Abstract
Circoviruses lack an autonomous DNA polymerase and are dependent on the replication machinery of the host cell for de novo DNA synthesis. Accordingly, the viral DNA needs to cross both the plasma membrane and the nuclear envelope before replication can occur. Here we report on the subcellular distribution of the beak and feather disease virus (BFDV) capsid protein (CP) and replication-associated protein (Rep) expressed via recombinant baculoviruses in an insect cell system and test the hypothesis that the CP is responsible for transporting the viral genome, as well as Rep, across the nuclear envelope. The intracellular localization of the BFDV CP was found to be directed by three partially overlapping bipartite nuclear localization signals (NLSs) situated between residues 16 and 56 at the N terminus of the protein. Moreover, a DNA binding region was also mapped to the N terminus of the protein and falls within the region containing the three putative NLSs. The ability of CP to bind DNA, coupled with the karyophilic nature of this protein, strongly suggests that it may be responsible for nuclear targeting of the viral genome. Interestingly, whereas Rep expressed on its own in insect cells is restricted to the cytoplasm, coexpression with CP alters the subcellular localization of Rep to the nucleus, strongly suggesting that an interaction with CP facilitates movement of Rep into the nucleus.
Circoviruses are animal viruses that have small (∼2-kb), covalently closed, circular, single-stranded DNA (ssDNA) genomes encapsidated within nonenveloped icosahedral virions (8). Members of the family Circoviridae are divided into genera based on their specific genome organization and host range. Porcine circovirus type 1 (PCV1), Porcine circovirus type 2 (PCV2) (12), and Beak and feather disease virus (BFDV) are the only formally recognized members of the genus Circovirus. In recent years, a number of novel circovirus-like viruses have been identified in nonpsittacine avian species. These include Columbid circovirus (34), Goose circovirus (34), Canary circovirus (29), and Duck circovirus (14), which have all been tentatively classified as members of the genus. All of these viruses possess ambisense genomes with two major open reading frames (ORFs) carried on opposite strands of the replicative double-stranded DNA (dsDNA) intermediate (33). These encode the replication-associated protein (Rep) and capsid protein (CP) from the virion and complementary strands, respectively (25, 27) (23, 26).
Circoviruses are dependent on the replication machinery of the host cell for de novo DNA synthesis (26). Although Rep is required to initialize viral replication, continuation of the process is dependent upon cellular enzymes expressed during S phase and commences only after the host cell has passed through mitosis (32). Since DNA synthesis occurs exclusively in the nucleus, the viral DNA needs to cross both the plasma membrane and the nuclear envelope before a productive infection can be established. The strict size limitations associated with the transport of molecules across the nuclear envelope exclude diffusion as a possible mechanism for the entry of the viral genome into the nucleus (11). Nuclear import of macromolecules is generally facilitated by protein-lined aqueous channels known as nuclear pore complexes (7). However, transport through the nuclear pore complexes is signal mediated, which necessitates the involvement of karyophilic proteins in the active nuclear import of DNA molecules (16). Protein-mediated nuclear transport of viral genomes has in fact been suggested for several DNA viruses (5, 19, 37).
In the case of the plant-infecting geminiviruses, which are thought to share the same mode of replication as the circoviruses, viral DNA transport is mediated by the CP and the nuclear shuttle protein (NSP) or the movement protein (MP), depending on the particular geminivirus species (19, 28, 30). Both the CP and NSP are actively targeted to the nucleus and are able to shuttle between the nucleus and the cytoplasm (18). The capsid protein of PCV2 has similarly been shown to localize to the nucleus (20, 21). The intracellular localization of the PCV CP is directed by a bipartite nuclear localization signal (NLS) situated at the N terminus of the protein (20). The karyophilic nature of this protein suggests that the CP of circoviruses may, like the geminivirus CP, be involved in DNA translocation.
In order to gain insight into the role of the CP in the life cycle of circoviruses, we have investigated the physical interactions of the BFDV CP with the viral DNA and with Rep. We used recombinant BFDV proteins expressed in insect cells, as no cell culture system exists for BFDV. The intracellular localization of the BFDV CP is shown to be directed by a bipartite NLS situated at the N terminus of the protein. Moreover, we have mapped a DNA binding region to the N terminus of the protein that falls within a region containing three potential NLSs. Interestingly, we also found that the nuclear localization of Rep in insect cells is CP dependent, strongly implying that Rep and CP of BFDV interact.
MATERIALS AND METHODS
Virus, cells, and sera.
All bacterial plasmids were maintained in Escherichia coli DH5α (Invitrogen). Recombinant baculoviruses were cultivated in Spodoptera frugiperda Sf-21 cells grown in TC-100 insect medium (Highveld Biological) supplemented with 10% fetal bovine serum, 50 μg/ml neomycin, 69.2 μg/ml penicillin G, and 100 μg/ml streptomycin. BFDV-positive sera used for Western blotting and cytochemistry were collected from African gray parrots showing clinical signs of the disease (15).
Generation of recombinant baculoviruses.
The 747-nucleotide (nt) fragment of the BFDV genome comprising the putative capsid protein open reading frame (nucleotides 1234 to 1980 of the genome [GenBank accession no. AY450443]) was amplified by PCR using primers wtCP-F (5′ GCGGCCGCATGCTGTGGGGCACCTCTAACTGC 3′) and wtCP-R (5′ CTCGAGTCTTTATTAAGTACTGGGATTG 3′) (sequences engineered to create endonuclease restriction sites are indicated in boldface type). The PCR product was ligated with the pGEM-T Easy plasmid (Promega), resulting in plasmid pGM.wtCP.
