Abstract
Epstein-Barr virus is an orally transmitted human herpesvirus that infects epithelial cells and establishes latency in memory B lymphocytes. Movement of virus between the two cell types is facilitated by changes in amounts of an envelope glycoprotein, gp42, which are effected by interaction of gp42 with HLA class II in a B cell. Here we used the differential ability of virus to bind to CD21-positive B cells and CD21-negative epithelial cells, which is also influenced by levels of gp42, to determine that the majority of virus shed in saliva is derived from an HLA class II-negative cell.
Epstein-Barr virus (EBV) is an orally transmitted human herpesvirus that infects more than 90% of the human population. Although the majority of primary infections are asymptomatic the virus can cause infectious mononucleosis, and long-term carriage can be associated with B-lymphocyte and epithelial-cell malignancies (16). These malignancies are reflective of the predominant tropisms of the virus. Current models of infection propose that productive replication in epithelial cells of the oropharynx facilitates access to B cells in Waldeyer's ring (15, 18) where the virus can establish latency in long-lived memory B cells. Sporadic terminal differentiation of these B cells into plasmablasts triggers onset of productive replication to initiate another cycle of infection in the same or in a new host (8).
During this cycle of infection, ease of access of virus to a B cell and an epithelial cell can be driven by the cell type of origin because of differences in the proteins used for entry into each (1). Entry of virus into a B cell is initiated by attachment of virus glycoprotein gp350/220 to the complement receptor type 2, CD21 (13, 17). Attachment to a CD21-negative epithelial cell can be initiated by binding of a complex of two glycoproteins, gHgL, to an as-yet-unidentified receptor, gHgLR (12, 14). Penetration of virus into a B cell requires glycoprotein gB and a complex of three proteins, gHgL and gp42 (5, 19). It is triggered by an interaction between gp42 and HLA class II (4, 10). Penetration of an HLA class II-negative epithelial cell requires gB and a two-part gHgL complex lacking gp42 (5, 20). It is triggered by a direct interaction between gHgL and a receptor which may be the same gHgLR that mediates attachment (2). To accommodate these differences, the EBV virion contains both two-part gHgL complexes and three-part gHgLgp42 complexes with mutually exclusive functions. Only three-part gHgLgp42 complexes mediate B cell infection, and only two-part complexes lacking gp42 mediate epithelial infection (20). The presence of gp42 blocks the interaction with gHgLR (2).
In an HLA class II-positive B cell, some three-part gHgLgp42 complexes are lost to an interaction with HLA class II which targets them to a degradative pathway. The ratio of gHgLgp42 to gHgL is thereby reduced. In an HLA class II-negative epithelial cell, this does not happen, and the ratio of gHgLgp42 to gHgL is higher (1). This results in a switch of tropism, with B-cell-derived virus being slightly more infectious for an epithelial cell and epithelial-cell-derived virus being as much as 2 logs more infectious for a B cell. It suggests a model in which virus replicating in epithelial cells in the oropharynx is strongly and rapidly targeted to the B-cell compartment at the beginning of the cycle of infection, whereas virus made in a differentiating B cell has a small increase in its ability to infect epithelial cells either for amplification in the existing host or, if shed directly from differentiating cells into saliva, for infection of epithelial cells in a new host.
We recently found that levels of gp42 in virus impact not only penetration but also attachment of virus to a gHgLR-expressing epithelial cell (2). Virus derived from HLA class II-positive B cells bound to gHgLR almost as well as it bound to CD21. In contrast, virus derived from an HLA class II-negative epithelial cell bound to CD21 as well as did the B-cell-derived virus but, because of a higher load of gp42, bound poorly to gHgLR. This difference suggested that the relative ability of virus to bind to CD21 or gHgLR might be used to determine whether virus in saliva originates from an HLA class II-positive or -negative cell.
