Abstract
Hyperactivation of Ras is one of the most common abnormalities in acute myeloid leukemia. In experimental models, Ras inhibits myeloid differentiation, which is characteristic of leukemia; however, the mechanism through which it disrupts hematopoiesis is poorly understood. In multipotent FDCP-mix cells, Ras inhibits terminal neutrophil differentiation, thereby indefinitely extending their proliferative potential. Ras also strongly promotes the sensitivity of these cells to granulocyte-macrophage colony-stimulating factor (GM-CSF). Using this model, we have dissected the signaling elements downstream of Ras to determine their relative contribution to the dysregulation of hematopoiesis. Cells expressing Ras mutants selectively activating Raf (Ras*T35S) or phosphatidylinositol 3-kinase (Ras*Y40C) did not significantly affect differentiation or proliferative capacity, whereas Ras*E37G (which selectively activates RalGEFs) perpetuated proliferation and blocked neutrophil development in a manner similar to that of Ras. Correspondingly, expression of constitutively active versions of these effectors confirmed the overriding importance of Ral guanine nucleotide exchange factors. Cells expressing Ras demonstrated hyperactivation of Ral, which itself was able to exactly mimic the phenotype of Ras, including hypersensitivity to GM-CSF. Conversely, dominant negative Ral promoted spontaneous neutrophil development. Ral, in turn, appears to influence differentiation through multiple effectors. These data show, for the first time, the importance of Ral in regulating differentiation and self-renewal in hematopoietic cells.
The 21-kDa guanine nucleotide-binding Ras proteins function as membrane-bound molecular switches linking the transduction of extracellular signals, via receptor and nonreceptor tyrosine kinases, to downstream effectors regulating diverse processes such as differentiation, proliferation, and cell cycle progression (7). Direct activation of Ras via point mutation is among the most common molecular abnormalities in hematopoietic malignancy, with an incidence of around 30% in myeloid leukemia and preleukemia (9, 25, 40, 58). Hyperactivation of Ras in leukemia also arises as a result of constitutive upstream signals arising from other common abnormalities such as those involving the Flt3 receptor (22, 46) and the Bcr-Abl fusion (24, 90). Conversely, Ras also becomes constitutively active as a result of loss of GTPase-activating protein (GAP) activity as seen in neurofibromatosis patients, who lack Nf1 Ras-GAP expression. These patients are at high risk of developing juvenile myelomonocytic leukemia (JMML) (65). Ras mutations also arise at high frequency in the preleukemic condition known as myelodysplastic syndrome (68), characterized by disorders of development resulting in hyperplastic marrow and peripheral blood cytopenia. The association of hyperactive Ras with both leukemia and preleukemia suggests that Ras signaling plays an important role during hematopoietic dysregulation and leukemic transformation.
Activated Ras genes are most commonly associated with promotion of proliferation of a variety of cell types. However, in the context of myeloid leukemia it has long been established that there are no differences in the proliferative capacity of leukemic cells compared to their normal counterparts. The accumulation of malignant cells is instead due to the failure of cells to undergo terminal differentiation and apoptosis (5). There is evidence from both in vitro and in vivo models that Ras influences the development of monocytic, erythroid, and granulocytic cells. This is substantiated by observations of primary mouse and human cells, where Ras appears to promote monocytic differentiation (10, 21), as well as to oppose the differentiation of erythroid progenitors (17, 88). In addition, neutrophilic leukocytosis has been observed with K-Ras transgenic mice (12), implying that Ras also affects the development of granulocytic cells.
Another commonly observed effect of Ras on myeloid hematopoietic cells is the capacity to reduce the requirements for exogenous growth factors (55, 73). This may arise in part from the fact that many hematopoietic cytokine receptors act through Ras, as previously reported for granulocyte-colony-stimulating factor (G-CSF), granulocyte-macrophage CSF (GM-CSF), and interleukin 3 (IL-3) receptors (18, 39, 72). Cytokines are known to influence the development of hematopoietic cells (37), with dysregulated cytokine signaling identified as a common feature of acute myeloid leukemia, ranging from mutational activation of growth factor receptor, as exemplified by the Flt3 receptor (28, 59, 89), to hyperresponsiveness to cytokine, such as the enhanced response to GM-CSF seen with JMML (23).
We have previously studied the effects of Ras on the multipotent factor-dependent cell line, FDCP-mix. These cells possess a nonleukemic phenotype, have a normal karyotype, are factor dependent, and can be directed to undergo multilineage development in response to cytokines (77). By changing growth conditions (serum and cytokines), it is possible to grow this cell line under self-renewing conditions, such that they retain their multipotent state; alternatively, they may be directed to differentiate to form erythroid or myeloid cells. In these cells, Ras selectively inhibits granulocytic differentiation of neutrophils, giving rise to continued proliferation of these partly differentiated cells. In addition, Ras also selectively increases the cells' sensitivity to GM-CSF, suggesting a link between the heightened response to this cytokine and an inhibition of differentiation (16). This phenotype therefore closely resembles that seen with JMML, which also commonly arises from hyperactivation of Ras (58).
As described above, the effects of Ras on myeloid development have been well documented; however, the mechanism by which it elicits these changes is poorly understood. Ras interacts with multiple downstream effectors via a highly conserved amino-terminal loop domain (switch I), encompassing amino acid residues 32 to 40 (83). The three best-established effector families are the Raf serine-threonine kinases, phosphatidylinositol 3-kinases (PI3-kinases), and the Ral guanine nucleotide exchange factor (RalGEF) family (54). In the context of myelopoiesis, activation of the mitogen-activated protein kinase (MAPK) cascade via Raf has been implicated in both disruption of differentiation and changes in cytokine dependency (20, 21, 88). PI3-kinase activity has been linked with both differentiation and inhibition of apoptosis (14, 20, 50) with demonstration of the synergistic requirement for Raf-mediated activation in leukemogenesis (6). Comparatively little has been reported regarding the role of RalGEFs in hematopoiesis, although they have been associated with both Jak/STAT signaling and the promotion of survival following growth factor withdrawal (33, 55). This family of effectors acts as GEFs for the Ral GTPases. Two highly similar Ral proteins, RalA and RalB, associate with a variety of downstream effectors (for a review, see reference 26): Sec5 and Exo84 during formation of the Exocyst complex, which participates in delivering vesicles to the plasma membrane (60, 61); the actin-binding protein, filamin (67); the ZO-1-associated nucleic acid-binding protein (designated ZONAB) (29), and Ral-binding protein 1 (RalBP1), which mediates GAP activity towards the Rho family GTPases Rac and Cdc42 (27).
Here, we show for the first time that Ral is a critical developmental regulator in hematopoietic cells. Ras promotes the activity of this GTPase through RalGEF family members. Further, activation of Ral is both necessary and sufficient to mimic the inhibition of neutrophil development imposed by Ras and is also responsible for promoting the sensitivity of these cells to GM-CSF. The data demonstrate an as-yet-unidentified role for Ral in controlling hematopoietic differentiation.
MATERIALS AND METHODS
Cell culture.
Culturing of FDCP-mix (clone A4) cells (77) under self-renewing conditions was carried out in Iscove's modified Dulbecco's medium (Sigma) supplemented with 20% batch-tested horse serum (GibcoBRL) and 10% WEHI-3B conditioned medium and maintained at 37°C with 5% CO2. Under these conditions, >95% of cells exhibited an immature blast cell morphology. Analysis of the effect of ectopically expressed genes on differentiation was carried out as follows. Mid-logarithmic-phase cells were harvested, washed in Hanks buffered saline solution containing 25 mM HEPES buffer (GibcoBRL), and resuspended to 2 × 104 cells/ml in 5 ml of differentiation medium: Iscove's modified Dulbecco's medium containing 20% batch-tested fetal calf serum, 1,000-U/ml human G-CSF, and 50-U/ml mouse GM-CSF (R&D Systems). Cells were subcultured if necessary in the same medium. Automated May-Grunwald-Giemsa staining (Bayer) of cytospins (20,000 cells) was carried out at day 7 (or as indicated), and morphological assessments were made under blind conditions. Cell proliferation was determined by viable cell counts as assessed by trypan blue exclusion, as previously described (4).
Sensitivity to cytokines and inhibitors.
To assess cytokine sensitivity, mid-logarithmic-phase cells were harvested and washed twice in 20 ml Hanks buffered saline solution. These were resuspended to 2 × 104 cells/ml in 5 ml differentiation medium (as above but excluding cytokines). Recombinant mouse IL-3 (R&D Systems) at a final specific activity of 0 to 1,000 U/ml, human G-CSF (0 to 10,000 U/ml), or mouse GM-CSF (0 to 500 U/ml) was added. GM-CSF hyperresponsiveness was assessed in the presence of constant G-CSF (50 U/ml), since GM-CSF alone did not support the viability of these cells. Cumulative proliferative capacity was determined by cell counts at day 5. The effect of the Rac1 inhibitor (NSC23766; Calbiochem) was assessed by addition at the start of the 7-day differentiation assay. The effect of PI3-kinase and MEK inhibitors was studied on FDCP-mix Ras* cells precultured for 7 days under differentiating conditions. These were exposed to LY294002 or PD98059 (Sigma) or U0126 (Cell Signaling Technology). Morphological and proliferative end points were scored 3 days later as described above.
cDNA constructs.
