Abstract
Hepatitis C virus (HCV) infection is frequently associated with the development of hepatocellular carcinomas and non-Hodgkin's B-cell lymphomas. Previously, we reported that HCV infection causes cellular DNA damage and mutations, which are mediated by nitric oxide (NO). NO often damages mitochondria, leading to induction of double-stranded DNA breaks (DSBs) and accumulation of oxidative DNA damage. Here we report that HCV infection causes production of reactive oxygen species (ROS) and lowering of mitochondrial transmembrane potential (ΔΨm) in in vitro HCV-infected cell cultures. The changes in membrane potential could be inhibited by BCL-2. Furthermore, an inhibitor of ROS production, antioxidant N-acetyl-l-cysteine (NAC), or an inhibitor of NO, 1400W, prevented the alterations of ΔΨm. The HCV-induced DSB was also abolished by a combination of NO and ROS inhibitors. These results indicated that the mitochondrial damage and DSBs in HCV-infected cells were mediated by both NO and ROS. Among the HCV proteins, core, E1, and NS3 are potent ROS inducers: their expression led to DNA damage and activation of STAT3. Correspondingly, core-protein-transgenic mice showed elevated levels of lipid peroxidation and oxidatively damaged DNA. These HCV studies thus identified ROS, along with the previously identified NO, as the primary inducers of DSBs and mitochondrial damage in HCV-infected cells.
Chronic liver injury by hepatitis C virus (HCV) leads to inflammation and liver regeneration over a period of time and induces cirrhosis, thus paving the way for events proceeding to liver neoplasia (5). Oxidative stress, imposed either directly by the virus or by the host-immune response, is a potentially important pathogenic mechanism in hepatitis C virus disease as well as other chronic liver diseases to initiate and promote multistage carcinogenesis (9, 28). Oxidants not only kill target cells but may also overwhelm cellular antioxidant defenses of neighboring cells, leading to damage of DNA, producing 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG), which is a typical form of oxidatively damaged DNA (27). Indeed, elevated levels of 8-oxodG has been reported in chronic hepatitis C patients (22). Liver tissues from HCV-infected patients show evidence of lipid peroxidase-protein adducts (19), and antioxidant therapy alleviates the extent of liver injury in chronic hepatitis C (17). Patients with chronic hepatitis C often exhibit increased production of tumor necrosis factor-α (TNF-α), a cytokine that can produce oxidative stress by stimulating the release of reactive oxygen species (ROS), such as superoxide radical and hydrogen peroxide (24). In addition, HCV uses the host endoplasmic reticulum (ER) as the primary site of envelope glycoprotein biogenesis and may thus induce the ER stress response (1). Mild ER stress inhibits the initiation of protein synthesis and slows cell growth, while extreme or prolonged ER stress can lead to apoptosis (23). Therefore, HCV may induce liver damage and hepatocellular carcinoma by a combination of mitochondrial damage and ER damage through ROS production.
Previously, we reported that HCV infection, through the core and NS3 proteins, induced NO, which, in turn, caused double-stranded DNA breaks (DSBs) (30). NO inactivates the mitochondrial respiratory chain enzymes and triggers blockade of the electron transfer chain and disruption of the mitochondrial transmembrane potential (14, 33). It may also increase electron leakage, followed by hypergeneration of endogenous ROS, leading to subsequent activation of caspases and apoptosis (33, 43). NS3 has been reported to induce ROS through NADPH oxidase (4, 41). Viral proteins may also directly activate ROS production without being mediated through NO, as we have shown that HCV E1 protein does not induce NO, but causes DSBs, presumably through the production of ROS (30). These combined effects of viral proteins may synergistically induce ROS. ROS play an important role, both as regulators of transcription factors and inhibitors of protein tyrosine phosphatases (10), as oxidative stress can trigger STAT3 tyrosine phosphorylation and its nuclear translocation (2, 6). We reasoned that HCV-induced ROS might participate in oxidative DNA/lipid damage and their downstream signal transduction pathways, leading to cancer.
These observations led us to examine whether HCV infection induces ROS. We studied the formation of endogenous ROS in HCV-infected B cells and the biological consequences of ROS production. Our studies demonstrated that ROS play a critical role in HCV pathogenesis.
MATERIALS AND METHODS
Mice.
Mice expressing the HCV core protein of genotype 1b under the control of the human elongation factor (EF) 1a promoter were generated (29) and bred at the USC transgenic mouse facility.
Establishment of NS3/4A inducible cell lines.
TRNS3/4A cell, which expresses HCV NS3/4A under an inducible Tet-On promoter, was established by transfection of pTRE-Tight-NS3/4A into U-2 OS cells and selected under G418 and puromycin. A cDNA fragment comprising nucleotides 3417 to 5474 (corresponding to amino acids 1027 to1711) of the HCV Con1 strain (genotype 1b) was amplified by PCR from pUC-Replicon (26) by using the sense primer 5′-GCACGAATTCACCATGGCGCCTATTACGGCCTACTCCCAACAGAC-3′ (the EcoRI site is underlined; the engineered translation initiation codon is double underlined) and the reverse primer 5′-GCTGTCTAGATTAGCACTCTTCCATCTCATCGAACTCCCGGTAA-3′ (the XbaI site is underlined; the engineered ochre stop codon is double underlined). PCR products were cloned into the EcoRI-XbaI sites of pTRE-Tight (CLONTECH) to yield the expression construct pTRE-NS3-4A. This construct allows expression of the NS3-4A proteins under the transcriptional control of a reverse tetracycline-controlled transactivator-dependent promoter. The constitutively reverse tetracycline-controlled transactivator-expressing, U-2 OS (human osteosarcoma) Tet-On cell line (32) (CLONTECH) was cotransfected with pTRE-NS3-4A and linearized puromycin selection marker (CLONTECH). G418 and puromycin double-resistant clones (TRNS3/4A cells) were isolated, screened for tightly regulated HCV NS3 gene expression, and characterized by Western blot analysis in the absence and presence of doxycycline (Sigma). Cloned TRNS3/4A cells were cultured in McCoy's 5A medium (Invitrogen) with 10% fetal bovine serum, 200 μg/ml G418, and 0.5 μg/ml of puromycin. To induce NS3 expression, the cell was treated with 1 μg/ml of doxycycline (Sigma) and incubated for 48 h.
Cells and electroporation.
Raji cells were obtained from ATCC and cloned before uses. The cells were maintained in RPMI medium 1640 (Invitrogen). The HCV (genotype 2B) used for Raji cell infection in this study was obtained from the supernatant of an HCV-producing B-cell line (SB cells) established from an HCV-infected splenic tumor (39). Typically, the supernatant (5 ml culture fluid) contained 1 × 105 viral RNA copies and was used for infection of Raji cells at a multiplicity of infection of 1. Approximately 80% of cells could be infected under this condition (37). For experiments involving BCL-2 overexpression, the empty vector or BCL-2 expression vector (kindly provided by Brian Lever from McMaster University) was transfected into Raji cells by electroporation using a Bio-Rad Gene Pulser at 300 V and 0.975 μF. After electroporation, the cells were allowed to recover in the complete medium and cultured for 2 days. Then, the transfected cells were selected in the presence of G418 (GIBCO BRL) at 2 mg/ml.
