Abstract
Due to difficulties in cell culture propagation, the mechanisms of hepatitis C virus (HCV) entry are poorly understood. Here, postbinding cellular mechanisms of HCV entry were studied using both retroviral particles pseudotyped with HCV envelope glycoproteins (HCVpp) and the HCV clone JFH-1 propagated in cell culture (HCVcc). HCVpp entry was measured by quantitative real-time PCR after 3 h of contact with target cells, and HCVcc infection was quantified by immunoblot analysis and immunofluorescence detection of HCV proteins expressed in infected cells. The functional role of clathrin-mediated endocytosis in HCV entry was assessed by small interfering RNA-mediated clathrin heavy chain depletion and with chlorpromazine, an inhibitor of clathrin-coated pit formation at the plasma membrane. In both conditions, HCVpp entry and HCVcc infection were inhibited. HCVcc infection was also inhibited by pretreating target cells with bafilomycin A1 or chloroquine, two drugs known to interfere with endosome acidification. These data indicate that HCV enters target cells by clathrin-mediated endocytosis, followed by a fusion step from within an acidic endosomal compartment.
Hepatitis C virus (HCV) infects about 170 million people around the world. Despite the importance of HCV as a human pathogen, little is known about its cell biology. The virus was identified and cloned more than 15 years ago (7), but the lack of a robust system allowing for the production of HCV in cell culture has hampered for many years functional studies on HCV infection.
In recent years, two major advances have made it possible to investigate HCV entry. A first advance has been the production of infectious retroviral particles pseudotyped with HCV envelope glycoproteins (3, 14, 24). Using this system of HCV pseudoparticles (HCVpp), observations on receptor usage and the facilitating role of high-density lipoprotein during entry were reported (3, 24, 55). A second major advance has been the recent development of a cell culture model for HCV (30, 56, 59). This system allows for the production of virus that can be efficiently propagated in cell culture (HCVcc). Therefore, the cell entry of HCV can now be investigated in the context of an infectious cycle.
HCV belongs to the Hepacivirus genus in the Flaviviridae family, which also includes the Flavivirus and Pestivirus genera (31). The HCV genome encodes three structural proteins, capsid protein C and envelope glycoproteins E1 and E2, which are associated in the form of a heterodimer (13). Several cellular proteins were reported to interact in vitro with isolated E2. These putative receptors include the tetraspanin CD81 (42), the scavenger receptor class B type I (SR-BI) (50), the lectins L-SIGN and DC-SIGN (18, 32), the asialoglycoprotein receptor (49), and heparan-sulfate proteoglycans (2). The low-density lipoprotein receptor was also proposed as a candidate receptor (1). The importance of CD81 and SR-BI in HCV entry was confirmed with HCVpp (3, 4, 24) as well as with HCVcc for CD81 (30, 56). Beyond receptor binding, virtually nothing is known about molecular and cellular mechanisms used by HCV during cell entry.
According to postbinding mechanisms of entry, enveloped viruses fall into two main types. Some viruses deliver their genome to the cytosol of target cells by fusing their envelope with the plasma membrane, while other ones enter by endocytosis. For many enveloped viruses that enter cells by endocytosis, an activation step occurs in endosomes which leads to the fusion of the viral envelope with the membrane of the endosome and the delivery of the viral genome into the cytosol. The acidic pH of endosomes is thought to play an essential role in triggering this fusion event, which is catalyzed by a viral envelope glycoprotein. Therefore, the pH sensitivity is often considered a good indication of entry by endocytosis. The best documented mode of endocytosis is the clathrin-mediated pathway. However, recent studies have revealed a surprising variety of endocytic routes (10, 39, 51).
Previous reports have indicated a pH dependency for HCVpp entry (4, 24), suggesting that HCV would enter cells by endocytosis. However, the functional importance of endocytosis in HCV infection has not been directly tested so far, and the endocytic pathway used by the virus has not been identified. In this paper, we report on postbinding cellular mechanisms of HCV entry. Using both a newly developed HCVpp entry assay and the JFH1-based HCVcc model, we confirmed the pH sensitivity of HCV entry and established the functional importance of clathrin-mediated endocytosis in HCV entry.
MATERIALS AND METHODS
Chemicals.
Dulbecco's modified Eagle's medium (DMEM), phosphate-buffered saline (PBS), OptiMEM, HEPES, and goat and fetal calf sera (FCS) were purchased from Invitrogen. Hoechst dye 33342 was from Molecular Probes. Chlorpromazine was from Alexis. Bafilomycin-A1 and Mowiol 3-88 were from Calbiochem. ExGen500 was purchased from Euromedex. All other chemicals were from Sigma.
Antibodies.
Rat monoclonal antibody (MAb) 3/11 (17) and mouse MAb A4 (15) were produced in vitro by using a MiniPerm apparatus (Heraeus) as recommended by the manufacturer. Anti-NS3 (486D39) MAb was kindly provided by J. F. Delagneau (Bio-Rad, France). Mouse anti-E2 MAb AP33 (9) was kindly provided by A. H. Patel (Institute of Virology, Glasgow, United Kingdom). Mouse anti-clathrin heavy chain (CHC) MAb was purchased from BD Biosciences. Goat anti-actin polyclonal antibody and mouse anti-simian virus 40 (SV40) T-antigen MAb (Pab101 [sc-147]) were from Santa Cruz. Anti-green fluorescent protein (GFP) MAb was from Roche. Alexa594-conjugated or Alexa555-conjugated goat anti-mouse secondary antibody was from Molecular Probes.
Cell culture.
293T human embryo kidney cells (HEK 293T), PLC/PRF/5 human hepatoma cells (ATCC CRL-8024), and Huh-7 human hepatoma cells were grown in Dulbecco's modified essential medium supplemented with glutamax and 10% fetal bovine serum.
Production of HCVpp.