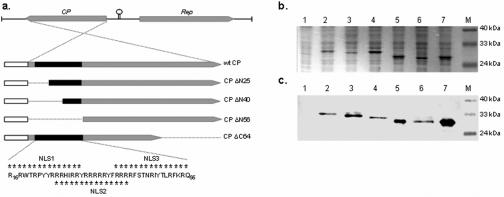
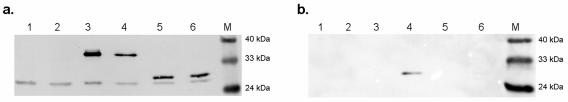
N-terminal-truncated CP ORFs were generated by amplifying various lengths of the wild-type (wt) CP with primer ΔN25-F (5′ GCGGCCGC ATGCACATCAGGCGATACC 3′; 25-amino-acid N-terminal deletion), ΔN40-F (5′ GCGGCCGCATGCGCTTCTCAACCAATAG 3′; 40-amino-acid N-terminal deletion), or ΔN56-F (5′ GCGGCCGCATGAAACGCCAATTCAAATTCC 3′; 56-amino-acid N-terminal deletion) used in conjunction with primer wtCP-R. The PCR products were ligated with plasmid pGEM-T Easy, resulting in recombinant plasmids pGM.CP-ΔN25, pGM.CP-ΔN40, and pGM.CP-ΔN56, respectively. In addition to the wild-type and N-terminal-truncated CP ORFs, a fifth CP ORF, in which the 64 carboxy-terminal residues were removed, was constructed. This was achieved by digesting plasmid pGM.wtCP with PstI and religating the larger fragment, yielding plasmid pGM.CP-ΔC64. The integrity of the ORFs was verified by sequence analysis. Figure 1a shows the truncated ORFs in relation to the wt CP.
FIG. 1.
(a) Schematic representation of the gene structure of BFDV and the capsid protein derivatives used in this study. The CPs (gray boxes) had a His6 affinity tag and the TEV protease recognition site (white boxes) fused to their N termini. The section of CP corresponding to the putative NLSs is indicated by a solid black box. Residues constituting the potential bipartite NLSs are indicated by asterisks. Thin black lines represent the deleted sections in each of the truncated derivatives. Insect cells were either mock infected or infected with the indicated recombinant baculoviruses. Cell lysates were fractionated 60 h p.i. on a 12% SDS-polyacrylamide gel (b) followed by Western blotting using Tetra-His mouse monoclonal IgG1 (c). Lanes 1, mock-infected cells; lanes 2, wt CP; lanes 3, CP ΔN25; lanes 4, CP ΔN40; lanes 5, CP ΔN56; lanes 6, CP ΔC64; lanes 7, CAT; lanes M, molecular size markers.
A Bac-to-Bac baculovirus expression system (Invitrogen) was used to express the various forms of the BFDV capsid protein. The NotI-XhoI fragment was excised from each of the respective plasmids and ligated with pFastBac HT donor plasmid that had been linearized using the same enzymes. The pFastBac HT donor plasmid is designed to express a His6 sequence and the tobacco etch virus (TEV) proteinase cleavage site fused to the N terminus of the gene of interest. E. coli DH10Bac cells (Gibco BRL), containing a helper vector which facilitates the transposition of the recombinant fragment into the baculovirus shuttle vector (bacmid), were transformed with recombinant donor plasmids. Sf-21 cells were transfected with purified recombinant bacmid DNA by use of Cellfectin (Invitrogen) according to the manufacturer's protocols. After 4 days of incubation, the culture media of the transfected cells were collected and stored at 4°C.
Expression and purification of recombinant proteins.
High-titer stocks of viruses were prepared by a single passage of infecting Sf-21 cells with a multiplicity of infection (MOI) of 0.1. Recombinant proteins were extracted from cells infected at an MOI of 5. Infected cells were harvested 60 h postinfection (p.i.), suspended in cell lysis buffer (10 mM Tris-HCl, pH 7.8, 50 mM KH2PO4, 300 mM NaCl, 1 mM β-mercaptoethanol, 40 mM imidazole), and subsequently lysed by sonication. Cell debris was removed by centrifuging the lysate at 10,000 × g for 30 min. The supernatant was further clarified by passing it through a 0.45-μm acetate filter (Osmonics) before loading it onto a HisTrap HP affinity chromatography column (Amersham Biosciences). The column was washed with 10 column volumes washing buffer (40 mM Tris-HCl, pH 7.5, 20% glycerol, 100 mM KCl, 1 mM β-mercaptoethanol, 40 mM imidazole), and proteins were eluted in 5 column volumes elution buffer (40 mM Tris-HCl, pH 7.5, 20% glycerol, 100 mM KCl, 300 mM imidazole). Purified proteins were dialyzed against dialysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM KCl, 1 mM dithiothreitol, 10% glycerol) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
SDS-PAGE and Western blotting.
Samples were diluted in sample treatment buffer, heated to 80°C for 10 min, and subjected to electrophoresis on 12% denaturing SDS-polyacrylamide gels. Proteins were stained with PageBlue 83 (BDH Chemicals Ltd.). The purity and concentration of the recombinant protein were determined by densitometric analysis using GeneTools software (Syngene, Synoptics Ltd.). For Western blotting, proteins were transferred onto nitrocellulose membranes (NitroBind; Osmonics Inc.) by using a Trans-Blot semidry transfer cell (Bio-Rad). The membrane was soaked in a 5% (wt/vol) bovine serum albumin (BSA) solution for 1 h before incubation with Tetra-His mouse monoclonal immunoglobulin G1 (IgG1) (1:1,000; QIAGEN) or sera from BFDV-infected psittacines (1:500). Bound antibodies were revealed using polyclonal rabbit anti-IgY sera (1:500; University of Cape Town) followed by the appropriate alkaline phosphatase anti-rabbit conjugate (1:1,000; Sigma) or alkaline phosphatase anti-mouse conjugate, depending on the primary antibody used. After equilibration in Tris-buffered saline, bound antibodies were detected with Nitro Blue Tetrazolium-5-bromo-4-chloro-3-indolylphosphate-toluidine salt substrate (Roche).
