Abstract
We have identified dihydroxythiophenes (DHT) as a novel series of human immunodeficiency virus type 1 (HIV-1) integrase inhibitors with broad antiviral activities against different HIV isolates in vitro. DHT were discovered in a biochemical integrase high-throughput screen searching for inhibitors of the strand transfer reaction of HIV-1 integrase. DHT are selective inhibitors of integrase that do not interfere with virus entry, as shown by the inhibition of a vesicular stomatitis virus G-pseudotyped retroviral system. Moreover, in quantitative real-time PCR experiments, no effect on the synthesis of viral cDNA could be detected but rather an increase in the accumulation of 2-long-terminal-repeat cycles was detected. This suggests that the integration of viral cDNA is blocked. Molecular modeling and the structure activity relationship of DHT demonstrate that our compound fits into a two-metal-binding motif that has been suggested as the essential pharmacophore for diketo acid (DKA)-like strand transfer inhibitors (Grobler et al., Proc. Natl. Acad. Sci. USA 99:6661-6666, 2002.). This notion is supported by the profiling of DHT on retroviral vectors carrying published resistance mutations for DKA-like inhibitors where DHT showed partial cross-resistance. This suggests that DHT bind to a common site in the catalytic center of integrase, albeit with an altered binding mode. Taken together, our findings indicate that DHT are novel selective strand transfer inhibitors of integrase with a pharmacophore homologous to DKA-like inhibitors.
The introduction of highly active antiretroviral therapy (HAART) in the mid-90s of the last century led to a dramatic improvement in the treatment of patients infected with human immunodeficiency virus (HIV) in industrially advanced countries (41, 44, 53). HAART has effectively transformed the HIV/AIDS disease from an invariably lethal infection to a chronic, but still mortal, illness. Current HAART regimens involve the combined oral administration of inhibitors against the two viral enzymes reverse transcriptase and HIV protease. Only recently, a novel antiviral principle in the form of the gp41-based fusion inhibitor T20 was introduced into salvage therapy of HAART patients (16, 47). However, despite these advances in therapy, substantial unmet medical needs in HIV treatment remain: in an increasing number of patients, HAART therapy ultimately fails due to the emergence of HIV variants that are resistant to currently used drugs (37, 43, 54); many side effects and long-term toxicities of current drugs impair the patient's quality of life (for information see the Guidelines for the Use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents at http://aidsinfo.nih.gov) (10); high pill burden and complicated dosing lead to low compliance (1, 12). Thus, the identification of improved agents that are designed to interrupt alternative stages in the viral life cycle represents a fundamental challenge in HIV research.
The integration of viral DNA into the cellular chromosome is a key step in the viral replication cycle, ensuring the stable maintenance of the viral genome in the host organism (13, 35, 55). Therefore, this essential reaction, which is catalyzed by HIV's third enzyme, integrase (IN), represents an attractive but so-far-unexploited target for therapeutic intervention (3, 15, 56). IN of HIV-1 subtype B isolates is a 32-kDa enzyme (288 amino acid residues) encoded together with the reverse transcriptase and the protease by the pol gene of HIV and is generated during virion maturation by proteolytic processing of the gag-pol precursor. IN is composed of three essential domains, with the central catalytic domain (residues 50 to 212) containing a triad of invariant carboxylate residues, D64, D116, and E152 (the so-called D,D-35E motif). The latter are involved in the coordination of divalent metal ions and are therefore required for catalysis. Whereas structural studies of IN reveal a single binding site for Mg2+ (formed by D64 and D116) (23, 38), there is also evidence from related phosphotransferases like avian sarcoma virus, RNase H, and type II restriction endonucleases that suggests that IN might possess a second metal binding site (formed by D63 and E152) which might be occupied in the presence of the DNA substrate (7, 14, 34).
Crystal structures of various integrase subdomains have been published (11, 24), but the active site has turned out to be of a highly flexible nature, thus hampering structural approaches for drug design (4, 36).
The integration of newly synthesized viral DNA into the host chromosome is a multistep process (3, 51). Initially, IN recognizes the long-terminal repeat (LTR) of the retro-transcribed viral DNA, leading to the assembly of various viral and cellular proteins in a preintegration complex. Subsequently, IN performs endonucleolytic processing of the 3′ ends of both strands via recognition of an absolutely conserved CA dinucleotide and specific cleavage of the terminal GT dinucleotide downstream, thereby generating two recessed 3′-OH ends (3′ processing). The latter serve as nucleophiles in the following strand transfer (ST) step, where, in a transesterification reaction, host DNA is cleaved and concomitantly and covalently linked to the viral genome. Finally, this irreversible incorporation of the HIV cDNA into the host chromosome is completed by DNA gap repair, most likely with the help of cellular repair enzymes.
Despite the identification of diverse classes of IN inhibitors in simple oligonucleotide-based assays (reviewed in reference 48), most of these compounds do not exhibit antiviral activity or are too toxic in cell culture (33). Other compounds lack specificity and have been abandoned (19). The first antiviral series of compounds that have been proven to act unambiguously via selective inhibition of IN were the 1,3-diketo acid (DKA) inhibitors identified in a strand transfer assay employing enzyme preassembled on oligonucleotides (26). It has been suggested that the prebinding of IN to viral DNA in a donor substrate complex configures an active site conformation that is not accessible in assays initiated with free enzyme. DKAs selectively bind to this complex, thereby competing with the target (or host) DNA substrate during the strand transfer reaction (18). From previous studies, it has been postulated that binding to and inhibition of IN by DKA is achieved by the interaction of the critical DKA pharmacophore with two divalent metal ions in the active site of IN (25). Consistent with this proposed mechanism, mutations that confer resistance to DKA map to the integrase active site adjacent to amino acids involved in the coordination of divalent metal ions. It has been reported that resistance mutations reduce IN enzymatic activity and, hence, the replicative fitness of affected viruses (20, 27, 28).
As expected from their novel mode of action, DKA-like inhibitors were also shown to be effective against clinical isolates that were resistant to reverse transcriptase and protease inhibitors (PIs) (28). Consequently, S-1360, a triazole analogue of DKA developed by Shionogi & Co. and GlaxoSmithKline, was the first integrase strand transfer inhibitor (INSTI) to enter clinical trials, but the development was stopped during phase I/II (Yoshinaga et al., Abstr. 9th Conf. Retrovir. Opportun. Infect., abstr. 8, 2002) (5). Subsequently, Merck reported on a novel series of potent INSTIs replacing the 1,3-diketo acid moiety by an isosteric 8-hydroxy-1,6-naphthyridine core that showed improved metabolic stability (59). In vivo efficacy of the naphthyridine carboxamide L-870,812 (50% inhibitory concentration [IC50] was 40 nM in a strand transfer assay) was reported in simian-human immunodeficiency virus-infected rhesus macaques (29), and a congener, L-870,810 (IC50 = 8 nM), moved into clinical trials, where it provided proof of concept in antiretroviral therapy-experienced and antiretroviral therapy-naïve patients (S. Little et al., Abstr. 12th Conf. Retrovir. Opportun. Infect., abstr. 161, 2005). However, recently the development of L-870,810 was discontinued in favor of MK-0518, which presumably represents another member of the naphthyridine carboxamide series characterized by an improved pharmacokinetic profile (J. O. Morales-Ramirez et al., Abstr. 10th Europ. AIDS Clin. Soc., abstr. LBPS 1/6, 2005) (17). Whether a second clinical development compound named GS-9137/JTK-313 belongs to the structurally related class of 4-oxoquinoline integrase inhibitors is presently not clear due to a lack of published data (50).
In the present study, we describe the identification and characterization of dihydroxythiophenes (DHT), a novel series of selective INSTIs with antiviral activity in cell culture. Biochemical and antiviral activities of DHT were profiled against those of recently described INSTIs, i.e., Merck's DKA and the former developmental compounds L-870,810 and S-1360. We furthermore demonstrate that DHT possess a pharmacophore that is homologous to the DKA motif, which suggests that DHT are mechanistically related. However, as DHT are only partially cross-resistant to previously described INSTIs, their binding mode to the IN active site is probably not identical.
MATERIALS AND METHODS
Cells and viruses.
Unless otherwise stated, cells were grown in RPMI 1640 medium (GIBCO) supplemented with 10% heat-inactivated fetal calf serum (hi-FCS), 2 mM l-glutamine (GIBCO), 100 μg/ml streptomycin (GIBCO), and 100 U/ml penicillin G (GIBCO) at 37°C in a 5% CO2-humidified atmosphere.
C8166, H9, and MT4 cell lines were obtained from the European Collection of Cell Cultures. The U1 cell line (described in reference 21) was kindly provided by T. M. Folks (CDC, Atlanta, Ga.).
Peripheral blood mononuclear cells (PBMCs) were isolated from HIV-1-negative donors by density gradient centrifugation, grown in RPMI 1640 medium supplemented with 15% fetal calf serum (FCS), and activated with 90 μg/ml phytohemagglutinin (Murex Biotech Ltd., Dartford, United Kingdom) and 40 U/ml interleukin-2 (Roche, Basel, Switzerland) for 3 days before infection.
For the preparation of primary monocytes/macrophages, PBMCs of HIV-1-negative donors were isolated by density gradient centrifugation as described above and seeded into tissue culture plates in RPMI medium supplemented with 5% hi-FCS and 10% hi-human serum antibody (Sigma-Aldrich). After 2 h, the supernatant was discarded and adherent cells were washed extensively and incubated for 4 days in the presence of the same medium. Subsequently, cells were detached from the plates and seeded into 96-well plates at 3 × 104 cells/well. After an additional incubation period of 3 days, cell monolayers were infected in the absence of human serum antibody.