Glycoprotein gp42 binds many but not all HLA class II alleles (6). Individuals expressing different alleles then presumably lose three-part gHgLgp42 complexes at levels consistent with the alleles that they express. To control for this, lymphoblastoid cell lines (LCL) were derived from six healthy donors by transforming their T-cell-depleted blood lymphocytes (11) with virus in their own saliva, which was obtained by use of a Salivette (Starstedt, Fisher Scientific International), centrifuged to remove cells, and clarified by filtration through a 0.8-μm filter. Virus was harvested from each individual LCL after induction with phorbol esters and sodium butyrate (9) and concentrated by centrifugation (12). LCL-derived virus and virus from saliva of the same donor were bound for 1 h on ice to EBV-negative CD21-positive Akata B cells or CD21-negative gHgLR-positive AGS epithelial cells (2). DNA was isolated with a QIAamp DNA blood minikit (QIAGEN Sciences) for real-time quantitative PCR (RQ-PCR) as described elsewhere (3) with primers that amplified the BamHI K region of EBV (5′-GGATGCGATTAAGGACCTTGTT-3′ and 5′-CGTCAAAGCTGCACACAGTCA-3′; base coordinates 109677 and 109753, respectively; GenBank accession no. VO1555) and/or the cellular C-reactive protein (CRP) (5′-CTTGACCAGCCTCTCTCATGC-3′ and 5′-TGCAGTCTTAGACCCCACCC-3′; base coordinates 132705 and 132605, respectively; GenBank accession no. AL445528). The BamHI K probe (5′-CAAAGCCCGCTCCTACCTGCAATATCA-3′) was labeled with 6-carboxyfluorescein, and the CRP probe (5′-TTTGGCCAGACAGGTAAGGGCCACC-3′) was labeled with VIC (PE Applied Biosystems). Serial dilutions of DNA from IB4, a B-cell line containing five copies of EBV per cell (7), served as a standard, and EBV copy number per sample was normalized to the amount of CRP DNA. Low levels of antibodies to gp350/220 are found in the saliva of a minority of donors and might have reduced levels of B-cell binding (21). We checked for this possible confounder by determining how much saliva-derived virus was able to bind to CD21. For each donor, the amount was between 76 and 90% of that added, indicating little or no effect of antibody.
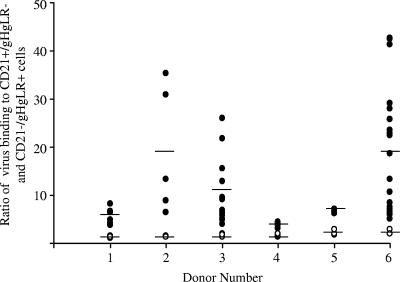
On average, slightly more LCL-derived virus bound to B cells than to epithelial cells, although the ratio of binding was never higher than 3 and the variability of binding in repeated assays was low (Table 1). There was considerably more variability in binding of saliva-derived virus in repeat assays done with saliva collected on different days (Fig. 1; not all donors shed virus in saliva in every collection). However, on average, for each donor, virus in saliva was significantly better able to bind to B cells than to epithelial cells, as summarized in Table 1.
TABLE 1.
Average ratio of binding of LCL- and saliva-derived virus to CD21+ gHgLR− EBV− Akata B cells and CD21− gHgLR+ EBV− AGS epithelial cells
| Donor | Binding ratio (no. of expts)a
|
t test P valuec | |
|---|---|---|---|
| LCL-derived virus | Saliva-derived virusb | ||
| 1 | 1.42 ± 0.22 (4)c | 5.72 ± 1.73 (6) | <0.05 |
| 2 | 1.44 ± 0.18 (4) | 18.98 ± 13.20 (5) | <0.05 |
| 3 | 1.69 ± 0.33 (3) | 10.19 ± 6.40 (15) | <0.05 |
| 4 | 1.58 ± 0.29 (4) | 3.66 ± 0.59 (5) | <0.001 |
| 5 | 2.27 ± 0.61 (3) | 6.71 ± 0.44 (3) | <0.001 |
| 6 | 2.31 ± 0.55 (3) | 18.09 ± 12.37 (21) | <0.001 |
Virus binding was measured by RQ-PCR as in Fig. 1.
Fresh saliva was collected for each experiment.
P values represent differences between LCL-derived and saliva-derived viruses for each donor.
FIG. 1.
Ratio of binding of LCL-derived virus (open symbols) and saliva-derived virus (closed symbols) from the same donor to CD21+ gHgLR− EBV− Akata B cells or CD21− gHgLR+ EBV− AGS epithelial cells. The amount of virus bound per cell was determined by dividing the RQ-PCR amplification of the BamHI K region of EBV DNA by the RQ-PCR amplification of cellular CRP. The average ratios for LCL and saliva virus binding for each donor are represented by the horizontal bars.
Previously, the relative amounts of gH and gp42 in B-cell-derived or epithelial-cell-derived virus had been determined by binding each to EBV-negative Akata B cells, staining with monoclonal antibody CL59 to gH or F-2-1 to gp42, and analyzing by flow cytometry (1). Most donors did not shed enough virus in saliva to repeat this assay. However, virus concentrated by centrifugation on four occasions from one donor who did had a higher ratio of gp42 to gH than did virus from the LCL derived from the same individual (Table 2).
TABLE 2.
Ratios of gp42 to gH in LCL-derived virus and saliva-derived virus from the same donor
| Expt | gp42/gH ratioa
|
Relative increase in gp42 | |
|---|---|---|---|
| LCL-derived virus | Saliva-derived virusb | ||
| 1 | 0.60 | 1.3 | 2.17 |
| 2 | 0.04 | 0.11 | 2.75 |
| 3 | 0.14 | 0.66 | 4.72 |
| 4 | 0.19 | 0.76 | 4.00 |
Measured by flow cytometry of virus bound to B cells and stained with a monoclonal antibody to gp42 or gH. Ratios are calculated from the percent of cells binding virus that with each antibody, to gp42 or gH, had a fluorescent signal above that of the isotype control.