Ras effector loop domain mutants and PI3-kinase* (K227E) (both provided by Julian Downward, London, United Kingdom) were expressed in pLXSN. The c-Raf1 catalytic domain was expressed in the pM5 retroviral vector (56). RalGEFs (Michael A. White, Dallas, Tex., and Johannes L. Bos, The Netherlands) and constitutively active and inhibitory RalA constructs (Chris J. Marshall, London and David Wynford-Thomas, Cardiff, Wales) were all subcloned into the retroviral expression vector pBABE-puro. RalB, RalA Q72L, RalAQ72L ΔCAAX, and the RalA effector loop domain mutant (RalAQ72L D49E, RalAQ72L D49N, and RalAQ72L ΔN11) constructs, all in pBABE-puro, were kindly provided by Christopher Counter (Duke University Medical Center).
Retrovirus infection.
FDCP-mix-Ras* and neomycin control lines were created as previously described (16). The remaining lines were created via the generation of replication-defective amphotropic retrovirus by transient transfection of the Phoenix-Ampho packaging line (American Type Culture Collection). This was subsequently used to infect the parental FDCP-mix line as described below. Non-tissue-culture 24-well plates were treated with 15 μg of RetroNectin (TaKaRa Biochemicals) per well and incubated at room temperature for 2 h. RetroNectin was replaced by 1% bovine serum albumin, and incubation was carried out for 30 min for blocking. The blocking solution was replaced by 400 μl of retroviral supernatant together with 105 cells at semilogarithmic growth phase in 1.5 ml of culture medium. This was incubated overnight at 37°C with 5% CO2, after which the infection procedure was repeated using fresh retroviral supernatant. Following infection, antibiotic selection was carried out in the presence of puromycin (2 μg/ml) or neomycin (800 μg/ml).
Activation assays and Western blotting.
Cells were lysed in 50 mM Tris (pH 7.4), 1% NP-40, 15% glycerol, 200 mM NaCl, 5 mM MgCl2, 5 mM NaF, 1 μM leupeptin, 0.1 μM aprotinin, and 1 mM phenylmethylsulfonyl fluoride. Detergent-insoluble material was removed by centrifugation (16,000 × g at 4°C for 20 min). Activation sensitive pull-down assays were carried out using the binding domains of RalBP1 (amino acids 397 to 518) and PAKα (amino acids 1 to 252) (both were kindly provided by Chris J. Marshall, London, United Kingdom), which specifically interact with active GTP-bound forms of Ral and Rac/Cdc42 proteins, respectively, essentially as described previously (71, 86). Glutathione S-transferase-binding domain-coupled beads were prepared by mixing at 4°C for 1 h, while the pull-down was carried out by mixing with the whole-cell lysate for 45 min. GTP proteins were eluted using 2× reducing sample buffer, boiled for 5 min at 95°C, and assessed by Western blotting (12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis). Comparative total protein content was determined by parallel Western blotting of the whole-cell lysates. Immunoprobing was carried out using the following antibodies: Ras G12V-sensitive monoclonal (Calbiochem), pMek1/2 (#2354), pErk1/2 (#4695), and pAkt/PKB (#9271), all from Cell Signaling Technology and RalA (BD Transduction Laboratories), Rac1 (Upstate Biotechnology), and Cdc42 (Santa Cruz Biotechnology).
RESULTS
Activation of Ral guanine nucleotide dissociation stimulator (RalGDS) mediates Ras-induced inhibition of differentiation.
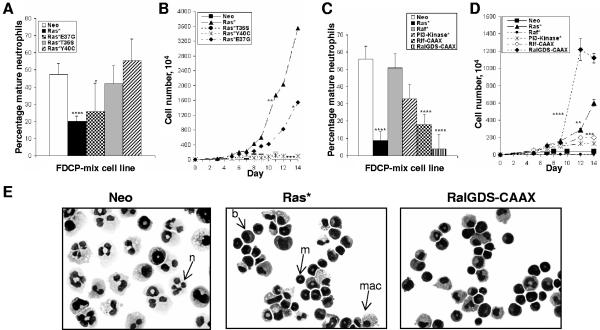
FDCP-mix cells undergo either self-renewal or differentiation, depending on the presence of hematopoietic growth factors. In the presence of G-CSF plus GM-CSF, these cells undergo predominantly neutrophil development, culminating in terminal differentiation and growth arrest. Expression of mutant Ras (Ras*) strongly inhibits the capacity of these cells to undergo terminal neutrophil development under these conditions (Fig. 1A and E). To determine which downstream targets of Ras influenced the development of these cells, we repeated these experiments using cells expressing effector mutants of Ras. Selective activation of Raf (Ras*T35S) or PI3-kinase (Ras*Y40C) did not significantly affect differentiation capacity; these cells consequently underwent terminal neutrophil development and growth arrest. In contrast, Ras*E37G (selectively activating RalGEFs-phospholipase Cε [PLCε]) inhibited neutrophil development (Fig. 1A), with concomitant increase in proliferation capacity (Fig. 1B). This observation was not due to the relative overexpression of Ras*E37G, as demonstrated by Western blotting (Fig. 2A); indeed, normalized expression of Ras*E37G was marginally lower than that of Ras*T35S or Ras*Y40C. Therefore, the Ras*E37G mutant is sufficient to replicate the antidifferentiation phenotype resulting from activation of Ras.
FIG. 1.
Differentiation and proliferation of FDCP-mix cells in the presence of Ras loss of function and constitutively active effector mutants. Morphological scoring and proliferative expansion of FDCP-mix cells in the presence of Ras loss-of-function mutants (A and B) and constitutively active mutants (C and D). Cells were seeded at 2 × 104/ml in medium containing G-CSF plus GM-CSF, and differentiation was assessed following May-Grunwald-Giemsa staining. Significant differences versus FDCP-mix-Neo cells were defined as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001. Error bars represent standard deviations from the mean of at least three independent experiments. (E) Morphology (magnification, ×400) after 7 days under differentiating conditions. Control cultures (Neo) are dominated by large segmented mature neutrophils (n). Cells expressing Ras* either fail to differentiate (b) or give rise to immature neutrophils with doughnut-shaped nuclei characteristic of mouse metamyelocytes (m); these cells retain proliferative capacity (16). Macrophage differentiation is unaffected (mac).
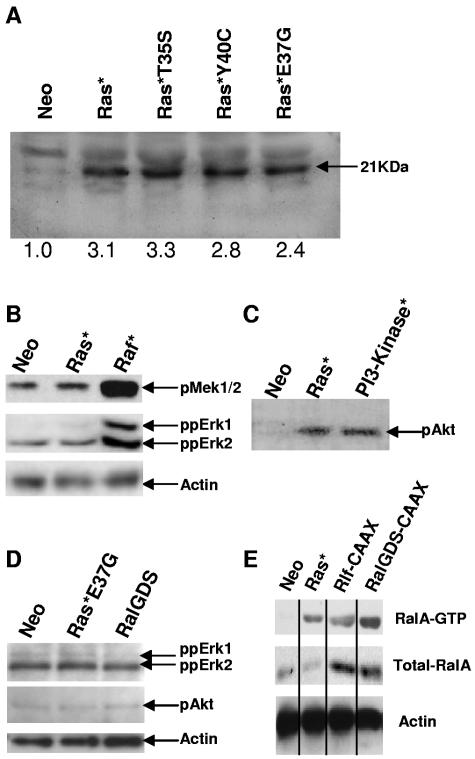
FIG. 2.
Expression and activity of effector mutants of Ras and constitutively active downstream targets of Ras. (A) Mutant Ras expression was probed using a mutant H-Ras-specific antibody. (B) Raf activity was investigated using the phosphorylation-specific antibodies to MEK1/2 and ERK1/2; (C) PI3-kinase activity was assessed using a pAkt/PKB antibody. (D) The effect of constructs activating RalGDS/Ral on Erk1/2 and pAkt/PKB activity was similarly determined. (E) Ral activity was determined using the GTP-sensitive probe as described in Materials and Methods. Results are representative of two independent experiments.
The Ras*E37G effector loop domain mutant has been previously shown to interact with a group of guanine nucleotide exchange factors for the Ral GTPase (e.g., RalGDS and Rlf) (84), as well as the phosphoinositide-specific PLCε (15, 45, 75). However, PLCε RNA is undetectable in FDCP-mix cells, as assessed by reverse transcription-PCR (data not shown), suggesting that RalGEFs were the principal target in this context.