Immunocytochemistry.
For indirect immunofluorescence, Sf-21 cells were seeded onto glass slides and infected with recombinant baculovirus at an MOI of 0.1. A recombinant virus expressing the chloramphenicol acetyltransferase protein (CAT) was used for comparative purposes. The cells were fixed and permeabilized at 60 h p.i., blocked in 5% BSA, and incubated with Tetra-His mouse monoclonal IgG1 (1:250; QIAGEN) for 3 h. After being washed, cells were incubated with secondary fluorescein isothiocyanate-conjugated anti-mouse IgG (1:500; Sigma). Cellular membranes and nuclei were stained with 1% Evans blue (BDH Chemicals Ltd.) and 10 μg/ml 4,6-diamidino-2-phenylindole (DAPI) (Sigma), respectively. Fluorescence was observed with a Nikon Diaphot inverted epifluorescence microscope. Images were captured using a Zeiss Axiocam digital camera system.
DNA binding assay.
The ability of each variant of the BFDV CP to bind single- and double-stranded DNA was assessed by electrophoretic mobility shift analysis. Purified protein was diluted to 0.5 μg/ml in binding buffer (100 mM Tris-HCl, pH 8, 300 mM KCl, 25 mM MgCl2, 20% glycerol, 500 μg/ml BSA) and added to 100 ng of M13mp18 single-stranded DNA (Amersham Biosciences), pUC18 plasmid DNA (New England Biolabs Inc.), or pBFDV plasmid DNA. The pBFDV plasmid DNA consisted of the entire BFDV genome (GenBank accession no. AY450443) cloned into the pGEM-T Easy plasmid (15). Plasmids were purified from E. coli DH5α by use of a QIAprep Spin miniprep kit (QIAGEN) according to the manufacturer's instructions. To confirm that the observed retardation of the DNA fragment was indeed caused by bound proteins, duplicate samples were treated with 1 μg/ml proteinase K (Roche). The samples were incubated at 37°C for 1 h, after which they were subjected to electrophoresis on 0.8% agarose gels. DNA was stained with 0.5 mg/ml ethidium bromide and visualized by UV illumination.
Coexpression of BFDV Rep and CP.
The 870-nt fragment of the BFDV genome comprising the Rep open reading frame (nucleotides 130 to 999 of the genome [GenBank accession no. AY450443]) was amplified by PCR using primers wtRep-F (5′ AGATCTATGCCGTCCAAGGAGGGATCTG 3′) and wtRep-R (5′ AAGCTTCTAATAATTGATGGGGTGGGCGAG 3′). The PCR product was ligated with the pGEM-T Easy plasmid, resulting in plasmid pGM.wtRep. The BglII/HindIII fragment was excised from the plasmid pGM.wtRep and ligated with the pFastBac HT donor plasmid that had been linearized using the same enzymes. An extended fragment, including the His6 affinity tag coding regions and TEV protease recognition site, was amplified by PCR using primers FB.HT-F (5′ AGATCTATGTCGTACCATCACCATCAC CATC 3′) and FB.HT-R (5′ AGATCTTTATGATCCTCTAGTACTTCTCGA 3′). The PCR product was ligated with pGEM-T Easy plasmid, resulting in plasmid pGM.HisRep. The 1,085-nt fragment was excised from the plasmid pGM.HisRep by using BglII and placed under control of the polyhedron promoter of the pFastBac Dual donor plasmid that had been linearized with BamHI, yielding plasmid pFBD.HisRep/O.
Plasmid pGM.wtCP was modified to include an XhoI restriction endonuclease recognition site at the 5′ end of CP, yielding plasmid pGM.wtCP-XhoI. This was achieved by PCR mutagenesis using primers wtCP-F-XhoI (5′ CTCGAGATGCTGT GGGCACCTCTAAC 3′) and wtCP-R. The XhoI-XhoI fragment of 747-nt was excised and placed under control of the p10 promoter of plasmid pFBD.HisRep/O, resulting in plasmid pFBD.HisRep/wtCP.
A similar strategy was used to generate a recombinant donor plasmid containing a His6-tagged CAT (Invitrogen) and BFDV wt CP. Recombinant baculoviruses expressing the various proteins were produced as described above.
Immunoprecipitation assay.
An immunoprecipitation assay was used to confirm that CP and Rep directly interact when coexpressed in insect cells. Insect cells were seeded into 25-cm3 culture dishes and infected with recombinant baculovirus at an MOI of 0.1. Infected cells were harvested 60 h p.i., suspended in phosphate-buffered saline (pH 7.4), and lysed by sonication. Proteins were immunoprecipitated using 1 μg Tetra-His mouse monoclonal IgG1 as previously described (31). The proteins were subsequently analyzed by SDS-PAGE and Western blotting as described above.
RESULTS
Expression of recombinant BFDV protein.
The capsid proteins of circoviruses contain a high proportion of basic amino acids at their N termini (27). It is known that the nuclear localization of PCV CP is directed by two NLSs located in this part of the protein. We were interested in determining whether the amino-terminal region of the BFDV capsid protein was similarly involved in nuclear localization. Three potential partially overlapping bipartite NLSs, positioned between residues 16 and 56 of the BFDV CP (Fig. 1), were identified using the online computer program PROSITE (http://www.expasy.org/prosite).