Detection of ROS and mitochondrial membrane potential.
Cells were labeled simultaneously with dihydroethidium (HE; Molecular Probes), which is oxidized by ROS to become ethidium (Eth) and emits red fluorescence, and 3,3′-dihexyloxacarbocyanine iodide [DiOC6(3), Molecular Probes], which is incorporated into mitochondria in nonlinear dependence on mitochondrial membrane potential (ΔΨm) and emits within the spectrum of green light. Conversion of HE to ethidium is carried out by superoxide anions. Once oxidized, ethidium is free to intercalate into DNA in the nucleus, where it emits fluorescence at 605 nm. The cells were labeled with both HE and DiOC6(3) for 15 min and analyzed immediately by fluorescence-activated cell sorter.
Immunoblotting.
Exponentially growing cells were harvested using a lysis buffer containing 50 mM Tris-HCl (pH 8), 150 mM NaCl, 2 mM phenylmethylsulfonyl fluoride, 1% Nonidet P-40 plus a recommended dose of complete protease inhibitor mixture tablets (Roche Diagnostics). Proteins were resolved by electrophoresis in sodium dodecyl sulfate-polyacrylamide gels (Invitrogen) and transferred onto nitrocellulose membranes (Immobulon-P; Millipore). The following primary antibodies were used for immunoblotting procedures: monoclonal anti-human BCL-2 (Santa Cruz Biotechnology) and anti-STAT3 and anti-STAT3 phosphotyrosine-705 (Cell Signaling Technology). The proteins were detected using peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin antibodies. Immunoreactivity was visualized by an enhanced chemiluminescence detection system (Amersham).
Ligation-mediated PCR.
Ligation-mediated PCR was performed as previously described (31) in the presence or absence of ROS inhibitor (NAC or superoxide dismutase [SOD]), nitric oxide inhibitor (1400W), or irreversible inhibitor for caspase-3, caspase-7, and caspase-10 (Z-Asp[methyl ester {OME}]-Glu[OME]-Val[OME]-Asp[OME]-fluoromethylketone [Z-DEVD-FMK]; MP Biomedicals, Irvine, CA].
Immunostaining.
STAT3 localization was determined by staining with anti-STAT3 monoclonal antibody, followed by detection with an appropriate fluorescein isothiocyanate-conjugated secondary antibody.
Transfection and luciferase assay.
The STAT3 reporter gene construct pLucTKS3, containing the human C-reactive-protein gene acute-phase response element (cAPRE), which includes seven copies of the STAT3-binding site, and a luciferase reporter gene, was a generous gift from Richard Jove of H. Lee Moffitt Cancer Center and Research Institute, University of South Florida College of Medicine (Tampa Bay, Florida) (42). Huh7 cells were grown on six-well plates and transfected with pLucTKS3, pRL, a Renilla luciferase expression plasmid, and dominant-negative STAT3 mutants. Transfection was performed using FuGENE 6 transfection reagent (Roche Diagnostics) or Gene Pulser II (Bio-Rad). After 48 h, cells were lysed and assayed for luciferase activities using a dual luciferase reporter assay system (Promega). Firefly luciferase activities were normalized to the internal control Renilla luciferase activity.
Measurement of lipid peroxidation products.
Appropriate amounts of cell culture (2 × 107 to 4 × 107 cells) or tissue homogenates (200 mg liver tissue) were prepared by sonication and stored at −70°C with 5 mM butylated hydroxytoluene (Sigma). For cells expressing viral proteins, cell lysates were prepared at 72 h after transfection. 4-Hydroxyalkenals and malondialdehyde were measured in the homogenates using a commercial assay (LPO-586; OXIS International Inc., Portland, OR). Protein concentration was determined by the Bradford assay (Bio-Rad).
Detection of 8-oxodG.
Cell or tissue lysates (100 μl) were incubated with 100 μg/ml hyaluronidase for 1 h at 37°C. The samples were then heated to 95°C for 5 min, cooled rapidly on ice, and digested for 2 h with 10 U of nuclease P1 (United States Biological, Swampscott, MA) at 37°C, followed by incubation with 2 U of alkaline phosphatase at 37°C for 1 h. The prepared samples were assayed using a commercial 8-oxodG-specific competitive enzyme-linked immunosorbent assay kit (OXIS Research).
Statistical analysis.
Statistical analysis of the data was performed by χ2 test. P values of <0.05 were considered to be statistically significant.
RESULTS
HCV induces ROS and reduces mitochondrial membrane potential.
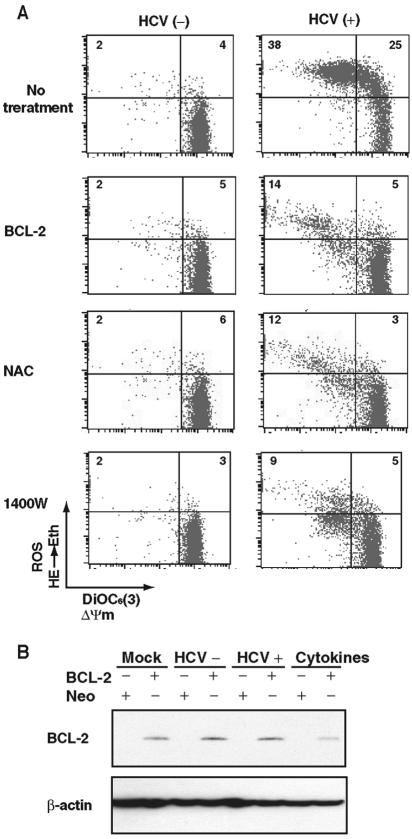
To understand the mechanism of HCV-induced cell damage, we measured mitochondrial membrane potential and ROS production, since HCV infection induces nitric oxide (NO) production (30), which in turn may disrupt electron transport in mitochondria and damages mitochondria, leading to an outburst of ROS (7). For this purpose, Raji cells were infected with HCV or UV-inactivated HCV; mitochondrial membrane potential and ROS levels were determined by using DiOC6(3) and HE, respectively, at 12 days postinfection. The results showed that HCV infection caused a significant increase in ROS levels in the cells (Fig. 1A, top panel). Simultaneously, the mitochondrial membrane potential (ΔΨm) decreased in the HCV-infected cells (Fig. 1A, upper left quadrants). To understand the mechanism of ROS induction and the decrease of ΔΨm in the HCV-infected cells, we first used an inhibitor of executor of apoptosis, BCL-2, during HCV infection. BCL-2 substantially reduced the extents of reduction of ΔΨm and increase of ROS in HCV-infected cells (Fig. 1A), which is consistent with the previous reports that BCL-2 expression normalizes ΔΨm and ROS production (38, 40). The expression of BCL-2 was confirmed by immunoblotting (Fig. 1B). Significantly, treatment with an ROS inhibitor (NAC) or an inducible nitric oxide synthase (iNOS) inhibitor (1400W) also prevented the production of ROS and reduction of mitochondrial membrane potential in HCV-infected cells (Fig. 1A). These results indicated that HCV infection reduces mitochondrial membrane potential through the production of both ROS and NO.