Most of the HIV-1 and HIV-2 strains used were kindly provided by H. von Briesen (Georg-Speyer-Haus, Frankfurt/Main, Germany). The HIV-1 Ba-L isolate was obtained from TEBU GmbH (Offenbach, Germany).
Chemistry.
Antiviral compounds were synthesized in the chemical department of Bayer HealthCare Pharma Research and dissolved in dimethyl sulfoxide at a 10 mM final concentration (f.c.). The compounds DHT-1 to -9 are accessible through standard methods (data not shown). The reference compounds DKA and L-870,810 that were used in this study were described earlier (52, 2).
Integrase strand transfer assay.
A commercially available electrochemiluminescence-based assay was employed for the assessment of IN strand transfer activity (BioVeris Europe [formerly IGEN Europe], Witney, United Kingdom) (described in reference 57) essentially according to the manufacturer's instructions. HIV-1 IN (prepared according to a method previously described in reference 32) was preassembled at 37°C for 30 min on biotinylated donor DNA oligonucleotides coupled to paramagnetic streptavidin-coated M-280 Dynabeads (Dynal, Norway). Subsequently, the resulting enzyme donor DNA beads were preincubated in 384-well microplates (Corning), with or without compounds at different concentrations, for 30 min at room temperature. Samples were mixed with ruthenium(bpy)32+-conjugated target DNA substrate (final concentration of 3.1 nM) and incubated for 90 min at room temperature in the presence of 8.5 mM MgCl2. Then, the strand transfer reaction was stopped by the addition of EDTA (final concentration, 25 mM) and electrochemiluminescence of samples was analyzed on an M series 384 reader (BioVeris Europe, Whitney, United Kingdom). The underlying method is based on the capture of the magnetic beads on an electrode surface in the flow cell and photodetection of product-dependent, ruthenium-emitted light upon excitation.
Integrase 3′ processing assay.
The 3′ processing assays were performed according to the method described previously (39), with minor modifications. As a substrate, a 21-mer double-stranded DNA oligonucleotide consisting of oligonucleotide A (5′-GTGTGGAAAATCTCTAGCAGT-3′) and oligonucleotide B (5′-ACTGCTAGAGATTTTCCACAC-3′) corresponding to the terminal 21 nucleotides of the U5 HIV LTR was used. Oligonucleotide A was 5′ end labeled with [γ-32P]ATP using T4 polynucleotide kinase (Invitrogen Life Technologies) and annealed to oligonucleotide B. For compound testing, 2.5 pmol HIV-1 integrase, 0.1 pmol double-stranded oligonucleotides, 25 mM morpholinepropanesulfonic acid (pH 7.2), 7.5 mM MnCl2, 14.3 mM 2-mercaptoethanol, and 0.1 mg/ml bovine serum albumin were incubated for 30 min at 37°C in the absence or presence of various concentrations of inhibitors. The reaction was stopped by adding the same volume of stop buffer containing 98% deionized formamide, 10 mM EDTA (pH 8.0), 0.05% xylene cyanol blue, and 0.05% bromophenol blue. Samples were then heated for 5 min at 95°C and loaded on a 20% acrylamide gel containing 7 M urea in Tris-borate-EDTA. Autoradiography and quantification were performed by exposing the dry gel on a phosphorimager (Fuji).
p24 enzyme-linked immunosorbent assay.
The level of HIV p24 antigen in the culture supernatants was assessed by an enzyme-linked immunosorbent assay using the Vironostika HIV-1 antigen kit (bioMerieux, The Netherlands) according to the manufacturer's instructions.
HIV replication assays.
The antiviral activity of compounds was assessed by a cytoprotection assay based on C8166 cells infected by the HIV-1 isolate D117II (49). Briefly, C8166 cells were batch infected with HIV-1 D117II and immediately distributed into 96-well plates at 5,000 cells per well in the presence or absence of test compounds at various concentrations. After 6 days of incubation, cell survival was detected by incubation of treated cells with the fluorogenic dye alamarBlue from Biosource International (Camarillo, CA) according to the manufacturer's instructions. Anti-HIV activities of compounds were expressed as 50% effective concentrations (EC50s). The cytotoxicity of compounds was determined in parallel by alamarBlue staining of uninfected samples.
Antiviral activities of compounds against some primary isolates were tested on PBMCs. PBMCs were infected with different isolates of HIV-1 and HIV-2 as indicated, distributed into 96-well plates at 4 × 104 cells per well in the presence or absence of test compounds at various concentrations, and incubated for 4 to 7 days without subcultivating. After that, cell-free supernatants were collected and tested for p24 antigen (described above). The anti-HIV activities of compounds were expressed as EC50s. The cytotoxicity of compounds was determined in parallel by alamarBlue staining according to the manufacturer's instructions.
Antiviral activities of compounds against the R5-tropic HIV-1 isolate Ba-L were assessed on primary monocytes/macrophages. Briefly, monolayers of monocytes/macrophages were infected in the presence or absence of compounds at various concentrations. Fresh medium, with or without compound, was added at days 3 and 11 postinfection (p.i.). On day 17 p.i., cell-free supernatants were collected and tested for p24 antigen (described above). The cytotoxicity of compounds was determined in parallel by alamarBlue staining according to the manufacturer's instructions.
Time of addition experiment.
MT4 cells were resuspended in ice-cold RPMI medium and preincubated with HIV-1 (D117-II) at a high multiplicity of infection (MOI) (an MOI of >1) on ice for 40 min to allow attachment of virus to cells. Samples were then washed twice with ice-cold medium to remove unbound virus. Subsequently, cells were resuspended in 37°C warm medium and immediately (at time zero [t0]) distributed in 96-well plates (1.3 × 104 cells/well). DHT-2 was added at t0 or at various time points thereafter (t = 1, 3, 8, and 27 h) at a final concentration of 15 μM. Reference compounds included were T20 (fusion inhibitor, f.c. 5 μM), efavirenz (EFV) (nonnucleoside reverse transcriptase inhibitor [NNRTI], f.c. 250 nM), DKA (INSTI, f.c. 5 μM), and saquinavir (SQV) (PI, f.c. 500 nM). Thirty-six hours postinfection, aliquots from each sample were collected and analyzed for virion production by p24 antigen capture as described above.
U1 cell assay.
Persistently infected U1 cells (described in reference 21) were seeded at 4 × 104 cells per well in RPMI medium, and test compounds were added at different concentrations. At the same time, HIV-1 production was induced by the addition of 10 ng/ml phorbol myristate acetate (PMA) and cells were incubated for 48 h at 37°C and 5% CO2. Cell-free supernatants were then collected and tested for p24 antigen (described above). The anti-HIV activities of compounds were expressed as EC50s.
HIV vector transduction assays.
The HIV-1-based lentiviral vector system that was used was originally described by Jarmy et al. (31). The production of pseudotyped viral vectors was performed by transient cotransfection of HEK293T cells with the plasmids pczVSV-G and pGJ3-luci essentially as described earlier. Briefly, HEK293T cells grown in Dulbecco's modified Eagle's medium (GIBCO; supplemented with 2 mM l-glutamine [GIBCO] 100 μg/ml streptomycin [GIBCO] and 100 U/ml penicillin G [GIBCO]) were transfected with the same amount of each plasmid using Lipofectamine 2000 as transfection reagent (Invitrogen Life Technologies) and incubated for 7 h at 37°C. After that, the medium was exchanged and incubation continued for an additional 16 h. At that point, sodium butyrate was added at a final concentration of 10 mM. Supernatants were harvested after a final incubation of 24 h.
Infection of HEK293 cells with vector-containing supernatants was performed essentially as described elsewhere (31). Briefly, HEK293 cells were mixed with HIV vector preparation and seeded at 5 × 104 cells per well in 96-well luminometer plates (Greiner) in the presence or absence of compounds at different concentrations. Twenty-four hours later, medium was removed and 100 μl of a 1:1 mix of lysis-buffer (3% Triton X-100, 11.5% glycerol, 2 mM dithiothreitol, 25 mM Na2HPO4, 25 mM Tris/Cl [pH 7.8]) and luciferase substrate (20 mM tricine [pH 7.8], 2.7 MgSO4, 0.1 mM EDTA, 33.3 mM dithiothreitol, 270 μM coenzyme A, 470 μM luciferin, 530 μM ATP) was added. Light emission was recorded with an in-house-developed camera system (Bayer HealthCare AG). The cytotoxicity of compounds (50% cytotoxic concentration [CC50]) was determined in parallel by alamarBlue staining according to the manufacturer's instructions.
In some experiments, vector transduction was performed using particles with authentic HIV-1 envelope. These HIV vectors were produced as described above, with the exception of cotransfecting pGJ3-luci and pEnvIIIB (kindly provided by C. Jassoy) instead of pczVSV-G. The resulting nonpseudotyped vector particles were used to infect H9 cells for 48 h in the absence or presence of test compounds. An assessment of luciferase activity in transduced cells was performed as described above.
Site-directed mutagenesis.
In vitro mutagenesis of integrase was performed with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions using the plasmid pGJ3-luci as a template. The introduction of mutations was verified by DNA sequencing.
Real-time PCR.