To test whether these potential NLSs were functional, we constructed a series of truncated capsid proteins, sequentially deleting N-terminal portions between residues 1 and 56 (Fig. 1a). Insect cells were infected with recombinant baculoviruses expressing the individual BFDV CP variants. Recombinant proteins CP ΔN25 (with a 25-amino-acid N-terminal deletion) and CP ΔC64 were expressed at levels similar to that of the wt CP (Fig. 1b). However, expression of CP ΔN40 and CP ΔN56 was notably higher than expression of the wt CP. Densitometric analysis revealed that at least 24% of the total cellular protein consisted of CP ΔN40, representing on average a 2.5-fold increase in expression level from that of wt CP. The integrity of the recombinant proteins was confirmed by Western blotting (Fig. 1c).
BFDV capsid protein is localized to the nucleus.
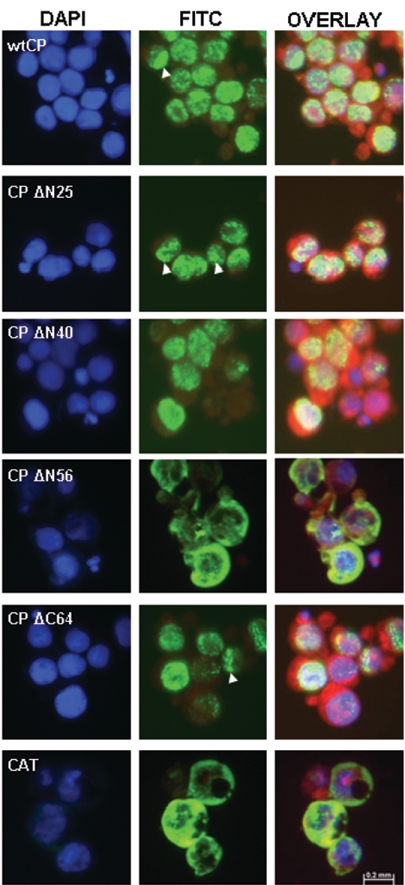
Figure 2 shows the subcellular localization of the respective recombinant proteins as determined by indirect immunofluorescence. Full-length CP was observed exclusively in the nuclei of infected cells. Fluorescence was generally distributed evenly throughout the nucleus but occasionally appeared to be punctiformly associated with the perinuclear region. Partial deletion of the putative NLSs did not significantly alter the localization of the recombinant proteins, with CP ΔN25 and CP ΔN40 similarly restricted to the nucleus. However, removal of the 56 amino-terminal residues completely abolished translocation of the recombinant protein into the nucleus, resulting in the uniform distribution of CP throughout the cytoplasm. The subcellular distribution of CP ΔN56 closely resembled that of CAT. A construct carrying a specific deletion of the 64 carboxy-terminal residues clearly exhibited nuclear import, confirming that the karyophilic activity of the CP is associated with the N-terminal portion of the protein.
FIG. 2.
Subcellular distribution of BFDV capsid protein. Insect cells were infected with recombinant baculovirus and probed with a monoclonal antibody directed against the N-terminal His6 affinity tag. Antibody binding was followed by a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (green). DAPI stain was used to define nuclei (blue), while membranes were visualized with Evans blue (red). Merged images (overlay) are also shown. Arrowheads indicate specific examples of the punctate perinuclear distribution of the relevant recombinant proteins.
BFDV capsid protein binds DNA.
It is very likely that the small size of the BFDV proteome has selectively favored protein multifunctionality. Coat protein-mediated nuclear transport of DNA has been described for various geminivirus species (19, 28), and we accordingly decided to investigate whether, besides its role in viral DNA encapsidation, BFDV CP might also bind DNA in the nucleus. To test this hypothesis, we examined the affinity of viral CP for both dsDNA and ssDNA molecules.
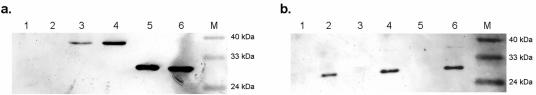
His-tagged recombinant proteins were purified by affinity chromatography under nondenaturing conditions by use of a HisTrap HP kit (Amersham Biosciences). The yields of purified recombinant protein varied between 9.5 and 12 mg/106 cells depending on the CP variant used, with purity approaching 85% for all preparations (data not shown). The ability of the BFDV CP to bind DNA was assessed by its ability to retard the electrophoretic movement of ssDNA and dsDNA within an agarose gel. Purified wild-type protein strongly retarded the movement of plasmid DNA containing the full-length BFDV genome (Fig. 3a). This interaction appeared to be sequence nonspecific, since pUC18 vector DNA was equally affected by the addition of the purified protein (Fig. 3b). There was also an apparent lack of sequence specificity in CP-ssDNA interactions in that BFDV CP bound the ssDNA of M13 phage (Fig. 3c). Treatment of reaction mixtures with proteinase K completely abolished the binding in all cases, indicating that the retardation was indeed caused by the formation of a protein-DNA complex. Gel mobility shift analysis of dsDNA incubated with increasing amounts of the purified protein suggested that the interaction was cooperative in nature. The reaction appeared to be completely saturated at a protein concentration of 0.5 μg protein per 1 ng DNA (Fig. 3d).
FIG. 3.
