Abstract
The Rep proteins of the adeno-associated virus (AAV) are required for viral replication in the presence of adenovirus helper functions and as yet poorly characterized cellular factors. In an attempt to identify such factors, we purified Flag-Rep68-interacting proteins from human cell lysates. Several polypeptides were identified by mass spectrometry, among which was ANP32B, a member of the acidic nuclear protein 32 family which takes part in the formation of the template-activating factor I/Set oncoprotein (TAF-I/Set) complex. The N terminus of Rep was found to specifically bind the acidic domain of ANP32B; through this interaction, Rep was also able to recruit other members of the TAF-I/Set complex, including the ANP32A protein and the histone chaperone TAF-I/Set. Further experiments revealed that silencing of ANP32A and ANP32B inhibited AAV replication, while overexpression of all of the components of the TAF-I/Set complex increased de novo AAV DNA synthesis in permissive cells. Besides being the first indication that the TAF-I/Set complex participates in wild-type AAV replication, these findings have important implications for the generation of recombinant AAV vectors since overexpression of the TAF-I/Set components was found to markedly increase viral vector production.
Adeno-associated virus type 2 (AAV-2 hereafter for brevity) is a small, nonpathogenic human virus originally discovered in adenovirus-infected cells (13). Indeed, it has become progressively evident that, for its productive replication, AAV-2 requires a series of still poorly characterized cellular functions that are triggered by the concomitant infection of the host cell with adenovirus, as well with other viruses, or by cell treatment with a vast series of chemical and physical agents possessing genotoxic activity (reviewed in reference 4). In the absence of helper functions, AAV-2 establishes a latent infection by integrating into a specific sequence of human chromosome 19q13.3 (19, 20, 23, 32).
Growing interest in AAV-2 molecular biology over the last few years has been fostered by the progressive recognition of its outstanding properties when used as a gene delivery vector in vivo. AAV-2 vectors transduce postmitotic cell tissues at high efficiency in vivo, including myocardium, skeletal muscle, brain, and retina. In these tissues, vector-driven transgene expression persists for very long periods of time, possibly for the whole lives of treated animals, without inducing inflammation or an immune response (9). Despite early success in the application of AAV-2 vector technology, several obstacles continue to hamper further development. In particular, the production of large quantities of AAV-2 vectors required for in vivo application in large animals and humans would greatly benefit from the possibility of increasing vector yield per infected cell. This possibility, however, remains limited by our incomplete understanding of the molecular mechanisms and the cellular factors required for vector replication.
The linear, single-stranded DNA (ssDNA) genome of AAV-2 is ∼4.7 kb long and is flanked at both ends by inverted terminal repeats that can fold into stable T-shaped hairpins, thus providing a free 3′-OH end that serves as an origin of DNA replication. The virus contains two open reading frames: Rep, which produces four partially overlapping polypeptides (Rep78, Rep68, Rep52, and Rep40), and Cap, which encodes the capsid proteins (VP1, VP2, and VP3).
The two largest isoforms of Rep, Rep78 and Rep68, are necessary for AAV-2 replication (16, 30), for site-specific integration (2, 38), and for transcriptional regulation of viral and cellular promoters (21). Rep binds a specific DNA sequence named the Rep binding site, which is present also in the inverted terminal repeats. Upon binding at the Rep binding site, Rep nicks nearby DNA at the specific terminal resolution site (14, 16, 31). While Rep40 and Rep52 do not bind and nick DNA or prove proficient for AAV-2 replication, they do still contain an ATP binding site and retain helicase activity (15, 34). From the structural point of view, the recently solved three-dimensional structures of the Rep68 endonuclease and helicase domains suggest strong functional similarities with large T antigen, the replicator protein of simian virus 40 (SV40) (12, 17).
AAV-2 replicates through a strand displacement mechanism that was first proposed by Tattersall and Ward (40). According to this model, newly replicated genomic ssDNA is produced from double-stranded DNA (dsDNA) intermediates by the concerted action of Rep and cellular factors. The minimal requirements for AAV-2 replication in vivo are either Rep78 or Rep68 and a minimal subset of adenovirus helper functions: E1, E2A, E4, and the VA1 RNA (11). As an alternative, it has been proposed that AAV-2 replication might use ssDNA as a template (43). So far, it has not been possible to reconstitute AAV-2 replication in vitro entirely with purified cellular proteins, although the ssDNA binding protein replication protein A, the proliferating nuclear antigen, and replication factor C have been shown to be required (29). In particular, replication protein A binds Rep78 and Rep68 and enhances their DNA binding and endonuclease activities (37).
It appears likely that, besides its intrinsic biochemical properties, most of the functions of Rep inside the cell are carried out in conjunction with cellular proteins. In fact, previous evidence has shown that Rep interacts with different factors, such as the transcriptional coactivator PC4 (44), the cell cycle regulator Rb (3), the nonhistone chromosomal protein HMGB1 (6), and protein kinase A (7). These interactions have been shown to regulate various aspects of the AAV-2 life cycle. Nevertheless, a comprehensive identification of Rep-containing protein complexes in vivo (i.e., in mammalian cell culture) has so far not been tackled.
Here we describe the results of a proteomic approach aimed at identifying cellular partners of AAV-2 Rep. This approach, which was based on the characterization of proteins physically binding to Flag-tagged Rep68 in vivo, brought about the initial identification of acidic nuclear protein 32B (ANP32B) as an interacting factor of Rep. ANP32B, together with ANP32A and ANP32E, belongs to the ANP32 family. Members of this family share an N-terminal globular domain that contains one or more leucine-rich repeats, involved in protein-protein interactions (18), and an extended acidic C-terminal domain. These proteins are highly expressed in tissues or cell types that undergo active proliferation (24, 42) or at particular stages of the development of the rat brain (28). ANP32A and ANP32B form a protein complex that also includes the two splicing isoforms (α and β) of the template-activating factor I/Set oncoprotein (TAF-I/Set) complex (5, 33), which belongs to the Nap-1 family of histone chaperones.
In this paper, we demonstrate that AAV-2 Rep physically and functionally interacts with the TAF-I/Set complex through the specific binding of its ANP32B component. The ANP32A and ANP32B subunits are required for AAV-2 replication, and in a consistent manner, ectopic expression of TAF-I/Set components increases the levels of AAV-2 replication intermediates, as well as the titer of recombinant AAV-2 (rAAV-2) vector preparations. This is the first functional indication that the TAF-I/Set complex has a role in AAV-2 DNA replication in vivo.
MATERIALS AND METHODS
Plasmids, cell culture, and small interfering RNA (siRNA).
Open reading frames of Rep68 and Rep40 were PCR amplified from pHisRep68 and cloned as a ClaI/KpnI fragment into pFlag-CMV2 (Sigma, St. Louis, Mo.) to obtain Flag-Rep68 and Flag-Rep40. Plasmid pHisRep68, a derivative of pET-16b (Novagen, Milwaukee, Wis.) used for the expression of N-terminally His-tagged Rep68, was obtained from M. Linden (Mt. Sinai School of Medicine, New York, N.Y). Plasmids expressing hemagglutinin (HA)-tagged ANP32A, ANP32B, and TAF-Iβ were constructed by PCR cloning into the pCGN vector (39). ANP32BΔC expresses residues 1 to 147 of wild-type ANP32B from the same backbone. The pDG helper plasmid for AAV-2 vector production and pTR-UF5, expressing green fluorescent protein (GFP) under the control of the cytomegalovirus promoter, were kind gifts of J. A. Kleinschmidt and N. Muzyczka, respectively. Human embryonic kidney (HEK293) cells were cultured in Dulbecco's modified Eagle's medium with Glutamax (Life Technologies, Inc.) supplemented with 10% fetal bovine serum (Life Technologies, Inc.) and gentamicin (100 μg/ml) at 37°C in a humidified 93% air-7% CO2 incubator. DNA transfections were performed by the standard calcium phosphate coprecipitation method. RNA interference (RNAi) with ANP32A and ANP32B was performed against the target sequences 5′-GAAGAAGAGCTTGGTGAAGAAGA-3′, corresponding to nucleotides (nt) 673 to 685 of the ANP32 mRNA (NM_006305), and 5′-GAAGAGGAGTTTGATGAAGAAGA-3′, corresponding to nt 559 to 671 of the ANP32B mRNA (NM_006401). Synthetic 21-nt double-stranded RNA oligonucleotides were purchased by Dharmacon (Chicago, IL).