FIG. 1.
(A) HCV-induced changes in mitochondrial membrane potential ΔΨm and ROS production in Raji cells. To measure mitochondrial membrane potential and ROS production, cells were incubated with DiOC6(3) and HE, respectively, at 37°C for 15 min. An experiment representative of four experiments is shown. In some experiments, the cells were treated with different inhibitors during virus infection as indicated. For BCL-2 expression, the cells were stably transfected with the BCL-2 expression plasmids before HCV infection. The numbers in each quadrant represent percentages of total cell population. (B) BCL-2 expression was confirmed by immunoblotting. β-Actin served as a loading control.
Core, E1, and NS3 induce ROS.
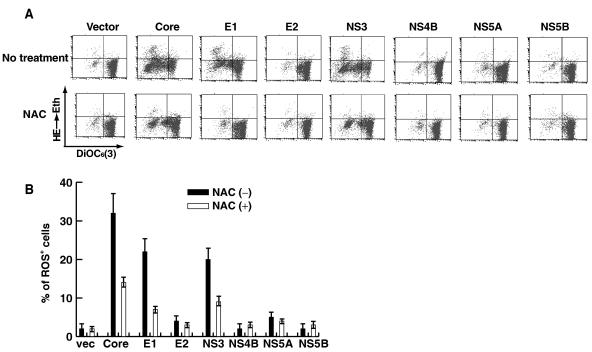
We have previously shown that HCV-induced NO production was mediated through core and NS3 proteins (30). To determine which viral gene products are responsible for ROS production, we examined ROS levels in Raji cells expressing individual viral proteins by transiently transfecting with an individual-protein-expressing plasmid. The results showed that, among all the viral proteins examined, core, E1, and NS3 proteins induced enhanced ROS production (Fig. 2A, upper panels, and B). Correspondingly, mitochondrial membrane potential was also reduced by the expression of these three proteins. The expression of these viral proteins was confirmed by immunoblotting (data not shown; see reference 30). The ROS inhibitor NAC substantially reduced viral-protein-induced ROS production (Fig. 2A, lower panels, and B) and restored mitochondrial membrane potential (Fig. 2A). These results indicated that intracellular expression of HCV core, E1, and NS3 proteins induces ROS and causes mitochondrial damage. Significantly, the reductions of ΔΨm induced by core and NS3 were only partially restored by NAC, consistent with the findings that these two proteins also induced NO (30), which may independently contribute to the damage of mitochondrial membrane. In contrast, the E1-induced ΔΨm reduction was almost completely reversed by the NAC treatment. Previously, we showed that E1 did not induce NO production (30).
FIG. 2.
(A) HCV core, E1, and NS3 induce ROS. Raji cells were transfected with plasmids expressing the individual viral proteins in the presence or absence of NAC and analyzed as explained in the legend to Fig. 1. (B) Quantitative measurement of viral-protein-induced ROS in the presence or absence of NAC from panel A. vec, vector.
HCV-induced ROS generate DSBs.
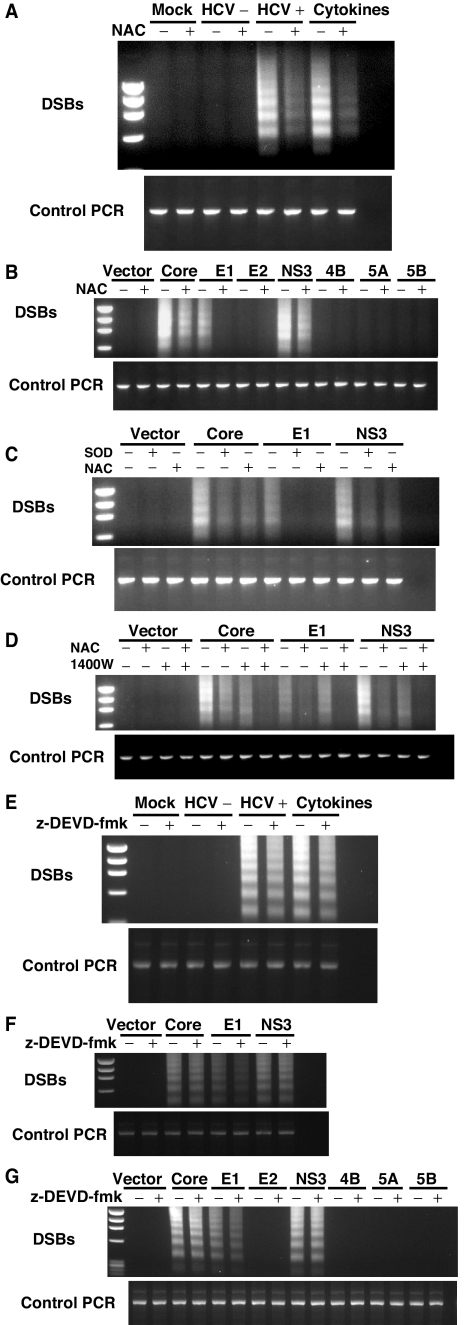
Previously, we demonstrated that HCV infection induces DSBs by excessive NO production induced by core and NS3 (30). Interestingly, E1 protein did not induce NO, and yet it caused DSBs (30). The finding that E1 induced ROS production (Fig. 2) suggested that ROS production may contribute to HCV-induced DSBs. Furthermore, ROS have been reported to induce DNA damage directly (44). We therefore analyzed the effects of the ROS inhibitor NAC on the extent of DSBs in HCV-infected cells. When HCV-infected cells were treated with NAC, there was a significant decrease in DSB formation (Fig. 3A), suggesting that HCV-induced ROS induce DSBs (Fig. 3A). Similarly, DSBs induced by a combination of various cytokines (TNF-α, gamma interferon, and interleukin-1β [IL-1β]) was also inhibited by NAC, as previously reported (30). These results indicated that HCV-induced DSBs were largely caused by ROS.
FIG. 3.