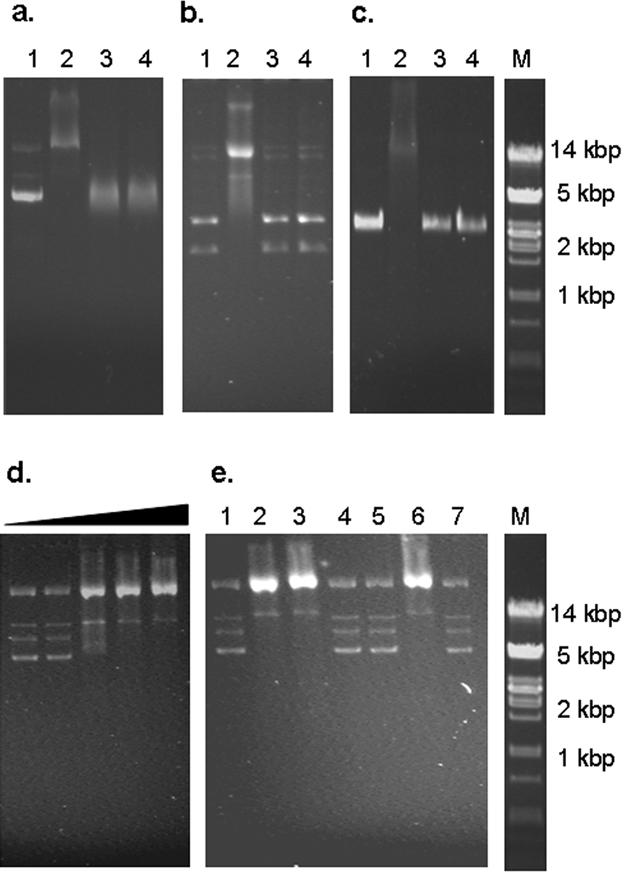
Electrophoretic mobility shift analysis of the interaction of recombinant BFDV CP with various DNA samples. (a) Plasmid preparations containing the BFDV genome, (b) plasmid preparations of pUC18, and (c) phage M13 ssDNA DNA samples were incubated in the absence of protein (lane 1), in the presence of purified wt CP (lane 2), and in the presence of purified CAT (lane 3). To confirm that the observed retardation of the DNA fragment was indeed caused by bound proteins, 1 μg/ml proteinase K was added to the samples containing purified CP (lane 4). (d) Increasing amounts of His-tagged CP were incubated with BFDV DNA. Lane 1, DNA only; lane 2, 10 ng; lane 3, 20 ng; lane 4, 50 ng; lane 5, 500 ng. (e) The interaction of each of the truncated variants of the BFDV CP with BFDV DNA was assessed in a similar manner. Lane 1, DNA only, lane 2, wt CP; lane 3, CP ΔN25; lane 4, CP ΔN40; lane 5, CP ΔN56; lane 6, CP ΔC64; lane 7, CAT. Lanes M, molecular size markers.
Mutational analyses of several DNA binding proteins have indicated that DNA binding regions often overlap with other functional domains (3, 4, 24). To test whether the NLSs and the DNA binding region of the BFDV CP overlap to any extent, the abilities of the truncated CP variants to bind double-stranded plasmid DNA containing the full-length BFDV genome were assayed (Fig. 3e). Deletion of either the first 25 or the last 64 residues did not noticeably alter the DNA binding activity of BFDV CP, with CP ΔN25 and CP ΔC64 exhibiting binding activities similar to that of the full-length protein. However, the removal of an additional 15 N-terminal residues (CP ΔN40) completely abolished this DNA binding ability. This is in contrast to its subcellular localization characteristics, with CP ΔN40 being strictly localized to the nucleus.
Interaction between BFDV CP and Rep.
It has been suggested recently that subcellular distribution of PCV CP is determined by an interaction with the viral Rep (26). We have shown here that this is unlikely to be the case with BFDV, since the CP of this virus does not depend on Rep for nuclear entry. In contrast to the CP, Rep does not contain any recognizable NLSs. In order to determine whether BFDV Rep and CP do interact to any extent, we coexpressed these proteins in insect cells (Fig. 4).
FIG. 4.
Coexpression of BFDV Rep and CP. Insect cells were either mock infected or infected with the indicated recombinant baculoviruses. Cell lysates were analyzed on a 12% SDS-polyacrylamide gel followed by Western blotting with (a) monoclonal antibodies directed against the His6 affinity tag fused to the N terminus of Rep or (b) sera from BFDV-infected psittacines. Lanes 1, mock-infected cells; lanes 2, wt CP; lanes 3, Rep; lanes 4, Rep plus CP; lanes 5, CAT; lanes 6, CAT plus wt CP; lanes M, molecular size markers.
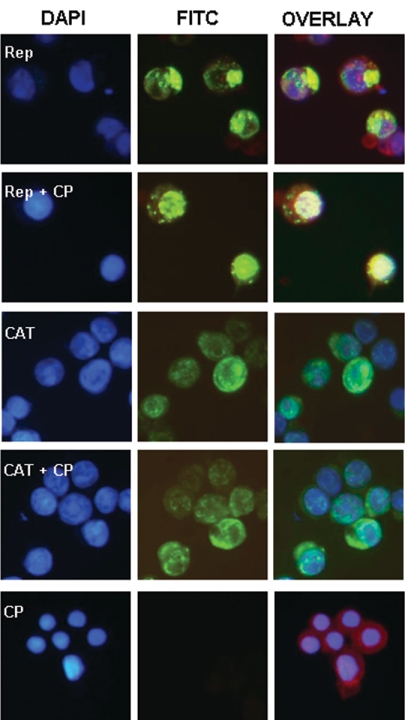
Indirect immunofluorescence assays of cells infected with recombinant baculovirus expressing only Rep clearly showed that the protein was localized exclusively in the cytoplasm, forming distinctive compact aggregates (Fig. 5, first row). When cells were infected with recombinant virus expressing both Rep and CP, the subcellular distribution of Rep changed markedly, with the protein now being predominantly nuclear localized (Fig. 5, second row). The proposed interaction appeared to be sequence specific, since coexpression of CP and CAT did not result in the relocation of CAT to the nucleus (Fig. 5, third and fourth rows).
FIG. 5.
Subcellular distribution of BFDV Rep in the absence and presence of CP. Insect cells were infected with recombinant baculovirus and probed with monoclonal antibodies directed against the His6 affinity tag fused to the N terminus of Rep and CAT. Monoclonal antibody probing was followed by a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (green). DAPI stain was used to define nuclei (blue), while membranes were visualized with Evans blue (red). Merged images (overlay) are also shown.