Antibodies.
Anti-Rep polyclonal rabbit antiserum was a kind gift from J. Kleinschmidt. Mouse monoclonal anti-Flag M2 antibody, mouse monoclonal anti-tubulin, and Flag M2 agarose-conjugated beads were purchased from Sigma (St. Louis, Mo.). Rat monoclonal anti-HA high-affinity (3F10) antibody was purchased from Roche Diagnostics. Mouse monoclonal anti-Ku70 antibody, mouse monoclonal anti-Ku80 antibody, and mouse monoclonal antibody against the catalytic subunit of the DNA-PK complex (DNA-PKcs) were purchased from NeoMarkers (Fremont, Calif.). The anti-ANP32A (I1PP2A, C-18) and anti-TAF-I (I2PP2A, E-15) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). The anti-ANP32B (PHAPI2a) antibody was purchased from Abcam (Cambridge, United Kingdom).
Protein purification and identification.
Thirty-six hours after transfection, ∼6 × 108 HEK293 cells were washed once in phosphate-buffered saline (PBS) and lysed on ice in lysis buffer (20 mM HEPES [pH 7.9], 150 mM NaCl, 0.5% NP-40, 1 mM dithiothreitol, protease inhibitor cocktail). The cell extract was sonicated once and then centrifuged for 15 min at 14,000 rpm at 4°C. An aliquot of the cleared extract was kept as input, while the rest was incubated with 100 μl of packed and preequilibrated Flag M2 agarose beads for 4 h at 4°C. Beads were rinsed twice in lysis buffer and then washed in the same buffer three times. Immunocomplexes were eluted by adding 500 μg/ml Flag peptide (Sigma, St. Louis, Mo.) in lysis buffer. The eluate was concentrated by standard trichloroacetic acid precipitation and resuspended in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) protein loading buffer. Proteins were then subjected to 10% SDS-PAGE and then stained with zinc stain by following the indications provided by the producer (Bio-Rad, Hercules, CA). Mass spectrometry-based protein identification was performed as previously described (45).
Coimmunoprecipitations and micrococcal nuclease (MNase) treatment.
Thirty-six hours after transfection, HEK293 cells were washed once in PBS and lysed on ice in 1 ml/dish of lysis buffer. Cleared cell extracts were incubated with preequilibrated Flag M2 agarose beads on a rotating wheel for 4 h at 4°C. Beads were rinsed twice with 1 ml of lysis buffer and then washed in the same buffer three times. MNase (Sigma, St. Louis, Missouri) treatment was performed essentially as described in reference 22. Proteins were eluted in 1× Laemmli buffer, boiled, and subjected to 10% SDS-PAGE.
In vitro pull-down assays.
The Sp6 in vitro coupled transcription-translation kit (Promega, Madison, Wisconsin) was used to radiolabel proteins with [35S]Met by following the protocol provided by the manufacturer. Production of recombinant HisRep68 and pull-down assays were previously described in reference 25.
AAV-2 replication assay.
Low-molecular-weight DNA was isolated essentially as described in reference 11. Samples were digested overnight at 37°C with DpnI to remove input bacterial plasmids, run on a DNA 0.7% agarose Tris-acetate-EDTA gel, and blotted. A 1-kb XhoI restriction fragment of pTR-UF5, radiolabeled with [α-32P]dCTP, was used as a probe in standard Southern blotting hybridizations.
For the gene-silencing experiments, synthetic siRNAs were transfected at a final concentration of 100 nM in HEK293 cells (3 × 105 cells were plated 24 h earlier in a 60-mm culture dish) by using the Gene Silencer reagent (San Diego, California) according to the manufacturer's instructions. Thirty-six hours after siRNA transfection, the medium was changed and the cells were transfected by the calcium phosphate method. Newly replicated viral genomes were collected 36 h after DNA transfection.
rAAV-2 vector production.
Thirty-six hours after transfection, about 3 × 107 HEK293 cells were scraped and collected in lysis buffer (50 mM HEPES [pH 7.6], 150 mM NaCl) and subjected to three freeze-thaw cycles to release the virions. The lysate was centrifuged to remove cell debris, and the supernatant was precipitated by adding 0.33 volume of saturated (NH4)2SO4. After 10 min of incubation on ice, samples were centrifuged for 30 min at 14,000 rpm and then the pellet was discarded and the supernatant was further precipitated by addition of 0.66 volume of saturated (NH4)2SO4 and incubation for 10 min on ice. Finally, the centrifuged supernatant was dialyzed overnight against PBS at 4°C.
To titrate encapsidated AAV-2 genomes, samples were first digested with DNase I for 1 h at 37°C, boiled for 2 min at 95°C to inactivate the enzyme, and incubated for 1 h at 56°C with proteinase K, which was further inactivated for 5 min at 95°C.
DNase- and proteinase K-digested samples were quantified by competitive PCR with a pair of primers that amplify a 243-bp fragment of the cytomegalovirus promoter present in pTR-UF5, together with scalar dilutions of a 223-bp competitor (47).
rAAV-2-GFP vectors obtained as described above were used to transduce HEK293 cells. Seventy-two hours posttransduction, fluorescence microscopy images of transduced cells were taken with an LSM510 Meta confocal microscope (Zeiss).
RESULTS
Proteomic analysis of Flag-Rep68 protein complexes.
To purify Rep-interacting proteins, we used a mammalian expression vector encoding the open reading frame of Rep68 fused with an N-terminal Flag tag. This epitope-tagged version of Rep68 was active in AAV-2 replication in vivo and showed a nuclear localization similar to that of the unmodified wild-type protein (data not shown; see also reference 37). Extracts from HEK293 cells transfected with Flag-Rep68, as well as from mock-transfected cells, were immunoprecipitated with M2 Flag antibody conjugated to agarose beads. Affinity-purified Flag-Rep68 protein and copurifying cellular factors were subsequently eluted with an excess of Flag peptide, subjected to 10% SDS-PAGE, and stained with zinc stain (Fig. 1).
FIG. 1.