Pseudotyped particles were produced as described previously (3). Plasmids were kindly provided by D. Lavillette, B. Bartosch, and F.-L. Cosset (INSERM U412, Lyon, France). Briefly, 293T cells were cotransfected with a murine leukemia virus (MLV)-based transfer vector encoding luciferase (37), a murine leukemia virus Gag-Pol packaging construct, and an envelope glycoprotein-expressing vector, phCMV-E1E2 (3), using Exgen 500 as recommended by the manufacturer. The phCMV-G, phCMV-RD114, phCMV-A, and phCMV-HA expression vectors encode the vesicular stomatitis virus G protein (VSV-G), the feline endogenous virus RD114 glycoprotein, the MLV 10A1 envelope glycoprotein (A-MLV), and the fowl plague virus H7 hemagglutinin (HA), respectively (48). The phCMV-NA expression vector for influenza virus neuraminidase (NA) was a kind gift from F.-L. Cosset. Pseudotyped particles harboring VSV-G, RD114, A-MLV, or hemagglutinin (HA) and NA envelope glycoproteins on murine leukemia virus cores (VSV-Gpp, RD114pp, A-MLVpp, and HA/NApp, respectively) were used as controls. Supernatants containing the pseudotyped particles were harvested 48 h after transfection, filtered through 0.45-μm-pore-sized membranes, and incubated with 300 U/ml DNase I (Roche) for 30 min at 37°C to remove excess plasmid DNA. Pseudotyped particle stocks were kept at 4°C and used within 1 week after production. The luciferase-based HCVpp infection assay was as previously described (37).
PCR-based entry assay.
Confluent cell monolayers grown in 12-well plates were infected with 600 μl of DNase-treated pseudoparticles in DMEM supplemented with 10% FCS and 20 mM HEPES. The multiplicity of infection was about 1 (range, 0.5 to 1.5). The infection was enhanced by spinoculation, as previously reported for human immunodeficiency virus (36). Plates were enclosed in plastic bags and centrifuged at 1,200 × g for 2 h at 37°C and then incubated for 1 h in a 5% CO2 incubator at 37°C. After 3 h of contact, the cells were washed three times with PBS. Total DNA was extracted using the Wizard SV Genomic DNA Purification System (Promega) according to the manufacturer's instructions. Each sample of DNA was purified from two pooled wells and was dissolved into 200 μl of DNase-free water.
Quantification of early reverse-transcribed viral DNA was performed by quantitative PCR assay. Primers were designed in the U3 region of the long terminal repeat (LTR) of MLV with the help of the Primer3 software (45). The sequences of the forward and reverse U3 primers were 5′-CCATCAGATGTTTCCAGGCT-3′ and 5′-GCGACTCAGTCTATCGGAGG-3′, respectively. DNA was amplified in glass capillaries using a LightCycler instrument (Roche Diagnostics, Meylan, France). The PCR mix contained 4 μl of sample DNA and 16 μl of FastSTART DNA Master SYBR Green I (Roche Diagnostics), 4 mM MgCl2, and 0.5 μM of each primer. The PCR amplification was performed for 40 cycles of 15 s at 95°C, 5 s at 60°C, and 7 s at 72°C. The specificity of the amplicon was determined by using melting curve analysis, which displayed only one peak in agreement with a specific amplicon.
The standard curves for quantification of the U3 LTR region of MLV were constructed with 10-fold serial dilutions ranging from 105 to 100 copies of phCMV-Luc plasmid. The standard curve was also used to assess PCR efficiency by examining its slope, which was consistently between −3.45 and −3.3 for each experiment. Individual samples were evaluated in duplicate. Absolute quantification of the viral load was evaluated by using the albumin gene as an internal control gene, as described previously (19). The conditions used to amplify the albumin gene were identical to those used for the U3 LTR region. The infection was scored as the ratio of U3 LTR DNA mean copy number over albumin DNA mean copy number. Background values obtained from parallel infections with pseudoparticles containing no envelope protein were subtracted. Data are presented as the percentage of infection relative to control conditions. Using this assay, HCVpp preparations usually yielded titers between 105 and 106 infectious units/ml on PLC/PRF/5 cells.
Production of HCVcc.
To generate genomic HCV RNA, the plasmid pJFH1 (56) was linearized at the 3′ end of the HCV cDNA by XbaI digestion. Following treatment with mung bean nuclease, the linearized DNA was then used as a template for in vitro transcription with the MEGAscript kit from Ambion. In vitro-transcribed RNA was delivered to Huh-7 cells by electroporation as described previously (25). Viral stocks were obtained by harvesting cell culture supernatants at 1 week posttransfection. Secondary viral stocks were obtained by additional amplifications on naïve Huh-7 cells. Infectious titers of viral stocks were estimated between 105 and 106 infectious units per ml, based on immunofluorescent detection of infected foci following infection of Huh7 cells with serial dilutions of viral stocks.
Other viruses.
The recombinant Sindbis virus expressing HCV envelope protein E1 was previously described (16). Stocks of SV40 virus were previously described (58). The recombinant Sendai virus (SeV) expressing a green fluorescent protein (GFP) was previously described (28). A purified stock of recombinant SeV was kindly provided by Laurent Roux, University of Geneva.
HCVcc infection.
For HCVcc infection assays, Huh-7 cells grown in 24-well plates were infected for 2 h at 37°C with HCVcc or control viruses. Each HCVcc stock was concentrated between 5 and 10 times by ultrafiltration using Vivaspin cartridges (molecular weight cutoff, 300,000) in order to get 20 to 40% infected cells. For experiments using small interfering RNA (siRNA), equal numbers of CHC siRNA-treated cells and control siRNA-treated cells were infected. For experiments using chlorpromazine and inhibitors of endosomal acidification, cells were preincubated with inhibitors for 30 min before infection, and the drugs were added to the infection and culture media up to the end of the experiment. In some control experiments, drugs were added at 2 h postinfection (hpi). Infections were scored by immunoblot analysis or by indirect immunofluorescence microscopy at 30 hpi (HCVcc and SV40), 16 hpi (SeV), or 5 hpi (Sindbis virus).
RNA interference.
Subconfluent cultures of PLC/PRF/5 or Huh-7 cells in 6-well plates were transfected twice with 80 pmol of synthetic double-stranded siRNA (Dharmacon) complexed with 4 μl of oligofectamine (Invitrogen) in a total volume of 1 ml of OptiMEM for 6 h. The interval between both siRNA transfections was 48 h. Cells were trypsinized 24 h after the second siRNA transfection and plated in 12-well plates for the HCVpp entry assay, or they were plated in 24-well plates for infection with HCVcc and infected 24 h after trypsinization. Just before infection, extra wells of cells treated with each siRNA were counted to ensure that equal numbers of cells were infected. Relative CHC levels were analyzed by immunoblotting equal amounts of cell lysates. The films were scanned and quantified with the Image J software. The CHC target sequence was UAAUCCAAUUCGAAGACCAAU (34). The control siRNA was originally designed to knock down the green fluorescent protein; its target sequence is GCUGACCCUGAAGUUCAUC.
Indirect immunofluorescence microscopy.