The direct interaction between Rep and CP was further confirmed by immunoprecipitation experiments. Crude lysates of infected insect cells were immunoprecipitated with a monoclonal antibody directed against the His6 affinity tag fused to the N terminus of Rep and analyzed by Western blotting (Fig. 6). Tetra-His mouse monoclonal IgG1 coimmunoprecipitated CP when it was expressed together with Rep. CP was, however, not immunoprecipitated when expressed either on its own or together with tagged CAT. These results are consistent with the observations made using the indirect immunofluorescence assays. Taken together, they suggest that the BFDV CP may be responsible for relocating Rep from the cytoplasm into the nucleus of infected cells through a direct protein-protein interaction.
FIG. 6.
Immunoprecipitation of BFDV Rep and CP expressed in insect cells. Proteins were immunoprecipitated with Tetra-His mouse monoclonal IgG1. Samples were analyzed on a 12% SDS-polyacrylamide gel followed by Western blotting with (a) monoclonal antibodies directed against the His6 affinity tag fused to the N terminus of Rep or (b) sera from BFDV-infected psittacines. Lanes 1, mock-infected cells; lanes 2, BFDV CP; lanes 3, Rep; lanes 4, Rep plus CP; lanes 5, CAT; lanes 6, CAT plus CP; lanes M, molecular size markers.
DISCUSSION
The BFDV capsid protein typically has a high proportion of arginine residues concentrated at the N terminus of the protein. The concentrated localization of these amino acids is known to inhibit efficient mRNA translation in both prokaryotic and eukaryotic expression systems (1) and often indicates the presence of NLSs (35). In accordance with this, three partly overlapping potential bipartite NLSs were identified between positions 16 and 56 at the N terminus of the BFDV CP. To test whether these potential NLSs were functional, we expressed, in addition to the wild-type CP, a series of CP variants, each lacking specific residues potentially involved in effective nuclear localization. Our results show that the BFDV CP is indeed localized to the nucleus following its expression and that the nuclear localization is directed by one or more of the NLSs situated within the 56 N-terminal residues of the protein. This is in accordance with previously published data on the subcellular localization of PCV CP, where two NLS motifs (12R-H-R-P-R-S-H and 34H-R-Y-R-W-R-R-K) situated in the first 41 residues of the protein were identified as being essential for nuclear targeting of the protein (20). It is important to note that although the NLSs are positioned similarly in both proteins, their exact compositions differ.
In 90% of karyophilic proteins for which both NLS and DNA binding regions have been identified, these regions actually overlap (6). This seems to be the case for the capsid proteins of circoviruses as well. Here we have shown that BFDV CP is capable of binding both ssDNA and dsDNA in a cooperative manner. The DNA binding region was mapped to the N terminus of the protein and falls within the region containing the three potential NLSs. The ability of the BFDV CP to bind DNA, coupled with the karyophilic nature of this protein, strongly suggests that it may be responsible for nuclear targeting of the viral genome. This is similar to the role of CP in the related geminiviruses, where CP-mediated nuclear transport of maize streak virus DNA is a prerequisite for the establishment of a productive infection of maize plants (2). The exact structure and possible mechanisms regulating the formation of such a nucleoprotein complex are, however, unknown.
The replication-associated protein of all circoviruses is derived from the virion-sense ORF and is an absolute requirement for viral replication (23). In this study, we demonstrated that the BFDV Rep expressed on its own in insect cells is restricted to the cytoplasm. This is in stark contrast to the PCV Rep, which has recently been shown to localize to the nucleus (9, 26). It is our contention that the nuclear localization of the BFDV Rep is facilitated by an interaction with the CP, which is responsible for trafficking it across the nuclear membrane. This is the converse of what seems to occur in the related PCV, where it is Rep that apparently facilitates trafficking of CP into the nucleus (26). Our hypothesis is strongly supported by subcellular localization data on individually expressed and coexpressed BFDV proteins. That it is a direct protein-protein interaction which is involved in this process is evident from our immunoprecipitation data. An analogous interaction between the CP and Rep of the geminivirus mung bean yellow mosaic India virus has recently been described (22). Rather than being implicated in nuclear trafficking, though, the mung bean yellow mosaic India virus Rep-CP interaction results in both the down-regulation of replication initiation activity and a general decrease in ssDNA accumulation and viral replication. Whether the BFDV CP-Rep interaction has a similar inhibitory effect on the activity of Rep remains to be determined.
It is becoming increasingly clear that the involvement of CPs in the life cycles of circoviruses and other ssDNA viruses may go far beyond their role in encapsidation. It has been established recently that the subcellular localization of PCV2 CP in permissive cells is dependent on the specific stage of infection (26). In addition to this, contrasting localization patterns have been observed for PCV nonpermissive cells compared to those observed for permissive cells. Despite the ability of the virus to accumulate within the cytoplasm of dendritic cells, there was no evidence of viral replication in these cells (10). Meerts et al. (26) have subsequently shown that in certain instances CP and Rep are nuclear localized in macrophages but that the localization is substantially delayed, resulting in a significant decrease in viral replication. A partial barrier to nuclear entry of Rep (and, by association, CP) may account for either the complete lack of or the substantial delay in viral replication.
The ability of the virus to replicate within an infected cell, however, does not necessarily guarantee the establishment of a productive infection. Replication of PCV has been shown to occur in certain human cells, but these infections were found to be nonproductive. In contrast to localization in macrophages, PCV CP is localized to the nucleus in infected human and primate cell lines, where it aggregates punctiformly (13). Despite the presence of viral DNA in the supernatant from PCV-infected human cells, no virus particles could be detected, suggesting that viral assembly may be disrupted in these cells. The exact way in which viral assembly is perturbed is, however, unclear.