(A) HCV- or (B) HCV protein-induced DSBs. DSBs were detected by linker-ligation PCR as previously reported (31). HCV-infected cells were analyzed 12 days after infection. The viral-protein-expressing cells were analyzed 5 days after transfection. Some experiments were performed in the presence of ROS and NO inhibitors, respectively. The positive and negative controls used are indicated. Cytokines (TNF-α, gamma interferon, and IL-1β) were used as positive controls for double-stranded DNA breaks. HCV+, HCV; HCV−, UV-inactivated HCV. (B) ROS inhibitor (NAC) prevented viral-protein-induced ROS. (C) NAC or SOD treatment completely inhibited E1-induced DSBs, but partially inhibited core or NS3-induced DSBs. (D) NAC and iNOS inhibitor (1400W) together completely inhibited viral-protein-induced DSBs. (E to G) Caspase inhibitor (Z-DEVD-FMK) did not inhibit viral-protein-induced DSBs.
ROS induced by core, E1, and NS3 contribute to DSBs.
Since core and NS3 induced NO (30), while core, E1, and NS3 induced ROS (Fig. 2), we next examined the relative contributions of these viral proteins to DSBs. We transiently expressed individual viral proteins into Raji cells in the presence or absence of ROS inhibitor. Among all of the HCV proteins, core, E1, and NS3 expression induced DSBs in Raji cells (Fig. 3B), in agreement with the finding that all three proteins induced ROS (Fig. 2). The presence of NAC completely inhibited DSBs induced by E1 but only partially reduced DSBs induced by core and NS3 (Fig. 3B). In addition, free radical scavenger SOD gave a similar pattern of inhibition (Fig. 3C), indicating that E1-induced DSBs was mediated entirely by ROS, but the core- and NS3-induced DSBs were only partially dependent on ROS. When both ROS and iNOS inhibitors (NAC and 1400W) were used, the core- and NS3-induced DSBs were completely blocked (Fig. 3D). These results were consistent with our previous report that the core- and NS3-induced NO contributed to DSBs (30). Therefore, E1-induced DSBs were caused by ROS alone, whereas core- and NS3-induced DSBs were caused by a combination of NO and ROS.
Caspase inhibitor did not significantly inhibit HCV-induced DSBs.
Since DSBs can also be generated by cellular apoptosis, we performed the virus infection in the presence of a caspase inhibitor (Z-DEVD-FMK). The results showed that the caspase inhibitor had only a very minor inhibitory effect on the DSBs associated with virus infection (Fig. 3E). Furthermore, the E1-induced DSBs were only partially inhibited by the caspase inhibitor. In contrast, caspase inhibitor did not inhibit core- or NS3-induced DSBs (Fig. 3F and G). These results indicated that the HCV-induced ROS contributed to the cascade of events leading to DSB generation independently of caspase activation. These results indicate that core- and NS3-induced DSBs were not mediated through caspase activation, but E1-induced DSBs were partially mediated through caspase activation.
HCV induces tyrosine phosphorylation and nuclear translocation of STAT3.
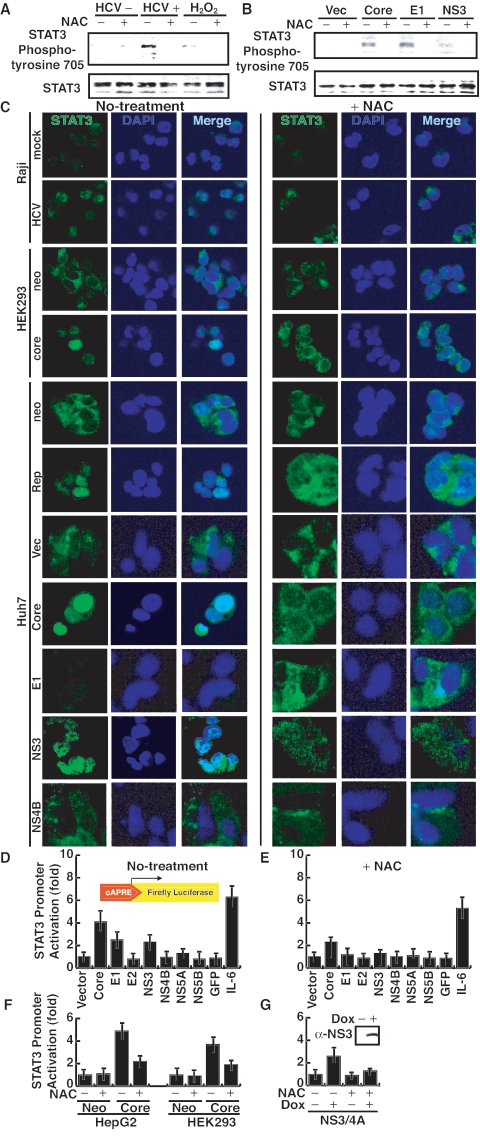
To understand the mechanism of DSB formation caused by HCV-induced ROS, we studied the cascade of events following HCV infection. We first examined the status of STAT3 tyrosine phosphorylation in HCV-infected cells, since ROS have been reported to activate STAT3 (2, 6). Nuclear lysates were immunoprecipitated with a polyclonal anti-STAT3 antibody and then immunoblotted with an antiphosphotyrosine antibody, since following activation, STAT3 is phosphorylated at tyrosine residue 705 and translocated into the nucleus, where it binds cognate DNA sequences as dimers (37). Furthermore, ROS have been reported to prevent dephosphorylation of STAT3 (13). We found that HCV infection induced an enhanced level of phosphorylated STAT3, similar to the H2O2 treatment, in Raji cells (Fig. 4A). Similarly, the expression of core, E1, and NS3 activated STAT3 phosphorylation (Fig. 4B). The overall STAT3 level remained unchanged. In addition, an HEK293 cell line stably expressing HCV core protein also has an elevated level of phosphorylated STAT3 protein (data not shown). All of the induced STAT3 phosphorylation was abolished by treatment with the ROS inhibitor NAC (Fig. 4A and B).
FIG. 4.
(A and B) HCV infection constitutively activates STAT3 tyrosine phosphorylation. Immunoblot analysis of STAT3 protein in cell extracts infected with HCV or transfected with individual HCV proteins. Cellular lysates were immunoprecipitated with anti-STAT3 polyclonal serum and immunoblotted with antiphosphotyrosine monoclonal antibody. Vec, vector. (C) Immunostaining of STAT3 under different conditions. Cells were stained with anti-STAT3 antibody (green) and DAPI (4′,6′-diamidino-2-phenylindole) (blue). neo, neomycin resistance gene transfected; mock, mock infected. (D and E) Transcriptional activation of STAT3 by individual HCV proteins in the absence of NAC (D) or in the presence of NAC (E) by using a luciferase reporter assay under the STAT3-regulated promoter. Huh7 cells were transfected with a STAT3 reporter gene construct (pLucTKS3), an internal control plasmid (pRL-null), and various viral-protein constructs. To serve as a control, a control plasmid (pLucTK) (vector) was used instead of the reporter gene construct. Forty-eight hours after transfection, cells were lysed and analyzed for luciferase activity. Vertical bars represent standard deviations (n = 4). GFP, green fluorescent protein. (F) Core stable transformants (HepG2 and HEK293 cells) versus neomycin resistance gene-transfected (Neo) cells in the luciferase assay. (G) Inducible (by doxycycline [Dox]) expression of NS3 in a stable transformant NS3 expression was confirmed by Western blotting using an anti-NS3 antibody (inset). Doxycycline was added in the culture supernatant of the cell line.
