 4B). More importantly, the observed solubility of PIC indicates that the transmembrane region was lost in the course of PIC generation. These findings may suggest the involvement of γ-secretase-dependent intramembrane cleavage. In fact, peptide aldehydes like MG132 that have been reported to block γ-secretase activity, in addition to the proteasome, caused the accumulation of both PΔE and PIC in cells (Fig. 4A) (22). We therefore investigated whether the generation of PIC out of RPTPκ is dependent on the activity of PS, the catalytic subunit of the γ-secretase complex. To address this, we analyzed the processing of the endogenous phosphatase in embryonic fibroblasts derived from PS1/PS2 wild-type and knockout mice (Fig. 4C, left) (16). Indeed, PΔE accumulates in PS1−/− PS2−/− fibroblasts, whereas it becomes further processed in wild-type cells. Alternatively, the treatment of wild-type cells with the γ-secretase inhibitor DAPT led to PΔE accumulation. Notably, concomitant with the increase in PΔE, even in the presence of specific proteasomal inhibitors like lactacystin and epoxomycin, the generation of PIC was abolished upon interference with PS activity, i.e., in PS1−/− PS2−/− fibroblasts or in wild-type cells treated with DAPT (Fig. 4C, middle and right). Moreover, direct PS-dependent conversion of PΔE to PIC was observed with a previously established in vitro assay for PS-dependent ICD generation (23, 40; data not shown). RPTPκ S3 cleavage by γ-secretase also operates in invasive renal carcinoma cells as the addition of the γ-secretase-specific inhibitors DAPT and L-685458 resulted in accumulation of PΔE (Fig. 4D). Additionally, RPTPκ was transfected into HEK293 cells stably expressing wild-type PS1 or the dominant-negative mutant PS1 D385N (23). As expected, RPTPκ PΔE accumulated in cells expressing PS1 D385N (Fig. 4E). These results conclusively demonstrate the involvement of PS1-dependent γ-secretase in PΔE cleavage, resulting in the release of PIC from the membrane.
FIG. 4.
The S2 cleavage product PΔE is subject to PS1-dependent processing, leading to generation of PIC. (A) Specific inhibition of the proteasome results in accumulation of an additional RPTPκ-specific isoform (PIC) that is slightly smaller than PΔE. RPTPκ was immunoprecipitated from HEK293 cells treated with the peptide aldehyde MG132 (5 μM) and the specific proteasomal inhibitor lactacystin (Lac, 5 μM) for 16 h. Note the accumulation of PIC upon lactacystin treatment and of both PΔE and PIC upon incubation with MG132. For comparison, PΔE generation was induced by TFP. −, no treatment. (B) PIC is a soluble RPTPκ isoform. HEK293 cells were treated with or without (−) lactacystin (Lac) for 16 h, and cellular proteins were separated into membrane (P100) and cytosolic (S100) fractions. (C) Accumulation of the ADAM product PΔE and lack of PIC in the absence of PS (PIC is marked by asterisks). RPTPκ was immunoprecipitated from untreated (−) PS1+/+/PS2+/+ and PS1−/−/PS2−/− fibroblasts or PS1+/+/PS2+/+ fibroblasts treated (+) with the γ-secretase inhibitor DAPT (2 μM) for 16 h. Additionally, cells were incubated with 5 μM lactacystin (Lac, middle panel) or 5 μM epoxomycin (Epo, right panel) for 16 h. Note the accumulation of PΔE upon inhibition of PS activity (left) concomitantly with the lack of stabilized PIC (middle and right). (D) PS-mediated processing of PΔE in renal carcinoma cells. 768-O cells grown to confluence were treated with dimethyl sulfoxide or the γ-secretase inhibitors DAPT (2 μM) and L-685458 (5 μM) for 8 h. (E) HEK293 cells expressing wild-type PS1 or PS1 D385N were transiently transfected with HA-tagged RPTPκ. (F) PS-mediated cleavage of RPTPμ. PS1+/+PS2+/+ and PS1−/−PS2−/− fibroblasts were incubated with or without (−) epoxomycin (5 μM) for 12 h and endogenously expressed RPTPμ was immunoprecipitated. RPTPμ shows a characteristic precursor band at 200 kDa and its P subunit at 100 kDa (5). Note that the accumulation of PIC strictly depends on PS activity upon inhibition of the proteasome. Abbreviations: α, anti; B, blotting; IP, immunoprecipitation.
To test whether related RPTPs undergo PS-dependent intramembrane proteolysis, we analyzed the endogenously expressed MAM family member RPTPμ in PS1+/+ PS2+/+ and PS1−/− PS2−/− fibroblasts treated with and without epoxomycin. As shown in Fig. 4F, the absence of PS activity results in RPTPμ PΔE accumulation, whereas PS activity is required for the generation of RPTPμ PIC upon inhibition of the proteasome. Then does RPTPμ also become cleaved in a preceding proteolytic step? RPTPμ is processed at S2 in response to TFP treatment, yielding a product that corresponds to PΔE (data not shown). Together, our results demonstrate the regulation of members of the MAM family of RPTPs by γ-secretase-mediated intramembrane proteolysis.
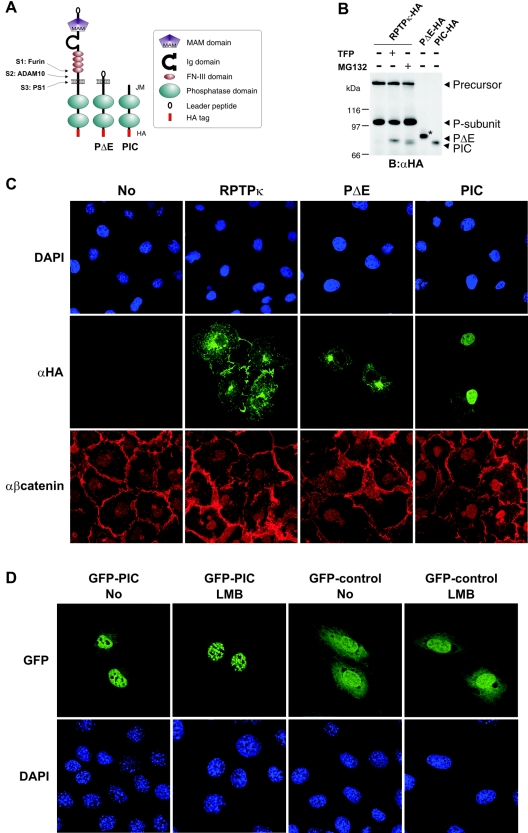
PIC is transported into the cell nucleus.