It should be noted that the behavior of the BFDV proteins in insect cells may not necessarily reflect the functionality of the proteins during a natural infection. Although most recombinant proteins expressed in insect cells are generally correctly processed, targeted to their appropriate cellular locations, and in most cases remain functionally active (17, 36), slight differences in the inherent characteristics of specific cell types can dramatically influence their behavior. While the relevance of our findings to the natural state of affairs remains to be determined, this behavior is strongly reminiscent of the behavior of the PCV proteome during productive infections of permissive cells. Adaptation of BFDV to tissue-cultured psittacine cells would undoubtedly allow researchers to clear up most of the questions raised here. However, all attempts to do so have thus far been unsuccessful.
In summary, we have shown that the capsid protein of BFDV is actively localized to the nucleus when it is expressed in insect cells. The CP is most likely targeted to the nucleus by one or more of three bipartite nuclear localization signals located at the N terminus of the protein. Moreover, this portion of the protein also appears to contain a DNA binding region which facilitates the binding of CP to both single- and double-stranded DNA in a cooperative manner. It remains to be determined whether the NLS and the DNA binding region are functionally coupled or whether they act independently. In addition to the protein-DNA interaction, the results strongly suggest that CP directly interacts with Rep, enabling cotranslocation of the latter into the nucleus. The precise domains involved in this interaction and the possible impact of Rep-CP interaction on other aspects of the virus life cycle remain to be determined. Fine mapping of the regions involved will undoubtedly shed light on the precise roles of CP in the BFDV life cycle.
Acknowledgments
We thank Jacques Theron (University of Pretoria, South Africa) for his assistance with and insight into several aspects of the project. The careful proofreading of the manuscript by Antoinette van Schalkwyk, Dionne Shepherd, Eric van der Walt, and Darren Martin prior to submission is also greatly appreciated.
The project was partly funded by the National Research Foundation of South Africa.
REFERENCES
- 1.Alexandrova, N., J. Niklinski, V. Bliskovsky, G. A. Otterson, M. Blake, F. J. Kaye, and M. Zajac-Kaye. 1995. The N-terminal domain of c-Myc associates with alpha-tubulin and microtubules in vivo and in vitro. Mol. Cell. Biol. 15:5188-5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulton, M. I., H. Steinkellner, J. Donson, P. G. Markham, D. I. King, and J. W. Davies. 1989. Mutational analysis of the virion-sense genes of maize streak virus. J. Gen. Virol. 70:2309-2323. [DOI] [PubMed] [Google Scholar]
- 3.Braem, C. V., K. Kas, E. Meyen, M. Debiec-Rychter, W. J. M. Van de Ven, and M. L. Voz. 2002. Identification of a karyopherin α2 recognition site in PLAG1, which functions as a nuclear localization signal. J. Biol. Chem. 277:19673-19678. [DOI] [PubMed] [Google Scholar]
- 4.Bruening, W., P. Moffett, S. Chia, G. Heinrich, and J. Pelletier. 1996. Identification of nuclear localization signals within the zinc fingers of the WT1 tumor suppressor gene product. FEBS Lett. 393:41-47. [DOI] [PubMed] [Google Scholar]
- 5.Clever, J., M. Yamada, and H. Kasamatsu. 1991. Import of simian virus 40 virions through nuclear pore complexes. Proc. Natl. Acad. Sci. USA 88:7333-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cokol, M., R. Nair, and B. Rost. 2000. Finding nuclear localization signals. EMBO Rep. 1:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbett, A. H., and P. A. Silver. 1997. Nucleocytoplasmic transport of macromolecules. Microbiol. Mol. Biol. Rev. 61:193-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fauquet, C. M., M. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball. 2005. Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Academic Press, London, United Kingdom.
- 9.Finsterbusch, T., T. Steinfeldt, R. Caliskan, and A. Mankertz. 2005. Analysis of the subcellular localization of the proteins Rep, Rep′ and Cap of porcine circovirus type 1. Virology 343:36-46. [DOI] [PubMed] [Google Scholar]
- 10.Gilpin, D. F., K. McCullough, B. M. Meehan, F. McNeilly, I. McNair, L. S. Stevenson, J. C. Foster, J. A. Ellis, S. Krakowka, B. M. Adair, and G. M. Allan. 2003. In vitro studies on the infection and replication of porcine circovirus type 2 in cells of the porcine immune system. Vet. Immunol. Immunopathol. 94:149-161. [DOI] [PubMed] [Google Scholar]
- 11.Greber, U. F. 1998. Delivery of animal virus into the nucleus, p. 89-114. In L. Seymour, A. Kabanov, and P. Felgner (ed.), Self-assembling complexes for gene delivery: from chemistry to clinical trial. Wiley, Sussex, United Kingdom.