To substantiate the data obtained with effector mutants, we repeated these experiments using constitutively active effectors of Ras. Consistent with the Ras effector mutant data, constitutively active forms of the RalGEFs (RalGDS and Rlf) were able to mimic the effects of Ras* under differentiating conditions (Fig. 1C). Interestingly, RalGDS-CAAX was more effective in inhibiting differentiation than Ras* itself. As expected, both these constructs concomitantly promoted proliferation under differentiating conditions (Fig. 1D). Also in agreement with the Ras effector mutant data, there was no difference in differentiation or proliferative capacity between control cells and those expressing Raf*. PI3-kinase* did demonstrate a modest antidifferentiation effect, but this did not achieve significance in this series of experiments.
As no significant difference in the development or proliferative capacity was observed with the Raf* or PI3-kinase*-expressing FDCP-mix cells, the integrity of these constructs was confirmed by Western blotting. As expected, expression of Raf* strongly promoted MEK1/2 and extracellular signal-regulated kinase 1 (ERK1) and ERK2 phosphorylation (Fig. 2B). Interestingly, Ras* itself was only marginally effective in this regard. This may indicate that activation of these targets is normally limited by endogenous levels of expression of Raf in these cells; alternatively, it might also be the case that simultaneous activation of other effectors by Ras* suppresses activation of MEK/ERK. We also confirmed the phosphorylation of Akt-protein kinase B (PKB) in the Ras* and PI3-kinase*-FDCP-mix cell lines, confirming activation of this pathway. Neither Ras*E37G nor RalGDS-CAAX gave rise to significant activation of either Akt or ERK (Fig. 2D). Taken together, these data confirmed the integrity of the constructs employed and indicated that constitutive Raf and PI3-kinase signaling appears to have little influence on neutrophil development of FDCP-mix cells. Furthermore, these data for the first time suggest a significant role for RalGEFs in regulating neutrophil differentiation.
The activity of RalGEFs in mediating the inhibition of neutrophil differentiation suggests that the modulation of Ral activity may be intrinsic to the observed phenotype. We therefore assessed RalA activity using the activation-sensitive glutathione S-transferase-RalBP1-RalBD probe. The level of Ral-GTP was elevated in FDCP-mix cells expressing Ras*, Rlf-CAAX, and RalGDS-CAAX (Fig. 2D). This observation demonstrates that Ras as well as the RalGEF molecules (Rlf and RalGDS) results in activation of Ral in these cells, which is consistent with a role for Ral in mediating the effects of Ras.
Active Ral is necessary and sufficient for dysregulated myelopoietic development.
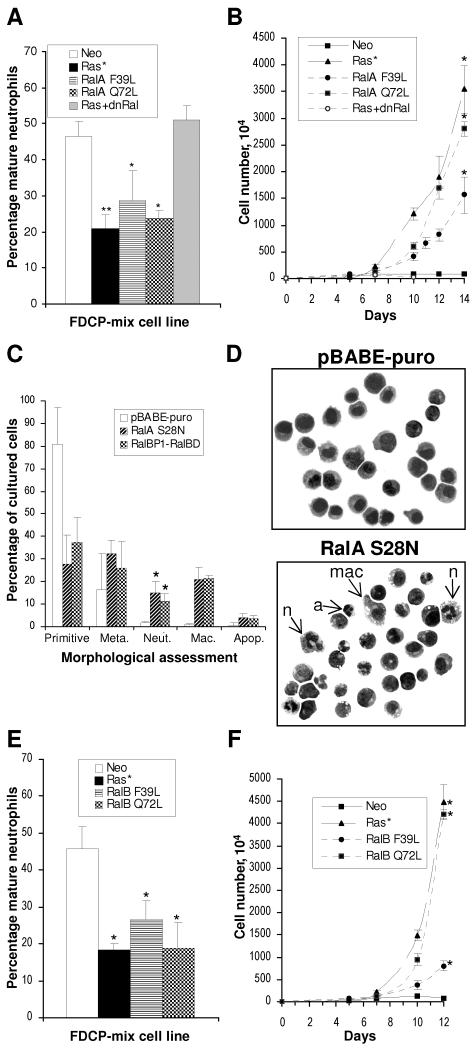
The above data indicate a correlation between active Ral and dysregulated hematopoiesis in FDCP-mix cells. To substantiate whether Ral is necessary and/or sufficient to inhibit neutrophil differentiation, we examined the effect of activated and dominant negative versions of RalA. The constitutively active Ral mutants used were that which is insensitive to Ral-GAP activity (mutated at Q72L) and the “fast-cycling” mutation (F39L) (52, 53, 70). Both RalA mutants examined mimicked the blocked development and increased proliferative capacity demonstrated by FDCP-mix-Ras* (Fig. 3A). This was also associated with increased proliferative capacity (Fig. 3B). The effects of dominant negative RalA constructs on the parental FDCP-mix cells substantiated these observations. Expression of dominant negative Ral (RalBP1-RalBD) in conjunction with Ras* abrogated the capacity of Ras* to inhibit differentiation and promote differentiation (Fig. 3A and B). Expression of dominant negative Ral constructs alone (RalBP1-RalBD or RalA S28N) promoted spontaneous differentiation of FDCP-mix cells grown under self-renewing conditions (IL-3) compared to the empty vector control line (Fig. 3C and D). Interestingly, these constructs also promoted monocyte differentiation. Therefore, constitutive inhibitory RalA is able to compromise the self-renewing capacity of FDCP-mix cells, resulting in spontaneous development. Recent work has demonstrated functional differences between the roles of RalA and RalB in the transformation of human cells (51). We therefore examined the possibility that there may also be differences in the capacity of these proteins to affect hematopoietic differentiation by repeating these experiments with the corresponding constitutively active RalB mutants. In this context, however, we found RalB to be just as effective as RalA (Fig. 3E and F).
FIG. 3.
Effect of RalA and RalB mutants on the development of FDCP-mix cells. (A and B) The effect of constitutively active mutants (F39L and Q72L) of RalA on the development of FDCP-mix cells and the effect of dominant negative RalA (RalBP1-RalBD) on FDCP-mix cells coexpressing Ras* are shown; conditions are as described in the legend to Fig. 1. (C) Spontaneous differentiation under self-renewing conditions of FDCP-mix cells expressing dominant negative RalA constructs alone. (D) Corresponding morphology of cells grown under self-renewing conditions is shown. Control cells (pBABE-puro) demonstrate a uniform blast cell appearance, while RalA S28N cells exhibit a variety of differentiated forms including mature neutrophils (n), macrophages (mac)−, and disintegrating forms of segmented neutrophils (a). Morphology was scored as described in Materials and Methods. (E and F) Effect of constitutively active mutants of RalB on FDCP-mix cells; conditions are as described in the legend to Fig. 1. Significant differences versus FDCP-mix-Neo cells are indicated as follows: *, P < 0.05; **, P < 0.01. Error bars represent standard deviations from the mean of at least three independent experiments.
Together, these data suggest that activation of RalA (or that of RalB) directly inhibits neutrophil differentiation and that its activity is necessary to maintain FDCP-mix cells in a self-renewing state.
Like Ras*, both RalGDS-CAAX and RalA F39L enhance GM-CSF sensitivity.
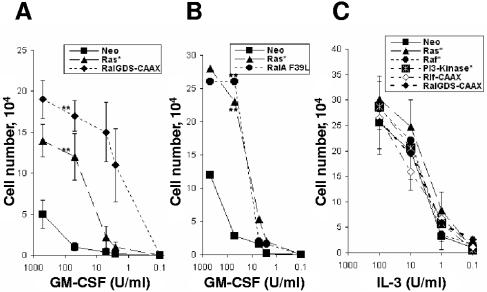
Previously, it was shown that the block in neutrophil development in this model was associated with an increased sensitivity to GM-CSF and that the block in development was dependent upon the continued presence of this cytokine (16). To establish the possible role of the downstream effectors of Ras in augmenting cytokine responsiveness, the growth of the aforementioned cell lines was examined by dose-response assays. We observed that expression of RalGDS-CAAX promoted hyperresponsiveness to GM-CSF in a manner similar to that seen with cells expressing Ras* (P < 0.01) (Fig. 4A). Similarly, cells expressing constitutively active Ral (RalA F39L) also replicated the increase in GM-CSF sensitivity seen in Ras* (Fig. 4B). In contrast, the absolute requirement of all these cell lines for exogenous IL-3 remained, with no significant difference in the dose-response data between Ras loss-of-function mutants (data not shown) or constitutively active mutants versus control cells (Fig. 4C). The data suggest that expression of Ras* selectively increased the sensitivity to GM-CSF and that both increased sensitivity to GM-CSF and differentiation arrest are mediated via the Ral pathway.