The day before transduction, HEK293T cells were seeded in 24-well plates at 2 × 105 cells per well. Transduction with wild-type (wt) vector was carried out with 4.2 × 106 50% tissue culture infective dose/ml. The vector was added to the cells in the presence of inhibitor. After 1.5 h of incubation at 37°C, cells were washed with phosphate-buffered saline and fresh medium with the respective inhibitor was added. At different time points after transduction (0 to 48 h), cells were harvested and total DNA was extracted using a DNeasy tissue kit (QIAGEN, Hilden, Germany). DNA samples were quantified by spectrophotometry at an optical density of 260 nm and adjusted to the same concentration. Real-time PCRs were performed essentially as described previously (8, 42). For the quantification of late reverse transcripts, the primers MH531 (5′-TGTGTGCCCGTCTGTTGTG-3′), MH532 (5′-GAGTCCTGCGTCGAGAGAGC-3′), MH535 (5′-AACTAGGGAACCCACTGCTTAAG-3′), and the TaqMan probe LRT-P (5′-[FAM]-CAGTGGCGCCCGAACAGGGA-[TAMRA]-3′) were used. For the quantification of 2-LTR circles, the primer MH535, AB536-1 (5′-TGTCTTTGGGAGTGAATTAGC-3′), and the TaqMan probe MH603 (5′-[FAM]-ACACTACTTGAAGCACTCAAGGCAAGCTTT-[TAMRA]-3′) were employed. The quantification of integrated provirus was carried out in a two-step PCR amplification (8, 42). For the outer PCR, the primers Alu (5′-TGCTGGGATTACAGGCGTGAG-3′) and Q-Alu-F-out (5′-GCTAACTAGGGAACCCACTGCTTA-3′) were used. The sequence of the primer used for quantification was, for Q-Alu-F-in, 5′-AGCTTGCCTTGAGTGCTTCAA-3′, for Q-Alu-R-in, 5′-TGACTAAAAGGGTCTGAGGGATCT-3′, and for TaqMan probe, 5′-(FAM)-TTACCAGAGTCACACAACAGACGGGCA-(TAMRA)-3′. Each experiment contained a standard curve of the amplicon being measured run in duplicate, ranging from 10 to 105 copies plus negative controls that lacked a template. For quantification of late reverse transcripts and 2-LTR circle PCR, standard curves were generated by the dilution of pGJ3-Luci DNA with matching sequences. In all cases, the DNA standard was diluted into 250 ng of uninfected cellular DNA to match the cellular samples. The Alu copy number standard DNA was generated as described recently (8). Reaction mixtures contained 1× Platinum pPCR SuperMix UDB (Invitrogen Life Technologies); 300 nM forward primer, 300 nM reverse primer, and 200 nM probe primer; and 250 ng of template DNA in a 50-μl volume. After initial incubation at 50°C for 2 min and 95°C for 10 min, 40 cycles of amplification were carried out at 95°C for 30 s, followed by 1 min 30 s at 60°C. Reactions were analyzed by using the ABI Prism 7700 sequence detection system (PE-Applied Biosystems).
Modeling.
The alignment was performed with Sybyl 6.9.2 (Tripos, Inc.) on three-dimensional structures generated using Concord with the MMFF94s force field. The alignment pattern used was the seven-atom motif [C:C(:CC(any) = O)O] describing the β-keto-hydroxy fragment.
RESULTS
DHT are potent strand transfer inhibitors of HIV integrase.
In an effort to identify inhibitors for strand transfer activity of IN, we performed a high-throughput screening of a low-molecular-weight compound library. The oligonucleotide-based assay that was used (Origen assay; BioVeris) basically resembled the ST assay reported by Hazuda et al. (26) involving preassembled IN. Among the tested compounds, dihydroxythiophenes demonstrated potent IN strand transfer inhibition, whereby DHT-1 and DHT-2 were the most active class members (IC50s of 0.03 μM and 0.02 μM, respectively) as shown in Table 1. The IC50 values of reference inhibitors for strand transfer were determined in parallel in this assay and were found to be 0.012 μM, 0.13 μM, and 0.001 μM for DKA, S-1360, and L-870,810, respectively.
TABLE 1.
Activity of DHT and reference INSTI in ST and 3′ processing assays
Results show the mean value of at least six separate experiments.
Results show the mean value of at least two separate experiments.
ND, not determined.
Besides the ST reaction, integrase also catalyzes the preceding 3′ processing of HIV LTRs in vitro and in vivo, with both reactions mediated by different regions of the same active site. Therefore, we asked to what extent DHT were able to inhibit 3′ processing using an oligonucleotide-based in vitro assay. Reference compounds for strand transfer reactions, like DKA, S-1360, and L-870,810, are known to be only moderate-to-weak inhibitors for 3′ processing, leading to IC50 values of 4 μM, 15 μM, and 4 μM, respectively (Table 1). Likewise, DHT-2, with an IC50 value of 12.5 μM, preferentially affects ST reaction versus 3′ processing.
DHT inhibit HIV replication in cell culture.
Although numerous inhibitors have been described which were identified in in vitro IN assays, only a few of them were reported to exhibit antiviral activities in cell culture. Therefore, we investigated the potential of DHT to inhibit HIV replication in a cell culture model of acute infection. Interestingly, as shown in Table 2, DHT showed potent antiviral activity in an HIV replication assay based on HIV-1-infected (D117II isolate) C8166 cells with an EC50 for DHT-2 of 0.3 μM. Activities of reference INSTIs were assessed in parallel and were found to be in the same range for DKA (EC50, 0.1 μM), and S-1360 (EC50, 0.3 μM), whereas L-870,810's antiviral activity was in the nanomolar range (EC50, 0.002 μM). The cytotoxicity of DHT-2 on C8166 cells, expressed as a CC50 value, was ∼25 μM. Thus, DHT-2 has a selectivity index of ∼83.
TABLE 2.
Antiviral activity and cytotoxicity of DHT in different cell culture systems
| Compound | HIV isolate | Target cell | Readout | Remarka | EC50 (μM) | CC50 (μM) |
|---|---|---|---|---|---|---|
| DHT-1 | D117II | C8166 | Cytoprotection | HIV-1/B/X4 | 0.4 | >12.5 |
| DHT-2 | D117II | C8166 | Cytoprotection | HIV-1/B/X4 | 0.3 | 25 |
| DKA | D117II | C8166 | Cytoprotection | HIV-1/B/X4 | 0.1 | 50 |
| S-1360 | D117II | C8166 | Cytoprotection | HIV-1/B/X4 | 0.3 | 25 |
| L-870,810 | D117II | C8166 | Cytoprotection | HIV-1/B/X4 | 0.002 | 3.1 |
| AZT | D117II | C8166 | Cytoprotection | HIV-1/B/X4 | 0.04 | NDd |
| DHT-1 | IIIB | MT4 | Cytoprotection | HIV-1/B/X4 | 0.4 | >12.5 |
| EFV | IIIB | MT4 | Cytoprotection | HIV-1/B/X4 | 0.009 | ND |
| DHT-2 | D34 | PBMC | p24 | HIV-1/B/X4 | 0.4 | 12.5 |
| AZTb | D34 | PBMC | p24 | HIV-1/B/X4 | 0.008 | ND |
| DHT-2 | GE98002 | PBMC | p24 | HIV-1/B/X4 | 0.2 | 12.5 |
| DHT-2 | ZA97005 | PBMC | p24 | HIV-1/C/X4 | 0.8 | ND |
| DHT-2 | TZ97002 | PBMC | p24 | HIV-1/A/X4 | 0.4 | ND |
| DHT-2 | TH92001 | PBMC | p24 | HIV-1/E/X4 | 0.2 | ND |
| DHT-2 | D-194 | PBMC | p24 | HIV-2 | 0.7 | ND |
| AZT | D-194 | PBMC | p24 | HIV-2 | 0.01 | ND |
| DHT-1 | Ba-L | M/Mc | p24 | HIV-1/B/R5 | 0.2 | 12.5 |
| AZT | Ba-L | M/M | p24 | HIV-1/B/R5 | 0.003 | ND |
Specification of HIV isolate with regard to HIV-1 or -2, tropism (X4 or R5), and clade (A, B, C, or E).
Reference compounds like AZT were included in each assay, but for clarity reasons, EC50 values are given for only D34-infected PBMCs.
M/M, monocyte/macrophage.
ND, not determined.
We also analyzed the activity of DHT on primary target cells like PBMCs and monocytes/macrophages and against different HIV-1 and HIV-2 isolates. As shown in Table 2, DHT proved to be broadly effective against all tested viral isolates, including variants of clades A, B, C, and E and of different tropisms (X4 and R5). Moreover, DHT were also active against HIV-2 with similar potencies (EC50 of 0.7 μM). As expected from their postulated mode of action, DHT were also shown to be effective against viral variants that were resistant to reverse transcriptase and protease inhibitors (data not shown).
Mode-of-action studies to confirm that DHT are authentic inhibitors of HIV IN.
In order to prove that the antiviral activity of DHT was indeed mediated by the inhibition of HIV integrase, various mode-of-action studies were performed.
Time-of-addition experiment.
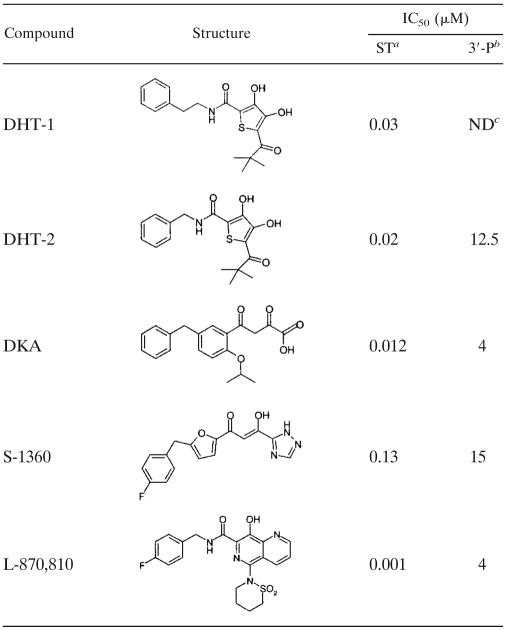
We carried out a time-of-addition experiment to investigate at which stage during the viral replication cycle intervention by DHT takes place (Fig. 1). Cells were infected at high multiplicities of infection (MOI, >1) to synchronize HIV replication, and antivirals, including reference compounds with known modes of action, were added at different time points postinfection, ranging from 0 to 27 h. As expected, the fusion inhibitor T20 already lost its activity when added 1 h after infection. In contrast, the protease inhibitor SQV retained most of its inhibitory effect even when added 28 h after infection, whereas the addition of the NNRTI EFV could only be postponed for up to ∼3 to 8 h without losing activity. As expected for the reference strand transfer inhibitor DKA, a loss of activity could be observed starting at 8 h p.i. A similar profile was found for DHT when tested in this assay: the compound remained active during the early steps of viral entry but lost its inhibitory effect when added 8 h p.i.