After having shown PS1-mediated conversion of RPTPκ PΔE to PIC, we sought to demonstrate a relationship between this cleavage step and the subcellular localization of both isoforms. To this end, we took advantage of recombinant PΔE and PIC. The PΔE construct codes for the cytoplasmic and transmembrane part plus a four-amino-acid extracellular stalk directly fused to the leader peptide (Fig. 5A). PIC corresponds to the intracellular portion plus two N-terminal residues (773 and 774) derived from the putative transmembrane region. When expressed in COS-7 cells, γ-secretase-derived PIC comigrated with the ectopically expressed version (Fig. 5B), which was detected by immunofluorescence predominantly in cell nuclei (Fig. 5C, right panel). Thus, once generated, soluble PIC can localize to cell nuclei. In contrast, RPTPκ-specific immunoreactivity was not observed in the nucleus upon the expression of RPTPκ and PΔE (Fig. 5C). This suggests that the relative amount of proteolysis-derived PIC was below the detection limit of our immunofluorescence analysis.
FIG. 5.
Exogenously expressed PIC localizes to the nucleus. (A) Schematic representation of ADAM-derived, transmembrane PΔE- and PS1-derived soluble PIC. All constructs used in this study were HA tagged at the C terminus. Ig, immunoglobulin. (B) Comparison of PS1-derived and recombinant PIC starting at residue 773 at the membrane-cytoplasm interface. COS-7 cells were transfected with HA-tagged RPTPκ and treated with (+) or without (−) TFP or MG132 to induce PΔE and PIC generation. Additionally, recombinant PΔE and PIC constructs were expressed in these cells. For comparison, only 1/5 of PΔE and 1/10 of PIC were transfected. The increase in size of recombinant PΔE (marked by an asterisk) is due to glycosylation (data not shown). B, blotting; α, anti. (C) COS-7 cells were transfected with the indicated HA-tagged RPTPκ isoforms, fixed and immunostained by using anti-HA antibody and AlexaFluor 488-labeled secondary antibody (green). Endogenous β-catenin was detected with a polyclonal antibody to β-catenin and AlexaFluor 546-labeled secondary antibody (red). Nuclear staining with DAPI (4′,6′-diamidino-2-phenylindole) is shown in the upper panels. Note the detection of PIC in the nucleus by confocal microscopy. Due to overexpression, transmembrane PΔE localizes predominantly in the endoplasmic reticulum and Golgi complex. α, anti. (D) Accumulation of PIC in nuclear bodies after the inhibition of CRM1-dependent nuclear export. NIH 3T3 cells were transfected with either GFP-PIC (left panels) or GFP control vector (right panels) and treated or not treated with leptomycin B (LMB) at 25 ng/ml for 3 h. After fixation, cells were stained with DAPI (4′,6′-diamidino-2-phenylindole) and observed with a Leica confocal microscope. Note the specific accumulation of GFP-PIC in nuclear bodies upon leptomycin B treatment.
We have previously found that the RPTPκ cytoplasmic part binds to β-catenin (11). β-Catenin is a multifunctional protein that combines the features of a structural component at sites of cell-cell contact with those of a transcriptional coactivator of LEF/TCF transcription factors (26). Tyrosine phosphorylation of β-catenin by EGF receptor (EGFR), ErbB2, c-Met, and Src can lead to a decrease of membane-bound β-catenin and, concomitantly, to an increase in the cytoplasmic pool (25, 36). In light of these reports and the fact that RPTPκ associates with β-catenin (Fig. 6A), we first analyzed the localization of β-catenin upon the overexpression of RPTPκ, PΔE, and PIC. As shown in Fig. 5C (lower panel), the localization of β-catenin is not markedly changed upon the overexpression of either RPTPκ isoform under basal conditions, i.e., without treatment of ligands that induce tyrosine phosphorylation of β-catenin.
To further examine nuclear localization of PIC, we fused GFP at the PIC N terminus (residue 773) and expressed it in NIH 3T3 fibroblasts. Again, although GFP localizes in either compartment, GFP-PIC was predominantly detected in the nucleus (Fig. 5D). Moreover, when the cells were treated with leptomycin B, an inhibitor of CRM1-dependent nuclear export (48), the nuclear/cytoplasmic ratio of GFP-PIC fluorescence further increased and the fusion protein accumulated in small granular structures inside the nucleus. In contrast, localization of control GFP was unaffected by the export inhibitor (Fig. 5D, right panel). This suggests that PIC has a leptomycin B-sensitive nuclear export signal sequence and that it is transported in and out of the nucleus. Future studies will elucidate whether these nuclear structures are so-called promyelocytic leukemia protein bodies (known also as nuclear body, nuclear domain 10, or promyelocytic leukemia protein oncogenic domains), which form granular structures of 250 to 500 nm in diameter and are present in the nuclei of most cells (2).
PIC dephosphorylates the coactivator β-catenin and increases TCF-mediated transcription.