Infected cells were processed for immunofluorescent detection of viral proteins as previously described (44). Nuclei were stained by a 1-min incubation in PBS containing 1 μg/ml Hoechst dye 33342. Coverslips were mounted on glass slides using Mowiol and observed with a Zeiss Axiophot equipped with a ×40 magnification, 1.3 numerical aperture oil immersion lens. Fluorescent signals were collected with a Princeton cooled charged device using specific fluorescence excitation and emission filters. Images were processed using Adobe Photoshop software. For quantification, images of 10 randomly picked areas of each coverslip were recorded. Cells labeled with anti-E2 MAb AP33 were counted as infected cells. The total number of cells was obtained from Hoechst-labeled nuclei. The infections were scored as the ratio of infected cells to total cells.
Immunoblotting.
Cells were lysed in 50 mM Tris-Cl buffer, pH 7.5, containing 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), and protease inhibitors, for 30 min on ice. Cells were collected, and the nuclei were pelleted. Protein concentration in the postnuclear supernatants was determined by the bicinchoninic acid method as recommended by the manufacturer (Sigma), using bovine serum albumin as the standard. Five micrograms of total protein was separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Hybond-ECL; Amersham) by using a Trans-Blot apparatus (Bio-Rad). The proteins of interest were revealed with specific primary antibodies followed by donkey anti-goat immunoglobulin G (IgG), goat anti-mouse IgG, or anti-rat IgG conjugated to peroxidase (Jackson Immunoresearch) as well as enhanced chemiluminescence detection (Amersham) as recommended by the manufacturer.
RESULTS
Real-time PCR-based HCVpp entry assay.
In order to study HCVpp entry, we first tried to use a luciferase-based assay (37). However, our attempts were quite ineffective, due to drug toxicity and differences in cell proliferation under various experimental conditions, over the 2-day culture period that is necessary for luciferase expression. To overcome these difficulties, we set up a new assay for HCVpp entry, derived from the method originally designed by Mothes et al. for studying retrovirus entry (33). It is based on the quantification by PCR of the cDNA synthesized in target cells during HCVpp entry. In order to quantify early events during HCVpp entry, we designed primers in the U3 region of the LTR, which is the first part of the viral genome to be retrotranscribed during the entry of a retroviral particle (21).
During the development of this assay, we observed that HCVpp infection yielded signals about three times higher in PLC/PRF/5 cells than in Huh-7 cells. This effect was observed with the PCR-based assay as well as with the luciferase-based assay (data not shown). Therefore, most of the experiments were carried out in PLC/PRF/5 cells.
Since HCVpp infection is sensitive to agents that neutralize the pH of endosomes (4, 24), we tested whether we could detect this pH sensitivity with the PCR-based assay in order to confirm that it indeed reflects this entry pathway. The importance of endosome acidification was studied with bafilomycin A1, which is a specific inhibitor of endosomal proton-ATP pumps. PLC/PRF/5 cells were preincubated for 30 min with various concentrations of bafilomycin A1 and infected in the presence of the inhibitor. Results (Fig. 1A) show that the amounts of retroviral DNA recovered from infected cells dramatically decreased for HCVpp and VSV-Gpp in a dose-dependent manner. In contrast, the synthesis of A-MLVpp-derived retroviral DNA was not affected by 20 or 50 nM bafilomycin A1, and it was reduced at 100 nM. Although significant, this reduction was much smaller than that observed for HCVpp and VSV-Gpp at the same concentration. A similar partial inhibition of A-MLVpp infection with 100 nM bafilomycin A1 was previously reported (4). Similar inhibitions of HCVpp and VSV-Gpp infection were observed with Huh-7 cells (data not shown). Inhibition of HCVpp and VSV-Gpp entry, but not of A-MLVpp entry, was also observed in the presence of chloroquine or NH4Cl in PLC/PRF/5 cells as well as in Huh-7 cells (data not shown). The results obtained with these different inhibitors of endosomal acidification confirmed that the PCR-based entry assay recapitulates HCVpp entry up to the fusion step.
FIG. 1.
PCR-based HCVpp entry assay. (A) PLC/PRF/5 cells were pretreated for 30 min with 20, 50, or 100 nM bafilomycin A1 or left untreated. The cells were then infected with HCVpp, VSV-Gpp, or A-MLVpp in the presence of the drug. After 2 h of spinoculation and an additional hour of incubation, total DNA was purified and retroviral DNA was quantified by real-time PCR. Mean values for the controls with no drug were 1.2 equivalent genomes per cell (EG/cell) for HCVpp, 1.5 EG/cell for VSV-Gpp, and 1.3 EG/cell for A-MLVpp infections. The background value obtained from cells infected with pseudotyped particles produced in the absence of envelope proteins corresponded to 0.03 EG/cell. Results were expressed as the percentage of entry in control cells treated with no drug. (B) PLC/PRF/5 cells were infected by spinoculation with different dilutions of the same HCVpp stock. Cells were either immediately lysed to extract DNA or cultured for 3 days to allow for luciferase expression. Retroviral DNA was quantified by real-time PCR (MLV DNA), and infection was quantified by luciferase assay (Luc). Mean values for the undiluted HCVpp infection were 1.3 EG/cell and 6.5 × 106 relative light units (RLU), and the background values obtained from cells infected with pseudotyped particles produced in the absence of envelope proteins corresponded to 0.018 EG/cell and 3.0 × 102 RLU. Results were expressed as the percentage of entry in cells infected with undiluted HCVpp stock (1:1).
We next examined whether the levels of retroviral DNA synthesis after 3 h of contact predicted the levels of luciferase expression 3 days later. PLC/PRF/5 cells were infected with different dilutions of an HCVpp stock, and they were then either immediately extracted to purify and quantify retroviral DNA or cultured for 3 days and assayed for luciferase activity. Each assay displayed dose dependency, and interassay comparisons revealed a clear correlation between the level of MLV DNA synthesis and the subsequent level of luciferase expression (Fig. 1B). These results indicate that the levels of DNA synthesis at 3 h accurately quantitate the levels of HCVpp entry.
HCVpp entry is clathrin dependent.