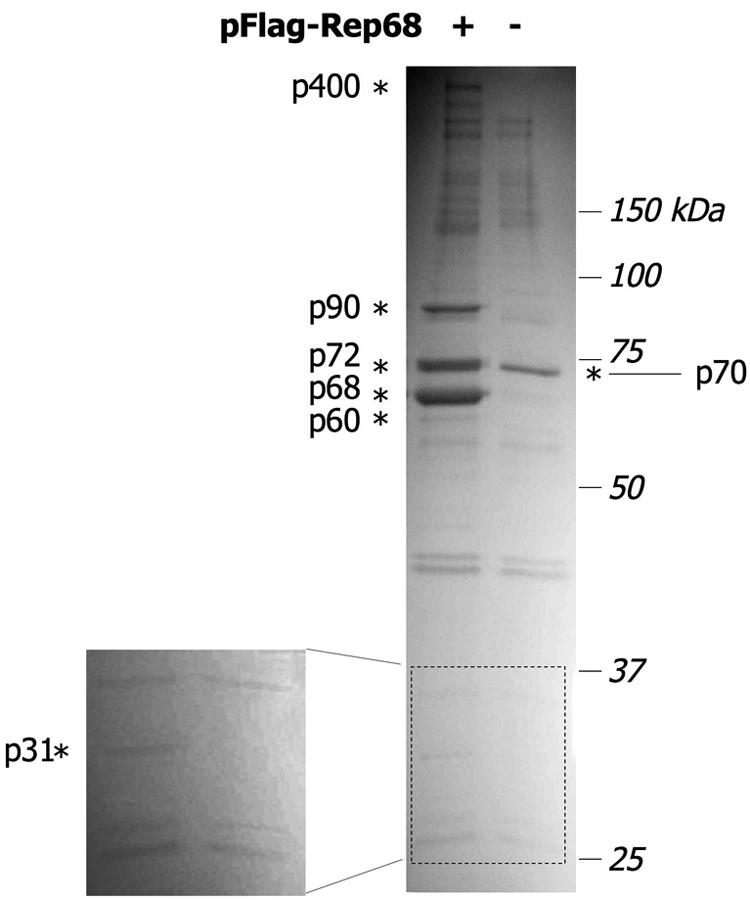
Identification of Rep-interacting proteins. Flag-immunoprecipitated material from Flag-Rep68-transfected and mock-transfected HEK293 cells was subjected to 10% SDS-PAGE, followed by zinc staining. Protein bands present exclusively in the sample transfected with Flag-Rep68 are marked by asterisks and named after their apparent molecular masses in the gel. The enlarged box shows the presence of a specific band migrating at 31 kDa (p31). These bands were excised from the gel and identified by electrospray tandem mass spectrometry (see text).
Compared to mock-transfected cells, six major protein bands were apparent only in the sample from Flag-Rep68-transfected cells, which were named p400, p90, p72, p68, p60, and p31, according to their relative molecular masses. These bands are indicated by asterisks in Fig. 1 and in the enlarged box showing the lower portion of the gel. An additional protein band (p70) was considered in the samples from mock-transfected cells, which migrated slightly faster than p72 in the complex eluted from the Flag-Rep68 cell extract. These seven bands were excised, and the corresponding proteins were identified by mass spectrometry.
Electrospray tandem mass spectrometry analysis of peptides obtained by trypsin digestion (Table S1 in the supplemental material) led to the identification of p400 as DNA-PKcs (accession no. NP_008835), p90 as Ku80 (accession no. NP_066964), p72 as Ku70 (accession no. NP_001460), p68 as AAV-2 Rep68 (accession no. 040500), p60 as heat shock protein 60 (accession no. NP_002147), and p31 as ANP32B (accession no. NP_006392). The band migrating at about 70 kDa in the control lane was identified as protein arginine methyltransferase 5 (accession no. NP_006100). Heat shock protein 60, a member of the heat shock protein family, is very abundant in the cell and is known to be prone to unspecific protein binding. Thus, it was not considered for further analysis.
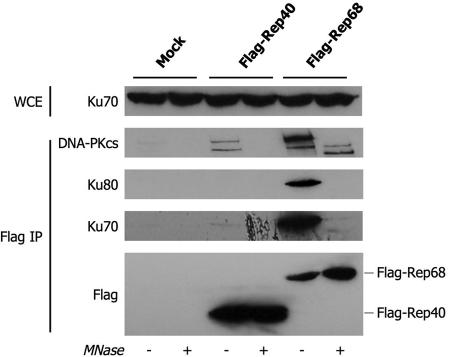
DNA-PKcs, Ku80, and Ku70 are the three components of the human DNA-PK complex involved in repairing DNA double-strand breaks by nonhomologous end joining (for a recent review, see reference 1). Previous evidence from our and other laboratories has suggested a possible role for the DNA-PK complex in the processing of AAV-2 genomes upon cell infection (8, 35, 36, 47). Since the Ku70-Ku80 heterodimer binds DNA ends and structural DNA elements with very high affinity, albeit with very low specificity, we were concerned about the possibility that the interaction between these proteins and Rep might be unspecific and that it might be mediated by DNA fragments possibly attached to Rep. To rule out this possibility, we performed standard coimmunoprecipitations between Flag-Rep68 and the components of the DNA-PK complex in the presence of MNase, in order to remove any intervening DNA that might bridge these interactions (22). Flag-Rep68, but not Flag-Rep40, strongly interacted with Ku70, Ku80, and DNA-PKcs. However, MNase treatment almost completely abolished these interactions (Fig. 2). This observation indicates that binding between Rep and the DNA-PK complex is not direct but requires DNA. The rest of this work therefore focused on the interaction between Rep68 and p31, which we identified as human ANP32B.
FIG. 2.
Coimmunoprecipitation of Flag-Rep40 and Flag-Rep68 with Ku70, Ku80, and DNA-PKcs. Lysates of HEK293 cells transfected with the indicated plasmids were immunoprecipitated (IP) with an anti-Flag antibody. The immunoprecipitated material was cleared with MNase for 30 min at 37°C to eliminate contaminating DNA, as indicated. Samples were then analyzed by Western blotting with antibodies against endogenous subunits of the DNA-PK complex (Ku70, Ku80, and DNA-PKcs). Western blot assays of input whole-cell lysates (WCE) for Ku70 are also shown.
The N terminus of Rep68 specifically binds the acidic domain of ANP32B.
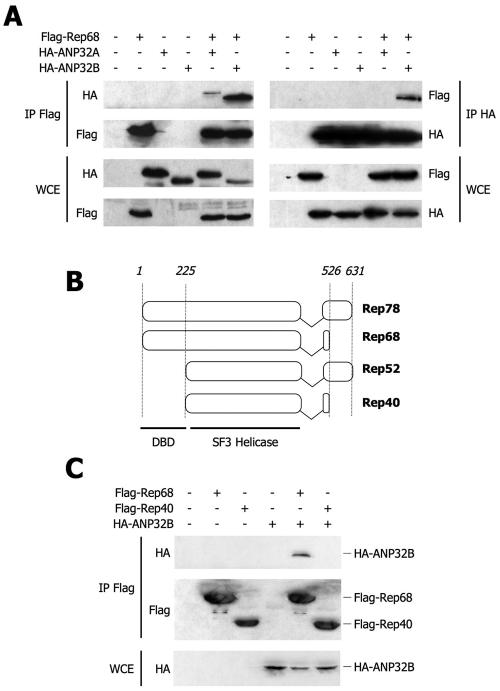
ANP32B is a member of the ANP32 family, which, in humans, also includes the closely related homologue protein ANP32A. To confirm that ANP32B is indeed an in vivo partner for Flag-Rep68 and to establish whether the latter can distinguish between different members of the ANP32 family, we performed coimmunoprecipitation experiments after cell transfection with flagged Rep and HA-tagged ANP32A or ANP32B. By using an anti-Flag antibody, we found that HA-ANP32B and, only to a much lesser extent, HA-ANP32A coimmunoprecipitated with Flag-Rep68 (Fig. 3A, left side). Control Western blot assays of the same immunoprecipitates and the input cell extracts showed that this difference in binding could not be attributed to different levels of expression of the two ANP32 constructs. These results were further corroborated by the observation that, by inverting the Flag and HA antibodies and by using the former for immunoprecipitation and the latter for Western blot assays, HA-ANP32B, but not HA-ANP32A, effectively pulled down Flag-Rep68 (Fig. 3A, right side).