FIG. 1.
Time-of-addition experiment. MT-4 cells were infected with HIV-1 (D117/II) at an MOI of >1, and test compounds were added at different time points after infection (0, 1, 3, 8, or 27 h). The production of viral p24 Ag was assessed 36 h after infection. Concentrations of compounds were as follows: T20, 5 μM; DKA, 5 μM; EFV, 0.25 μM; SQV, 0.5 μM; and DHT-2, 15 μM. OD450, optical density at 450 nm; OD620, optical density at 620 nm.
This result strongly suggests that DHT are inhibiting an early but postentry step of the viral life cycle, which is in accordance with its postulated anti-integrase activity. However, the time resolution of this experiment does not allow the exclusion of an interaction of DHT with the reverse transcriptase of HIV.
Persistently infected cells.
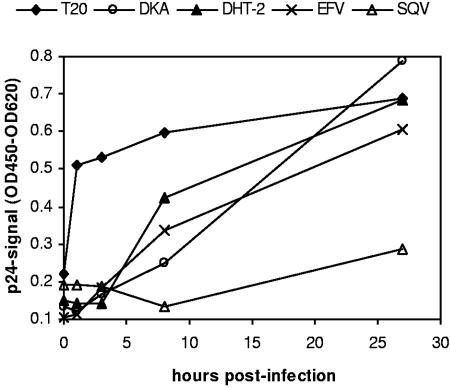
To investigate whether DHT exert an additional effect during late postintegration steps of the viral life cycle, DHT-2 was evaluated for virus production on persistently infected U1 cells. This cell line bears stably integrated HIV provirus with a defective LTR, which can be induced to synthesize viral particles by the addition of PMA. Control compounds included in the experiment were the NRTI zidovudine (AZT), which showed no effect at concentrations from 0.012 to 0.750 μM, and the PI SQV, which strongly reduced the production of mature HIV particles by more than 95% at its highest concentration of 0.750 μM (Fig. 2). No dose-dependent inhibition of the HIV yield was detectable for DHT-2 when tested at concentrations from 0.12 to 7.5 μM, which suggests that DHT do not target late steps of viral replication.
FIG. 2.
Effect of DHT-2 on production of HIV-1 by persistently infected U1 cells. U1 cells were treated with PMA to induce HIV-1 production in the absence or presence of AZT, SQV, or DHT-2 and incubated for 48 h. The amount of p24 in the supernatants is expressed as the percentage of untreated control. Note that the upper concentration range (a) is given for AZT and SQV, while the lower range (b) is given for DHT-2.
HIV vector transduction.
To further underline the interference with early postentry steps of HIV replication, we evaluated DHT in a replication-incompetent lentiviral vector system (described in reference 31). HIV-1-derived vectors pseudotyped with the vesicular stomatitis virus G (VSV-G) envelope protein carried a luciferase reporter gene that is activated after successful integration of the lentiviral genome into HEK293 target cell DNA. Thus, this system represents an extremely helpful tool for the characterization of inhibitors for reverse transcription and integration under single-cycle conditions.
As depicted in Table 3, we found that DHT-1 exerted potent inhibition of lentiviral transduction with an EC50 of 0.12 μM, which is in the same range as the EC50 for the former developmental compound S-1360 (EC50, 0.3 μM). However, higher potencies were observed for DKA and L-870,810, with EC50 values of 0.03 and 0.007 μM, respectively. These values are in good accordance with the graduated potencies of DHT and reference compounds in the biochemical strand transfer assay and in the replication assay described above and suggest that antiviral activity of DHT in cellular systems is indeed coupled to the inhibition of integrase strand transfer activity. Moreover, due to the lack of gp120/gp41 in the retroviral particles, an interference of DHT with HIV env-mediated entry steps can be excluded.
TABLE 3.
Activity of DHT and reference compounds using different HIV vector systems
| Compound | EC50 (μM)
|
CC50 (μM)a | ||
|---|---|---|---|---|
| VSV-G-pseudo typed vectora | VSV-G-pseudotyped vectora (RT mut)c | Nonpseudo typed vectorb | ||
| DHT-1 | 0.12 | 0.18 | 0.09 | >25 |
| DKA | 0.03 | NDd | 0.06 | >25 |
| S-1360 | 0.3 | ND | ND | >25 |
| L-870,810 | 0.007 | ND | 0.004 | >1 |
| T-20 | >25 | ND | 0.12 | ND |
| NVP | 0.06 | >10 | 0.05 | ND |
Results from using HEK293 as target cells (see Materials and Methods).
Results from using H9 as target cells (see Materials and Methods).
Vector carries a double mutation, K103N/Y181C, in RT
ND, not determined.
To further corroborate this independence of HIV env, we also investigated the activity of DHT on viral transduction of nonpseudotyped vector particles. Whereas the gp41-targeting fusion inhibitor T20 was inactive in VSV-G-pseudotyped HIV vectors and active in nonpseudotyped HIV vectors, EC50 values for DHT remained essentially unchanged in the presence or absence of env (EC50s of 0.12 and 0.09 μM, respectively). It should be mentioned that, accordingly, DHT did not appear to bind to gp120 in vitro as analyzed by surface plasmon resonance experiments using immobilized recombinant gp120 (data not shown).
We then asked whether the introduction of key mutations against marketed NNRTIs would have any influence on the antiviral activity of DHT. Therefore, variants of vector particles were generated bearing the K103N/Y181C double mutation in the RT open reading frame. The introduction of these mutations caused a marked increase in the EC50 of nevirapine (NVP) and EFV (data not shown) and led to resistance indices of >120. In contrast, no reduction of DHT potency was observed on K103N/Y181C particles. This finding substantiates their lack of cross-resistance to current NNRTIs (see above).
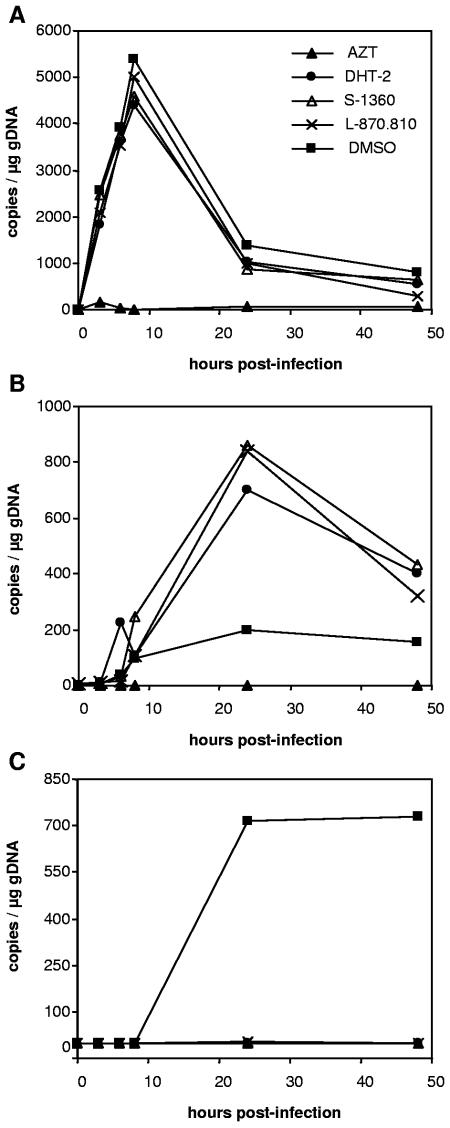
Real-time PCR.
To investigate in detail at which stage during early postentry HIV is affected by DHT, we followed the fate of viral nucleic acids in HIV-infected cells. HEK293T cells were transduced by HIV vectors ensuring single-cycle infection conditions. Subsequently, DNA extracts of HEK293T cells transduced in the absence or presence of compounds were analyzed by a set of quantitative PCRs (Fig. 3). Using different specific primers and probes, we followed the synthesis of late reverse transcripts, 2-LTR circles, and proviral integration over a period of 48 h p.i. Blocking reverse transcription with AZT results in a reduced formation of 2-LTR circles and integrated provirus. In contrast, neither DHT-2 nor the reference INSTIs S-1360 and L-870,810 decreased the production of late transcripts but they induced a significant accumulation of 2-LTR circles. Moreover, the integration of proviral DNA in the presence of either reference INSTIs or DHT-2 was severely reduced to background level. In summary, these results strongly support that the antiviral activity of DHT is a direct consequence of its effect on proviral integration.
FIG. 3.
Effect of DHT on HIV DNA during HIV vector transduction. 293T cells were transduced with HIV-1 vectors at a high MOI in the absence or presence of inhibitors (AZT, DHT-2, S-1360, L-870.810, and dimethyl sulfoxide [DMSO]). Cells were harvested, and DNA was extracted at different time points after infection. Samples were analyzed by quantitative PCR for late reverse transcripts (A), 2-LTR circles (B), and integrated proviral DNA (C).
Structure-activity relationship of DHT.
To analyze the structure-activity relationship (SAR) of DHT, analogs of DHT-1, with modifications at positions R1, R2, and R3, were synthesized and tested for activity in the strand transfer assay and against HIV replication (Table 4).