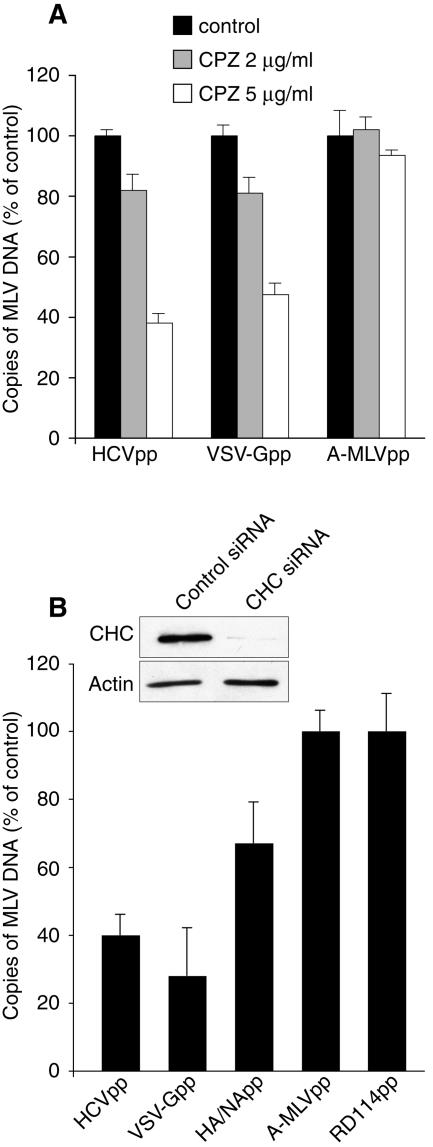
The pH dependency of HCVpp entry implies that HCVpp fusion does not occur at the plasma membrane but from within an intracellular acidic compartment, most probably an endosome. To assess the role of clathrin-mediated endocytosis in HCVpp entry, we first examined the inhibitory effect of chlorpromazine. Chlorpromazine causes clathrin lattice to assemble on endosomal membranes and at the same time prevents the assembly of coated pits at the plasma membrane (57). Chlorpromazine inhibited the entry of HCVpp in PLC/PRF/5 cells in a dose-dependent manner but had minimal effect on A-MLVpp (Fig. 2A). As expected for a clathrin-dependent virus (53), chlorpromazine also inhibited VSV-Gpp entry (Fig. 2A). At 5 μg/ml chlorpromazine, HCVpp entry was inhibited by about 60% and VSV-Gpp entry by about 50%. The use of higher chlorpromazine concentrations resulted in cell detachment during spinoculation of Huh-7 or PLC/PRF/5 cells. This toxicity precluded the use of 10 μg/ml chlorpromazine, a concentration that strongly inhibited bovine viral diarrhea virus infection in MDBK cells (29).
FIG. 2.
Clathrin-mediated entry of HCVpp. (A) PLC/PRF/5 cells were pretreated for 30 min with 2 or 5 μg/ml chlorpromazine (CPZ) or were left untreated. The cells were then infected with HCVpp, VSV-Gpp, or A-MLVpp by spinoculation in the presence of the drug. (B) PLC/PRF/5 cells were transfected twice with CHC siRNA or control siRNA, as explained in Materials and Methods. Two days after the second siRNA transfection, cells were spinoculated with HCVpp, VSV-Gpp, HA/NApp, RD114pp, or A-MLVpp. The cellular content of retroviral DNA was quantified by real-time PCR as a measure of entry and is expressed as a percentage of entry in untreated control cells (A) or in cells treated with control siRNA (B). The inset in panel B shows an immunoblot of the relative CHC and actin contents in siRNA-treated cells.
To further confirm that HCVpp entry requires an active pathway of clathrin-mediated endocytosis, we also knocked down the clathrin heavy chain (CHC) using siRNA technology. Following two successive transfections of siRNA, CHC levels were down-regulated to about 20% of controls or less (Fig. 2B). PLC/PRF/5 cells treated with CHC or control siRNA were infected with HCVpp, and entry was quantified by quantitative PCR. For comparison, the entry of retroviral particles pseudotyped with envelope glycoproteins of VSV (VSV-Gpp) or influenza virus (HA/NApp), two viruses known to enter by clathrin-mediated endocytosis, were also quantified. We also monitored the entry of retroviral particles pseudotyped with envelope glycoproteins of two retroviruses known to enter by a nonendocytic route (A-MLVpp and RD114pp). VSV-Gpp entry was strongly inhibited in CHC siRNA-treated cells (Fig. 2B). Similar levels of inhibition were achieved for HCVpp, whereas HA/NApp entry was significantly less inhibited than that of HCVpp and VSV-Gpp, in keeping with the documented dual-entry pathway of influenza virus by both clathrin-mediated and clathrin-independent endocytosis (46, 52). In contrast, the entry of RD114pp or A-MLVpp was not affected by CHC depletion (Fig. 2B), as expected for particles coated with envelope glycoproteins of pH-independent retroviruses. Similar results were obtained with Huh-7 cells (data not shown).
The inhibition of HCVpp entry by chlorpromazine or by CHC depletion strongly suggests that HCVpp enters PLC/PRF/5 cells by clathrin-mediated endocytosis.
HCVcc infection is clathrin dependent.
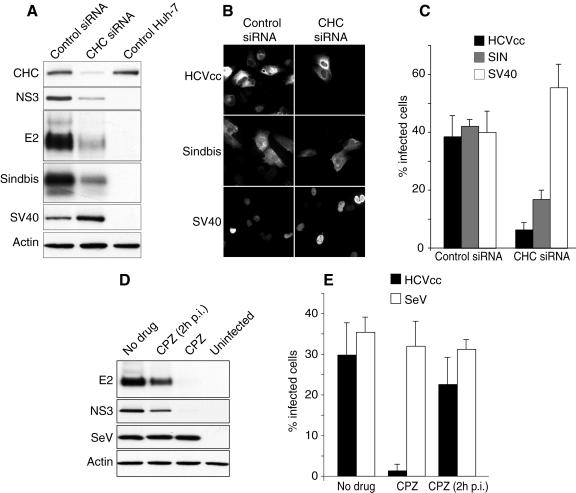
Recently, it has become possible to generate infectious HCV particles in cell culture (HCVcc), allowing the study of HCV entry with infectious viral particles. Preliminary experiments indicated that HCVcc does not infect PLC/PRF/5 cells, in contrast to HCVpp (C. Wychowski, unpublished observation). The reason for this difference is not yet understood, but it may involve a postentry restriction in PLC/PRF/5 cells. Therefore, we used Huh-7 cells to test whether the infection by HCVcc is dependent on clathrin-mediated endocytosis. Huh-7 cells were transfected twice with CHC or control siRNA, infected with HCVcc for 2 h at 37°C, and cultured for 30 h to allow for expression of viral proteins. The cells were then lysed and analyzed by immunoblotting or fixed and processed for immunofluorescence. As shown in Fig. 3A, the expression levels of E2 and NS3 were significantly lower at 30 hpi in CHC siRNA-treated cells than in cells treated with control siRNA or untreated cells. Similar results were obtained at 48 hpi (data not shown).