FIG. 3.
The N terminus of Rep68 specifically binds ANP32B in vivo. (A) Coimmunoprecipitation experiments. Lysates from HEK293 cells transfected with the combinations of plasmids indicated at the top were immunoprecipitated (IP) with an anti-Flag antibody and Western blotted with an anti-HA antibody or immunoprecipitated with an anti-HA antibody and Western blotted with an anti-Flag antibody, as indicated. Immunoprecipitates were treated with MNase for 30 min at 37°C. Western blot assays of the input whole-cell lysate (WCE) are also shown as a measurement of the total input material. (B) Schematic representation of the Flag-tagged isoforms of Rep used in panel C. Residues 1 to 224 of Rep contain the DNA binding domain (DBD); Rep40 is the shortest naturally occurring isoform of Rep, encompassing residues 225 to 526, which still include an SF3 viral helicase domain. (C) Binding of Rep68 and Rep40 to ANP32B by immunoprecipitation. Lysates from HEK293 cells were transfected with the indicated combinations of plasmids to test whether Flag-Rep40 was able to interact with HA-ANP32B. Western blot assays of input whole-cell lysate (WCE) are also shown as a measurement of the input material.
Given that ANP32A and ANP32B are 70% identical in terms of amino acidic sequence, the results of these experiments strongly indicate that Flag-Rep68 specifically interacts only with ANP32B, a conclusion that agrees well with the results obtained in the proteomic analysis.
Next, we set out to determine which region of Rep68 mediated the interaction with ANP32B. Upon productive AAV-2 infection, four different isoforms of Rep are produced through the use of alternative start sites and alternative splicing (Fig. 2B). Rep40 corresponds to residues 225 to 526 of Rep68 and is the shortest natural isoform produced by AAV-2 (16); while it retains helicase activity, it lacks the DNA binding domain, which is located in the N terminus of full-length Rep (Fig. 3B). As shown in Fig. 3C, binding of HA-ANP32B to Flag-Rep40 was not detected under the same experimental conditions in which the protein indeed interacted with Flag-Rep68. This result clearly indicates that the first 224 N-terminal residues of Rep are necessary for the interaction with ANP32B.
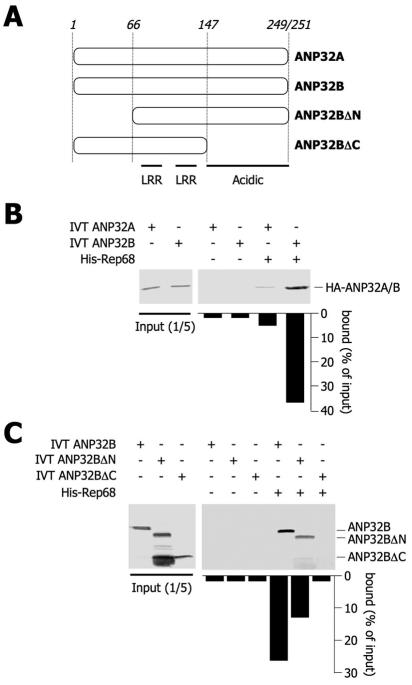
As a logical follow-up of these in vivo experiments and to demonstrate that this protein-protein interaction can also take place in vitro, 35S-labeled, transcribed-translated ANP32A and ANP32B were incubated with recombinant HisRep68 bound on beads. 35S-labeled proteins retained on the resin were quantified and measured as a percentage of the initial input (Fig. 4B). The Ni-nitrilotriacetic acid (NTA) column alone did not display any detectable unspecific binding to ANP32A or to ANP32B. Instead, we found that HisRep68 was able to retain 36.5% of 35S-labeled ANP32B. In contrast, only 5% of the initial ANP32A was retained by HisRep68, a clear indication that Rep68 is able to discriminate between different ANP32 family members and, more in general, between proteins that possess long stretches of acidic residues.
FIG. 4.
Rep68 binds the C terminus of ANP32B but not ANP32A in vitro. (A) Schematic representation of functional domains of ANP32 proteins and of the mutant proteins used in panels B and C. LRR, leucine-rich repeats. (B) In vitro pull-down experiments. In vitro-transcribed-translated (IVT), [35S]methionine-labeled ANP32 proteins were incubated with recombinant His-Rep68 bound on Ni-NTA beads. The levels of bound proteins were measured as the quantity of radioactivity that was retained on the Ni-NTA beads (as counted by PhosphorImager) and expressed as the percentage of the input. (C) Same as panel B, but deletion mutant forms of ANP32B were used in in vitro pull-down assays with His-Rep68 as indicated. The same experiments were independently performed at least three times, and superimposable results were obtained.
To map the ANP32B domain responsible for the interaction with Rep68, we constructed two deletion mutant proteins (Fig. 4A). ANP32BΔN lacks the first 66 residues of ANP32B, while ANP32BΔC is devoid of the C-terminal region of ANP32B, which is rich in acidic residues. Both deletion mutant proteins still possess two centrally located leucine-rich repeats. By in vitro pull-down assays, we observed that the acidic region of ANP32B was necessary for the protein-protein interaction with Rep68, since ANP32BΔC was not competent for binding (Fig. 4C). ANP32BΔN could still interact with HisRep68 in vitro, albeit at a lower level than the wild-type protein.
Together, the results of these in vitro experiments show that HisRep68 binds the C-terminal acidic domain of ANP32B.
ANP32A and ANP32B are required for AAV-2 replication.
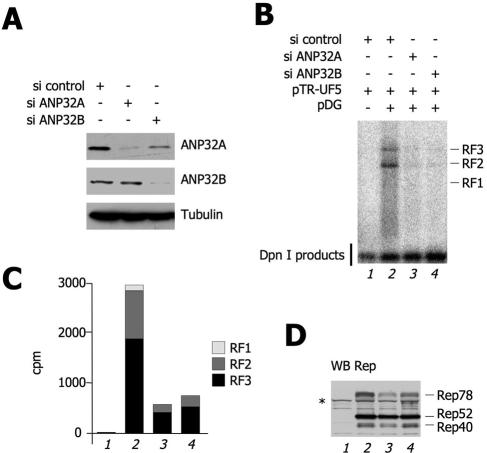
The Rep protein, which binds the AAV-2 origin and possesses multiple enzymatic functions, is essential for AAV-2 DNA replication. The observation that Rep68 physically interacts with ANP32B raises the question of whether this protein might have a role in regulating Rep activities. To test the requirement for endogenous ANP32 proteins in AAV-2 replication, we performed a series of AAV-2 DNA replication assays with HEK293 cells in which expression of ANP32A or ANP32B was down-regulated by RNAi. Two specific siRNA oligonucleotides were designed which could discriminate between these two highly homologous proteins. Both siRNAs were able to silence >85% of the expression of their respective targets from 48 h after transfection onward, as assessed by Western blot analysis (Fig. 5A). After 36 h from the beginning of siRNA treatment, cells were transfected with an AAV-2 vector (pTR-UF5) together with the pDG plasmid, which provides Rep, Cap, and a minimal subset of adenovirus helper genes in trans (11). Low-molecular-weight DNA was then extracted at 72 h and digested with DpnI to remove unreplicated input plasmid, and the presence of AAV-2 replication intermediates was assessed by Southern blotting. As expected, in control siRNA-transfected cells and in the absence of the replication functions provided by the pDG plasmid, no rAAV-2 replication products could be visualized by autoradiography (Fig. 5B). Instead, when pDG was cotransfected together with pUF5, three major bands corresponding to AAV-2 dsDNA replication intermediates were apparent (replicative forms 1, 2 and 3, designated RF1, RF2, and RF3, respectively, in Fig. 5B). In cells in which the expression of ANP32B had been silenced, AAV-2 DNA replication was almost completely impaired. To our surprise, however, the same outcome was also evident when the ANP32A form was knocked down. Radioactivity associated with each of the AAV-2 replication intermediate bands were quantified, and the results are plotted in Fig. 5C. Inhibition of AAV-2 DNA replication by the anti-ANP32A and -ANP32B siRNAs were clearly not attributable to changes in Rep protein levels (Fig. 5D).