FIG. 4.
GM-CSF hypersensitivity in FDCP-mix cells is mediated via the Ral pathway. Dose-response curves with GM-CSF (A and B) and to IL-3 (C) are shown. Cell growth was assessed after 5 days of culture with the indicated concentrations of cytokines. Significant difference versus FDCP-mix-Neo cells is indicated as follows: **, P < 0.01. Error bars represent standard deviations from the mean of at least three independent experiments.
Role of downstream effectors of Ral in the inhibition of neutrophil differentiation.
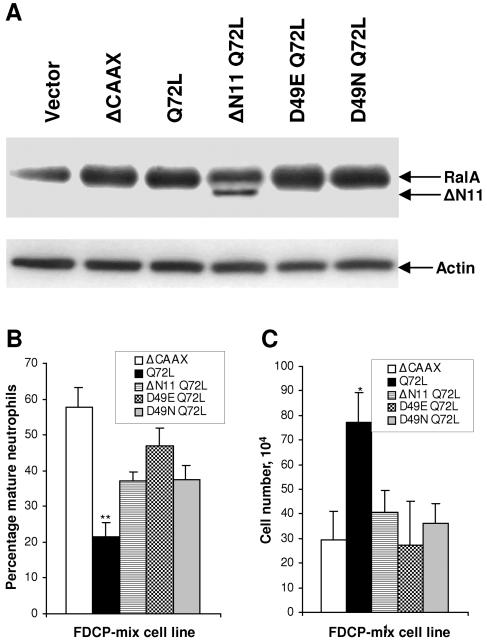
Mutants of Ral which have impaired capacity to activate effectors downstream of Ral have been previously described; deletion of the N-terminal amino acids (RalAΔN11) reduces phospholipase D1 binding (41), the RalAD49E mutant is defective in binding Sec5 and Exo84 (60, 61), and the RalAD49N mutant has diminished affinity for RalBP1 (11). We examined the phenotype conferred by constitutively active (RalAQ72L) versions of these mutants to indicate the contribution of these Ral effectors to the dysregulation of hematopoiesis. A mutant lacking the biologically essential CAAX motif acted as the negative control in these experiments. Each mutant appeared to be expressed at similar levels against the relatively high endogenous levels of RalA in these cells (Fig. 5A). Compared to RalAQ72L, we found that each of the effector mutants compromised the activity of constitutively active RalA in the inhibition of neutrophil differentiation (Fig. 5B). Correspondingly, none of the effector mutants retained any capacity to promote the proliferation of these cells under differentiating conditions (Fig. 5C). In common with RalAQ72L, these mutants did not affect the proliferative rate of these cells under self-renewing conditions (data not shown). The simplest interpretation of these data is that multiple effectors of Ral contribute towards its ability to inhibit differentiation and promote self-renewal of hematopoietic cells.
FIG. 5.
Role of downstream effectors of Ral in the inhibition of neutrophil differentiation. (A) The expression of RalA effector loop domain mutants in FDCP-mix cells by immunoblotting is shown; the corresponding expression of actin is also shown as a loading control. Morphological scoring after 7 days under differentiating conditions (B) and corresponding proliferative expansion (C) of FDCP-mix cells in the presence of RalA effector loop domain mutants (conditions are as described in the legend to Fig. 1) is shown. Significant differences versus FDCP-mix-RalAQ72L ΔCAAX cells are indicated as follows: *, P < 0.05; **, P < 0.01. Error bars represent standard deviations from the mean of three independent experiments.
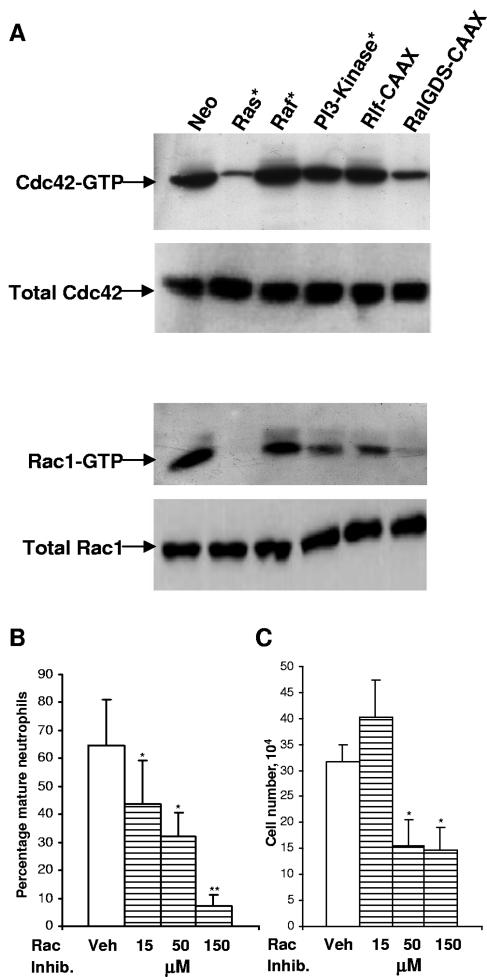
Given that activation of one of these Ral effectors, RalBP1, has been previously associated with inhibition of differentiation (32), we investigated the individual contribution of this effector to the observed inhibition of differentiation. Activation of RalBP1 functions as a GAP towards the Rho-related proteins Rac and Cdc42. Hyperactivation of Ral by Ras should therefore result in negative regulation of Rac1 and Cdc42 activity if these proteins are in fact a target of Ral in this context. We therefore examined the active-protein levels via binding-domain pull-down assays. This analysis revealed that whereas control FDCP-mix cells (Neo) expressed these Rho-related proteins in their active (GTP-bound) state, the lines expressing Ras* and RalGDS-CAAX were almost completely deficient in GTP-bound Rac1 and Cdc42 (Fig. 6A). The comparative difference in expression of these GTP-bound proteins between the Rlf-CAAX and RalGDS-CAAX lines showed that the latter effector more closely mimicked the effects of Ras*. Interestingly, we also observed that PI3-kinase* weakly reduced GTP-bound levels of Rac1 and Cdc42. These observations correlate with the weak inhibitory effect on neutrophil development described above (Fig. 1C) and suggest a degree of cross talk from this effector.
FIG. 6.
Expression and activity of Rac1 and Cdc42 and effect of inhibition of Rac1. After 7 days in the presence of G-CSF plus GM-CSF, normalized protein from whole-cell lysates of the indicated FDCP-mix cell line was assayed. A total of 10% of the lysate was assessed for total protein levels by Western blotting; the remainder was immunoprecipitated with PAK-Rac-binding domain. Immunoprobing was carried out using the Cdc42 polyclonal and Rac1 monoclonal antibodies. (A) The results shown are representative of two independent experiments. (B) The effect of the Rac1 inhibitor, NSC23766, on neutrophil differentiation; conditions are as described in the legend to Fig. 1. (C) Corresponding effect of the Rac1 inhibitor, NSC23766, on cell growth after 7 days in culture. No significant cell death was observed in any of the conditions used (<5% of cells). A significant difference versus vehicle alone treated FDCP-mix cells is indicated as follows: *, P < 0.05; **, P < 0.01. Error bars represent standard deviations from the mean of three experiments.
Recently, a highly selective cell-permeable inhibitor for Rac1, NSC23766, has become available (31); we therefore examined the influence of Rac1 inhibition on FDCP-mix development. NSC23766 strongly inhibited neutrophil differentiation in a dose-dependent manner (Fig. 6B), however, at higher doses, it was also antiproliferative (Fig. 6C). Nevertheless, the degree of inhibition at low doses was at least consistent with a role for Rac1 inactivation in mediating part of the antidifferentiation effects of Ras.
PI3-kinase activity is required to maintain the differentiation block imposed by Ras.