FIG. 3.
Clathrin-mediated entry of HCVcc. (A to C) Huh-7 cells were treated with CHC or control siRNA and then infected with HCVcc, a recombinant Sindbis virus expressing HCV E1, or SV40. (A) Cells were lysed at 5 hpi (Sindbis) or 30 hpi (HCVcc and SV40), and the cell lysates were analyzed by immunoblotting with antibodies to E2, NS3 (HCVcc), E1 (Sindbis), T-antigen (SV40), CHC, and actin. (B) Cells were fixed and processed for immunofluorescent detection of E2 (HCVcc), E1 (Sindbis), or T antigen (SV40). (B) For each virus, the fields presented contain similar numbers of CHC or control siRNA-treated cells. (C) Percentage of infected cells in CHC and control siRNA-treated cells. (D and E) Huh-7 cells were pretreated for 30 min and then infected for 2 h with HCVcc in the presence or the absence of chlorpromazine (CPZ; 5 μg/ml). Cells infected in the absence of drug were cultured with no drug, or chlorpromazine was added 2 hpi (CPZ 2hpi). Infection was analyzed (D) by immunoblotting with antibodies to E2, NS3, GFP (SeV), and actin or (E) by immunofluorescent detection of infected cells using an anti-E2 MAb (HCVcc) or by GFP fluorescence (SeV). Results are presented as percentages of infected cells.
In comparison, the infection of siRNA-treated Huh-7 cells with Sindbis virus was also monitored. Sindbis virus enters cells by clathrin-mediated endocytosis (6). In order to facilitate the detection of Sindbis virus infection, we used a recombinant Sindbis virus driving the expression of HCV envelope protein E1 in infected cells (16). The infection was detected by immunoblotting cell lysates with anti-E1 (HCV) MAb A4. Note that in these experiments E1 is a transgene that is used as a marker and does not play any role in recombinant Sindbis virus entry. As expected, CHC depletion also had an inhibitory effect on Sindbis virus infection (Fig. 3A).
As a control, we infected siRNA-treated cells with SV40, which enters by clathrin-independent endocytosis in Huh-7 cells (12). Cells were lysed at 30 hpi, and the levels of T antigen were visualized by immunoblotting. In contrast to HCV and Sindbis virus, SV40 infection was not inhibited in CHC-depleted cells. Instead, we consistently observed in cells treated with CHC siRNA higher levels of T-antigen expression than in cells treated with control siRNA (Fig. 3A). Similar results were obtained with other unrelated control siRNAs (data not shown), thus excluding an inhibitory effect of the control siRNA on SV40 infection or T-antigen expression. This apparent up-regulation of SV40 infection upon CHC depletion was not further investigated in this study.
To verify that the lower levels of HCV proteins observed by immunoblotting resulted from an inhibition of the infection and not from a reduced expression of HCV proteins, infected cells were analyzed by immunofluorescence microscopy. The expression levels of HCV, Sindbis virus, or SV40 proteins in individual cells appeared similar in CHC siRNA-treated cells and in controls (Fig. 3B and data not shown). The number of cells infected by HCVcc was reduced in CHC siRNA-treated cells by about 80% (Fig. 3C). Similar results were obtained when HCVcc infection was scored with an anti-C MAb (data not shown). The number of Sindbis virus-infected cells was also reduced in CHC siRNA-treated cells (by 60%), whereas SV40-infected cells were present in increased numbers (Fig. 3C). These data confirmed the inhibitory role of CHC depletion on HCVcc infection.
The role of clathrin-mediated endocytosis in HCV infection was further confirmed with chlorpromazine. When Huh-7 cells were pretreated with chlorpromazine for 30 min and then infected with HCVcc, the expression levels of E2 and NS3 were dramatically reduced (Fig. 3D) and the number of infected cells was very low (Fig. 3E). In contrast, when Huh-7 cells were infected in the absence of chlorpromazine and the drug was added 2 hpi, the expression levels of HCV proteins E2 and NS3 were only slightly reduced (Fig. 3D), and the number of infected cells was similar to that in control untreated cells (Fig. 3E).
As a control, we assessed the impact of chlorpromazine on the infection of Huh-7 cells by Sendai virus (SeV). SeV is a paramyxovirus which enters by direct fusion at the plasma membrane. To facilitate detection, we used a recombinant SeV expressing GFP (28). Chlorpromazine treatment had no effect on the levels of GFP expression (Fig. 3D) or the number of SeV-infected cells (Fig. 3E). These results indicate that the inhibitory effect of chlorpromazine on HCVcc infection primarily results from an inhibition of an early step of the infection, most probably corresponding to entry.
Taken together, the data obtained with CHC depletion and chlorpromazine treatment indicate that HCVcc entry is dependent on an active pathway of clathrin-mediated endocytosis.
HCVcc infection is pH dependent.
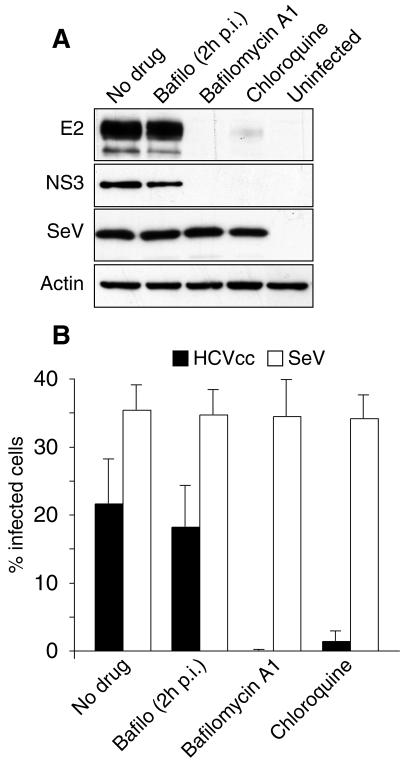
Since both HCVpp entry and HCVcc infection assays indicated a functional role for clathrin-mediated endocytosis in HCV entry, we also verified that HCVcc infection is indeed dependent on acidic endosomal pH. Huh-7 cells were pretreated for 30 min, infected for 2 h with HCVcc, and cultured for 30 h in the presence of 50 nM bafilomycin A1 or 20 μM chloroquine. These experimental conditions were chosen because they had a strong inhibitory effect on HCVpp entry in PLC/PRF/5 cells and on BVDV infection in MDBK cells (29). HCVcc infection was quantified by immunoblotting. As shown in Fig. 4A, the cellular levels of both E2 and NS3 at 30 hpi were dramatically reduced in the presence of each inhibitor compared to levels for untreated control cells infected with the same amounts of HCVcc. When the cells were infected and bafilomycin A1 was added to infected cells at 2 hpi, the levels of E2 and NS3 were similar to those in the controls, indicating that the drug had no inhibitory effect on HCVcc infection at a postentry step. In contrast, bafilomycin A1 and chloroquine had no effect on the levels of GFP expression mediated by recombinant SeV infection (Fig. 4A).