Fresh saliva was collected for each experiment.
Our results, overall, indicate that virus in saliva generally carries larger amounts of gp42 than B-cell-derived virus, suggesting that, in aggregate, it is derived from an HLA class II-negative cell. This is consistent with recent observations of EBV in tonsil epithelium of healthy carriers (15) and with a model in which amplification of virus in an HLA class II-negative epithelial cell is part of the normal cycle of persistence of EBV.
Acknowledgments
This work was supported by Public Health Service grant DE016669 from the National Institute of Dental and Craniofacial Research and P20RR01874 from the National Center for Research Resources.
REFERENCES
- 1.Borza, C. M., and L. M. Hutt-Fletcher. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594-599. [DOI] [PubMed] [Google Scholar]
- 2.Borza, C. M., A. J. Morgan, S. M. Turk, and L. M. Hutt-Fletcher. 2004. Use of gHgL for attachment of Epstein-Barr virus to epithelial cells compromises infection. J. Virol. 78:5007-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gan, Y.-J., B. I. Razzouk, T. Su, and J. W. Sixbey. 2002. A defective, rearranged Epstein-Barr virus genome in EBER-negative and EBER-positive Hodgkin's disease. Am. J. Pathol. 160:781-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haan, K. M., W. W. Kwok, R. Longnecker, and P. Speck. 2000. Epstein-Barr virus entry utilizing HLA-DP or HLA-DQ as a coreceptor. J. Virol. 74:2451-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haan, K. M., S. K. Lee, and R. Longnecker. 2001. Different functional domains in the cytoplasmic tail of glycoprotein gB are involved in Epstein-Barr virus induced membrane fusion. Virology 290:106-114. [DOI] [PubMed] [Google Scholar]
- 6.Haan, K. M., and R. Longnecker. 2000. Coreceptor restriction within the HLA-DQ locus for Epstein-Barr virus infection. Proc. Natl. Acad. Sci. USA 97:9252-9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King, W., A. L. Thomas-Powell, N. Raab-Traub, M. Hawke, and E. Kieff. 1980. Epstein-Barr virus RNA. V. Viral RNA in a restringently infected, growth-transformed cell line. J. Virol. 36:506-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laichalk, L. L., and D. A. Thorley-Lawson. 2005. Terminal differentiation into plasma cells initiated the replicative cycle of Epstein-Barr virus in vivo. J. Virol. 79:1296-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lake, C. M., and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus that lacks glycoprotein gN is impaired in assembly and infection. J. Virol. 74:11162-11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, Q. X., M. K. Spriggs, S. Kovats, S. M. Turk, M. R. Comeau, B. Nepom, and L. M. Hutt-Fletcher. 1997. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J. Virol. 71:4657-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, Q. X., S. M. Turk, and L. M. Hutt-Fletcher. 1995. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 69:3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molesworth, S. J., C. M. Lake, C. M. Borza, S. M. Turk, and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus gH is essential for penetration of B cell but also plays a role in attachment of virus to epithelial cells. J. Virol. 74:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nemerow, G. R., C. Mold, V. K. Schwend, V. Tollefson, and N. R. Cooper. 1987. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J. Virol. 61:1416-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oda, T., S. Imai, S. Chiba, and K. Takada. 2000. Epstein-Barr virus lacking glycoprotein gp85 cannot infect B cells and epithelial cells. Virology 276:52-58. [DOI] [PubMed] [Google Scholar]
- 15.Pegtel, D. M., J. Middeldorp, and D. A. Thorley-Lawson. 2004. Epstein-Barr virus infection in ex vivo tonsil epithelial cell cultures of asymptomatic carriers. J. Virol. 78:12613-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 17.Tanner, J., J. Weis, D. Fearon, Y. Whang, and E. Kieff. 1987. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping and endocytosis. Cell 50:203-213. [DOI] [PubMed] [Google Scholar]
- 18.Tugizov, S. M., J. W. Berline, and J. M. Palefsky. 2003. Epstein-Barr virus infection of polarized tongue and nasopharyngeal epithelial cells. Nat. Med. 9:307-314. [DOI] [PubMed] [Google Scholar]
- 19.Wang, X., and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J. Virol. 72:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, X., W. J. Kenyon, Q. X. Li, J. Mullberg, and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao, Q.-Y., M. Rowe, A. J. Morgan, C. K. Sam, U. Prasad, H. Dang, Y. Zeng, and A. B. Rickinson. 1991. Salivary and serum IgA antibodies to the Epstein-Barr virus glycoprotein gp340: incidence and potential for virus neutralization. Int. J. Cancer 48:45-50. [DOI] [PubMed] [Google Scholar]