The presence of two PTP domains in PIC and its nuclear localization raise an interesting question: is PIC a nuclear protein tyrosine phosphatase? Our investigations of the proteolytic processing suggest the existence of membrane-bound, cytoplasmic and nuclear pools of the RPTPκ-derived catalytic domains. As mentioned above, the RPTPκ-binding protein β-catenin can also be detected in these subcellular fractions and tyrosine phosphorylation of the protein has been causally linked to its transition from the membrane to the cytoplasm. However, tyrosine phosphorylation of β-catenin has also been implicated in modulation of its transcriptional coactivator activity by stimulating association with the basal transcription factor TATA-binding protein (TBP) in the nucleus (37). This report prompted us to analyze the effects of overexpression of RPTPκ, PΔE, and PIC on tyrosine phosphorylation of β-catenin. Although association of β-catenin with RPTPκ has been reported (11) (Fig. 6A), dephosphorylation of the protein by RPTPκ has not yet been demonstrated in the cellular context.
We therefore started our analysis by activating endogenous RPTPκ in cells and by analyzing the effects on β-catenin tyrosine phosphorylation. A major obstacle in the field of RPTP research is the lack of cognate ligands with the ability to modulate RPTP activity. The experiment shown in Fig. 2H addressed the induction of RPTPκ S2 cleavage by the affinity-purified anti-RPTPκEC antibody, which is directed against the extracellular portion of the phosphatase (Fig. 2D). Here we asked whether this antibody, possibly by inducing aggregation of the phosphatase at the cell surface, also increases its catalytic activity. EGF stimulation induces the association of EGFR with β-catenin in MDA-MB-468 breast carcinoma cells, resulting in the tyrosine phosphorylation of β-catenin (15). Although the treatment of these cells with anti-RPTPκEC antibody did not affect the phosphorylation of EGFR, it led to a marked decrease in the β-catenin tyrosine phosphorylation induced by EGFR (Fig. 6B). This observation suggests that anti-RPTPκEC antibody activates the phosphatase. This activation clearly correlated with the amount of antibody added to the cell medium (Fig. 6C). Conversely, silencing the expression of RPTPκ by siRNA increased EGF-induced phosphorylation of β-catenin (Fig. 6D). In contrast, treatment with TFP, an RPTPκ shedding inducer (Fig. 2E), decreased the EGF-induced phosphorylation of β-catenin (Fig. 6E). We interpret these results as an indication that the induction of S2 cleavage accompanies RPTP activation. In fact, all stimuli shown in Fig. 2, i.e., cell density (Fig. 2A and data not shown), TFP (Fig. 2E), and anti-RPTPκEC antibody treatment (Fig. 2H), lead to a reduction in β-catenin tyrosine phosphorylation.
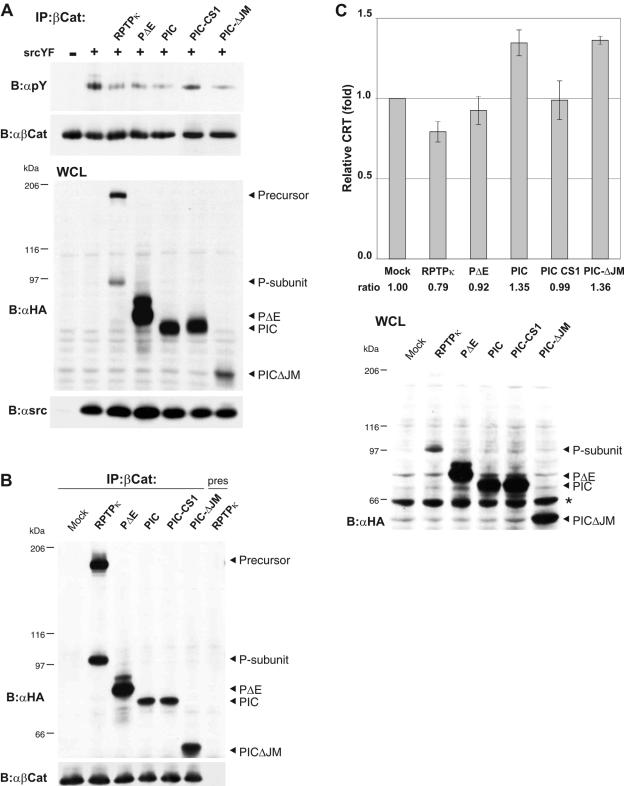
We next asked whether the catalytic activity of the membrane-proximal PTP domain accounts for the dephosphorylation of β-catenin. We therefore transfected HEK293 cells with a constitutive active mutant of Src (SrcYF) to induce phosphorylation of the protein (Fig. 6F, upper panel). This phosphorylation was reduced by coexpression of RPTPκ but not by a mutant version with a transition of the membrane-proximal catalytic cysteine to alanine. These results conclusively identify β-catenin as a target of RPTPκ in living cells and reveal the importance of the membrane-proximal PTP domain in the dephosphorylation reaction.
We then asked whether PIC expression also decreases Src-induced tyrosine phosphorylation of β-catenin or whether the isoforms differentially affect phosphorylation. We also included a catalytically inactive version of PIC, PIC-CS1 (inactivation of the membrane-proximal PTP domain), and a PIC deletion mutant that is devoid of the juxtamembrane part. This sequence connects the transmembrane region with the first PTP domain and has been implicated in binding to β-catenin (11). First, wild-type phosphatase, PΔE, and PIC dephosphorylated the protein with comparable efficiencies (Fig. 7A). Second, PIC-CS1 did not affect phosphorylation. Third, PICΔJM effectively dephosphorylated β-catenin under these conditions. Accordingly, RPTPκ, PΔE, PIC, PIC-CS1, and PICΔJM coprecipitate with β-catenin (Fig. 7B). The fact that PICΔJM still binds to β-catenin is surprising as it suggests that primary binding of β-catenin to RPTPκ may rely on the PTP domains. Collectively, we interpret these results as an indication that PIC participates in dephosphorylation of β-catenin and that the RPTPκ P subunit is an active phosphatase that does not require cleavage at S2 or S3 in order to become active. In line with these findings goes the observation that γ-secretase inhibitors did not affect tyrosine phosphorylation of β-catenin (data not shown).