TABLE 4.
SAR of DHT in integrase strand transfer and HIV replication assays
IC50 in integrase strand transfer assay.
EC50 in HIV replication assay based on D117II-infected C8166 cells.
We observed that methylation of one (DHT-3 and DHT-4) or both (DHT-5) hydroxy groups at R2 and R3 of the thiophene led to a total loss of activity in both the biochemical IN strand transfer assay and the HIV replication assay. Thus, both OH groups represent essential structural elements of this class of IN inhibitors that are similar to the chelating diketo function of DKA inhibitors. In contrast, the modulation of the activity of DHT could be achieved by modification of the R1 position, with a benzyl substituent (DHT-2) being the most favorable residue tested so far in both assay systems. N methylation at the amide nitrogen as present in compound DHT-6 led to a reduced activity in both assays, underlining the importance of the coplanarity of the pharmacophore.
Three-dimensional-alignment of the DHT pharmacophore to DKA and naphthyridine carboxamides.
For DKA-like inhibitors, a two-metal-binding model has been proposed in which the diketo acid pharmacophore coordinates two divalent metal ions in the catalytic site of the integrase donor substrate complex, thereby competing with the target DNA (25).
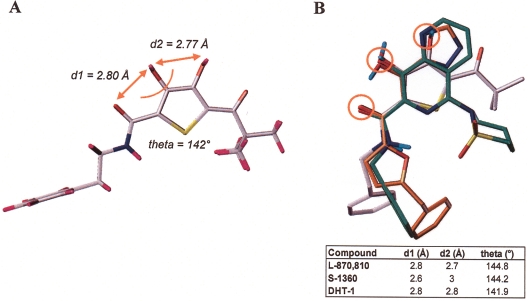
To examine whether the mode of action of DHT might also involve an interaction with active site metal ions, a three-dimensional model of DHT-1 was generated (shown in Fig. 4A) and employed as a template to align its β-keto-dihydroxythiophene pharmacophore both to the DKA-like S-1360 and the naphthyridine L-870,810. The overlay of low-energy conformers for all three compounds, as depicted in Fig. 4B, indicates that the spatial orientation of the DHT functionalities is in good agreement with the positioning of essential residues in S-1360 and in the naphthyridine. Moreover, as reported for DKA and naphthyridines and as shown in Fig. 4B, the central pharmacophore of DHT is almost planar. Distances (d1 and d2) and angles between residues putatively involved in chelating active site metal ions were calculated and were found to be in the same range for all three inhibitors: d1 = 2.6 to 2.8 Å; d2 = 2.7 to 3.0 Å; theta = 142 to 145°. It is of note that these values are in good agreement to the data of Grobler et al. (25) describing the two-metal-binding motif for DKA-like inhibitors: d1 = 2.70 Å; d2 = 2.99 Å; theta = 142°.
FIG. 4.
Three-dimensional-structure of DHT and alignment to reference INSTI. (A) Three-dimensional model of DHT-1. Values for distances d1 and d2 and the angle theta are given. (B) Overlay of DHT-1 (in gray), S-1360 (in orange), and L-870,810 (in green). Values for distances d1 and d2 and angle theta shown in panel A are listed in the table. Circles mark residues putatively involved in chelating active site metal ions.
The more distal parts of DHT are significantly less congruent to the respective parts of S-1360 and the naphthyridine due to the relatively flexible side chains. A strict constraint alignment was not performed for these parts, since there is no experimental information available for deciding the required side chain conformation.
DHT are only partially cross-resistant to DKA and naphthyridine carboxamides.
In order to compare DHT in more detail to known classes of INSTIs, we profiled DHT on IN carrying resistance mutations for S-1360, DKA, and L-870,810 in a cellular system. It has been reported that the introduction of some key resistance mutations induced by INSTIs, e.g., N155S, severely impairs replicative fitness of mutant strains and so complicates their phenotypic characterization. Therefore, our analysis was performed with the help of replication-incompetent HIV vector particles carrying defined modifications in the IN open reading frame.
A panel of vector particles containing recently described DKA resistance mutations and combinations thereof was constructed and examined for the particles' susceptibilities to DHT-1 and -2, DKA, S-1360, and to the recently reported naphthyridine carboxamide L-870,810 (Table 5). For DKA a loss of susceptibility on modified vector particles carrying S153Y (1-fold), T66I/S153Y (9-fold), or S153Y/N155S (47-fold) was in good agreement with published data generated on mutant HIV isolates (27, 29). When we profiled the two other reference INSTIs for these three mutants we observed that S-1360 was broadly cross-resistant (4-, 14-, and 25-fold, respectively), whereas resistance of L-870,810 was restricted to S153/N155S (15-fold). In contrast, neither DHT-1 nor DHT-2 exhibited a significant loss of efficacy with any of these mutants. However, activity of all IN inhibitors, including DHT, was strongly reduced on mutant vectors carrying the substitutions T66I/N155S (17- to 25-fold) and T66I/S153Y/N155S (more than 33-fold), providing further evidence for DHT being authentic inhibitors of HIV integrase.
TABLE 5.
Activities of DHT and reference INSTIs against HIV vectors containing various site-directed mutations in integrase
| Mutation | Reduction (n-fold) in susceptibility of mutants to:
|
|||||
|---|---|---|---|---|---|---|
| DHT-1 | DHT-2 | DKA | S-1360 | L-870,810 | AZT | |
| S153Y | 2 | 2 | 1 | 4 | 2 | 1 |
| T66I/S153Y | 1 | 1 | 9 | 14 | 3 | 1 |
| S153Y/N155S | 2 | 2 | 47 | 25 | 15 | 1 |
| T66I/N155S | 17 | 25 | 94 | 14 | 190 | 1 |
| T66I/S153Y/N155S | >33 | >43 | >125 | >25 | >286 | 1 |
| T66I | 0a | 0b | 1 | 5 | 1 | 1 |
| N155H | 1 | 1 | 1 | 5 | 26 | 1 |
| N155S | 1 | 1 | 10 | 15 | 30 | 1 |
| Q148K | 1 | 1 | 9 | 17 | 21 | 1 |
| V1511 | 1 | 1 | 5 | 17 | 36 | 1 |
Mutant with fourfold-higher susceptibility than the wt.
Mutant with sixfold-higher susceptibility than the wt.
Another striking observation was made with regard to the role of the T66I single mutation: whereas sensitivities of DKA, L-870,810, and S-1360 were not, or only weakly, decreased in the presence of T66I, it clearly was associated with a hypersensitivity (four- to sixfold) to DHT. However, the T66I mutation made a crucial contribution to the resistance of vector particles to DHT observed in T66I/N155S (17- to 25-fold) and T66I/S153Y/N155S (more than 33-fold) mutants relative to N155S (1-fold) and S153Y/N155S (2-fold), respectively.
We also analyzed DHT for cross-resistance against the recently described N155H mutation raised in vivo against L-870,812, a congener of L-870,810. Interestingly, besides L-870,810 (26-fold), only S-1360 (6-fold) exhibited low cross-resistance, whereas both DKA and DHT retained their activities completely. Finally, DHT were profiled against a panel of modified vectors carrying single mutations which have been reported to confer resistance to the DKA-like inhibitor S-1360 (i.e., Q148K, V151I, and N155S). In this case, only DHT remained fully active, while DKA (5- to 10-fold) and L-870,810 (21- to 36-fold) were moderately to strongly cross-resistant.
It is of note that the naphthyridine carboxamide L-870,810, a bioisosteric homolog of DKA, showed a substantial overlap in its cross-resistance profile to DKA-like compounds. However, the decrease of sensitivity against L-870,810 in the presence of T66I/S153Y (3-fold) and S153Y/N155S (15-fold) was less severe than that observed for DKA (9- and 47-fold, respectively) and S-1360 (14- and 25-fold, respectively). Moreover, the naphthyridine resistance mutation N155H induced no (onefold for DKA) or only weak resistance (sixfold for S-1360) against DKA-like inhibitors.
In summary, each INSTI, including DHT, showed its distinct resistance profile on our panel of engineered HIV vectors carrying mutations against DKA, S-1360, or naphthyridine carboxamides, although a certain overlap was present. DHT showed the most restricted profile in the underlying collection of mutants, whereas S-1360 was cross-reactive, to different extents, against all tested variants.
DISCUSSION
A huge number of compounds have been reported to inhibit HIV integrase activity in vitro in oligonucleotide-based assays (48), but to date, only DKA and its homologs, the naphthyridine carboxamides, proved to be unambiguous, well-characterized integrase inhibitors that reached clinical development status. The assessment of a second clinical development compound named GS-9137/JTK-313 is not yet possible due to a lack of published data.
Here we describe the identification, characterization, and profiling of DHT, a novel series of HIV INSTIs that are active against HIV replication, with a pharmacophore homologous to the previously described DKA-like and naphthyridine compounds.
Our experimental data clearly indicate that DHT are selective inhibitors for HIV IN strand transfer with a potency intermediate to Merck's naphthyridine L-870,810, their DKA, and Shionogi's former developmental compound, S-1360. As has been reported for other INSTIs, the activity of DHT in 3′ processing assays was markedly lower (27). The observation that both activities of IN could be affected differentially by inhibitors, together with results from mutagenesis studies, substantiates the hypothesis that strand transfer and 3′ processing are mediated by distinct subregions or different structural confirmations of the same catalytic site (9, 40).