FIG. 4.
pH dependency of HCVcc infection. Huh-7 cells were pretreated for 30 min and then infected with HCVcc or SeV in the presence of 50 nM bafilomycin A1 or 20 μM chloroquine or in the absence of any inhibitor of endosomal acidification. Cells infected in the absence of drug were cultured with no drug, or bafilomycin A1 was added 2 hpi (Bafilo 2hpi). HCVcc infection was analyzed (A) by immunoblotting with antibodies to E2, NS3, GFP (SeV) and actin or (B) by immunofluorescent detection of infected cells using an anti-E2 MAb (HCVcc) or by GFP fluorescence (SeV). Results are presented as percentages of infected cells.
HCVcc infection was also scored by immunofluorescence microscopy using anti-E2 and anti-C antibodies. No infected cells were observed in cells pretreated and infected in the presence of 50 nM bafilomycin A1 (Fig. 4B and data not shown). In contrast, cells that were infected with no drug and incubated with 50 nM bafilomycin A1 from 2 hpi up to 30 hpi displayed a percentage of infected cells similar to that of controls. Chloroquine also dramatically reduced the number of infected cells (Fig. 4B). In contrast, neither bafilomycin A1 nor chloroquine treatments had any impact on the number of SeV-infected cells (Fig. 4B).
Together with the results from immunoblot analysis, these results indicate the HCVcc entry is sensitive to agents that neutralize the acidic pH of cellular endosomes.
DISCUSSION
To examine the entry of HCV in target cells, we have used two complementary approaches. First we have developed a new assay based on the use of HCVpp to specifically study early entry steps mediated by HCV envelope glycoproteins. This assay is based on the quantification of retroviral DNA synthesis, which occurs soon after the fusion of the retroviral particle with a cellular membrane (21). Presumably, this assay is only dependent on the entry steps mediated by the heterodimer E1E2 (binding, endocytosis, and fusion) and on the activity of the reverse transcriptase of the HCVpp retroviral core. Second, we made use of the recently developed JFH-1 infectious clone to investigate in parallel the entry pathway of HCVcc in the context of an infectious cycle.
Both assays yielded similar conclusions on the mechanisms of HCV entry. Our results indicate that HCVpp and HCVcc enter cells by clathrin-mediated endocytosis. The partial inhibition observed in cells transfected with CHC siRNA is probably due to the incomplete depletion of clathrin from the cells after siRNA treatment rather than to the existence of an alternative entry pathway for HCV, since a similar behavior was observed for Sindbis virus and VSV-Gpp, two controls for clathrin-mediated viral entry (6, 53). HCVcc infection was also found to be sensitive to agents that interfere with the acidic pH of endosomes. Similar findings on the pH dependency of HCVcc entry were recently reported by Tscherne et al. with a recombinant HCVcc clone expressing luciferase (54). This is consistent with an entry mediated by clathrin-mediated endocytosis, since clathrin-coated vesicles deliver their content into endosomes with an acidic content.
Many other viruses are known to hijack endocytic pathways. The sensitivity to lysosomotropic agents has often been considered evidence for an entry by endocytosis. However, viruses insensitive to lysosomotropic agents may also enter cells by endocytosis (12, 20, 33, 40). Several endocytic pathways were recently identified in addition to the clathrin-mediated pathway (10, 39, 51). These newly discovered pathways differ in the type of vesicular carrier formed at the plasma membrane to carry out the internalization step and also differ in the type of intracellular compartments to which the internalized material is transported. Some of them carry ligands to the classical acidic early and late endosomes, which are also reached by endocytic ligands originating from clathrin-coated vesicles, while other ones deliver their ligands to compartments with neutral pH, such as caveosomes, endoplasmic reticulum, or the Golgi complex (35, 39). Not surprisingly, it appears that viruses have evolved to adapt to these different entry routes. Several enveloped and nonenveloped viruses have been reported to use various alternative endocytic pathways, whether they are sensitive or not to acidic pH. SV40 enters through caveola-mediated internalization in CV-1 cells (40). Echovirus 1 and rotavirus are internalized by clathrin- and caveola-independent pathways, which require dynamin function and are sensitive to the cholesterol-sequestering drug methyl-β-cyclodextrin (41, 47). Lymphocytic choriomeningitis virus is endocytosed into cells by noncoated vesicles and reaches an acidic intracellular compartment where a process of pH-dependent fusion occurs (5). Influenza viruses can be endocytosed both by clathrin-independent and clathrin-dependent mechanisms into the same cell (46, 52).
During experiments involving CHC depletion, we made use of SV40 as a control for clathrin-independent endocytosis. SV40 was recently reported to infect Huh-7 cells by a unique route involving neither clathrin-mediated endocytosis nor the internalization of caveolae, which are absent from Huh-7 cells (12). In Huh-7 cells, the endocytosis of SV40 virions occurs through small uncoated vesicles that are formed at the plasma membrane in a dynamin-independent manner, and they are sensitive to cholesterol-sequestering drugs (12). Interestingly, we found that the depletion of clathrin heavy chain results in an increased SV40 infection, possibly by enhancing its endocytic pathway. It is well established that many regulatory factors have opposite effects on clathrin-mediated and on caveolae/lipid raft-mediated endocytosis (38). We can therefore hypothesize that the inhibition of clathrin-mediated endocytosis might induce compensatory mechanisms up-regulating alternative endocytic pathways in order to control the homoeostasis of the plasma membrane. Similarly, up-regulation of dynamin-independent fluid-phase uptake has been reported in cells expressing dominant-negative dynamin mutants (11).