FIG. 5.
ANP32B and ANP32A are required for AAV-2 replication. (A) siRNA-induced knockdown of ANP32A and ANP32B. HEK293 cells were transfected with the indicated siRNA oligonucleotides. Forty-eight hours posttransfection, the endogenous levels of the ANP32A and ANP32B proteins were measured by Western blotting as indicated. A Western blot assay against tubulin is shown as a control. (B) Effect of ANP32A and ANP32B knockdown on AAV-2 DNA replication. HEK293 cells were pretreated either with control, anti-ANP32A, or anti-ANP32B siRNA, as indicated. Thirty-six hours after siRNA transfection, cells were cotransfected with various combinations of plasmids as indicated. Seventy-two hours posttransfection, low-molecular-weight DNA was extracted by the Hirt method and digested with DpnI to remove input bacterial plasmids. An AAV-2 DNA fragment was used in a Southern blot assay to visualize de novo rAAV-2 replication monomer, dimer, and tetramer intermediates, designated RF1, RF2, and RF3, respectively. The lowest band is a DpnI digestion product and serves as an internal transfection and loading control. The same experiment was repeated three times with similar results. (C) Quantification of the AAV-2 replicative forms of panel B. The intensities of the bands corresponding to RF1, RF2, and RF3 were quantified by PhosphorImager and plotted in the histogram as indicated. (D) Levels of expression of the Rep isoforms. Cell extracts from the experiment shown in panel A were analyzed by Western blotting (WB) with an anti-Rep antibody. The asterisk opposite Rep78 indicates an unspecific band also present in the sample not transfected with pDG.
Collectively, the results of this experiment clearly indicate that ANP32A and ANP32B are required for AAV-2 DNA replication.
The TAF-I/Set complex copurifies with Flag-Rep68 in vivo.
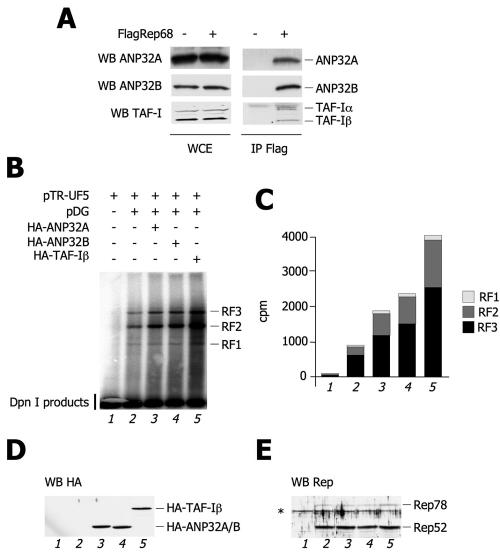
The decrease in AAV-2 replication levels caused by the silencing of ANP32A was in a sense unexpected, considering that, in contrast to ANP32B, this protein does not directly associate with Rep either in the in vitro pull-down assays or when overexpressed in coimmunoprecipitations (Fig. 3A and 4B). Nevertheless, previous observations indicate that ANP32B associates with ANP32A in a protein complex that also includes the histone chaperone TAF-I/Set (5). These findings raise the possibility that Rep, by specifically binding ANP32B, might also recruit ANP32A and TAF-I/Set. To assess the presence of other components of the TAF-I/Set complex not found in the proteomic analysis, as well as to confirm the identification of endogenous ANP32B as a protein partner of Rep68, we probed Flag-Rep68 immunoprecipitates with specific antibodies against endogenous ANP32A, ANP32B, and TAF-I. Immunoprecipitations were performed in the presence of MNase in order to rule out any possible role of DNA in mediating the physical interaction between the proteins under examination. Indeed, as shown in Fig. 5A, endogenous ANP32B was found to associate with Flag-Rep68. In addition, the Rep immunoprecipitate also contained ANP32A and the two splicing isoforms of TAF-I, TAF-Iα and TAF-Iβ (Fig. 6A).
FIG. 6.
Endogenous TAF-I/Set subunits associate with Flag-Rep68 and increase AAV-2 replication. (A) Rep coimmunoprecipitates with endogenous TAF-I/Set. Lysates of HEK293 cells transfected either with an empty vector plasmid or with Flag-Rep68 were immunoprecipitated (IP) with an anti-Flag antibody. MNase-treated immunoprecipitates were probed with specific antibodies against endogenous subunits of the Set/TAF-I complex (ANP32A, ANP32B, and TAF-I). Western blot (WB) assays of input whole-cell lysates (WCE) are also shown. (B) Levels of AAV-2 DNA replication after overexpression of TAF-I/Set members. The indicated combinations of plasmids were cotransfected into HEK293 cells. Thirty-six hours posttransfection, samples were treated as described in the legend to Fig. 5B. The same experiment was repeated three times with similar results. (C) Quantification of the AAV-2 replicative forms of panel B. The intensities of the bands corresponding to RF1, RF2, and RF3 were quantified by PhosphorImager and plotted in the histogram as indicated. (D) Levels of expression of TAF-I/Set proteins. Cell extracts from the experiment shown in panel B were analyzed by Western blotting with an anti-HA antibody. (E) Levels of expression of Rep. Cell extracts from the experiment shown in panel B were analyzed by Western blotting with an antibody against Rep.
Thus, these results indicate that Rep68, through its specific binding to ANP32B, becomes part of a larger set of proteins also containing all of the endogenous subunits of the TAF-I/Set complex.
The TAF-I/Set complex stimulates AAV-2 replication.
The observations reported above, together with the RNAi results, prompted us to consider whether overexpression of ANP32B, as well as of the other TAF-I/Set complex subunits, might have a positive effect on AAV-2 replication. To test this possibility, we performed AAV-2 replication assays by cotransfecting pTR-UF5 and pDG with or without expression vectors for HA-ANP32A, HA-ANP32B, and HA-TAF-Iβ (Fig. 6B). When any of these three proteins was cotransfected, a remarkable increase in the quantity of AAV-2 replication products was observed, which was particularly striking in the case of TAF-Iβ (Fig. 6B and quantifications in Fig. 6C, lanes 3, 4, and 5). In these experiments, we verified that the three HA-tagged proteins were expressed at comparable levels (Fig. 6D) and that they did not influence the overall amounts of the various Rep isoforms (Fig. 6E). Moreover, the ectopic expression of HA-ANP32A, HA-ANP32B, or HA-TAF-Iβ did not substantially affect the cell cycle profiles of transfected HEK293 cells (data not shown), thus ruling out the possibility that, under these experimental conditions, overexpression of these proteins had a relevant effect on the DNA replication of the host cell.