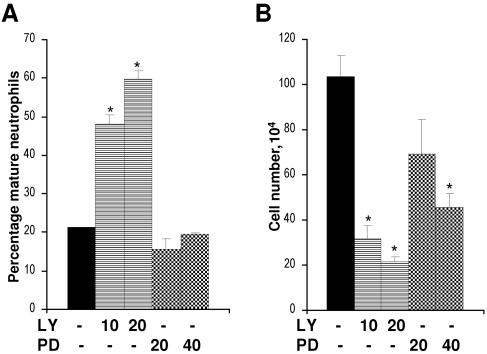
Experiments using activated effectors and effector mutants of Ras suggested that hyperactivation of PI3-kinase was not sufficient to block neutrophil differentiation; however, the data in Fig. 6A and 1C indicated that PI3-kinase activity did participate in this process. We therefore examined the requirement for both PI3-kinase activity, as well as MAPK activity, to maintain the differentiation block imposed by Ras. We treated differentiation-blocked FDCP-mix-Ras* cells with inhibitors to either PI3-kinase or MEK. Inhibition of PI3-kinase with LY294002 strongly promoted differentiation (Fig. 7A) and correspondingly inhibited proliferation (Fig. 7B), indicating that the maintenance of the differentiation-inhibited phenotype was dependent on PI3-kinase activity. In contrast, inhibition of MEK with PD98059 had no significant effect on the differentiation of these cells, despite the fact that it inhibited their proliferation (Fig. 7B). The MEK1/2 inhibitor, U0126, gave similar results (data not shown). These indicate that PI3-kinase activity is required to maintain the antidifferentiation phenotype elicited by Ras. Correspondingly, MAPK activity does not appear to play a role in inhibiting the differentiation of these cells. These data are consistent with the lack of influence of activated Raf and the corresponding effector mutant of Ras on neutrophil development (Fig. 1). They are also consistent with the fact that Ras itself does not constitutively promote MAPK activity in this context (Fig. 2).
FIG. 7.
Effect of inhibition of PI3-kinase and MEK on the differentiation of FDCP-mix Ras* cells. (A) Differentiation-blocked FDCP-mix Ras* cells (day 7 cells cultured under differentiating conditions at 2 × 105 cells/ml) were treated with inhibitors LY294002 (LY) and PD98059 (PD) at the indicated μM concentrations, their differentiated status was assessed 3 days later. (B) Cell growth was determined at the same time point. None of the experimental conditions resulted in significant cell death (<5% of cells). Significant differences versus untreated FDCP-mix-Ras* are indicated as follows: *, P < 0.05. Error bars represent standard deviations from the mean of three independent experiments.
DISCUSSION
Although there is some understanding of the processes which regulate hematopoietic differentiation at a transcriptional level, little is known of the intracellular events which regulate this developmental programming. These are influenced by environmental factors such as the presence of cytokines and stromal factors (3). Since many of these environmental signals are transduced via Ras proteins, it is perhaps not surprising that in cells expressing constitutively active Ras these signals become “distorted,” leading to a dysregulation of development. Previous work from our laboratory established that mutational activation of Ras selectively inhibited neutrophil development and concomitantly increased sensitivity to GM-CSF (16). In this study, we identify the downstream targets of Ras, which mediate these changes in developmental programming and cytokine responsiveness. We examined three well-characterized Ras effector proteins (Raf, PI3-kinase, and RalGEFs) and their related pathways (54). Studies using effector mutants of Ras indicated that it was RalGEF signaling which was shown to have the predominant role in the dysregulated development mediated by Ras. Although the use of these effector mutants allows the dissection of signaling activity in a relatively nonmanipulative manner (in that the expression level and activity of downstream targets are not artificially altered), the use of these mutants does not provide direct evidence of the interacting proteins (including as-yet-unidentified effectors). We therefore repeated these experiments using constitutively active Raf, PI3-kinase, and RalGEFs. Data from these experiments replicated the phenotypes of the corresponding effector loop domain mutants, both substantiating our initial observations and definitively identifying RalGEFs as the key effector in mediating this phenotype. Although the involvement of these RalGEFs or their putative effector, Ral, during hematopoiesis has not been previously reported, their ubiquitous expression has been identified within the relevant compartments including bone marrow, spleen, and thymus (2, 85).
The involvement of the RalGEFs implicated Ral signaling as being important for neutrophil development in FDCP-mix cells. Use of the activation-specific probe for Ral confirmed the association of increased Ral activity with the expression of Ras and RalGEFs. Furthermore, we also demonstrated that promoting the activity of Ral by using constitutively active mutants was itself sufficient to block neutrophil development. Conversely, expression of dominant negative Ral promoted differentiation of FDCP-mix cells even when cultured under self-renewing conditions. These data suggest that the activity of this GTPase is essential to maintain the self-renewing potential of these cells. Given that a universal property of leukemic blasts is their capacity for self-renewal, these data suggest that Ral activity may be an important factor in maintaining the self-renewing potential of leukemic cells. This is the first demonstration of a role for Ral in hematopoietic differentiation and self-renewal; however, Ral has been previously implicated as a developmental regulator in Drosophila melanogaster eye (57) and in inhibiting differentiation of PC12 cells, as defined by neurite outgrowth (32). In the context of cellular transformation, Ral activity has a long-established role (30, 80, 84); this role may be more significant in human cells (35, 69). The function of Ral activity has been assessed in a variety of contexts: a RalGDS knockout study has shown a role for Ral activity in inhibiting apoptotic death (34), and distinct functions for RalA and RalB have been identified, with RalB being required for suppression of apoptosis while RalA is required for anchorage-independent proliferation (13). RalA has also been reported to have a preferential affinity for exocyst complex (74), although it is possible that in the context of overexpression of Ral, differences in affinity may not be limiting. Recent work has also identified a dominant role for RalA in the transformation of human cells (51).
We have not so far established how Ral ultimately inhibits neutrophil development. Studies employing effector mutants of Ral suggest that this GTPase influences neutrophil development through a variety of effectors. Previous work has shown that all the established Ral effectors are able to influence differentiation. Increase in phospholipase D1 activity is closely associated with neutrophil differentiation (64, 66). The exocyst complex has not yet been directly linked with hematopoietic development, but it is associated with neurite outgrowth in neuronal and PC12 cells (47, 82). In both these instances, however, the increase in effector activity is associated with promoting rather than inhibiting differentiation. RalBP1 is thought to act as a GAP (or negative regulator) towards the Rho superfamily of Rac1/Cdc42 GTPases (43). These Rho family proteins have been previously associated with differentiation of epithelial cells (78), myoblasts (19, 36, 76), dendrites (49), and dorsal root ganglia (42, 81) and during PC12 neurite formation (87). It might therefore be predicted that increased RalBP1 activity would be associated with an inhibition of differentiation in these contexts; in the PC12 model, there is indeed evidence that RalGEFs inhibit differentiation by suppressing the activity of Rac and Cdc42 (32). Similarly, we present evidence that inhibition of Rac1 activity does suppress differentiation in FDCP-mix cells.
The surprisingly negligible contribution of the Raf and PI3-kinase effectors to hematopoietic dysregulation in this study may be due to the prevailing importance of the Ral pathway in this context. Alternatively, optimum activation of Raf and PI3-kinase may already be present in FDCP-mix cells; therefore, increasing their activity via Ras may be of little significance. In the context of the Raf effector pathway, it is interesting to note that, as with primary leukemias (38), the presence of activated Ras did not provoke constitutive activation of MAPK in FDCP-mix cells; even inhibition with MAPK inhibitors failed to influence the differentiation-inhibited status of FDCP-mix Ras* cells. These data, together with the lack of effect of constitutive activation of Raf, indicate that this effector is not playing a role in the regulation of neutrophil differentiation, although it may, as previously suggested (63), play a role in maintaining their proliferative capacity. In contrast to Raf, activation of PI3-kinase via the Ras*Y40C effector mutant or the constitutively active version of this kinase did provoke a modest antidifferentiation effect in these cells, implying that this effector may also participate in this process. These data were supported by the observation that inhibition of PI3-kinase activity reversed the effect of Ras on neutrophil differentiation, indicating that the maintenance of the antidifferentiated state was at least dependent on PI3-kinase activity. Since Ral has also been shown to be activated in a PI3-kinase-dependent manner in neutrophils (62), this may indicate that multiple effectors of Ras (RalGEFs and PI3-kinases) may converge on this GTPase. This possibility is also supported by the observation that expression of constitutively active PI3-kinase also suppressed Rac1 activation and, to a certain extent, Cdc42 activation. It has been suggested that PI3-kinase may participate in RalGDS activation by promoting its association with PI3-kinase-dependent kinase 1, which in turn appears to relieve autoinhibition of the catalytic domain of RalGDS (79).
A further observation of interest relates to the increased sensitivity to GM-CSF in cells expressing constructs that directly or indirectly activated Ral. We have previously shown that the influence of GM-CSF is crucial in maintaining the block in differentiation, in that its withdrawal leads to spontaneous terminal differentiation. These observations have interesting parallels with that of JMML. In JMML, 20 to 30% of cases are associated with loss of Nf1 Ras-GAP function, leading to hyperactivation of Ras. JMML-derived progenitor cells display a marked hypersensitivity towards GM-CSF, as shown by CFU-GM assays (23), with a similar phenotype demonstrated with Nf1−/− mouse cells (8, 48). While these studies strongly suggest that heightened sensitivity to GM-CSF in JMML is explicitly mediated via Ras (44), there is uncertainty as to the mechanism through which this arises. In Myb-transformed Nf1−/− murine cells, increased sensitivity appears to arise from autocrine production of GM-CSF that was dependent on Raf/MEK/ERK signaling (20). However, only around 50% of JMML patients demonstrate excessive GM-CSF production, despite the fact that they universally show increased sensitivity to this cytokine. In the model described here, we could find no evidence of autocrine production of this cytokine, also suggesting that additional and/or alternative mechanisms exist. In this context, it will be of interest to determine the activation state of Ral in Nf1−/− cells and whether their heightened response to GM-CSF is dependent on the activity of this GTPase.