Most interestingly, DHT showed broad antiviral activity against different laboratory and primary isolates of HIV on different target cells with selectivity indices between 31 and 83 and retained efficacy against strains carrying resistances for marketed reverse transcriptase inhibitors and PIs (data not shown). To investigate whether this antiviral activity of DHT in cell culture was indeed linked to the inhibition of integrase strand transfer, various mode-of-action studies were performed. Results from time-of-addition (TOA) studies and experiments with HIV vectors and persistently infected U1 cells point to an early mechanism of action for DHT. Furthermore, as DHT in TOA experiments start to lose activity from ∼8 h p.i. onwards and are inactive in VSV-G or HIV env-pseudotyped HIV vectors, we could exclude any intervention of DHT with early entry/fusion steps. As interference with early entry steps has been described for the first-generation IN inhibitors AR177 (19) and l-chicoric acid (46), exclusion of intervention with this step in particular is an important issue when characterizing cellular activity of novel IN inhibitors.
As the time resolution of TOA experiments in general does not allow the possibility of discriminating RT from IN steps, it was necessary to analyze the fate of viral nucleic acid in a set of quantitative PCRs on DNA purified from HIV vector-transduced cells in the absence or presence of DHT at various time points following infection. The profile of DHT was almost indistinguishable from that of reference INSTI, strongly supporting our hypothesis that the antiviral effect of DHT in cell culture is in fact due to the specific inhibition of the DNA strand transfer reaction. Recently, the quantification of viral nucleic acid species by real-time PCR was used to characterize two novel types of integrase inhibitors with a new mode of action, the pyranodipyrimidines and the styrylquinolines. For V-165, a pyranodipyrimidine prototype, only a moderate increase in the amount of 2-LTR circles was reported, whereas, in the case of the FZ41, a styrylquinoline prototype, no circular HIV DNA accumulated at all. For both classes, it was concluded that each compound interacts with IN and so interferes with the stability, the formation, and/or the nuclear import of pre-integration complexes in infected cells (6, 45). Therefore, both compounds have been classed as integrase-binding inhibitors (56). In contrast to V-165 and FZ-41, DHT markedly increased the formation of 2-LTR circles, which suggests that the inhibitory effect of DHT is most likely restricted to the strand transfer reaction.
It should be added that DHT did not show any activity in biochemical assays for HIV reverse transcriptase and HIV protease (data not shown). Moreover, when tested for activity against various bacterial and human topoisomerases, which are also involved in DNA transesterification reactions, DHT were inactive. These data further substantiate the specificity of DHT.
In order to get more insight into the molecular mechanism of the interaction of DHT with IN, we then analyzed their SAR. The chemical derivatization of DHT revealed that both OH groups linked to the thiophene are essential for their inhibitory activities, suggesting that these functionalities might be key elements of a pharmacophore that is isosteric to the DKA motif. Modifications of the metal-binding DKA pharmacophore has been described earlier in the case of the closely related analogs 5-CITEP and S-1360, where the carboxylate moiety of the pharmacophore was functionally replaced by a tetrazole and a triazole, respectively. (24, 25, 5) (Yoshinaga et al., Abstr. 9th Conf. Retrovir. Opportun. Infect., abstr. 8, 2002). In addition, efforts to reduce the electrophilic nature of the 1,3-diketone moiety resulted in the identification of the homologous 8-hydroxy-1,6-naphthyridine-7-carboxamides, with L-870,810 representing the first member providing proof of concept in HIV-infected patients (30, 58, 59) (S. Little et al., Abstr. 12th Conf. Retrovir. Opportun. Infect., abstr. 161, 2005). It has been reported that the coplanar conformation of the 1,3-diketo acid moiety is an important feature of the DKA pharmacophore, which was taken into consideration when designing the 8-hydroxy-(1,6)-naphthyridines (59). Interestingly, coplanarity of the pharmacophore was favorable also for DHT, underlining that the β-keto-dihydroxythiophene moiety is isosteric to the DKA motif.
Our notion that the DHT motif is mimicking the common pharmacophore of known INSTI was further supported by the molecular modeling of DHT and overlay to S-1360 and the naphthyridine L-870,810. This study demonstrated a good alignment of the spatial orientation and geometry of the essential chelating moieties of DHT compared to the reference compounds. Moreover, we were able to illustrate the coplanarity of the pharmacophore of both DHT and other DKA-like inhibitors discussed above. Thus, DHT fulfill essential criteria of the recently described two-metal-binding model postulated for DKA-like inhibitors (25). However, as demonstrated by the SAR of the DHT, the β-keto-dihydroxythiophene motif is necessary but not sufficient for the activity because less conserved pendant groups allow the modulation of the potency of the inhibitor. In agreement with this, the alignment of more distal parts of the DHT to other inhibitors in the molecular model appears to be less stringent.
With regard to L-870,810, it has been published that the alignment to DKA could be performed in forward and reverse orientations and it has been suggested that the latter is preferred during binding of the inhibitor to the IN active site (30) (see below). Whether DHT are also able to bind to the active site in a similar flipped orientation needs to be further investigated.
A two-metal-binding model has been postulated as the molecular basis for the inhibition of IN by the DKA-like motif in which the diketo acid pharmacophore coordinates two divalent metal ions in the catalytic site of the integrase, thereby blocking the target DNA site of the strand transfer complex (25). This strand transfer complex represents a unique conformation of the highly flexible IN active site required for the binding of INSTI and induced by the preceding binding of donor DNA (18). Due to the described structural similarities of the DHT pharmacophore to the common motif for INSTIs, we postulate that DHT block IN by interacting with active site metal ions in a similar manner. This notion was supported by competition experiments in which the increase of target DNA oligonucleotides led to an increase of the IC50 in the strand transfer assay (preliminary data not shown).
To date, the flexible nature of the IN active site in the absence of donor DNA has hampered the accurate cocrystallization of IN with INSTI. So we used a resistance profiling approach in order to obtain insight into how DHT might bind to the IN active site. Thus, compounds were profiled on a panel of engineered replication-incompetent HIV vectors carrying published mutations against DKA, S1360, and L-870,810. The use of this system facilitated the evaluation of mutants with combinations of active site mutations that are known to reduce viral fitness or are lethal when introduced into infectious virus (e.g., T66I/S153Y/N155S) (28). Of all of the tested INSTIs, the DHT exhibited the most restricted profile on the underlying panel of mutant vectors. In contrast to the other INSTIs, none of the single mutations and neither the T66I/S153Y nor the S153/N155S double mutation had any significant impact on the activity of DHT. However, their effectiveness was markedly impaired by the combinations T66I/N155S and T66I/S153Y/N155S, both leading to a strong cross-resistance to all reference INSTIs. Importantly, neither of these combinations have yet appeared in resistance selection experiments and, for the triple mutation, it has been reported that its introduction into an infectious molecular clone led to a nonviable mutant. Therefore, it has been concluded that, in these variants, the residual catalytic activity of IN is insufficient to allow viral replication (28).
It has been suggested that the DKA resistance-associated amino acids T66, S153, and N155 are not directly interacting with the inhibitor because they are generally not oriented into the active site (30). Rather, it has been assumed that these substitutions induce a shift of the adjacent metal-binding center formed by the active site residues D64, D116, and E152 that leads to an altered affinity or positioning of the complexed Mg2+ ions and to a decreased affinity of the inhibitor (30). Thus, our finding that resistance to DHT could so far be observed only on mutant vectors carrying lethal combinations of mutations may be explained by an altered mode of interaction of DHT to a common binding site of IN that does not allow a comparable shift of the metal binding center. The hypothesis that DHT exhibit an alternate binding mode to IN, though mechanistically related to other INSTI, is further supported by the observation that the T66I single substitution, although of little or no impact on the resistance of reference INSTI, surprisingly induced hypersensitivity to DHT. Drug hypersensitivity and resensitization have been described for certain RT-associated mutations (e.g., M184V and L74V in RT) and are most likely involved in altered substrate discrimination or excision (22). However, the biochemical mechanism underlying T66I hypersensitivity to DHT remains to be elucidated.
From our profiling data, it also became obvious that cross-resistance between the recently described INSTIs S-1360, DKA, and L-870,810 is incomplete. This indicates the presence of multiple binding sites for INSTI with a common pharmacophore within the IN catalytic center. Interestingly, an alternate binding mode for L-870,810 has been described by Hazuda et al. (30). L-870,810-resistant viruses that were selected in their study contained mutations V72I, F121Y, T125K, and V151I, which were not associated with cross-resistance to the mechanistically identical DKA. Vice versa, the authors showed that DKA resistance mutations (i.e., T66I, S153Y, M154I, and combinations thereof) had no significant effect on the effectiveness of L-870,810. When both sets of mutations were mapped in the IN active site, it turned out that distinct regions were affected. The authors concluded that the nonoverlap of resistance profiles was due to an unexpected reverse orientation of L-870,810 induced by favorable interactions of specific pendant substituents. Our experimental data for the resistance profiling of different INSTI is in good accordance with these findings; however, due to the nature of the analyzed mutants, the differences in the profiles of DKA and L-870,810 described here are less pronounced than those published by Hazuda et al. (30).
As we have not been successful in the identification of IN mutations mediating resistance against DHT up to now (mainly due to a severely impaired replicative fitness of DHT-treated isolates), a more detailed analysis of the interaction of DHT with the IN has been hampered. Therefore, it will be instructive to analyze whether the recently published naphthyridine resistance mutations at positions V72, F121, T125, F121, and V151 (30) might exert any influence on the activity of DHT.
It should also be mentioned that more recently, additional resistance mutations raised in vitro against the DKA L-708,906 have been reported by Fikkert et al. (20). The authors describe the emergence of DKA-resistant strains that, after 60 passages, accumulated mutations at T66I, L74M, and S230R that led to a 10-fold reduced susceptibility to L-708,906 and cross-resistance to S-1360. However, phenotypic profiling of viral variants with this resistance pattern against L-870,810 and against our DHT remains to be performed.