FIG. 7.
PIC is an active protein tyrosine phosphatase that promotes β-catenin-mediated transcription. (A) Dephosphorylation of β-catenin by PIC. HEK293 cells were transfected with empty vector (−) or constitutive active Src (SrcYF) (+) to induce tyrosine phosphorylation of β-catenin. All RPTPκ constructs used here are HA tagged at the C terminus. RPTPκ, PΔE, PIC, PIC-CS1, and PICΔJM were cotransfected, and tyrosine phosphorylation of β-catenin was analyzed (upper panel). Transfection controls are shown below. PIC-CS1 is a catalytically inactive version; PICΔJM is devoid of the juxtamembrane sequence. (B) Coprecipitation of PIC and PICΔJM with β-catenin. HEK293 cells were transfected with either empty vector (mock) or the indicated HA-tagged constructs, and β-catenin was immunoprecipitated. Preserum was used as a control (right lane). RPTPκ-derived constructs were detected with anti-HA antibody. (C) PIC enhances transcriptional activation of β-catenin, whereas RPTPκ suppresses it. HCT116 cells were cotransfected with β-catenin/TCF reporter constructs and either respective RPTPκ plasmids or empty vector (Mock). PIC-CS1 is a catalytically inactive PIC version that harbors a cysteine-to-alanine transition in the proximal PTP domain. Luciferase activity was determined using the dual luciferase kit (Promega), and the data were normalized to the cotransfected cytomegalovirus Renilla plasmid. Error bars represent the standard deviations of triplicate assays. The expression of the RPTPκ constructs is shown in the lower panel (an unspecific bovine serum albumin signal is marked by an asterisk). Abbreviations: α, anti; B, blotting; IP, immunoprecipitation.
Because tyrosine phosphorylation of β-catenin has been shown to modulate TCF-mediated transcription in the nucleus, we expressed the same isoforms in HCT116 colon carcinoma cells. HCT116 cells are characterized by activated β-catenin signaling, due to a stabilizing mutation in β-catenin. As expected, the expression of full-length RPTPκ suppressed the activity of a reporter gene placed under the control of a β-catenin- and TCF-sensitive promoter (Fig. 7C). This is in accordance with our data showing that RPTPκ is a negative regulator of β-catenin tyrosine phosphorylation (Fig. 6). In contrast to this observation, the expression of PIC as well as PICΔJM promoted β-catenin's transcriptional activity. Hence, this effect depends on the catalytic activity of PIC's proximal PTP domain since PIC-CS1 does not increase TCF-mediated transcription (Fig. 7C). Although RPTPκ, PΔE, and PIC share the ability to dephosphorylate β-catenin, they have different effects on TCF-mediated transcription. This indicates that PIC may target additional regulatory proteins of the transcription regulatory complex. We propose that RPTPκ and PIC serve opposing functions in the regulation of TCF-mediated transcription and suggest that RPTP intramembrane proteolysis constitutes a switch in the regulation of RPTPκ signaling.
DISCUSSION
Nuclear localization of some nonreceptor PTPs, including TC45 and SHP-1, has been observed. Both PTPs have been implicated in the dephosphorylation of transcription factors or direct activators of such proteins. For instance, TC45, derived through alternative splicing of T-cell PTP, functions as the nuclear tyrosine phosphatase of signal transducers and activators of transcription (44). In contrast, SHP-1 may regulate the transcriptional coactivator function of β-catenin in intestinal epithelial cells (9). This suggestion is backed by the fact that the expression of TCF/LEF target genes like the cyclin D1 gene and c-myc is increased in the jejunum and colon derived from allelic viable moth-eaten (mev/mev) mice, whose SHP-1 activity is strongly compromised (9).
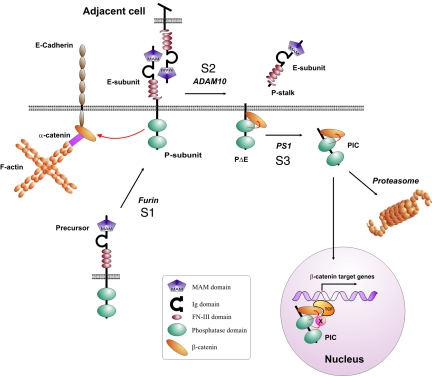
In this report, we suggest for the first time the nuclear localization of an RPTP. This is accomplished by sequential proteolytic processing by three proteases at three sites: S1, S2, and S3 (Fig. 8). While S1 cleavage by furin participates in RPTP maturation in the secretory pathway, cleavage at S2 by ADAM 10 results in the shedding of the phosphatase extracellular fragment. Finally, intramembrane proteolysis at S3 by γ-secretase releases PIC to the cytoplasm. In this proteolytic pathway, processing by ADAMs may play a major regulatory role. The ability of the anti-RPTPκEC antibody to activate S2 cleavage may suggest that trans-interactions between RPTPκ proteins that are expressed on adjacent cells lead to S2 processing (Fig. 8). Such a mechanism has been reported for ADAM 10-mediated processing of Notch and its ligand, Delta (24, 30), and ephrin-A2 and its receptor, Eph3A (14). A recent report from Saftig and colleagues further confirmed the role of ADAM 10 in the regulation of cell adhesion by demonstrating aggregate formation of ADAM 10-deficient cells, with increased N-cadherin-mediated adhesion being important (38). Moreover, ADAM 10 has been implicated in calcium signaling and the precursor of ADAM 10 has been found in association with calmodulin. Interestingly, calcium influx triggers the dissociation of calmodulin from ADAM 10, thereby leading to activation of the protease (31). We observed that the addition of calmodulin inhibitors, like TFP, induced S2 processing of RPTPκ. Accordingly, ADAM 10 is involved in RPTPκ processing induced by TFP. Moreover, ADAM 10 also mediates S2 cleavage in confluent fibroblast cultures, indicating that the activity of the protease is triggered by cell contact formation. This goes in line with the aforementioned reports. Notably, PMA failed to induce shedding of the phosphatase. An explanation for this unexpected finding could be that PMA selectively activates ADAM 17, a protease that is not involved in RPTPκ shedding in cells that have been used herein. Indeed, it is well accepted that different stimuli activate individual ADAMs. Because ADAM 10 has been implicated in cell density- and calcium-dependent shedding events, one cannot exclude the possibility that calcium signaling operates downstream of cell contact assembly.