To determine whether ROS-induced STAT3 phosphorylation affected subcellular localization of STAT3, we immunostained STAT3 in HCV-infected cells and cells expressing the various viral proteins. Most of STAT3 was found to be localized in the nucleus of HCV-infected cells, while most of it was formed in the cytoplasm of the mock-infected cells (Fig. 4C, top two panels). In the infected cells treated with NAC, STAT3 was found predominantly in the cytoplasm. In Huh7 cells transiently expressing core or E1, STAT3 was also localized mainly in the nucleus. However, in the NS3-expressing cells, some of STAT3 was found in the cytoplasm. One possibility is that the expression level of NS3 was too low. Another possibility is that NS3 protein caused substantial effects on other signaling pathways (12) which may affect the STAT3 nuclear translocation. In Huh7 cells harboring a subgenomic HCV replicon, which expresses HCV nonstructural proteins (NS3 through NS5B), STAT3 was also predominantly in the nucleus (Fig. 4C). In contrast, in Huh7 cells expressing the neomycin resistance or HCV NS4B protein or transfected with the vector plasmid, STAT3 was localized mainly in the cytoplasm (Fig. 4C). In the HEK293 cell line stably expressing the core protein, STAT3 was also found predominantly localized in the nucleus, in contrast to cells expressing the neomycin resistance gene. These results combined demonstrate that HCV infection constitutively activated STAT3, resulting in its nuclear translocation. This activation was mediated by core, E1, and NS3 proteins. When these cells were treated with NAC, most of STAT3 reverted back to the cytoplasm. Therefore, we concluded that the nuclear translocation of STAT3 induced by HCV proteins was mediated by ROS.
HCV-activated STAT3 enhances transcriptional activities of STAT3-regulated genes.
We next attempted to demonstrate that ROS induced by HCV infection or viral proteins can enhance the transcription of cellular genes regulated by STAT3. We performed a luciferase reporter assay using a STAT3-regulated reporter plasmid DNA in Huh7 and Raji cells expressing the various HCV proteins. The results showed that core, E1, and NS3 enhanced the luciferase activities by two- to fourfold in Huh7 cells, while none of the other viral proteins caused any increase (Fig. 4D). The amounts of the viral proteins expressed in the transfected cells were determined by immunoblotting (data not shown; see also reference 30). As a control, interleukin-6 (IL-6) treatment enhanced the STAT3-regulated luciferase activity by sixfold. When the cells were treated with NAC, the luciferase activity regulated by the STAT3-responsive element was reduced. In contrast, the IL-6-induced enhancement of the STAT3-regulated transcription activities was not inhibited by the NAC treatment (Fig. 4D and E), consistent with the previous report (45). The enhancement of luciferase activity was similarly observed in stable cell lines expressing core protein (Fig. 4F) or NS3 protein (Fig. 4G), or harboring an HCV subgenomic replicon (data not shown). These results further support the conclusion that HCV core, E1 and NS3 activate STAT3 through the production of ROS.
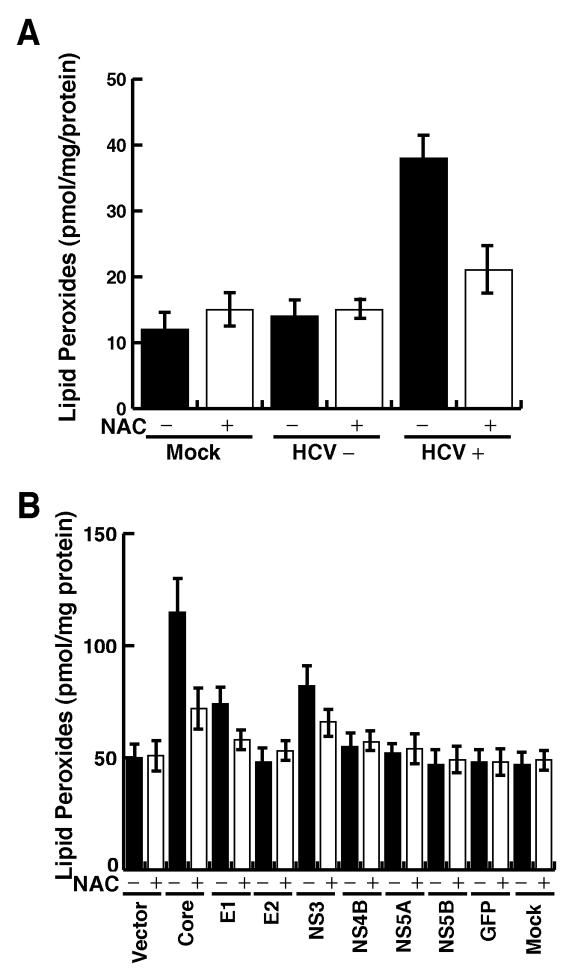
HCV enhanced lipid peroxidation.
ROS are known to cause lipid peroxidation (25), and HCV core protein has been shown to increase lipid peroxidation products in the cells (35). To determine whether the HCV-induced ROS production could lead to enhanced lipid peroxidation, we measured the total lipid peroxidation products in extracts from HCV-infected Raji cells. The results showed that HCV-infected cells had a significantly higher level of lipid peroxides than that in the uninfected counterpart (Fig. 5A). The expression of core, E1, and NS3 alone also resulted in a significant increase in the amounts of lipid peroxide products in Huh7 and HepG2 cells (Fig. 5B and data not shown). The ROS inhibitor NAC reduced the oxidation of lipids. These results suggest that core, E1, and NS3 protein expression can cause oxidative injury in both Raji and hepatocyte cell lines.
FIG. 5.
(A) HCV infection induces lipid peroxidation. Total cellular lipid peroxidation products 4-hydroxyalkenals and malondialdehyde were determined by colorimetric lipid peroxidation assay from cellular extracts of HCV-infected and uninfected Raji cells. HCV+, HCV; HCV−, UV-inactivated HCV; mock, mock infected. (B) Huh7 cells transfected with individual-viral-protein expression constructs were analyzed for lipid peroxides at 48 h posttransfection.