Although DHT are potent inhibitors of HIV in cell culture with an attractive mode of action, this compound class will not be further pursued due to unfavorable pharmacokinetic properties. However, in the absence of precise cocrystals of inhibitors bound to the IN active site, DHT and other INSTIs might help to further elucidate the molecular mechanism of the underlying strand transfer reaction.
In summary, we have presented DHT as a novel class of specific INSTIs that block HIV replication in cell culture. Although the DHT pharmacophore is homologous to the recently published DKA motif and fits into the two-metal-binding model, its cross-resistance to reference INSTIs is incomplete, suggesting that DHT have a different mode of interaction with a common binding site within the catalytic center of IN. Moreover, we have added further general evidence for the existence of multiple binding sites for structurally different but mechanistically related inhibitors in the IN active site recently proposed by Hazuda et al. (30).
Acknowledgments
We thank B. Bröhland, M. Kiessler, U. Schick, M. Kotzur, and A. Schommers for excellent technical assistance and G. Hewlett for critical reading of the manuscript. The support of C. Jassoy and H. von Briesen, who kindly provided us with reagents and HIV isolates, is also highly appreciated.
REFERENCES
- 1.Adam, B. D., E. Maticka-Tyndale, and J. J. Cohen. 2003. Adherence practices among people living with HIV. AIDS Care 15:263-274. [DOI] [PubMed] [Google Scholar]
- 2.Anthony, N. J., R. P. Gomez, S. D. Young, M. Egbertson, J. S. Wai, L. Zhuang, M. Embrey, L. Tran, J. Y. Melamed, H. M. Langford, J. P. Guare, T. E. Fisher, S. M. Jolly, M. S. Kuo, D. S. Perlow, J. J. Bennett, and T. W. Funk. 2002. Preparation of (poly)azanaphthalenylcarboxamides as HIV integrase inhibitors. PCT Int. Appl. WO 2002030930.
- 3.Anthony, N. J. 2004. HIV-1 integrase: a target for new AIDS chemotherapeutics. Curr. Top. Med. Chem. 4:979-990. [DOI] [PubMed] [Google Scholar]
- 4.Asante-Appiah, E., S. H. Seeholzer, and A. M. Skalka. 1998. Structural determinants of metal-induced conformational changes in HIV-1 integrase. J. Biol. Chem. 273:35078-35087. [DOI] [PubMed] [Google Scholar]
- 5.Billich, A. 2003. S-1360 Shionogi-GlaxoSmithKline. Curr. Opin. Investig. Drugs 4:206-209. [PubMed] [Google Scholar]
- 6.Bonnenfant, S., C. M. Thomas, C. Vita, F. Subra, E. Deprez, F. Zouhiri, D. Desmaele, J. D'Angelo, J. F. Mouscadet, and H. Leh. 2004. Styrylquinolines, integrase inhibitors acting prior to integration: a new mechanism of action for anti-integrase agents. J. Virol. 78:5728-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bujacz, G., J. Alexandratos, A. Wlodawer, G. Merkel, M. Andrake, R. A. Katz, and A. M. Skalka. 1997. Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity. J. Biol. Chem. 272:18161-18168. [DOI] [PubMed] [Google Scholar]
- 8.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 9.Calmels, C., V. R. de Soultrait, A. Caumont, C. Desjobert, A. Faure, M. Fournier, L. Tarrago-Litvak, and V. Parissi. 2004. Biochemical and random mutagenesis analysis of the region carrying the catalytic E152 amino acid of HIV-1 integrase. Nucleic Acids Res. 32:1527-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr, A., K. Samaras, A. Thorisdottir, G. R. Kaufmann, D. J. Chisholm, and D. A. Cooper. 1999. Diagnosis, prediction, and natural course of HIV-1 protease-inhibitor-associated lipodystrophy, hyperlipidaemia, and diabetes mellitus: a cohort study. Lancet 353:2093-2099. [DOI] [PubMed] [Google Scholar]
- 11.Chen, J. C., J. Krucinski, L. J. Miercke, J. S. Finer-Moore, A. H. Tang, A. D. Leavitt, and R. M. Stroud. 2000. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. USA 97:8233-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chesney, M. 2003. Adherence to HAART regimens. AIDS Patient Care STDs. 17:169-177. [DOI] [PubMed] [Google Scholar]
- 13.Chiu, T. K., and D. R. Davies. 2004. Structure and function of HIV-1 integrase. Curr. Top. Med. Chem. 4:965-977. [DOI] [PubMed] [Google Scholar]
- 14.Davies, J. F., II, Z. Hostomska, Z. Hostomsky, S. R. Jordan, and D. A. Matthews. 1991. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science 252:88-95. [DOI] [PubMed] [Google Scholar]
- 15.Debyser, Z., P. Cherepanov, B. Van Maele, E. De Clercq, and M. Witvrouw. 2002. In search of authentic inhibitors of HIV-1 integration. Antivir. Chem. Chemother. 13:1-15. [DOI] [PubMed] [Google Scholar]
- 16.De Clercq, E. 2005. Emerging anti-HIV drugs. Expert Opin. Emerg. Drugs 10:241-274. [DOI] [PubMed] [Google Scholar]
- 17.Embrey, M. W., J. S. Wai, T. W. Funk, C. F. Homnick, D. S. Perlow, S. D. Young, J. P. Vacca, D. J. Hazuda, P. J. Felock, K. A. Stillmock, M. V. Witmer, G. Moyer, W. A. Schleif, L. J. Gabryelski, L. Jin, I. Chen, J. D. Ellis, B. K. Wong, J. H. Lin, Y. M. Leonard, N. N. Tsou, and L. Zhuang. 2005. A series of 5-(5,6)-dihydrouracil substituted 8-hydroxy-[1,6]naphthyridine-7-carboxylic acid 4-fluorobenzylamide inhibitors of HIV-1 integrase and viral replication in cells. Bioorg. Med. Chem. Lett. 15:4550-4554. [DOI] [PubMed] [Google Scholar]
- 18.Espeseth, A. S., P. Felock, A. Wolfe, M. Witmer, J. Grobler, N. Anthony, M. Egbertson, J. Y. Melamed, S. Young, T. Hamill, J. L. Cole, and D. J. Hazuda. 2000. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc. Natl. Acad. Sci. USA 97:11244-11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Este, J. A., C. Cabrera, D. Schols, P. Cherepanov, A. Gutierrez, M. Witvrouw, C. Pannecouque, Z. Debyser, R. F. Rando, B. Clotet, J. Desmyter, and E. De Clercq. 1998. Human immunodeficiency virus glycoprotein gp120 as the primary target for the antiviral action of AR177 (Zintevir). Mol. Pharmacol. 53:340-345. [DOI] [PubMed] [Google Scholar]
- 20.Fikkert, V., B. Van Maele, J. Vercammen, A. Hantson, B. Van Remoortel, M. Michiels, C. Gurnari, C. Pannecouque, M. De Maeyer, Y. Engelborghs, E. De Clercq, Z. Debyser, and M. Witvrouw. 2003. Development of resistance against diketo derivatives of human immunodeficiency virus type 1 by progressive accumulation of integrase mutations. J. Virol. 77:11459-11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Folks, T. M., J. Justement, A. Kinter, C. A. Dinarello, and A. S. Fauci. 1987. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science 238:800-802. [DOI] [PubMed] [Google Scholar]
- 22.Frankel, F. A., B. Marchand, D. Turner, M. Götte, and M. A. Wainberg. 2005. Impaired rescue of chain-terminated DNA synthesis associated with L74V mutation in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 49:2657-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldgur, Y., F. Dyda, A. B. Hickman, T. M. Jenkins, R. Craigie, and D. R. Davies. 1998. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl. Acad. Sci. USA 95:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldgur, Y., R. Craigie, G. H. Cohen, T. Fujiwara, T. Yoshinaga, T. Fujishita, H. Sugimoto, T. Endo, H. Murai, and D. R. Davies. 1999. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc. Natl. Acad. Sci. USA 96:13040-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grobler, J. A., K. Stillmock, B. Hu, M. Witmer, P. Felock, A. S. Espeseth, A. Wolfe, M. Egbertson, M. Bourgeois, J. Melamed, J. S. Wai, S. Young, J. Vacca, and D. J. Hazuda. 2002. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc. Natl. Acad. Sci. USA 99:6661-6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hazuda, D. J., J. C. Hastings, A. L. Wolfe, and E. A. Emini. 1994. A novel assay for the DNA strand-transfer reaction of HIV-1 integrase. Nucleic Acids Res. 22:1121-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 28.Hazuda, D., W. Schleif, L. Gabryelski, J. Grobler, P. Felock, K. Stillmock, A. Espeseth, R. Danzeisen, R. Danovich, M. Miller, and M. Witmer. 2001. Resistance to integration inhibitors: evolution of active site mutations, relationship to fitness and enzyme co-factor utilization. Antivir. Ther. 6:S4. [Google Scholar]
- 29.Hazuda, D. J., S. D. Young, J. P. Guare, N. J. Anthony, R. P. Gomez, J. S. Wai, J. P. Vacca, L. Handt, S. L. Motzel, H. J. Klein, G. Dornadula, R. M. Danovich, M. V. Witmer, K. A. A. Wilson, L. Tussey, W. A. Schleif, L. S. Gabryelski, L. X. Jin, M. D. Miller, D. R. Casimiro, E. A. Emini, and J. W. Shiver. 2004. Integrase inhibitors and cellular immunity suppress retroviral replication in rhesus macaques. Science 305:528-532. [DOI] [PubMed] [Google Scholar]