FIG. 8.
Hypothetical model of the RPTP S1/S2/S3 cleavage mechanism and activation of β-catenin-mediated transcription. Furin-mediated cleavage constitutively yields two subunit RPTPκ proteins at the cell surface. Homophilic binding between RPTPκ proteins expressed in trans at high cell densities may induce S2 cleavage by ADAM 10, resulting in the release of the homophilic binding site into the cell supernatant. The remaining part, PΔE, is subject to γ-secretase/PS1-dependent intramembrane proteolysis that allows translocation of catalytically active PIC to the nucleus. Proteasomal destruction decreases the level of PIC in cells and may prohibit its nuclear import. PIC dephosphorylates the coactivator β-catenin and, unlike RPTPκ, increases TCF-mediated transcription. Note that PIC may additionally dephosphorylate transcriptional regulatory proteins associated with the β-catenin/TCF complex. Ig, immunoglobulin.
While there is an increased rate of cleavage at cell confluence, a higher rate of transcription of the RPTPκ gene has also been demonstrated (11). Consequently, higher RPTPκ protein levels have been observed at cell confluence (11). In accordance with this, RPTPκ accumulates at points of cell contact. The same has been reported for the MAM family member RPTPμ (12). Therefore, at high cell densities, the cleavage of MAM family RPTPs can be compensated easily by an increased rate of gene transcription. Our study suggests that S2 cleavage is part of a signaling, but also a degradation, pathway. Because MAM family RPTPs harbor cytoplasmic PTP domains, it is conceivable that the extracellular portions serve a sensor rather than a mechanical function in living tissues. If true, S2 cleavage would not affect the adhesive force between cells expressing RPTPκ in vivo; instead, it could lead to the relocalization of the PTP domains. Indeed, this study demonstrates that S2 cleavage forms the precondition for processing by γ-secretase and nuclear translocation of the PTP domains (see below).
The finding that TFP and anti-RPTPκEC antibody not only induce RPTPκ cleavage at S2 but also increase the catalytic activity of the phosphatase is interesting. Thus, there is a correlation between RPTPκ S2 cleavage and activation of the enzyme. However, direct inhibition of RPTP processing does not seem to interfere with RPTP activation (data not shown). Moreover, when overexpressed, the RPTPκ precursor, PΔE, and PIC dephosphorylated β-catenin equally well, indicating that the precursor is indeed an active phosphatase. It is therefore likely that cleavage accompanies RPTP activation, but cleavage does not seem to be causally related to it. Notably, TFP is the first compound identified so far that activates endogenous tyrosine phosphatase activity in cancer cells, thereby inhibiting tyrosine kinase-mediated signaling.
An increasing number of type I transmembrane proteins have been identified as substrates for γ-secretase-dependent intramembrane cleavage after ectodomain shedding, including Notch, APP, CD44, ErbB4, low-density lipoprotein receptor-related protein, and cadherins (20, 21). Some liberated ICDs translocate to the nucleus. For instance, the liberated Notch ICD interacts with the CSL (C promoter-binding factor) transcription factor and converts it from a transcriptional repressor to an activator, resulting in the expression of Notch target genes (21). The intracellular fragments of CD44 and ErbB4 have also been detected in the nucleus. Moreover, CD44 ICD can potentiate transactivation induced by the transcriptional coactivator CBP/p300 (33). In contrast, the nuclear function of ErbB4 remains undefined (32). Other γ-secretase substrates, like APP, cadherins, or Syndecan-3, have not been demonstrated to function in the nucleus. For example, the N-cadherin ICD binds to the transcription factor CBP in the cytoplasm to promote its proteasomal degradation, thereby inhibiting CRE-dependent transcription (28).
Our results reveal that PIC is a low-abundance protein and that its accumulation seems to be tightly regulated by both γ-secretase activity and proteasomal destruction. It should be noted at this point that the Notch ICD is also quite labile but plays an important role as transcriptional coactivator in the nucleus. In fact, the Notch ICD is hardly detectable from living tissues on Western blots (21). In our experiments, very minor amounts of endogenous PIC can indeed be detected with anti-RPTPκJM antibody in high-cell-density cultures without proteasomal inhibitors, but we noted that PIC accumulation was not always reproducible in these experiments (data not shown). Notably, the detection of PIC upon the overexpression of the RPTPκ precursor may simplify this issue. γ-Secretase has been implicated in maintaining membrane protein homeostasis, and the possibility that, in the membrane fraction, γ-secretase is analogous to the intracellular proteasome has been discussed. However, in most if not all cases, γ-secretase-mediated cleavage actually forms the precondition for the action of the proteasome, even for those ICDs for which a signaling function has been assigned (see above).