Despite the variety of approaches aimed at dysregulated Ras signaling in a clinical setting (1), none has proved particularly effective so far. Therefore, the identification of alternative pharmacological targets may be of significance. In the context of leukemia, such a target may be the small GTPase Ral.
Acknowledgments
We are grateful to Chris J. Marshall and associated laboratory members (Institute of Cancer Research, London, United Kingdom) for expert technical advice, assistance, and reagents supplied, including RalA F39L and the RalBP1- and PAKα-binding domain constructs. We thank Julian Downward (London Research Institute) for providing Ras effector loop and PI3-kinase* (K227E) mutant retroviral constructs, Michael A. White (University of Texas Southwestern Medical School) for RalGDS-CAAX, and Johannes L. Bos (University Medical Center Utrecht, Utrecht, The Netherlands) for Rlf-CAAX. We thank Ali Bounacer and David Wynford-Thomas (Cardiff University) for helpful discussions and the pBABE puro-RalA S28N construct and Christopher Counter (Duke University Medical Center) for the RalB and additional RalA constructs.
Part of this work was funded by grants from the Leukemia Research Fund of Great Britain. N.O. was also supported by grants from the Tom Owen Memorial Fund (College of Medicine, Cardiff, Wales), the British Journal of Hematology, and the Leukemia Research Fund Shalit Traveling Fellowship.
REFERENCES
- 1.Ahmadian, M. R. 2002. Prospects for anti-ras drugs. Br. J. Haematol. 116:511-518. [DOI] [PubMed] [Google Scholar]
- 2.Albright, C. F., B. W. Giddings, J. Liu, M. Vito, and R. A. Weinberg. 1993. Characterization of a guanine-nucleotide dissociation stimulator for a Ras-related GTPase. EMBO J. 12:339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander, W. S. 1998. Cytokines in hematopoiesis. Int. Rev. Immunol. 16:651-682. [DOI] [PubMed] [Google Scholar]
- 4.Altman, S. A., L. Randers, and G. Rao. 1993. Comparison of trypan blue dye exclusion and fluorometric assays for mammalian cell viability determinations. Biotechnol. Prog. 9:671-674. [DOI] [PubMed] [Google Scholar]
- 5.Andreeff, M. 1986. Cell kinetics of leukemia. Semin. Hematol. 23:300-314. [PubMed] [Google Scholar]
- 6.Blalock, W. L., P. M. Navolanic, L. S. Steelman, J. G. Shelton, P. W. Moye, J. T. Lee, R. A. Franklin, A. Mirza, M. McMahon, M. K. White, and J. A. McCubrey. 2003. Requirement for the PI3K/Akt pathway in MEK1-mediated growth and prevention of apoptosis: identification of an Achilles heel in leukemia. Leukemia 17:1058-1067. [DOI] [PubMed] [Google Scholar]
- 7.Boguski, M. S., and F. McCormick. 1993. Proteins regulating Ras and its relatives. Nature 366:643-654. [DOI] [PubMed] [Google Scholar]
- 8.Bollag, G., D. W. Clapp, S. Shih, F. Adler, Y. Y. Zhang, P. Thompson, B. J. Lange, M. H. Freedman, F. McCormick, T. Jacks, and K. Shannon. 1996. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat. Genet. 12:144-148. [DOI] [PubMed] [Google Scholar]
- 9.Bos, J. L. 1989. ras oncogenes in human cancer: a review. Cancer Res. 49:4682-4689. [PubMed] [Google Scholar]
- 10.Braun, B. S., D. A. Tuveson, N. Kong, D. T. Le, S. C. Kogan, J. Rozmus, M. M. Le Beau, T. E. Jacks, and K. M. Shannon. 2004. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc. Natl. Acad. Sci. USA 101:597-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantor, S. B., T. Urano, and L. A. Feig. 1995. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol. Cell. Biol. 15:4578-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan, I. T., J. L. Kutok, I. R. Williams, S. Cohen, L. Kelly, H. Shigematsu, L. Johnson, K. Akashi, D. A. Tuveson, T. Jacks, and D. G. Gilliland. 2004. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J. Clin. Investig. 113:528-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chien, Y., and M. A. White. 2003. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 4:800-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowburn, A. S., K. A. Cadwallader, B. J. Reed, N. Farahi, and E. R. Chilvers. 2002. Role of PI3-kinase-dependent Bad phosphorylation and altered transcription in cytokine-mediated neutrophil survival. Blood 100:2607-2616. [DOI] [PubMed] [Google Scholar]
- 15.Cullen, P. 2001. Ras Effectors: buying shares in Ras plc. Curr. Biol. 11:R342-344. [DOI] [PubMed] [Google Scholar]
- 16.Darley, R. L., and A. K. Burnett. 1999. Mutant RAS inhibits neutrophil but not macrophage differentiation and allows continued growth of neutrophil precursors. Exp. Hematol. 27:1599-1608. [DOI] [PubMed] [Google Scholar]
- 17.Darley, R. L., L. Pearn, N. Omidvar, M. Sweeney, J. Fisher, S. Phillips, T. Hoy, and A. K. Burnett. 2002. Protein kinase C mediates mutant N-Ras-induced developmental abnormalities in normal human erythroid cells. Blood 100:4185-4192. [DOI] [PubMed] [Google Scholar]
- 18.de Koning, J. P., A. A. Soede-Bobok, A. M. Schelen, L. Smith, D. van Leeuwen, V. Santini, B. M. Burgering, J. L. Bos, B. Lowenberg, and I. P. Touw. 1998. Proliferation signaling and activation of Shc, p21Ras, and Myc via tyrosine 764 of human granulocyte colony-stimulating factor receptor. Blood 91:1924-1933. [PubMed] [Google Scholar]
- 19.Deng, X., D. Z. Ewton, B. Pawlikowski, M. Maimone, and E. Friedman. 2003. Mirk/dyrk1B is a Rho-induced kinase active in skeletal muscle differentiation. J. Biol. Chem. 278:41347-41354. [DOI] [PubMed] [Google Scholar]
- 20.Donovan, S., W. See, J. Bonifas, D. Stokoe, and K. M. Shannon. 2002. Hyperactivation of protein kinase B and ERK have discrete effects on survival, proliferation, and cytokine expression in Nf1-deficient myeloid cells. Cancer Cell 2:507-514. [DOI] [PubMed] [Google Scholar]
- 21.Dorrell, C., K. Takenaka, M. D. Minden, R. G. Hawley, and J. E. Dick. 2004. Hematopoietic cell fate and the initiation of leukemic properties in primitive primary human cells are influenced by Ras activity and farnesyltransferase inhibition. Mol. Cell. Biol. 24:6993-7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dosil, M., S. Wang, and I. R. Lemischka. 1993. Mitogenic signalling and substrate specificity of the Flk2/Flt3 receptor tyrosine kinase in fibroblasts and interleukin 3-dependent hematopoietic cells. Mol. Cell. Biol. 13:6572-6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emanuel, P. D., L. J. Bates, R. P. Castleberry, R. J. Gualtieri, and K. S. Zuckerman. 1991. Selective hypersensitivity to granulocyte-macrophage colony-stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood 77:925-929. [PubMed] [Google Scholar]
- 24.Faderl, S., M. Talpaz, Z. Estrov, S. O'Brien, R. Kurzrock, and H. M. Kantarjian. 1999. The biology of chronic myeloid leukemia. N. Engl. J. Med. 341:164-172. [DOI] [PubMed] [Google Scholar]
- 25.Farr, C. J., R. K. Saiki, H. A. Erlich, F. McCormick, and C. J. Marshall. 1988. Analysis of RAS gene mutations in acute myeloid leukemia by polymerase chain reaction and oligonucleotide probes. Proc. Natl. Acad. Sci. USA 85:1629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feig, L. A. 2003. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 13:419-425. [DOI] [PubMed] [Google Scholar]
- 27.Feig, L. A., T. Urano, and S. Cantor. 1996. Evidence for a Ras/Ral signaling cascade. Trends Biochem. Sci. 21:438-441. [DOI] [PubMed] [Google Scholar]
- 28.Fenski, R., K. Flesch, S. Serve, M. Mizuki, E. Oelmann, K. Kratz-Albers, J. Kienast, R. Leo, S. Schwartz, W. E. Berdel, and H. Serve. 2000. Constitutive activation of FLT3 in acute myeloid leukaemia and its consequences for growth of 32D cells. Br. J. Haematol. 108:322-330. [DOI] [PubMed] [Google Scholar]