- 30.Hazuda, D. J., N. J. Anthony, R. P. Gomez, S. M. Jolly, J. S. Wai, L. H. Zhuang, T. E. Fisher, M. Embrey, J. P. Guare, M. S. Egbertson, J. P. Vacca, J. R. Huff, P. J. Felock, M. V. Witmer, K. A. Stillmock, R. Danovich, J. Grobler, M. D. Miller, A. S. Espeseth, L. X. Jin, I. W. Chen, J. H. Lin, K. Kassahun, J. D. Ellis, B. K. Wong, W. Xu, P. G. Pearson, W. A. Schleif, R. Cortese, E. Emini, V. Summa, M. K. Holloway, and S. D. Young. 2004. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. Proc. Natl. Acad. Sci. USA 101:11233-11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarmy, G., M. Heinkelein, B. Weissbrich, C. Jassoy, and A. Rethwilm. 2001. Phenotypic analysis of the sensitivity of HIV-1 to inhibitors of the reverse transcriptase, protease, and integrase using a self-inactivating virus vector system. J. Med. Virol. 64:223-231. [DOI] [PubMed] [Google Scholar]
- 32.Jenkins, T. M., A. Engelman, R. Ghirlando, and R. Craigie. 1996. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem. 271:7712-7718. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, A. A., C. Marchand, and Y. Pommier. 2004. HIV-1 integrase inhibitors: a decade of research and two drugs in clinical trial. Curr. Top. Med. Chem. 4:1059-1077. [DOI] [PubMed] [Google Scholar]
- 34.Kovall, R. A., and B. W. Matthews. 1999. Type II restriction endonucleases: structural, functional, and evolutionary relationships. Curr. Opin. Chem. Biol. 3:578-583. [DOI] [PubMed] [Google Scholar]
- 35.LaFemina, R. L., C. L. Schneider, H. L. Robbins, P. L. Callahan, K. LeGrow, E. Roth, W. A. Schleif, and E. A. Emini. 1992. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 66:7414-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, M. C., J. Deng, J. M. Briggs, and Y. Duan. 2005. Large-scale conformational dynamics of the HIV-1 integrase core domain and its catalytic loop mutants. Biophys. J. 88:3133-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Little, S. J., S. Holte, J. P. Routy, E. S. Daar, M. Markowitz, A. C. Collier, R. A. Koup, J. W. Mellors, E. Connick, B. Conway, M. Kilby, L. Wang, J. M. Whitcomb, N. S. Hellmann, and D. D. Richman. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347:385-394. [DOI] [PubMed] [Google Scholar]
- 38.Maignan, S., J. P. Guilloteau, Q. Zhou-Liu, C. Clement-Mella, and V. Mikol. 1998. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J. Mol. Biol. 282:359-368. [DOI] [PubMed] [Google Scholar]
- 39.Marchand, C., N. Neamati, and Y. Pommier. 2001. In vitro human immunodeficiency virus type 1 integrase assays. Methods Enzymol. 340:624-633. [DOI] [PubMed] [Google Scholar]
- 40.Marchand, C., X. Zhang, G. C. Pais, K. Cowansage, N. Neamati, T. R. Burke, Jr., and Y. Pommier. 2002. Structural determinants for HIV-1 integrase inhibition by beta-diketo acids. J. Biol. Chem. 277:12596-12603. [DOI] [PubMed] [Google Scholar]
- 41.Mocroft, A., B. Ledergerber, C. Katlama, O. Kirk, P. Reiss, A. d'Arminio Monforte, B. Knysz, M. Dietrich, A. N. Phillips, and J. D. Lundgren. 2003. Decline in the AIDS and death rates in the EuroSIDA study: an observational study. Lancet 362:22-29. [DOI] [PubMed] [Google Scholar]
- 42.O'Doherty, U., W. J. Swiggard, D. Jeyakumar, D. McGain, and M. H. Malim. 2002. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 76:10942-10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oette, M., R. Kaiser, M. Daumer, D. Akbari, G. Fatkenheuer, J. K. Rockstroh, J. Stechel, A. Rieke, S. Mauss, D. Schmaloer, K. Gobels, C. Vogt, M. Wettstein, and D. Haussinger. 2004. Primary drug-resistance in HIV-positive patients on initiation of first-line antiretroviral therapy in Germany. Eur. J. Med. Res. 9:273-278. [PubMed] [Google Scholar]
- 44.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, and S. D. Holmberg. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 45.Pannecouque, C., W. Pluymers, B. Van Maele, V. Tetz, P. Cherepanov, E. De Clercq, M. Witvrouw, and Z. Debyser. 2002. New class of HIV integrase inhibitors that block viral replication in cell culture. Curr. Biol. 12:1169-1177. [DOI] [PubMed] [Google Scholar]
- 46.Pluymers, W., N. Neamati, C. Pannecouque, V. Fikkert, C. Marchand, T. R. Burke, Jr., Y. Pommier, D. Schols, E. De Clercq, Z. Debyser, and M. Witvrouw. 2000. Viral entry as the primary target for the anti-HIV activity of chicoric acid and its tetra-acetyl esters. Mol. Pharmacol. 58:641-648. [DOI] [PubMed] [Google Scholar]
- 47.Pomerantz, R. J., and D. L. Horn. 2003. Twenty years of therapy for HIV-1 infection. Nat. Med. 9:867-873. [DOI] [PubMed] [Google Scholar]
- 48.Pommier, Y., A. A. Johnson, and C. Marchand. 2005. Integrase inhibitors to treat HIV-AIDS. Nat. Rev. Drug Discov. 4:236-248. [DOI] [PubMed] [Google Scholar]
- 49.Rubsamen-Waigmann, H., W. R. Willems, U. Bertram, and H. von Briesen. 1989. Reversal of HIV-phenotype to fulminant replication on macrophages in perinatal transmission. Lancet ii:1155-1156. [DOI] [PubMed] [Google Scholar]
- 50.Satoh, M., T. Motomura, T. Matsuda, K. Kondo, K. Ando, K. Matsuda, S. Miyake, and H. Uehara. 2005. 4-Oxoquinoline compounds and utilization thereof as HIV integrase inhibitors. PCT Int. Appl. WO 2005113508.
- 51.Van Maele, B., and Z. Debyser. 2005. HIV-1 integration: an interplay between HIV-1 integrase, cellular, and viral proteins. AIDS Rev. 7:26-43. [PubMed] [Google Scholar]
- 52.Wai, J. S., M. S. Egbertson, L. S. Payne, T. E. Fisher, M. W. Embrey, L. O. Tran, J. Y. Melamed, H. M. Langford, J. P. Guare, L. G. Zhuang, V. E. Grey, J. P. Vacca, M. K. Holloway, A. M. Naylor-Olsen, D. J. Hazuda, P. J. Felock, A. L. Wolfe, K. A. Stillmock, W. A. Schleif, L. J. Gabryelski, and S. D. Young. 2000. 4-aryl-2,4-dioxobutanoic acid inhibitors of HIV-1 integrase and viral replication in cells. J. Med. Chem. 43:4923-4926. [DOI] [PubMed] [Google Scholar]
- 53.Weiss, R. A. 2003. HIV and AIDS in relation to other pandemics. EMBO Rep. 4:S10-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wensing, A. M., D. A. van de Vijver, G. Angarano, B. Asjo, C. Balotta, E. Boeri, R. Camacho, M. L. Chaix, D. Costagliola, A. De Luca, I. Derdelinckx, Z. Grossman, O. Hamouda, A. Hatzakis, R. Hemmer, A. Hoepelman, A. Horban, K. Korn, C. Kucherer, T. Leitner, C. Loveday, E. MacRae, I. Maljkovic, C. de Mendoza, L. Meyer, C. Nielsen, E. L. Op de Coul, V. Ormaasen, D. Paraskevis, L. Perrin, E. Puchhammer-Stockl, L. Ruiz, M. Salminen, J. C. Schmit, F. Schneider, R. Schuurman, V. Soriano, G. Stanczak, M. Stanojevic, A. M. Vandamme, K. Van Laethem, M. Violin, K. Wilbe, S. Yerly, M. Zazzi, C. A. Boucher, and SPREAD Programme. 2005. Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: implications for clinical management. J. Infect. Dis. 192:958-966. [DOI] [PubMed] [Google Scholar]
- 55.Wiskerchen, M., and M. A. Muesing. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 69:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Witvrouw, M., V. Fikkert, J. Vercammen, B. Van Maele, Y. Engelborghs, and Z. Debyser. 2005. Identification of authentic inhibitors of HIV-integration. Curr. Med. Chem. Anti-Infective Agents 4:153-165. [Google Scholar]
- 57.Yan, H., T. C. Mizutani, N. Nomura, T. Takakura, Y. Kitamura, H. Miura, M. Nishizawa, M. Tatsumi, N. Yamamoto, and W. Sugiura. 2005. A novel small molecular weight compound with a carbazole structure that demonstrates potent human immunodeficiency virus type-1 integrase inhibitory activity. Antivir. Chem. Chemother. 16:363-373. [DOI] [PubMed] [Google Scholar]
- 58.Young, S. D., and the HIV Integrase Inhibitor Discovery Team. 2002. Discovery of a potent antiviral HIV integrase inhibitor with potential clinical utility. Antivir. Ther. 7:S3. [Google Scholar]
- 59.Zhuang, L., J. S. Wai, M. W. Embrey, T. E. Fisher, M. S. Egbertson, L. S. Payne, J. P. Guare, Jr., J. P. Vacca, D. J. Hazuda, P. J. Felock, A. L. Wolfe, K. A. Stillmock, M. V. Witmer, G. Moyer, W. A. Schleif, L. J. Gabryelski, Y. M. Leonard, J. J. Lynch, Jr., S. R. Michelson, and S. D. Young. 2003. Design and synthesis of 8-hydroxy-[1,6]naphthyridines as novel inhibitors of HIV-1 integrase in vitro and in infected cells. J. Med. Chem. 46:453-456. [DOI] [PubMed] [Google Scholar]