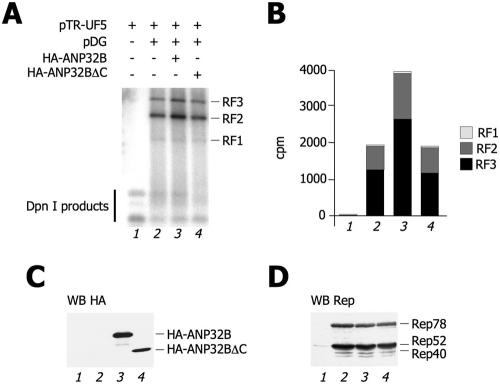
As a final control, we assessed the specificity of the increase in AAV-2 DNA replication after ANP32B protein overexpression. For this purpose, we performed an additional DNA replication assay in which we either transfected full-length HA-ANP32B or a C-terminally truncated form ANP32BΔC which does not bind Rep68 (Fig. 4C). As shown in Fig. 7A and quantified in Fig. 7B, while the former protein significantly increased the levels of AAV-2 replication intermediate, overexpression of the mutated protein was completely ineffective. Figure 7C and D show that, in these experiments, both proteins were expressed at comparable levels and did not alter the expression of the different Rep isoforms.
FIG. 7.
The effect of ANP32B on AAV-2 replication is dependent on Rep68 binding. (A) Levels of AAV-2 DNA replication after expression of wild-type or truncated ANP32B. The indicated combinations of plasmids were cotransfected into HEK293 cells. Thirty-six hours posttransfection, samples were treated as described in the legend to Fig. 4B. The same experiment was repeated three times with similar results. (B) Quantification of the AAV-2 replicative forms of panel A. The intensities of the bands corresponding to RF1, RF2, and RF3 were quantified by PhosphorImager and plotted in the histogram as indicated. (C) Levels of expression of ANP32B proteins. Cell extracts from the experiment shown in panel A were analyzed by Western blotting (WB) with an anti-HA antibody. (D) Levels of expression of Rep isoforms. Cell extracts from the experiment shown in panel A were analyzed by Western blotting with an anti-Rep antibody.
Taken together, these results clearly show that the TAF-I/Set complex positively regulates AAV-2 replication and that this effect is mediated by the specific interaction of Rep with ANP32B.
The TAF-I/Set complex increases rAAV-2 titers.
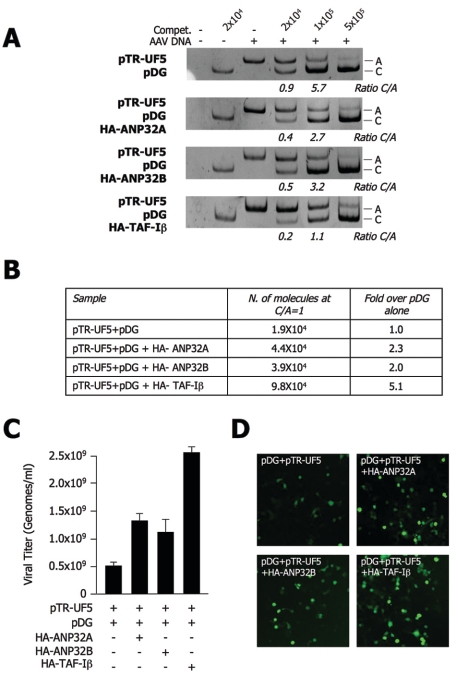
Having observed an increase in the production of dsDNA rAAV-2 genomes by overexpressing TAF-I/Set subunits, we considered whether this would result in an increase in the viral titers of rAAV-2 preparations. Therefore, we set up a series of rAAV-2 vector production experiments. In brief, HEK293 cells were transfected with pTR-UF5 and pDG with or without plasmids expressing HA-tagged TAF-I/Set components. Virus particles were released from producing cells by repeated freeze-thaw cycles and then fractionated by ammonium sulfate precipitation. Total protein levels were quantified for each preparation and normalized accordingly. Samples were first digested with DNase I to remove nonencapsidated genomes, and then capsid proteins were removed by proteinase K digestion. Finally, viral titers were determined by competitive quantitative PCR (47). The titer of encapsidated viral genomes was determined by comparing the PCR coamplification of constant quantities of the sample with increasing scalar amounts of a synthetic DNA molecule of a different size, acting as a competitor, whose concentration is known. The results of a representative competitive PCR titration are shown in Fig. 8A. The PCR amplification products were resolved by gel electrophoresis, and the ratio of competitor (C) to AAV-2 DNA amplification (A) products was evaluated by densitometric scanning. For each amplification, the C/A ratios were plotted against the input amount of competitor DNA and the line fitting the experimental data was calculated. According to the equation describing this line, the number of nascent DNA molecules was evaluated at a C/A ratio of 1 (Fig. 8B). As evaluated by at least three independent experiments, AAV-2 titers were significantly increased when the different component of the TAF-I/Set complex were cotransfected with pTR-UF5 and pDG (Fig. 8C). In particular, cotransfection of the pDG helper plasmid together with HA-TAF-Iβ resulted in a more-than-fivefold increase in vector production. The rAAV-2-GFP vector preparations retained full infectivity, since when equal volumes of vectors were used to transduce target HEK293 cells, the number of fluorescent cells and the intensity of fluorescence were proportional to the vector titer. Representative images of fluorescent HEK293 cells after transduction with the different vector preparations are shown in Fig. 8D.
FIG. 8.
TAF-I/Set subunits increase rAAV-2 titers. (A) Determination of rAAV-2 titers by competitive PCR. HEK293 cells were transfected with pTR-UF5 and the combinations of plasmids indicated to the left of each gel. After 36 h, rAAV-2 vectors were prepared as described in Materials and Methods. The DNA content of encapsidated AAV-2 genomes was quantified by competitive PCR. Viral DNA was added to scalar amounts of competitor (Compet.) DNA (as indicated at the top), followed by PCR amplification. Below the gels, the C/A ratios are reported. (B) Quantification of the abundance of AAV-2 genomes amplified in panel A. For each amplification, the C/A ratios were plotted against the input amount of competitor DNA and the line fitting the experimental data was calculated. According to its equation, the number of AAV-2 DNA molecules was evaluated at a C/A ratio of 1. (C) Quantification of AAV-2 titers. The histogram shows the rAAV-2 titers in HEK293 cells transfected with the plasmids indicated at the bottom. The results (mean ± standard deviation of at least three independent experiments) are reported as the number (N.) of AAV-2 genomes per milliliter. (D) Transduction efficiency of rAAV-2 vectors produced as described in panel A. Fresh HEK293 cells were transduced with the same volume of the rAAV-2-GFP vector preparations produced either with or without overexpressing components of the TAF-I/Set complex, as indicated. Fluorescence microscopy images of rAAV-2-GFP-transduced cells were taken after 72 h.
Together, these results indicate that overexpression of different components of the TAF-I/Set complex significantly improves the efficiency of rAAV-2 vector production. Of possible interest is the fact that, under the same experimental conditions, we could not observe any further increase upon the concomitant cotransfection of different TAF-I/Set member proteins together (data not shown).
DISCUSSION
Viral DNA replication essentially depends on protein factors provided by host cells. Small DNA virus genomes, such as SV40 and AAV-2, contain only two open reading frames, of which one codes for the proteins of the capsid and the other codes for a multifunctional protein essential for viral DNA replication. This is the case for SV40 T antigen and for AAV-2 Rep, which act by recognizing the viral origin of DNA replication, as well as providing several enzymatic activities (primarily DNA helicase). In both cases, all of the other molecular requirements are carried out through the recruitment, catalyzed by the viral replicative protein, of a vast array of proteins of cellular origin, including DNA polymerases and accessory factors. Despite much effort, the identities of many of these factors have remained elusive.