- 29.Frankel, P., A. Aronheim, E. Kavanagh, M. S. Balda, K. Matter, T. D. Bunney, and C. J. Marshall. 2005. RalA interacts with ZONAB in a cell density-dependent manner and regulates its transcriptional activity. EMBO J. 24:54-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frankel, P., M. Ramos, J. Flom, S. Bychenok, T. Joseph, E. Kerkhoff, U. R. Rapp, L. A. Feig, and D. A. Foster. 1999. Ral and Rho-dependent activation of phospholipase D in v-Raf-transformed cells. Biochem. Biophys. Res. Commun. 255:502-507. [DOI] [PubMed] [Google Scholar]
- 31.Gao, Y., J. B. Dickerson, F. Guo, J. Zheng, and Y. Zheng. 2004. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. USA 101:7618-7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goi, T., G. Rusanescu, T. Urano, and L. A. Feig. 1999. Ral-specific guanine nucleotide exchange factor activity opposes other Ras effectors in PC12 cells by inhibiting neurite outgrowth. Mol. Cell. Biol. 19:1731-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goi, T., M. Shipitsin, Z. Lu, D. A. Foster, S. G. Klinz, and L. A. Feig. 2000. An EGF receptor/Ral-GTPase signaling cascade regulates c-Src activity and substrate specificity. EMBO J. 19:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez-Garcia, A., C. A. Pritchard, H. F. Paterson, G. Mavria, G. Stamp, and C. J. Marshall. 2005. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell 7:219-226. [DOI] [PubMed] [Google Scholar]
- 35.Hamad, N. M., J. H. Elconin, A. E. Karnoub, W. Bai, J. N. Rich, R. T. Abraham, C. J. Der, and C. M. Counter. 2002. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 16:2045-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heller, H., E. Gredinger, and E. Bengal. 2001. Rac1 inhibits myogenic differentiation by preventing the complete withdrawal of myoblasts from the cell cycle. J. Biol. Chem. 276:37307-37316. [DOI] [PubMed] [Google Scholar]
- 37.Hoang, T. 2004. The origin of hematopoietic cell type diversity. Oncogene 23:7188-7198. [DOI] [PubMed] [Google Scholar]
- 38.Iida, M., M. Towatari, A. Nakao, H. Iida, H. Kiyoi, Y. Nakano, M. Tanimoto, H. Saito, and T. Naoe. 1999. Lack of constitutive activation of MAP kinase pathway in human acute myeloid leukemia cells with N-Ras mutation. Leukemia 13:585-589. [DOI] [PubMed] [Google Scholar]
- 39.Itoh, T., A. Muto, S. Watanabe, A. Miyajima, T. Yokota, and K. Arai. 1996. Granulocyte-macrophage colony-stimulating factor provokes RAS activation and transcription of c-fos through different modes of signaling. J. Biol. Chem. 271:7587-7592. [DOI] [PubMed] [Google Scholar]
- 40.Janssen, J. W., A. C. Steenvoorden, J. Lyons, B. Anger, J. U. Bohlke, J. L. Bos, H. Seliger, and C. R. Bartram. 1987. RAS gene mutations in acute and chronic myelocytic leukemias, chronic myeloproliferative disorders, and myelodysplastic syndromes. Proc. Natl. Acad. Sci. USA 84:9228-9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang, H., J. Q. Luo, T. Urano, P. Frankel, Z. M. Lu, D. A. Foster, and L. A. Feig. 1995. Involvement of Ral GTPase in v-Src-induced phospholipase-D activation. Nature 378:409-412. [DOI] [PubMed] [Google Scholar]
- 42.Jin, Z., and S. M. Strittmatter. 1997. Rac1 mediates collapsin-1-induced growth cone collapse. J. Neurosci. 17:6256-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jullien-Flores, V., O. Dorseuil, F. Romero, F. Letourneur, S. Saragosti, R. Berger, A. Tavitian, G. Gacon, and J. H. Camonis. 1995. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with Cdc42/Rac GTPase-activating protein activity. J. Biol. Chem. 270:22473-22477. [DOI] [PubMed] [Google Scholar]
- 44.Kalra, R., D. C. Paderanga, K. Olson, and K. M. Shannon. 1994. Genetic analysis is consistent with the hypothesis that NF1 limits myeloid cell growth through p21ras. Blood 84:3435-3439. [PubMed] [Google Scholar]
- 45.Kelley, G. G., S. E. Reks, J. M. Ondrako, and A. V. Smrcka. 2001. Phospholipase Cε: a novel Ras effector. EMBO J. 20:743-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiyoi, H., T. Naoe, Y. Nakano, S. Yokota, S. Minami, S. Miyawaki, N. Asou, K. Kuriyama, I. Jinnai, C. Shimazaki, H. Akiyama, K. Saito, H. Oh, T. Motoji, E. Omoto, H. Saito, R. Ohno, and R. Ueda. 1999. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood 93:3074-3080. [PubMed] [Google Scholar]
- 47.Lalli, G., and A. Hall. 2005. Ral GTPases regulate neurite branching through GAP-43 and the exocyst complex. J. Cell Biol. 171:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Largaespada, D. A., C. I. Brannan, N. A. Jenkins, and N. G. Copeland. 1996. Nf1 deficiency causes Ras-mediated granulocyte/macrophage colony stimulating factor hypersensitivity and chronic myeloid leukaemia. Nat. Genet. 12:137-143. [DOI] [PubMed] [Google Scholar]
- 49.Lee, A., W. Li, K. Xu, B. A. Bogert, K. Su, and F. B. Gao. 2003. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development 130:5543-5552. [DOI] [PubMed] [Google Scholar]
- 50.Lewis, J. L., S. B. Marley, M. Ojo, and M. Y. Gordon. 2004. Opposing effects of PI3 kinase pathway activation on human myeloid and erythroid progenitor cell proliferation and differentiation in vitro. Exp. Hematol. 32:36-44. [DOI] [PubMed] [Google Scholar]
- 51.Lim, K. H., A. T. Baines, J. J. Fiordalisi, M. Shipitsin, L. A. Feig, A. D. Cox, C. J. Der, and C. M. Counter. 2005. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell 7:533-545. [DOI] [PubMed] [Google Scholar]
- 52.Lin, R., S. Bagrodia, R. Cerione, and D. Manor. 1997. A novel Cdc42Hs mutant induces cellular transformation. Curr. Biol. 7:794-797. [DOI] [PubMed] [Google Scholar]
- 53.Lin, R., R. A. Cerione, and D. Manor. 1999. Specific contributions of the small GTPases Rho, Rac, and Cdc42 to Dbl transformation. J. Biol. Chem. 274:23633-23641. [DOI] [PubMed] [Google Scholar]
- 54.Marshall, C. J. 1996. Ras effectors. Curr. Opin. Cell Biol. 8:197-204. [DOI] [PubMed] [Google Scholar]
- 55.Matsuguchi, T., and A. S. Kraft. 1998. Regulation of myeloid cell growth by distinct effectors of Ras. Oncogene 17:2701-2709. [DOI] [PubMed] [Google Scholar]
- 56.McGlynn, A. P., R. A. Padua, A. K. Burnett, and R. L. Darley. 2000. Alternative effects of RAS and RAF oncogenes on the proliferation and apoptosis of factor-dependent FDC-P1 cells. Leuk. Res. 24:47-54. [DOI] [PubMed] [Google Scholar]
- 57.Mirey, G., M. Balakireva, S. L'Hoste, C. Rosse, S. Voegeling, and J. Camonis. 2003. A Ral guanine exchange factor-Ral pathway is conserved in Drosophila melanogaster and sheds new light on the connectivity of the Ral, Ras, and Rap pathways. Mol. Cell. Biol. 23:1112-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyauchi, J., M. Asada, M. Sasaki, Y. Tsunematsu, S. Kojima, and S. Mizutani. 1994. Mutations of the N-ras gene in juvenile chronic myelogenous leukemia. Blood 83:2248-2254. [PubMed] [Google Scholar]
- 59.Mizuki, M., R. Fenski, H. Halfter, I. Matsumura, R. Schmidt, C. Muller, W. Gruning, K. Kratz-Albers, S. Serve, C. Steur, T. Buchner, J. Kienast, Y. Kanakura, W. E. Berdel, and H. Serve. 2000. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the ras and STAT5 pathways. Blood 96:3907-3914. [PubMed] [Google Scholar]
- 60.Moskalenko, S., D. O. Henry, C. Rosse, G. Mirey, J. H. Camonis, and M. A. White. 2002. The exocyst is a Ral effector complex. Nat. Cell Biol. 4:66-72. [DOI] [PubMed] [Google Scholar]