We interpret our data as an indication that the low abundance and the short half life of PIC in cells serves important regulatory roles. For example, at a high cell density, the activation of ADAM 10 may lead to increased PΔE generation, which concomitantly results in higher levels of PIC generated by γ-secretase. If we assume that proteasomal activity is not affected by cell contact formation, increased PIC levels may facilitate its import into the nucleus. Although we showed nuclear staining of recombinant overexpressed PIC, specificity of the nuclear localization was demonstrated by the inhibition of a CRM1-dependent export. Under these conditions, the intensity of PIC nuclear staining further increased and the fragment became localized to nuclear bodies. Such granular domains have been implicated in gene regulation, DNA repair, tumor suppression, apoptosis, and proteolysis (2). Given the fact that endogenous PIC levels are low, visualization of the endogenous fragment with anti-RPTPκJM antibody in the nucleus is difficult, even upon inhibition of the proteasome. A simple explanation could be that anti-RPTPκJM antibody does not allow the detection of endogenous PIC. Although the abundance of nuclear PIC is low, it might be sufficient to fulfill a signaling function as transcriptional activator.
Contrary to all of the previously identified intracellular products generated by γ-secretase cleavage, PIC is an active protein tyrosine phosphatase. Our findings implicate RPTPκ and PIC in the dephosphorylation of β-catenin (Fig. 8). Although the role of β-catenin tyrosine phosphorylation in cell adhesion and cellular signaling is not yet clearly defined, it is assumed that the phosphorylation of E-cadherin-associated β-catenin by activated growth factor receptor tyrosine kinases and Src contributes to the disassembly of adherens junctions (35). In contrast, the activation of RPTPκ may lead to an accumulation of β-catenin at adherens junctions and, concomitantly, to a decrease in the cytoplasmic pool. Consequently, less cytoplasmic β-catenin may be available for nuclear translocation and activation of TCF-mediated transcription. The data shown in Fig. 7C may thus suggest that RPTPκ preferentially targets the cytoplasmic pool of β-catenin, causing its translocation to the membrane.
Recently, β-catenin tyrosine phosphorylation has been directly linked to the transcription of TCF target genes: phosphorylation of the protein may increase its association with the basal transcription factor TBP, thereby activating TCF-4-dependent transcription (37). For instance, in SW480 cells, a β-catenin phosphorylation mimic mutant (Tyr-654-Glu) showed increased binding to TBP compared to that of the wild-type protein. Accordingly, β-catenin Tyr-654-Glu increased TCF-4-mediated transcription to a higher extent in a gene reporter assay (approximately 200% in SW-480 cells and 100% in NIH 3T3 and MiaPaCa-2 cells). Along this line goes the observation that the protein tyrosine phosphatase SHP-1, as mentioned above, decreases the transcriptional activity from TCF-sensitive dihydrofolate reductase, c-myc, and cyclin D1 promoters by about 50% (9).
Although both RPTPκ and PIC isoforms dephosphorylated β-catenin with comparable efficiencies, they differentially modulated TCF-mediated transcription. The reason for this is unknown, but it is clear that the ability of PIC to dephosphorylate the transcriptional coactivator β-catenin cannot solely account for the observed increase in TCF-mediated transcription. Furthermore, whereas PIC was localized to granular nuclear structures, β-catenin was not (data not shown). This indicates that the function of PIC in these domains is independent from β-catenin. On the other hand, the deletion of PIC's juxtamembrane region does not impair either β-catenin dephosphorylation or TCF activation. Our data do not exclude the possibility that PIC has access to additional transcriptional regulators that are associated with the β-catenin/TCF complex (Fig. 8). Hence, the dephosphorylation of β-catenin-associated factors by PIC, but not by full-length RPTPκ, may account for the contrasting effects on TCF-mediated transcription. It is well accepted that additional factors are required to boost β-catenin's function as a transcriptional coactivator, and β-catenin does not contain the enzymatic activities that are necessary for modification of the chromatin structure. Instead, β-catenin recruits chromatin-modifying enzymes, such as p300/CBP and Brg-1 as well as other regulatory proteins, like pygopus, TIP49, and TBP, which then allow the recruitment of the basal transcription machinery (17). Given the fact that most of these proteins work in concert to tightly regulate TCF activation and that they are physically associated with each other, it is indeed possible that PIC targets components of this complex independently from β-catenin. This hypothesis is backed by the observation that PIC increases TCF-mediated transcription, while SHP-1, presumably by dephosphorylating nuclear β-catenin at tyrosine 654, decreases it. Furthermore, the mild effects of PIC overexpression on TCF activation (approximately 30%) may be explained, at least in part, by the absence of a particular factor that acts at the coactivator level and is required for efficient transcriptional induction by PIC. Although further studies are needed to clarify the regulation of TCF transcriptional activity by tyrosine phosphorylation in the nucleus, our study provides fundamental insights into the regulation of RPTPs by proteolytic processing. It will be interesting to analyze the contribution of these enzymes to biological processes that are modulated by ADAMs and γ-secretase, ranging from tumor growth to neurodegeneration.
Supplementary Material
Acknowledgments
We thank Stefan Hart for providing siRNAs directed against ADAM 10, 15, 17 and anti-ADAM 15 antibody, Bart De Strooper for PS knockout cells, Bert Vogelstein for pGL3-OT and pGL3-OF reporter constructs, and Markus Schmid for anti-RPTPμJM antibody. We also thank Ann-Katrin Ludwig, Uta Eichelsbacher, and Renate Gautsch for excellent technical assistance and Pjotr Knyazev and Sushil Kumar for critical readings of the manuscript.