In this work, we have identified several cellular proteins that associate with Rep (Fig. 1). Three of these proteins (DNA-PKcs, Ku80, and Ku70) constitute the DNA-PK complex that is involved in the repair of double-strand breaks by nonhomologous end joining. However, we also found that binding of Rep to this complex strictly depended on the presence of DNA since DNase treatment abolished the interaction. This observation does not rule out the possibility that the DNA-PK complex is indeed involved in Rep-mediated AAV-2 DNA processing, also considering that Ku binds rAAV-2 genomes in vivo in the absence of Rep (47). However, it is equally compatible with the possibility that the apparent interaction between Rep and DNA-PK might ensue as an artifact consequent to DNA bridging, as already described in other instances (see, among others, reference 22).
In our proteomic analysis, we could find ANP32B as a bona fide interactor of Rep68. We observed that ANP32B binds the amino terminus of Rep68, which is shared by the major Rep isoforms (Rep78 and Rep68) but not by the shorter ones (Rep52 and Rep40), which are not proficient in AAV-2 replication (30). In addition, we found that Flag-Rep68 preferentially binds ANP32B compared to ANP32A both in vitro and when ectopically expressed in vivo. The last observation underlines the high specificity of the interaction between the two proteins, also considering that ANP32A and ANP32B are highly homologous (>80% homology, >70% amino acid identity). In addition, both possess long stretches of acidic residues at their C termini which might be endowed with low-affinity binding to basic domains of other proteins. However, only the C terminus of ANP32B binds Rep.
Two complementary sets of experiments highlight the role of the Rep-ANP32B interaction in the context of AAV-2 DNA replication. First, when ectopically expressed in an AAV-2 replication assay, ANP32B significantly enhances the levels of AAV-2 replication, as concluded from the increase in all dsDNA viral replication intermediates. Second, in a consistent manner, AAV-2 DNA replication is remarkably impaired in cells in which the levels of endogenous ANP32B are down-regulated by RNAi.
While performing these functional experiments, we unexpectedly observed that the same functional effects were also obtained, respectively, by overexpressing ANP32A and by down modulating its expression with a specific siRNA. Thus, even though Rep only binds ANP32B in vitro, both this protein and ANP32A similarly affect AAV-2 DNA replication. These apparently puzzling results are explained by the fact that Rep, through its interaction with ANP32B, is able to recruit all of the endogenous subunits of the TAF-I/Set complex, which also includes ANP32A and the histone chaperone TAF-I. Accordingly, we were able to show that the ectopic expression of all known different subunits of the TAF-I/Set complex, and most notably of TAF-Iβ, caused a marked increase in AAV-2 DNA replication. The specificity of this effect is further highlighted by the observation that the overexpression of ANP32BΔC, which lacks the C-terminal domain and does not bind Rep, had no effect in these assays. Taken together, these results indicate that the TAF-I/Set complex acts at the level of AAV-2 DNA replication and that its function is required for this process to take place. Of interest, this notion is also indirectly strengthened by the observation that the TAF-I/Set components are phylogenetically conserved in Drosophila melanogaster Sf9 cells, which robustly support Rep78-dependent rAAV-2 DNA replication (41).
Our findings represent the first indication that the proteins belonging to the TAF-I/Set complex actively participate in AAV-2 replication. What might be the mechanism of action of these cellular factors? The TAF-I/Set complex binds nucleosomes and was originally shown to inhibit transcription by masking histones from being acetylated, hence the original name INHAT (inhibitor of acetyltransferase) (33). In contrast, however, other studies have suggested that the histone binding activity of the TAF-I chaperone is rather necessary for the activation of transcription from chromatinized templates in vitro (10). This last piece of evidence is further corroborated by the fact that TAF-I was originally discovered in an in vitro screening of cellular factors capable of promoting replication and transcription of the adenovirus genome when this was compacted with basic viral proteins into a structure called the “core” (26, 27). In this context, the likely activity carried out by TAF-I is to remodel this adenovirus core, thereby making the template DNA accessible to the transcription and replication apparatus. Consistent with this function, both ANP32A and the TAF-I/Set histone chaperone are present on adenovirus DNA during the first phases of viral infection (46). Taken together, these findings clearly suggest a role for the TAF-I complex in the replication of viral DNA genomes by the host cell machinery. Considering the known differences in molecular structure and basic molecular mechanisms between adenovirus and AAV-2, it might well be speculated that the TAF-I/Set complex regulates AAV-2 replication by changing the chromatin conformation of AAV-2 templates and rendering it more suitable for DNA replication. Our observation that all three AAV-2 DNA replication intermediates are equally affected by overexpressing or down-regulating the TAF-I/Set complex members is consistent with this hypothesis. In this context, it will also be interesting to understand whether the participation of TAF-I/Set in the replication of both AAV-2 and adenovirus might be somehow part of the helper function that the latter virus provides to AAV-2.
Finally, the increase in DNA replication caused by the overexpression of members of the TAF-I/Set complex, and in particular of TAF-Iβ, also results in a remarkable increase in rAAV-2 titers when analyzed in a standard AAV-2 vector production protocol. We believe that this observation has important implications for the gene therapy field, especially since most of the protocols for AAV-2 vector production still rely on transient cotransfection of large amounts of HEK293 cells with a plasmid corresponding to the vector and one or more plasmids expressing the AAV-2 proteins and different adenovirus helper functions. Thus, the possibility of increasing the titers of the viral preparations by ectopic expression of TAF-Iβ would result in a substantial improvement in the overall efficiency of the procedure.
Supplementary Material
Acknowledgments
We thank M. Linden, J. Kleinschmidt, and N. Muzyczka for kindly providing reagents. We are grateful to B. Boziglav and M. Dapas for excellent technical assistance and to S. Kerbavcic for editorial help.