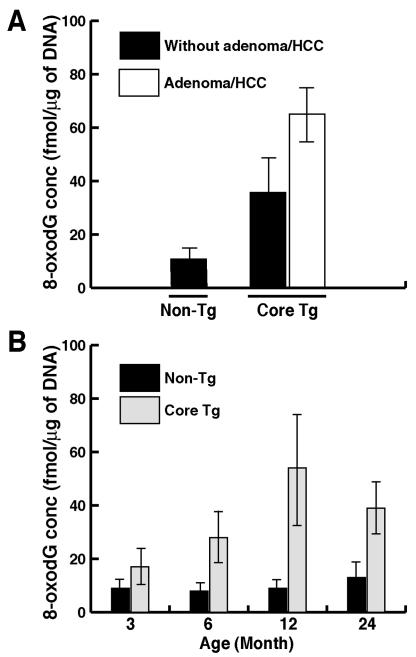
Core protein induces oxidative DNA damage in a murine model.
To determine whether the core protein can induce ROS and increase oxidative DNA damage in whole animal, we measured a hepatic oxidative DNA damage product (8-oxodG) in transgenic mice expressing HCV core protein. The 8-oxodG is thought to arise via hydroxyl-radical attack in the C-8 position of deoxyguanosine and is thus a specific marker for oxidative damage (20, 21). Livers of core-transgenic mice at the age of 12 months, even in the absence of adenoma or hepatocellular carcinoma (HCC), had a higher level of 8-oxodG (34 ± 11 [n = 9] versus 12 ± 4 fmol/μg of DNA [nontransgenic littermates, n = 14]). In the liver of transgenic mice with adenoma or HCC at age of 12 months, the 8-oxodG level was even higher (65 ± 20 fmol/μg of DNA [n = 4]), almost fivefold higher than that in nontransgenic mice (Fig. 6A). These results indicate that oxidative DNA damage was significantly higher in HCV core transgenic mice.
FIG. 6.
(A) Oxidative DNA damage in HCV core transgenic mice. The 8-oxodG in the livers of the core-transgenic (Core Tg) mice and age-matched nontransgenic littermates at 12 months of age with or without evidence of adenoma or HCC was measured. (B) Oxidative DNA damage as a function of age (n = 15). DNA was isolated from livers of transgenic mice or age-matched nontransgenic controls from 3 to 24 months of age.
To determine whether the increased production of 8-oxodG was due to the expression of core or merely reflected an age-associated accumulation, 8-oxodG levels in the liver at different age groups of transgenic and nontransgenic mice were examined. The results showed that 8-oxodG levels remained almost the same at different ages of nontransgenic mice (Fig. 6B). In contrast, the 8-oxodG level in the livers of core-transgenic animals increased with age (Fig. 6B), corresponding to the increased liver pathology (data not shown). These results indicate that the increases in 8-oxodG concentrations in the core-transgenic mice were not merely due to aging process, but more likely reflected the pathological changes of the liver.
DISCUSSION
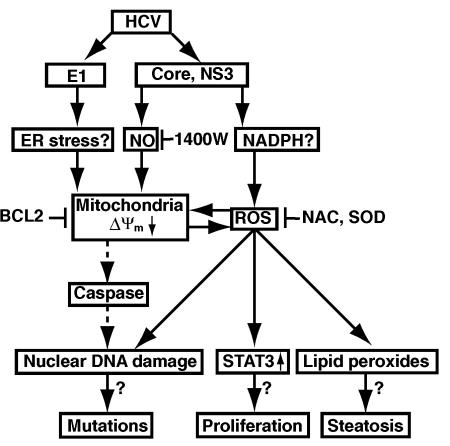
Our present study provides evidence that HCV alters mitochondrial ΔΨm, and increases ROS production. Treatment of the HCV-infected cells with ROS inhibitors effectively prevented mitochondrial damage and production of ROS. The production of ROS leads to oxidative DNA damage and lipid peroxidation in the HCV-infected cells. In addition, we showed that HCV-induced ROS activated STAT3, which may lead to the downstream proliferative responses, as STAT3 has been reported as an oncogene (2). Therefore, ROS, together with the previously reported NO (30), directly or indirectly play important roles in HCV pathogenesis. The multiple effects mediated through ROS may account for steatosis (through lipid peroxides) and oncogenesis (through DNA mutations and STAT3 activation) associated with HCV. Also, they can explain the acute hepatocyte damage (through the production of STAT3) induced by HCV. The cascade of these events is presented in Fig. 7.
FIG. 7.
Hypothetical pathways mediated by HCV-induced ROS.
ROS are generated as a consequence of mitochondrial electron transport caused by a small proportion of the electron flow interacting with oxygen molecules before reaching the cytochrome oxidase complex. The biochemical mechanisms behind the HCV-mediated mitochondrial damage remain to be fully investigated. Our data showed that E1 protein, together with core and NS3, are responsible for the ROS production in HCV-infected cells. Core and NS3 induce NO production (30), which is expected to cause mitochondrial membrane damage and decrease ΔΨm due to the opening of permeability transition pores followed by the depolarization of the inner mitochondrial membrane and massive ROS production (16). NO could also be activated by reacting with another free radical, superoxide (O2−), to form strong oxidant peroxynitrite anions (ONOO−), which are potent oxidants irreversibly inhibiting multiple respiratory complexes (complexes I, II, and IV) and aconitase, and activating proton leak and permeability transition pore (3). Downregulation of complex IV activity led to up-regulation of peroxynitrite anion, which results in increases in O2− and H2O2, which may contribute to NO-induced cell death (3). We initially hypothesized that HCV-induced NO disrupts mitochondrial electron transport, leading to an outburst of ROS. Indeed, core and NS3 activated the iNOS promoter, enhancing its expression (30). However, our data presented here showed that NO is not enough to account for the HCV-induced DNA damage, since the simultaneous application of inhibitors of both NO and ROS was required for complete abolition of HCV-induced DNA damage. Thus, core and NS3 proteins may activate other ROS sources to amplify ROS generation cycle. In contrast, the E1-induced lipid damage and DNA damage were mediated entirely through ROS production. The mechanism of E1-induced ROS production is still not clear. One possibility is that E1 induces stress response since it is synthesized in the ER, thus, leading to mitochondrial membrane damage. ER stress has been reported to activate CHOP and caspase-12, leading to apoptosis with activation of caspase (34, 46).
It should be noted that DNA damage could be an indirect result of apoptosis induced by HCV. However, we previously demonstrated that the HCV-infected cells devoid of apoptosis also showed enhanced DSBs (31). Furthermore, in this study, we showed that the caspase inhibitor Z-DEVD-FMK did not prevent HCV-induced DSBs. Therefore, the ROS-induced cascade of events are likely important for the pathogenesis and oncogenesis associated with HCV infection.