- 61.Moskalenko, S., C. Tong, C. Rosse, G. Mirey, E. Formstecher, L. Daviet, J. Camonis, and M. A. White. 2003. Ral GTPases regulate exocyst assembly through dual subunit interactions. J. Biol. Chem. 278:51743-51748. [DOI] [PubMed] [Google Scholar]
- 62.M'Rabet, L., P. J. Coffer, R. M. Wolthuis, F. Zwartkruis, L. Koenderman, and J. L. Bos. 1999. Differential fMet-Leu-Phe- and platelet-activating factor-induced signaling toward Ral activation in primary human neutrophils. J. Biol. Chem. 274:21847-21852. [DOI] [PubMed] [Google Scholar]
- 63.Muszynski, K. W., F. W. Ruscetti, G. Heidecker, U. Rapp, J. Troppmair, J. M. Gooya, and J. R. Keller. 1995. Raf-1 protein is required for growth factor-induced proliferation of hematopoietic cells. J. Exp. Med. 181:2189-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neri, L. M., R. Bortul, P. Borgatti, G. Tabellini, G. Baldini, S. Capitani, and A. M. Martelli. 2002. Proliferating or differentiating stimuli act on different lipid-dependent signaling pathways in nuclei of human leukemia cells. Mol. Biol. Cell 13:947-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niemeyer, C. M., M. Arico, G. Basso, A. Biondi, R. A. Cantu, U. Creutzig, O. Haas, J. Harbott, H. Hasle, G. Kerndrup, F. Locatelli, G. Mann, B. Stollmann-Gibbels, E. T. Veer-Korthof, E. van Wering, M. Zimmermann, et al. 1997. Chronic myelomonocytic leukemia in childhood: a retrospective analysis of 110 cases. Blood 89:3534-3543. [PubMed] [Google Scholar]
- 66.Ohguchi, K., S. Nakashima, Z. Tan, Y. Banno, S. Dohi, and Y. Nozawa. 1997. Increased activity of small GTP-binding protein-dependent phospholipase D during differentiation in human promyelocytic leukemic HL60 cells. J. Biol. Chem. 272:1990-1996. [DOI] [PubMed] [Google Scholar]
- 67.Ohta, Y., N. Suzuki, S. Nakamura, J. H. Hartwig, and T. P. Stossel. 1999. The small GTPase RalA targets filamin to induce filopodia. Proc. Natl. Acad. Sci. USA 96:2122-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Padua, R. A., B. A. Guinn, A. I. Al Sabah, M. Smith, C. Taylor, T. Pettersson, S. Ridge, G. Carter, D. White, D. Oscier, S. Chevret, and R. West. 1998. RAS, FMS and p53 mutations and poor clinical outcome in myelodysplasias: a 10-year follow-up. Leukemia 12:887-892. [DOI] [PubMed] [Google Scholar]
- 69.Rangarajan, A., S. J. Hong, A. Gifford, and R. A. Weinberg. 2004. Species- and cell type-specific requirements for cellular transformation. Cancer Cell 6:171-183. [DOI] [PubMed] [Google Scholar]
- 70.Reinstein, J., I. Schlichting, M. Frech, R. S. Goody, and A. Wittinghofer. 1991. p21 with a phenylalanine 28→leucine mutation reacts normally with the GTPase activating protein GAP but nevertheless has transforming properties. J. Biol. Chem. 266:17700-17706. [PubMed] [Google Scholar]
- 71.Sahai, E., M. F. Olson, and C. J. Marshall. 2001. Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 20:755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Satoh, T., M. Nakafuku, A. Miyajima, and Y. Kaziro. 1991. Involvement of ras p21 protein in signal-transduction pathways from interleukin 2, interleukin 3, and granulocyte/macrophage colony-stimulating factor, but not from interleukin 4. Proc. Natl. Acad. Sci. USA 88:3314-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scheele, J. S., D. Ripple, and M. Lubbert. 2000. The role of ras and other low molecular weight guanine nucleotide (GTP)-binding proteins during hematopoietic cell differentiation. Cell. Mol. Life Sci. 57:1950-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shipitsin, M., and L. A. Feig. 2004. RalA but not RalB enhances polarized delivery of membrane proteins to the basolateral surface of epithelial cells. Mol. Cell. Biol. 24:5746-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song, C., C. D. Hu, M. Masago, K. Kariya, Y. Yamawaki-Kataoka, M. Shibatohge, D. Wu, T. Satoh, and T. Kataoka. 2001. Regulation of a novel human phospholipase C, PLCε, through membrane targeting by Ras. J. Biol. Chem. 276:2752-2757. [DOI] [PubMed] [Google Scholar]
- 76.Sordella, R., W. Jiang, G. C. Chen, M. Curto, and J. Settleman. 2003. Modulation of Rho GTPase signaling regulates a switch between adipogenesis and myogenesis. Cell 113:147-158. [DOI] [PubMed] [Google Scholar]
- 77.Spooncer, E., C. M. Heyworth, A. Dunn, and T. M. Dexter. 1986. Self-renewal and differentiation of interleukin-3-dependent multipotent stem-cells are modulated by stromal cells and serum factors. Differentiation 31:111-118. [DOI] [PubMed] [Google Scholar]
- 78.Stappenbeck, T. S., and J. I. Gordon. 2000. Rac1 mutations produce aberrant epithelial differentiation in the developing and adult mouse small intestine. Development 127:2629-2642. [DOI] [PubMed] [Google Scholar]
- 79.Tian, X., G. Rusanescu, W. Hou, B. Schaffhausen, and L. A. Feig. 2002. PDK1 mediates growth factor-induced Ral-GEF activation by a kinase-independent mechanism. EMBO J. 21:1327-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Urano, T., R. Emkey, and L. A. Feig. 1996. Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 15:810-816. [PMC free article] [PubMed] [Google Scholar]
- 81.Vastrik, I., B. J. Eickholt, F. S. Walsh, A. Ridley, and P. Doherty. 1999. Sema3A-induced growth-cone collapse is mediated by Rac1 amino acids 17-32. Curr. Biol. 9:991-998. [DOI] [PubMed] [Google Scholar]
- 82.Vega, I. E., and S. C. Hsu. 2001. The exocyst complex associates with microtubules to mediate vesicle targeting and neurite outgrowth. J. Neurosci. 21:3839-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.White, M. A., C. Nicolette, A. Minden, A. Polverino, L. Vanaelst, M. Karin, and M. H. Wigler. 1995. Multiple ras functions can contribute to mammalian-cell transformation. Cell 80:533-541. [DOI] [PubMed] [Google Scholar]
- 84.White, M. A., T. Vale, J. H. Camonis, E. Schaefer, and M. H. Wigler. 1996. A role for the Ral guanine nucleotide dissociation stimulator in mediating Ras-induced transformation. J. Biol. Chem. 271:16439-16442. [DOI] [PubMed] [Google Scholar]
- 85.Wolthuis, R. M. F., B. Bauer, L. J. vantVeer, A. M. M. deVriesSmits, R. H. Cool, M. Spaargaren, A. Wittinghofer, B. M. T. Burgering, and J. L. Bos. 1996. RalGDS-like factor (Rlf) is a novel Ras and Rap 1A-associating protein. Oncogene 13:353-362. [PubMed] [Google Scholar]
- 86.Wolthuis, R. M. F., B. Franke, M. vanTriest, B. Bauer, R. H. Cool, J. H. Camonis, J. W. N. Akkerman, and J. L. Bos. 1998. Activation of the small GTPase Ral in platelets. Mol. Cell. Biol. 18:2486-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yasui, H., H. Katoh, Y. Yamaguchi, J. Aoki, H. Fujita, K. Mori, and M. Negishi. 2001. Differential responses to nerve growth factor and epidermal growth factor in neurite outgrowth of PC12 cells are determined by Rac1 activation systems. J. Biol. Chem. 276:15298-15305. [DOI] [PubMed] [Google Scholar]
- 88.Zhang, J., and H. F. Lodish. 2004. Constitutive activation of the MEK/ERK pathway mediates all effects of oncogenic H-ras expression in primary erythroid progenitors. Blood 104:1679-1687. [DOI] [PubMed] [Google Scholar]
- 89.Zheng, R., M. Levis, O. Piloto, P. Brown, B. R. Baldwin, N. C. Gorin, M. Beran, Z. Zhu, D. Ludwig, D. Hicklin, L. Witte, Y. Li, and D. Small. 2004. FLT3 ligand causes autocrine signaling in acute myeloid leukemia cells. Blood 103:267-274. [DOI] [PubMed] [Google Scholar]
- 90.Zou, X., and K. Calame. 1999. Signaling pathways activated by oncogenic forms of Abl tyrosine kinase. J. Biol. Chem. 274:18141-18144. [DOI] [PubMed] [Google Scholar]