In the Flaviviridae family, the entry of viruses of the Flavivirus and Pestivirus genera was examined before. Early electron microscopy studies suggested that the flavivirus West Nile virus enters cells through clathrin-coated vesicles (22). This mode of entry was recently confirmed by the use of a dominant-negative form of Eps15 as a way to functionally probe the clathrin-mediated pathway (8). This is consistent with numerous studies which have shown that the envelope glycoprotein E of flaviviruses undergoes a conformational transition under acidic conditions from a native dimeric form to a fusogenic trimeric form (23). Such an acid-triggered conformational transition has not been reported at the present time for the envelope proteins of pestiviruses or HCV.
The pestivirus BVDV also enters by clathrin-mediated endocytosis (26, 29). Consistent with this mode of entry, BVDV infection is sensitive to agents that neutralize the pH of endosomes. However, unlike flaviviruses, BVDV and HCVcc particles are stable and stay infectious when incubated at acidic pH before infection (26, 54). In similar conditions, flaviviruses, alphaviruses, and other enveloped viruses are inactivated, presumably because the pH triggers irreversible conformational changes in the envelope glycoproteins of these viruses similar to those associated with the fusion process during entry. This indicates that a pH drop is not sufficient to induce the conformational changes associated with the fusion process mediated by the envelope glycoproteins of pestiviruses and hepaciviruses, or that the changes in conformation triggered under acidic conditions are reversible in the absence of a cellular membrane. Such reversible conformational changes were reported for VSV (43).
It has been suggested that HCV and BVDV envelope glycoproteins must be primed to acquire a fusogenic conformation triggered by pH. This priming event can be mimicked to some extent by pretreating BVDV particles with reducing agents (26). Under reducing conditions, BVDV virions became pH sensitive, and their fusion to the plasma membrane was inducible at pH 5.
Our study suggests that, like flaviviruses and pestiviruses, HCV enters cells through clathrin-mediated endocytosis and fusion from within an acidic endosomal compartment. At the present time, we still do not know the exact nature of the endosomal compartment that is competent for HCV fusion. We also do not know whether the acidic pH of endosomes acts directly on the conformation of HCV envelope glycoproteins E1 and E2 to trigger fusion or whether it acts indirectly by activating other endosomal agents, which in turn would promote HCV fusion. It would be interesting to determine if HCV fusion requires a priming event to allow an acid-triggered conformational change of the E1E2 heterodimer or if the fusion may be directly induced by a pH drop, as is the case for flaviviruses. Tscherne et al. recently reported that HCVcc infection by direct fusion at the plasma membrane induced by a pH drop was very inefficient (54). However, it is not known if the blockade was at the fusion step or at a postfusion step. On the other hand, experiments carried out in vitro with HCVpp indicated that fusion to liposomes mediated by HCV E1E2 heterodimer is readily inducible at acidic pH (27). This suggests that a preliminary priming event would not be necessary for HCV E1E2-mediated fusion. It remains to be determined if conformational changes in E1E2 are associated with the fusion process and if similar findings are observed with HCVcc particles. Such questions could be addressed once it becomes possible to purify HCVcc particles for in vitro studies.
Acknowledgments
We thank Laetitia Corset and Anne Goffard for help with real-time PCR. We are grateful to J. F. Delagneau, A. Patel, and J. McKeating for providing us with antibodies, D. Lavillette and F.-L. Cosset for plasmids, and L. Roux and F. Lafont for the purified stock of SeV. Some data were generated with the help of the Imaging Core Facility of the Calmette campus.
This work was supported by a grant from the “Agence Nationale de Recherche sur le Sida et les Hépatites Virales” (ANRS) to Y.R. and C.W.E.B. was supported by a postdoctoral fellowship from the ANRS. T.W. was partly supported by a grant from the Ministry of Health, Labor, and Welfare of Japan, the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO), and Research on Health Sciences focusing on Drug Innovation from the Japan Health Sciences Foundation. J.D. is an international scholar of the Howard Hughes Medical Institute.
REFERENCES
- 1.Agnello, V., G. Ábel, M. Elfahal, G. B. Knight, and Q. X. Zhang. 1999. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth, H., C. Schäfer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. Von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003-41012. [DOI] [PubMed] [Google Scholar]
- 3.Bartosch, B., J. Dubuisson, and F.-L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F.-L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 5.Borrow, P., and M. B. Oldstone. 1994. Mechanism of lymphocytic choriomeningitis virus entry into cells. Virology 198:1-9. [DOI] [PubMed] [Google Scholar]
- 6.Carbone, R., S. Fre, G. Iannolo, F. Belleudi, P. Mancini, P. G. Pelicci, M. R. Torrisi, and P. P. Di Fiore. 1997. eps15 and eps15R are essential components of the endocytic pathway. Cancer Res. 57:5498-5504. [PubMed] [Google Scholar]
- 7.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 8.Chu, J. J., and M. L. Ng. 2004. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J. Virol. 78:10543-10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clayton, R. F., A. Owsianka, J. Aitken, S. Graham, D. Bhella, and A. H. Patel. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 76:7672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conner, S. D., and S. L. Schmid. 2003. Regulated portals of entry into the cell. Nature 422:37-44. [DOI] [PubMed] [Google Scholar]
- 11.Damke, H., T. Baba, A. M. van der Bliek, and S. L. Schmid. 1995. Clathrin-independent pinocytosis is induced in cells overexpressing a temperature-sensitive mutant of dynamin. J. Cell Biol. 131:69-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damm, E. M., L. Pelkmans, J. Kartenbeck, A. Mezzacasa, T. Kurzchalia, and A. Helenius. 2005. Clathrin- and caveolin-1-independent endocytosis: entry of simian virus 40 into cells devoid of caveolae. J. Cell Biol. 168:477-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummer, H. E., A. Maerz, and P. Poumbourios. 2003. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 546:385-390. [DOI] [PubMed] [Google Scholar]
- 15.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duvet, S., F. Chirat, A. M. Mir, A. Verbert, J. Dubuisson, and R. Cacan. 2000. Reciprocal relationship between alpha1,2 mannosidase processing and reglucosylation in the rough endoplasmic reticulum of Man-P-Dol deficient cells. Eur. J. Biochem. 267:1146-1152. [DOI] [PubMed] [Google Scholar]