This work was supported by grants from the Telethon Foundation Italy; from the FIRB program of the Ministero dell'Istruzione, Universita' e Ricerca, Italy; and from the Fondazione Cassa di Risparmio of Trieste, Italy.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bakkenist, C. J., and M. B. Kastan. 2004. Initiating cellular stress responses. Cell 118:9-17. [DOI] [PubMed] [Google Scholar]
- 2.Balague, C., M. Kalla, and W. W. Zhang. 1997. Adeno-associated virus Rep78 protein and terminal repeats enhance integration of DNA sequences into the cellular genome. J. Virol. 71:3299-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batchu, R. B., M. A. Shammas, J. Y. Wang, J. Freeman, N. Rosen, and N. C. Munshi. 2002. Adeno-associated virus protects the retinoblastoma family of proteins from adenoviral-induced functional inactivation. Cancer Res. 62:2982-2985. [PubMed] [Google Scholar]
- 4.Berns, K. I., and R. M. Linden. 1995. The cryptic life style of adeno-associated virus. Bioessays 17:237-245. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, C. M., I. E. Gallouzi, and J. A. Steitz. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costello, E., P. Saudan, E. Winocour, L. Pizer, and P. Beard. 1997. High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 16:5943-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Pasquale, G., and S. N. Stacey. 1998. Adeno-associated virus Rep78 protein interacts with protein kinase A and its homolog PRKX and inhibits CREB-dependent transcriptional activation. J. Virol. 72:7916-7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan, D., Y. Yue, and J. F. Engelhardt. 2003. Consequences of DNA-dependent protein kinase catalytic subunit deficiency on recombinant adeno-associated virus genome circularization and heterodimerization in muscle tissue. J. Virol. 77:4751-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flotte, T. R. 2004. Gene therapy progress and prospects: recombinant adeno-associated virus (rAAV) vectors. Gene Ther. 11:805-810. [DOI] [PubMed] [Google Scholar]
- 10.Gamble, M. J., H. Erdjument-Bromage, P. Tempst, L. P. Freedman, and R. P. Fisher. 2005. The histone chaperone TAF-I/SET/INHAT is required for transcription in vitro of chromatin templates. Mol. Cell. Biol. 25:797-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimm, D., A. Kern, K. Rittner, and J. A. Kleinschmidt. 1998. Novel tools for production and purification of recombinant adeno-associated virus vectors. Hum. Gene Ther. 9:2745-2760. [DOI] [PubMed] [Google Scholar]
- 12.Hickman, A. B., D. R. Ronning, R. M. Kotin, and F. Dyda. 2002. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus. Rep. Mol. Cell 10:327-337. [DOI] [PubMed] [Google Scholar]
- 13.Hoggan, M. D., N. R. Blacklow, and W. P. Rowe. 1966. Studies of small DNA viruses found in various adenovirus preparations: physical, biological, and immunological characteristics. Proc. Natl. Acad. Sci. USA 55:1467-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Im, D. S., and N. Muzyczka. 1989. Factors that bind to adeno-associated virus terminal repeats. J. Virol. 63:3095-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Im, D. S., and N. Muzyczka. 1992. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J. Virol. 66:1119-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Im, D. S., and N. Muzyczka. 1990. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 61:447-457. [DOI] [PubMed] [Google Scholar]
- 17.James, J. A., C. R. Escalante, M. Yoon-Robarts, T. A. Edwards, R. M. Linden, and A. K. Aggarwal. 2003. Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure (London) 11:1025-1035. [DOI] [PubMed] [Google Scholar]
- 18.Kobe, B., and A. V. Kajava. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11:725-732. [DOI] [PubMed] [Google Scholar]
- 19.Kotin, R. M., R. M. Linden, and K. I. Berns. 1992. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 11:5071-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotin, R. M., M. Siniscalco, R. J. Samulski, X. D. Zhu, L. Hunter, C. A. Laughlin, S. McLaughlin, N. Muzyczka, M. Rocchi, and K. I. Berns. 1990. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 87:2211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyostio, S. R., R. S. Wonderling, and R. A. Owens. 1995. Negative regulation of the adeno-associated virus (AAV) P5 promoter involves both the P5 Rep binding site and the consensus ATP-binding motif of the AAV Rep68 protein. J. Virol. 69:6787-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai, J. S., and W. Herr. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA 89:6958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linden, R. M., P. Ward, C. Giraud, E. Winocour, and K. I. Berns. 1996. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 93:11288-11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malek, S. N., A. I. Katumuluwa, and G. R. Pasternack. 1990. Identification and preliminary characterization of two related proliferation-associated nuclear phosphoproteins. J. Biol. Chem. 265:13400-13409. [PubMed] [Google Scholar]
- 25.Marcello, A., P. Massimi, L. Banks, and M. Giacca. 2000. Adeno-associated virus type 2 Rep protein inhibits human papillomavirus type 16 E2 recruitment of the transcriptional coactivator p300. J. Virol. 74:9090-9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto, K., K. Nagata, M. Ui, and F. Hanaoka. 1993. Template activating factor I, a novel host factor required to stimulate the adenovirus core DNA replication. J. Biol. Chem. 268:10582-10587. [PubMed] [Google Scholar]
- 27.Matsumoto, K., M. Okuwaki, H. Kawase, H. Handa, F. Hanaoka, and K. Nagata. 1995. Stimulation of DNA transcription by the replication factor from the adenovirus genome in a chromatin-like structure. J. Biol. Chem. 270:9645-9650. [DOI] [PubMed] [Google Scholar]
- 28.Mutai, H., Y. Toyoshima, W. Sun, N. Hattori, S. Tanaka, and K. Shiota. 2000. PAL31, a novel nuclear protein, expressed in the developing brain. Biochem. Biophys. Res. Commun. 274:427-433. [DOI] [PubMed] [Google Scholar]
- 29.Ni, T. H., W. F. McDonald, I. Zolotukhin, T. Melendy, S. Waga, B. Stillman, and N. Muzyczka. 1998. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J. Virol. 72:2777-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni, T. H., X. Zhou, D. M. McCarty, I. Zolotukhin, and N. Muzyczka. 1994. In vitro replication of adeno-associated virus DNA. J. Virol. 68:1128-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owens, R. A., M. D. Weitzman, S. R. Kyostio, and B. J. Carter. 1993. Identification of a DNA-binding domain in the amino terminus of adeno-associated virus Rep proteins. J. Virol. 67:997-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samulski, R. J., X. Zhu, X. Xiao, J. D. Brook, D. E. Housman, N. Epstein, and L. A. Hunter. 1991. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 10:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo, S. B., P. McNamara, S. Heo, A. Turner, W. S. Lane, and D. Chakravarti. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104:119-130. [DOI] [PubMed] [Google Scholar]
- 34.Smith, R. H., and R. M. Kotin. 1998. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J. Virol. 72:4874-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song, S., P. J. Laipis, K. I. Berns, and T. R. Flotte. 2001. Effect of DNA-dependent protein kinase on the molecular fate of the rAAV2 genome in skeletal muscle. Proc. Natl. Acad. Sci. USA 98:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song, S., Y. Lu, Y. K. Choi, Y. Han, Q. Tang, G. Zhao, K. I. Berns, and T. R. Flotte. 2004. DNA-dependent PK inhibits adeno-associated virus DNA integration. Proc. Natl. Acad. Sci. USA 101:2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stracker, T. H., G. D. Cassell, P. Ward, Y. M. Loo, B. van Breukelen, S. D. Carrington-Lawrence, R. K. Hamatake, P. C. van der Vliet, S. K. Weller, T. Melendy, and M. D. Weitzman. 2004. The Rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication. J. Virol. 78:441-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Surosky, R. T., M. Urabe, S. G. Godwin, S. A. McQuiston, G. J. Kurtzman, K. Ozawa, and G. Natsoulis. 1997. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J. Virol. 71:7951-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka, M., and W. Herr. 1990. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell 60:375-386. [DOI] [PubMed] [Google Scholar]
- 40.Tattersall, P., and D. C. Ward. 1976. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature 263:106-109. [DOI] [PubMed] [Google Scholar]
- 41.Urabe, M., C. Ding, and R. M. Kotin. 2002. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum. Gene Ther. 13:1935-1943. [DOI] [PubMed] [Google Scholar]
- 42.Walensky, L. D., D. S. Coffey, T. H. Chen, T. C. Wu, and G. R. Pasternack. 1993. A novel Mr 32,000 nuclear phosphoprotein is selectively expressed in cells competent for self-renewal. Cancer Res. 53:4720-4726. [PubMed] [Google Scholar]
- 43.Ward, P., and R. M. Linden. 2000. A role for single-stranded templates in cell-free adeno-associated virus DNA replication. J. Virol. 74:744-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weger, S., M. Wendland, J. A. Kleinschmidt, and R. Heilbronn. 1999. The adeno-associated virus type 2 regulatory proteins Rep78 and Rep68 interact with the transcriptional coactivator PC4. J. Virol. 73:260-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wysocka, J., M. P. Myers, C. D. Laherty, R. N. Eisenman, and W. Herr. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17:896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue, Y., J. S. Johnson, D. A. Ornelles, J. Lieberman, and D. A. Engel. 2005. Adenovirus protein VII functions throughout early phase and interacts with cellular proteins SET and pp32. J. Virol. 79:2474-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zentilin, L., A. Marcello, and M. Giacca. 2001. Involvement of cellular double-stranded DNA break binding proteins in processing of the recombinant adeno-associated virus genome. J. Virol. 75:12279-12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.