The livers of transgenic mice expressing the core protein display marked increases in lipid peroxides even before the development of HCC. The 8-oxodG may cause mutations in cellular DNA, resulting in increased mutation frequencies in these mice. This is consistent with our model of HCV oncogenesis, which proposes that the HCV-induced mutator phenotype (31) is at least partially responsible for tumor formation (Fig. 7). The ROS-induced STAT3 activation is expected to cause significant alteration of the cell growth properties, since the activated STAT3 molecule has been shown to stimulate cell growth (2). In addition, STAT3 is also the major inducer of acute-phase pathology of liver disease (36), including acute hepatitis associated with HCV infection. Constitutive activation of STAT3 has been shown to inhibit apoptosis (8), induce transformation by oncoproteins (18), and cause B-cell differentiation, particularly in the terminal differentiation of B cells into antibody-secreting plasma cells (11, 15). Thus, the STAT3-induced dysregulation of cellular responses may explain high incidence of hepatocellular carcinomas and lymphoproliferative diseases in hepatitis C patients.
In summary, the evidence presented here shows that HCV-induced ROS play a critical role in the establishment of acute and chronic liver diseases through the induction of DNA/lipid damage and STAT3 activation. ROS and/or iNOS inhibitors may provide therapeutic measures for HCV patients to protect the liver tissue against oxidative stress and chromosomal damage.
Acknowledgments
We thank Brian Leber from McMaster University for BCL-2 expression vectors, Richard Jove from University of South Florida College of Medicine for STAT3 reporters, Tatsuo Miyamura from National Institute of Infectious Disease, Tokyo, Japan, for the E1 expression vector, and Harold Soucier from University of Southern California for fluorescence-activated cell sorter analysis.
This project was supported by NIH research grants AI 40038 and CA108302.
REFERENCES
- 1.Benali-Furet, N. L., M. Chami, L. Houel, F. De Giorgi, F. Vernejoul, D. Lagorce, L. Buscail, R. Bartenschlager, F. Ichas, R. Rizzuto, and P. Paterlini-Brechot. 2005. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene 24:4921-4933. [DOI] [PubMed] [Google Scholar]
- 2.Bromberg, J. F., M. H. Wrzeszczynska, G. Devgan, Y. Zhao, R. G. Pestell, C. Albanese, and J. E. Darnell, Jr. 1999. Stat3 as an oncogene. Cell 98:295-303. [DOI] [PubMed] [Google Scholar]
- 3.Brown, G. C. 2001. Regulation of mitochondrial respiration by nitric oxide inhibition of cytochrome c oxidase. Biochim. Biophys. Acta 1504:46-57. [DOI] [PubMed] [Google Scholar]
- 4.Bureau, C., J. Bernad, N. Chaouche, C. Orfila, M. Beraud, C. Gonindard, L. Alric, J. P. Vinel, and B. Pipy. 2001. Nonstructural 3 protein of hepatitis C virus triggers an oxidative burst in human monocytes via activation of NADPH oxidase. J. Biol. Chem. 276:23077-23083. [DOI] [PubMed] [Google Scholar]
- 5.Butel, J. S. 2000. Viral carcinogenesis: revelation of molecular mechanisms and etiology of human disease. Carcinogenesis 21:405-426. [DOI] [PubMed] [Google Scholar]
- 6.Carballo, M., M. Conde, R. El Bekay, J. Martin-Nieto, M. J. Camacho, J. Monteseirin, J. Conde, F. J. Bedoya, and F. Sobrino. 1999. Oxidative stress triggers STAT3 tyrosine phosphorylation and nuclear translocation in human lymphocytes. J. Biol. Chem. 274:17580-17586. [DOI] [PubMed] [Google Scholar]
- 7.Cassina, A., and R. Radi. 1996. Differential inhibitory action of nitric oxide and peroxynitrite on mitochondrial electron transport. Arch. Biochem. Biophys. 328:309-316. [DOI] [PubMed] [Google Scholar]
- 8.Catlett-Falcone, R., T. H. Landowski, M. M. Oshiro, J. Turkson, A. Levitzki, R. Savino, G. Ciliberto, L. Moscinski, J. L. Fernandez-Luna, G. Nunez, W. S. Dalton, and R. Jove. 1999. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 10:105-115. [DOI] [PubMed] [Google Scholar]
- 9.Factor, V. M., D. Laskowska, M. R. Jensen, J. T. Woitach, N. C. Popescu, and S. S. Thorgeirsson. 2000. Vitamin E reduces chromosomal damage and inhibits hepatic tumor formation in a transgenic mouse model. Proc. Natl. Acad. Sci. USA 97:2196-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fialkow, L., C. K. Chan, D. Rotin, S. Grinstein, and G. P. Downey. 1994. Activation of the mitogen-activated protein kinase signaling pathway in neutrophils. Role of oxidants. J. Biol. Chem. 269:31234-31242. [PubMed] [Google Scholar]
- 11.Fornek, J. L., L. T. Tygrett, T. J. Waldschmidt, V. Poli, R. C. Rickert, and G. S. Kansas. 2006. Critical role for Stat3 in T-dependent terminal differentiation of IgG B cells. Blood 107:1085-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 13.Heffetz, D., I. Bushkin, R. Dror, and Y. Zick. 1990. The insulinomimetic agents H2O2 and vanadate stimulate protein tyrosine phosphorylation in intact cells. J. Biol. Chem. 265:2896-2902. [PubMed] [Google Scholar]
- 14.Hibbs, J. B., Jr., Z. Vavrin, and R. R. Taintor. 1987. l-Arginine is required for expression of the activated macrophage effector mechanism causing selective metabolic inhibition in target cells. J. Immunol. 138:550-565. [PubMed] [Google Scholar]
- 15.Hirano, T., K. Ishihara, and M. Hibi. 2000. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 19:2548-2556. [DOI] [PubMed] [Google Scholar]
- 16.Hortelano, S., B. Dallaporta, N. Zamzami, T. Hirsch, S. A. Susin, I. Marzo, L. Bosca, and G. Kroemer. 1997. Nitric oxide induces apoptosis via triggering mitochondrial permeability transition. FEBS Lett. 410:373-377. [DOI] [PubMed] [Google Scholar]
- 17.Houglum, K., A. Venkataramani, K. Lyche, and M. Chojkier. 1997. A pilot study of the effects of d-α-tocopherol on hepatic stellate cell activation in chronic hepatitis C. Gastroenterology 113:1069-1073. [DOI] [PubMed] [Google Scholar]
- 18.Ihara, S., K. Nakajima, T. Fukada, M. Hibi, S. Nagata, T. Hirano, and Y. Fukui. 1997. Dual control of neurite outgrowth by STAT3 and MAP kinase in PC12 cells stimulated with interleukin-6. EMBO J. 16:5345-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kageyama, F., Y. Kobayashi, T. Kawasaki, S. Toyokuni, K. Uchida, and H. Nakamura. 2000. Successful interferon therapy reverses enhanced hepatic iron accumulation and lipid peroxidation in chronic hepatitis C. Am. J. Gastroenterol. 95:1041-1050. [DOI] [PubMed] [Google Scholar]