This work was supported by a grant from the K. L. Weigand'sche Stiftung to S.L., Deutsche Forschungsgemeinshaft SFB415/TP-B9 to P.S., Interuniversity Attraction Poles Program P5/19 of the Belgian Federal Science Policy Office, and the European Union (APOPIS: LSHM-CT-2003-503330) to P.S. and D.H.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aicher, B., M. M. Lerch, T. Muller, J. Schilling, and A. Ullrich. 1997. Cellular redistribution of protein tyrosine phosphatases LAR and PTPσ by inducible proteolytic processing. J. Cell Biol. 138:681-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borden, K. L. B. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradykalnay, S. M., A. J. Flint, and N. K. Tonks. 1993. Homophilic binding of PTP mu, a receptor-type protein tyrosine phosphatase, can mediate cell-cell aggregation. J. Cell Biol. 122:961-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradykalnay, S. M., and N. K. Tonks. 1995. Protein tyrosine phosphatases as adhesion receptors. Curr. Opin. Cell Biol. 7:650-657. [DOI] [PubMed] [Google Scholar]
- 5.Campan, M., M. Yoshizumi, N. G. Seidah, M. E. Lee, C. Bianchi, and E. Haber. 1996. Increased proteolytic processing of protein tyrosine phosphatase μ in confluent vascular endothelial cells—the role of pc5, a member of the subtilisin family. Biochemistry 35:3797-3802. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, J., K. Wu, M. Armanini, N. Orourke, D. Dowbenko, and L. A. Lasky. 1997. A novel protein-tyrosine phosphatase related to the homotypically adhering κ and μ receptors. J. Biol. Chem. 272:7264-7277. [DOI] [PubMed] [Google Scholar]
- 7.De Strooper, B. 2003. Aph-1, Pen-2, and Nicastrin with presenilin generate an active gamma-Secretase complex. Neuron 38:9-12. [DOI] [PubMed] [Google Scholar]
- 8.Diaz-Rodriguez, E., A. Esparis-Ogando, J. C. Montero, L. Yuste, and A. Pandiella. 2000. Stimulation of cleavage of membrane proteins by calmodulin inhibitors. Biochem. J. 346:359-367. [PMC free article] [PubMed] [Google Scholar]
- 9.Duchesne, C., S. Charland, C. Asselin, C. Nahmias, and N. Rivard. 2003. Negative regulation of β-catenin signaling by tyrosine phosphatase SHP-1 in intestinal epithelial cells. J. Biol. Chem. 278:14274-14283. [DOI] [PubMed] [Google Scholar]
- 10.Fischer, O. M., S. Hart, A. Gschwind, N. Prenzel, and A. Ullrich. 2004. Oxidative and osmotic stress signaling in tumor cells is mediated by ADAM proteases and heparin-binding epidermal growth factor. Mol. Cell. Biol. 24:5172-5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs, M., T. Muller, M. M. Lerch, and A. Ullrich. 1996. Association of human protein tyrosine phosphatase κ with members of the armadillo family. J. Biol. Chem. 271:16712-16719. [DOI] [PubMed] [Google Scholar]
- 12.Gebbink, M., G. C. M. Zondag, G. M. Koningstein, E. Feiken, R. W. Wubbolts, and W. H. Moolenaar. 1995. Cell surface expression of receptor protein tyrosine phosphatase RPTP μ is regulated by cell-cell contact. J. Cell Biol. 131:251-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haass, C. 2004. Take five—BACE and the γ-secretase quartet conduct Alzheimer's amyloid β-peptide generation. EMBO J. 23:483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hattori, M., M. Osterfield, and J. G. Flanagan. 2000. Regulated cleavage of a contact-mediated axon repellent. Science 289:1360-1365. [DOI] [PubMed] [Google Scholar]
- 15.Hazan, R. B., and L. Norton. 1998. The epidermal growth factor receptor modulates the interaction of E-cadherin with the actin cytoskeleton. J. Biol. Chem. 273:9078-9084. [DOI] [PubMed] [Google Scholar]
- 16.Herreman, A., D. Hartmann, W. Annaert, P. Saftig, K. Craessaerts, L. Serneels, L. Umans, V. Schrijvers, F. Checler, H. Vanderstichele, V. Baekelandt, R. Dressel, P. Cupers, D. Huylebroeck, A. Zwijsen, F. Van Leuven, and B. De Strooper. 1999. Presenilin 2 deficiency causes a mild pulmonary phenotype and no changes in amyloid precursor protein processing but enhances the embryonic lethal phenotype of presenilin 1 deficiency. Proc. Natl. Acad. Sci. USA 96:11872-11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurlstone, A., and H. Clevers. 2002. T-cell factors: turn-ons and turn-offs. EMBO J. 21:2303-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, Y., P., H. Wang, P. D'Eustachio, J. M. Musacchio, J. Schlessinger, and J. Sap. 1993. Cloning and characterization of RPTPκ, a new member of the receptor protein tyrosine phosphatase family with a proteolytically cleaved cellular adhesion molecule-like extracellular region. Mol. Cell. Biol. 13:2942-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klarlund, J., K. 1985. Transformation of cells by an inhibitor of phosphatases acting on phosphotyrosine in proteins. Cell 41:707-717. [DOI] [PubMed] [Google Scholar]
- 20.Koo, E. H., and R. Kopan. 2004. Potential role of presenilin-1 regulated signaling pathways in sporadic neurodegeneration. Nat. Med. 10(Suppl.):S26-S33. [DOI] [PubMed] [Google Scholar]
- 21.Kopan, R., and M. X. Ilagan. 2004. Gamma-secretase: proteasome of the membrane? Nat. Rev. Mol. Cell Biol. 5:499-504. [DOI] [PubMed] [Google Scholar]
- 22.Kopan, R., E. H. Schroeter, H. Weintraub, and J. S. Nye. 1996. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl. Acad. Sci. USA 93:1683-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lammich, S., M. Okochi, M. Takeda, C. Kaether, A. Capell, A. K. Zimmer, D. Edbauer, J. Walter, H. Steiner, and C. Haass. 2002. Presenilin-dependent intramembrane proteolysis of CD44 leads to the liberation of its intracellular domain and the secretion of an Aβ-like peptide. J. Biol. Chem. 277:44754-44759. [DOI] [PubMed] [Google Scholar]