- 12.Hamel, A. L., L. L. Lin, and G. P. Nayar. 1998. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J. Virol. 72:5262-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hattermann, K., C. Roedner, C. Schmitt, T. Finsterbusch, T. Steinfeldt, and A. Mankertz. 2004. Infection studies on human cell lines with porcine circovirus type 1 and porcine circovirus type 2. Xenotransplantation 11:284-294. [DOI] [PubMed] [Google Scholar]
- 14.Hattermann, K., C. Schmitt, D. Soike, and A. Mankertz. 2003. Cloning and sequencing of duck circovirus (DuCV). Arch. Virol. 148:2471-2480. [DOI] [PubMed] [Google Scholar]
- 15.Heath, L., D. P. Martin, L. Warburton, M. Perrin, W. Horsfield, C. Kingsley, E. P. Rybicki, and A. L. Williamson. 2004. Evidence of unique genotypes of beak and feather disease virus in southern Africa. J. Virol. 78:9277-9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiscox, J. A. 2002. The nucleolus—a gateway to viral infection? Arch. Virol. 147:1077-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho, Y., H. R. Lo, T. C. Lee, C. P. Wu, and Y. C. Chao. 2004. Enhancement of correct protein folding in vivo by a non-lytic baculovirus. Biochem. J. 382:695-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, H., M. I. Boulton, K. J. Oparka, and J. W. Davies. 2001. Interaction of the movement and coat proteins of maize streak virus: implications for the transport of viral DNA. J. Gen. Virol. 82:35-44. [DOI] [PubMed] [Google Scholar]
- 19.Liu, H., M. I. Boulton, C. L. Thomas, D. A. Prior, K. J. Oparka, and J. W. Davies. 1999. Maize streak virus coat protein is karyophyllic and facilitates nuclear transport of viral DNA. Mol. Plant-Microbe Interact. 12:894-900. [DOI] [PubMed] [Google Scholar]
- 20.Liu, Q., S. K. Tikoo, and L. A. Babiuk. 2001. Nuclear localization of the ORF2 protein encoded by porcine circovirus type 2. Virology 285:91-99. [DOI] [PubMed] [Google Scholar]
- 21.Liu, Q., P. Willson, S. Attoh-Poku, and L. A. Babiuk. 2001. Bacterial expression of an immunologically reactive PCV2 ORF2 fusion protein. Protein Expr. Purif. 21:115-120. [DOI] [PubMed] [Google Scholar]
- 22.Malik, P. S., V. Kumar, B. Bagewadi, and S. K. Mukherjee. 2005. Interaction between coat protein and replication initiation protein of mung bean yellow mosaic India virus might lead to control of viral DNA replication. Virology 337:273-283. [DOI] [PubMed] [Google Scholar]
- 23.Mankertz, A., B. Mueller, T. Steinfeldt, C. Schmitt, and T. Finsterbusch. 2003. New reporter gene-based replication assay reveals exchangeability of replication factors of porcine circovirus types 1 and 2. J. Virol. 77:9885-9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matheny, C., M. L. Day, and J. Milbrandt. 1994. The nuclear localization signal of NGFI-A is located within the zinc finger DNA binding domain. J. Biol. Chem. 269:8176-8181. [PubMed] [Google Scholar]
- 25.Meehan, B. M., F. McNeilly, D. Todd, S. Kennedy, V. A. Jewhurst, J. A. Ellis, L. E. Hassard, E. G. Clark, D. M. Haines, and G. M. Allan. 1998. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J. Gen. Virol. 79:2171-2179. [DOI] [PubMed] [Google Scholar]
- 26.Meerts, P., G. Misinzo, F. McNeilly, and H. J. Nauwynck. 2005. Replication kinetics of different porcine circovirus 2 strains in PK-15 cells, fetal cardiomyocytes and macrophages. Arch. Virol. 150:427-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niagro, F. D., A. N. Forsthoefel, R. P. Lawther, L. Kamalanathan, B. W. Ritchie, K. S. Latimer, and P. D. Lukert. 1998. Beak and feather disease virus and porcine circovirus genomes: intermediates between the geminiviruses and plant circoviruses. Arch. Virol. 143:1723-1744. [DOI] [PubMed] [Google Scholar]
- 28.Palanichelvam, K., T. Kunik, V. Citovsky, and Y. Gafni. 1998. The capsid protein of tomato yellow leaf curl virus binds cooperatively to single-stranded DNA. J. Gen. Virol. 79:2829-2833. [DOI] [PubMed] [Google Scholar]
- 29.Phenix, K. V., J. H. Weston, I. Ypelaar, A. Lavazza, J. A. Smyth, D. Todd, G. E. Wilcox, and S. R. Raidal. 2001. Nucleotide sequence analysis of a novel circovirus of canaries and its relationship to other members of the genus Circovirus of the family Circoviridae. J. Gen. Virol. 82:2805-2809. [DOI] [PubMed] [Google Scholar]
- 30.Qin, S., B. M. Ward, and S. G. Lazarowitz. 1998. The bipartite geminivirus coat protein aids BR1 function in viral movement by affecting the accumulation of viral single-stranded DNA. J. Virol. 72:9247-9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theron, J., and L. H. Nel. 1997. Stable protein-RNA interaction involves the terminal domains of bluetongue virus mRNA, but not the terminally conserved sequences. Virology 229:134-142. [DOI] [PubMed] [Google Scholar]
- 32.Tischer, I., D. Peters, R. Rasch, and S. Pociuli. 1987. Replication of porcine circovirus: induction by glucosamine and cell cycle dependence. Arch. Virol. 96:39-57. [DOI] [PubMed] [Google Scholar]
- 33.Todd, D. 2000. Circoviruses: immunosuppressive threats to avian species: a review. Avian Pathol. 29:373-394. [DOI] [PubMed] [Google Scholar]
- 34.Todd, D., J. H. Weston, D. Soike, and J. A. Smyth. 2001. Genome sequence determinations and analyses of novel circoviruses from goose and pigeon. Virology 286:354-362. [DOI] [PubMed] [Google Scholar]
- 35.Whittaker, G. R., and A. Helenius. 1998. Nuclear import and export of viruses and virus genomes. Virology 246:1-23. [DOI] [PubMed] [Google Scholar]
- 36.Wu, X., C. Cao, Y. Xu, and X. Lu. 2004. Construction of a host range-expanded hybrid baculovirus of BmNPV and AcNPV, and knockout of cysteinase gene for more efficient expression. Sci. China C 47:406-415. [DOI] [PubMed] [Google Scholar]
- 37.Yamada, M., and H. Kasamatsu. 1993. Role of nuclear pore complex in simian virus 40 nuclear targeting. J. Virol. 67:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]