- 17.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner, J. P., R. J. Durso, R. R. Arrigale, G. P. Donovan, P. J. Maddon, T. Dragic, and W. C. Olson. 2003. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 100:4498-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gault, E., Y. Michel, A. Dehée, C. Belabani, J. C. Nicolas, and A. Garbarg-Chenon. 2001. Quantification of human cytomegalovirus DNA by real-time PCR. J. Clin. Microbiol. 39:772-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gianni, T., G. Campadelli-Fiume, and L. Menotti. 2004. Entry of herpes simplex virus mediated by chimeric forms of nectin1 retargeted to endosomes or to lipid rafts occurs through acidic endosomes. J. Virol. 78:12268-12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goff, S. P. 2001. Retroviridae: The retroviruses and their replication, p. 1871-1940. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott, Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 22.Gollins, S. W., and J. S. Porterfield. 1985. Flavivirus infection enhancement in macrophages: an electron microscopic study of viral cellular entry. J. Gen. Virol. 66:1969-1982. [DOI] [PubMed] [Google Scholar]
- 23.Heinz, F. X., K. Stiasny, and S. L. Allison. 2004. The entry machinery of flaviviruses. Arch. Virol. Suppl. 2004:133-137. [DOI] [PubMed] [Google Scholar]
- 24.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 26.Krey, T., H. J. Thiel, and T. Rümenapf. 2005. Acid-resistant bovine pestivirus requires activation for pH-triggered fusion during entry. J. Virol. 79:4191-4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavillette, D., B. Bartosch, D. Nourrisson, G. Verney, F.-L. Cosset, F. Penin, and E. I. Pécheur. 2006. Hepatitis C virus glycoproteins mediate low pH-dependent membrane fusion with liposomes. J. Biol. Chem. 281:3909-3917. [DOI] [PubMed] [Google Scholar]
- 28.Le Mercier, P., D. Garcin, S. Hausmann, and D. Kolakofsky. 2002. Ambisense Sendai viruses are inherently unstable but are useful to study viral RNA synthesis. J. Virol. 76:5492-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lecot, S., S. Belouzard, J. Dubuisson, and Y. Rouillé. 2005. Bovine viral diarrhea virus entry is dependent on clathrin-mediated endocytosis. J. Virol. 79:10826-10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wölk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 31.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: The viruses and their replication, p. 991-1042. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott, Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 32.Lozach, P. Y., H. Lortat-Jacob, A. de Lacroix de Lavalette, I. Staropoli, S. Foung, A. Amara, C. Houlès, F. Fieschi, O. Schwartz, J.-L. Virelizier, F. Arenzana-Seisdedos, and R. Altmeyer. 2003. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 278:20358-20366. [DOI] [PubMed] [Google Scholar]
- 33.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103:679-689. [DOI] [PubMed] [Google Scholar]
- 34.Motley, A., N. A. Bright, M. N. Seaman, and M. S. Robinson. 2003. Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 162:909-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nichols, B. 2003. Caveosomes and endocytosis of lipid rafts. J. Cell Sci. 116:4707-4714. [DOI] [PubMed] [Google Scholar]
- 36.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Op De Beeck, A., C. Voisset, B. Bartosch, Y. Ciczora, L. Cocquerel, Z. Keck, S. Foung, F.-L. Cosset, and J. Dubuisson. 2004. Characterization of functional hepatitis C virus envelope glycoproteins. J. Virol. 78:2994-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelkmans, L., E. Fava, H. Grabner, M. Hannus, B. Habermann, E. Krausz, and M. Zerial. 2005. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 436:78-86. [DOI] [PubMed] [Google Scholar]
- 39.Pelkmans, L., and A. Helenius. 2003. Insider information: what viruses tell us about endocytosis. Curr. Opin. Cell Biol. 15:414-422. [DOI] [PubMed] [Google Scholar]
- 40.Pelkmans, L., J. Kartenbeck, and A. Helenius. 2001. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 3:473-483. [DOI] [PubMed] [Google Scholar]
- 41.Pietiäinen, V., V. Marjomäki, P. Upla, L. Pelkmans, A. Helenius, and T. Hyypiä. 2004. Echovirus 1 endocytosis into caveosomes requires lipid rafts, dynamin II, and signaling events. Mol. Biol. Cell 15:4911-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 43.Puri, A., J. Winick, R. J. Lowy, D. Covell, O. Eidelman, A. Walter, and R. Blumenthal. 1988. Activation of vesicular stomatitis virus fusion with cells by pretreatment at low pH. J. Biol. Chem. 263:4749-4753. [PubMed] [Google Scholar]
- 44.Rouillé, Y., F. Helle, D. Delgrange, P. Roingeard, C. Voisset, E. Blanchard, S. Belouzard, J. McKeating, A. H. Patel, G. Maertens, T. Wakita, C. Wychowski, and J. Dubuisson. 2006. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 80:2832-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 46.Rust, M. J., M. Lakadamyali, F. Zhang, and X. Zhuang. 2004. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 11:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sánchez-San Martín, C., T. López, C. F. Arias, and S. López. 2004. Characterization of rotavirus cell entry. J. Virol. 78:2310-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandrin, V., and F. L. Cosset. 2006. Intracellular versus cell surface assembly of retroviral pseudotypes is determined by the cellular localization of the viral glycoprotein, its capacity to interact with Gag and the expression of the Nef protein. J. Biol. Chem. 281:528-542. [DOI] [PubMed] [Google Scholar]
- 49.Saunier, B., M. Triyatni, L. Ulianich, P. Maruvada, P. Yen, and L. D. Kohn. 2003. Role of the asialoglycoprotein receptor in binding and entry of hepatitis C virus structural proteins in cultured human hepatocytes. J. Virol. 77:546-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sieczkarski, S. B., and G. R. Whittaker. 2002. Dissecting virus entry via endocytosis. J. Gen. Virol. 83:1535-1545. [DOI] [PubMed] [Google Scholar]
- 52.Sieczkarski, S. B., and G. R. Whittaker. 2002. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J. Virol. 76:10455-10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun, X., V. K. Yau, B. J. Briggs, and G. R. Whittaker. 2005. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology 338:53-60. [DOI] [PubMed] [Google Scholar]
- 54.Tscherne, D. M., C. T. Jones, M. J. Evans, B. D. Lindenbach, J. A. McKeating, and C. M. Rice. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voisset, C., N. Callens, E. Blanchard, A. Op De Beeck, J. Dubuisson, and N. Vu-Dac. 2005. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J. Biol. Chem. 280:7793-7799. [DOI] [PubMed] [Google Scholar]
- 56.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, L. H., K. G. Rothberg, and R. G. Anderson. 1993. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J. Cell Biol. 123:1107-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wychowski, C., D. Benichou, and M. Girard. 1986. A domain of SV40 capsid polypeptide VP1 that specifies migration into the cell nucleus. EMBO J. 5:2569-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]