- 20.Kasai, H., and S. Nishimura. 1984. Hydroxylation of deoxyguanosine at the C-8 position by ascorbic acid and other reducing agents. Nucleic Acids Res. 12:2137-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasai, H., S. Nishimura, Y. Kurokawa, and Y. Hayashi. 1987. Oral administration of the renal carcinogen, potassium bromate, specifically produces 8-hydroxydeoxyguanosine in rat target organ DNA. Carcinogenesis 8:1959-1961. [DOI] [PubMed] [Google Scholar]
- 22.Kato, J., M. Kobune, T. Nakamura, G. Kuroiwa, K. Takada, R. Takimoto, Y. Sato, K. Fujikawa, M. Takahashi, T. Takayama, T. Ikeda, and Y. Niitsu. 2001. Normalization of elevated hepatic 8-hydroxy-2′-deoxyguanosine levels in chronic hepatitis C patients by phlebotomy and low iron diet. Cancer Res. 61:8697-8702. [PubMed] [Google Scholar]
- 23.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 24.Kizaki, M., A. Sakashita, A. Karmakar, C. W. Lin, and H. P. Koeffler. 1993. Regulation of manganese superoxide dismutase and other antioxidant genes in normal and leukemic hematopoietic cells and their relationship to cytotoxicity by tumor necrosis factor. Blood 82:1142-1150. [PubMed] [Google Scholar]
- 25.Laurent, A., C. Nicco, J. Tran Van Nhieu, D. Borderie, C. Chereau, F. Conti, P. Jaffray, O. Soubrane, Y. Calmus, B. Weill, and F. Batteux. 2004. Pivotal role of superoxide anion and beneficial effect of antioxidant molecules in murine steatohepatitis. Hepatology 39:1277-1285. [DOI] [PubMed] [Google Scholar]
- 26.Lee, K. J., J. Choi, J. H. Ou, and M. M. Lai. 2004. The C-terminal transmembrane domain of hepatitis C virus (HCV) RNA polymerase is essential for HCV replication in vivo. J. Virol. 78:3797-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis, J. G., and D. O. Adams. 1987. Inflammation, oxidative DNA damage, and carcinogenesis. Environ. Health Perspect. 76:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieber, C. S. 1997. Role of oxidative stress and antioxidant therapy in alcoholic and nonalcoholic liver diseases. Adv. Pharmacol. 38:601-628. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Z. X., H. Nishida, J. W. He, M. M. Lai, N. Feng, and G. Dennert. 2002. Hepatitis C virus genotype 1b core protein does not exert immunomodulatory effects on virus-induced cellular immunity. J. Virol. 76:990-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Machida, K., K. T. Cheng, V. M. Sung, K. J. Lee, A. M. Levine, and M. M. Lai. 2004. Hepatitis C virus infection activates the immunologic (type II) isoform of nitric oxide synthase and thereby enhances DNA damage and mutations of cellular genes. J. Virol. 78:8835-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machida, K., K. T. Cheng, V. M. Sung, S. Shimodaira, K. L. Lindsay, A. M. Levine, M. Y. Lai, and M. M. Lai. 2004. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc. Natl. Acad. Sci. USA 101:4262-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maul, R. S., and D. D. Chang. 1999. EPLIN, epithelial protein lost in neoplasm. Oncogene 18:7838-7841. [DOI] [PubMed] [Google Scholar]
- 33.Moncada, S., and J. D. Erusalimsky. 2002. Does nitric oxide modulate mitochondrial energy generation and apoptosis? Nat. Rev. Mol. Cell Biol. 3:214-220. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa, T., H. Zhu, N. Morishima, E. Li, J. Xu, B. A. Yankner, and J. Yuan. 2000. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature 403:98-103. [DOI] [PubMed] [Google Scholar]
- 35.Okuda, M., K. Li, M. R. Beard, L. A. Showalter, F. Scholle, S. M. Lemon, and S. A. Weinman. 2002. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology 122:366-375. [DOI] [PubMed] [Google Scholar]
- 36.Pahl, H. L., and P. A. Baeuerle. 1996. Activation of NF-κB by ER stress requires both Ca2+ and reactive oxygen intermediates as messengers. FEBS Lett. 392:129-136. [DOI] [PubMed] [Google Scholar]
- 37.Sasse, J., U. Hemmann, C. Schwartz, U. Schniertshauer, B. Heesel, C. Landgraf, J. Schneider-Mergener, P. C. Heinrich, and F. Horn. 1997. Mutational analysis of acute-phase response factor/Stat3 activation and dimerization. Mol. Cell. Biol. 17:4677-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimizu, S., Y. Eguchi, W. Kamiike, Y. Funahashi, A. Mignon, V. Lacronique, H. Matsuda, and Y. Tsujimoto. 1998. Bcl-2 prevents apoptotic mitochondrial dysfunction by regulating proton flux. Proc. Natl. Acad. Sci. USA 95:1455-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sung, V. M., S. Shimodaira, A. L. Doughty, G. R. Picchio, H. Can, T. S. Yen, K. L. Lindsay, A. M. Levine, and M. M. Lai. 2003. Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection. J. Virol. 77:2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Susin, S. A., N. Zamzami, M. Castedo, E. Daugas, H. G. Wang, S. Geley, F. Fassy, J. C. Reed, and G. Kroemer. 1997. The central executioner of apoptosis: multiple connections between protease activation and mitochondria in Fas/APO-1/CD95- and ceramide-induced apoptosis. J. Exp. Med. 186:25-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thoren, F., A. Romero, M. Lindh, C. Dahlgren, and K. Hellstrand. 2004. A hepatitis C virus-encoded, nonstructural protein (NS3) triggers dysfunction and apoptosis in lymphocytes: role of NADPH oxidase-derived oxygen radicals. J. Leukoc. Biol. 76:1180-1186. [DOI] [PubMed] [Google Scholar]
- 42.Turkson, J., T. Bowman, R. Garcia, E. Caldenhoven, R. P. De Groot, and R. Jove. 1998. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol. Cell. Biol. 18:2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ushmorov, A., F. Ratter, V. Lehmann, W. Droge, V. Schirrmacher, and V. Umansky. 1999. Nitric-oxide-induced apoptosis in human leukemic lines requires mitochondrial lipid degradation and cytochrome C release. Blood 93:2342-2352. [PubMed] [Google Scholar]
- 44.Yu, T. W., and D. Anderson. 1997. Reactive oxygen species-induced DNA damage and its modification: a chemical investigation. Mutat. Res. 379:201-210. [DOI] [PubMed] [Google Scholar]
- 45.Zhong, Z., Z. Wen, and J. E. Darnell, Jr. 1994. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264:95-98. [DOI] [PubMed] [Google Scholar]
- 46.Zinszner, H., M. Kuroda, X. Wang, N. Batchvarova, R. T. Lightfoot, H. Remotti, J. L. Stevens, and D. Ron. 1998. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12:982-995. [DOI] [PMC free article] [PubMed] [Google Scholar]