- 24.Lieber, T., S. Kidd, and M. W. Young. 2002. Kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 16:209-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lilien, J., J. Balsamo, C. Arregui, and G. Xu. 2002. Turn-off, drop-out: functional state switching of cadherins. Dev. Dyn. 224:18-29. [DOI] [PubMed] [Google Scholar]
- 26.Logan, C. Y., and R. Nusse. 2004. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20:781-810. [DOI] [PubMed] [Google Scholar]
- 27.Logeat, F., C. Bessia, C. Brou, O. Lebail, S. Jarriault, N. G. Seidah, and A. Israel. 1998. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 95:8108-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marambaud, P., P. H. Wen, A. Dutt, J. Shioi, A. Takashima, R. Siman, and N. K. Robakis. 2003. A CBP binding transcriptional repressor produced by the PS1/ɛ-cleavage of N-cadherin is inhibited by PS1 FAD mutations. Cell 114:635-645. [DOI] [PubMed] [Google Scholar]
- 29.McAndrew, P. E., A. Frostholm, R. A. White, A. Rotter, and A. H. M. Burghes. 1998. Identification and characterization of RPTP rho, a novel RPTP mu/kappa-like receptor protein tyrosine phosphatase whose expression is restricted to the central nervous system. Mol. Brain Res. 56:9-21. [DOI] [PubMed] [Google Scholar]
- 30.Mumm, J. S., E. H. Schroeter, M. T. Saxena, A. Griesemer, X. L. Tian, D. J. Pan, W. J. Ray, and R. Kopan. 2000. A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol. Cell 5:197-206. [DOI] [PubMed] [Google Scholar]
- 31.Nagana, O., and H. Saya. 2004. Mechanism and biological significance of CD44 cleavage. Cancer Sci. 95:930-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni, C. Y., M. P. Murphy, T. E. Golde, and G. Carpenter. 2001. γ-Secretase cleavage and nuclear localization of Erb4 receptor tyrosine kinase. Science 294:2179-2181. [DOI] [PubMed] [Google Scholar]
- 33.Okamoto, I., Y. Kawano, D. Murakami, T. Sasayama, N. Araki, T. Miki, A. J. Wong, and H. Saya. 2001. Proteolytic release of CD44 intracellular domain and its role in the CD44 signaling pathway. J. Cell Biol. 155:755-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Östman, A., Q. Yang, and N. K. Tonks. 1994. Expression of DEP-1, a receptor-like protein-tyrosine-phosphatase, is enhanced with increasing cell density. Proc. Natl. Acad. Sci. USA 91:9680-9684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pallen, C. J., and P. H. Tong. 1991. Elevation of membrane tyrosine phosphatase activity in density-dependent growth-arrested fibroblasts. Proc. Natl. Acad. Sci. USA 88:6996-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Moreno, M., C. Jamora, and E. Fuchs. 2003. Sticky business: orchestrating cellular signals at adherens junctions. Cell 112:533-548. [DOI] [PubMed] [Google Scholar]
- 37.Piedra, J., D. Martinez, J. Castano, S. Miravet, M. Dunach, and A. G. de Herreros. 2001. Regulation of β-catenin structure and activity by tyrosine phosphorylation. J. Biol. Chem. 276:20436-20443. [DOI] [PubMed] [Google Scholar]
- 38.Reiss, K., T. Maretzky, A. Ludwig, T. Tousseyn, B. de Strooper, D. Hartmann, and P. Saftig. 2005. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and β-catenin nuclear signalling. EMBO J. 24:742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sap, J., Y. P. Jiang, D. Friedlander, M. Grumet, and J. Schlessinger. 1994. Receptor tyrosine phosphatase R-PTP-κ mediates homophilic binding. Mol. Cell. Biol. 14:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sastre, M., H. Steiner, K. Fuchs, A. Capell, G. Multhaup, M. M. Condron, D. B. Teplow, and C. Haass. 2001. Presenilin-dependent γ-secretase processing of β-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep. 2:835-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seals, D. F., and S. A. Courtneidge. 2003. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 17:7-30. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi, S., K. Kasai, K. Hatsuzawa, N. Kitamura, Y. Misumi, Y. Ikehara, K. Murakami, and K. Nakayama. 1993. A mutation of furin causes the lack of precursor-processing activity in human colon carcinoma LoVo cells. Biochem. Biophys. Res. Commun. 195:1019-1026. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi, S., T. Nakagawa, K. Kasai, T. Banno, S. J. Duguay, W. J. M. Van de Ven, K. Murakami, and K. Nakayama. 1995. A second mutant allele of furin in the processing-incompetent cell line, LoVo. J. Biol. Chem. 270:26565-26569. [DOI] [PubMed] [Google Scholar]
- 44.ten Hoeve, J. M. de Jesus Ibarra-Sanchez, Y. Fu, W. Zhu, M. Tremblay, M. David, and K. Shuai. 2002. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol. Cell. Biol. 22:5662-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tonks, N. K., and B. G. Neel. 2001. Combinatorial control of the specificity of protein tyrosine phosphatases. Curr. Opin. Cell Biol. 13:182-195. [DOI] [PubMed] [Google Scholar]
- 46.Wang, H., Z. Lian, M. M. Lerch, Z. Chen, W. Xie, and A. Ullrich. 1996. Characterization of pcp-2, a novel receptor protein tyrosine phosphatase of the mam domain family. Oncogene 12:2555-2562. [PubMed] [Google Scholar]
- 47.Watanabe, H., R. Takahashi, Q. K. Tran, K. Takeuchi, K. Kosuge, H. Satoh, A. Uehara, H. Terada, H. Hayashi, R. Ohno, and K. Ohashi. 1999. Increased cytosolic Ca2+ concentration in endothelial cells by calmodulin antagonists. Biochem. Biophys. Res. Commun. 265:697-702. [DOI] [PubMed] [Google Scholar]
- 48.Zheng, B., Y. B. Ma, R. S. Ostrom, C. Lavoie, G. N. Gill, P. A. Insel, X. Y. Huang, and M. G. Farquhar. 2001. RGS-PX1, a GAP for GalphaS and sorting nexin in vesicular trafficking. Science 294:1939-1942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.