Abstract
Of 30 baculovirus genomes that have been sequenced to date, the only nonlepidopteran baculoviruses include the dipteran Culex nigripalpus nucleopolyhedrovirus and two hymenopteran nucleopolyhedroviruses that infect the sawflies Neodiprion lecontei (NeleNPV) and Neodiprion sertifer (NeseNPV). This study provides a complete sequence and genome analysis of the nucleopolyhedrovirus that infects the balsam fir sawfly Neodiprion abietis (Hymenoptera, Symphyta, Diprionidae). The N. abietis nucleopolyhedrovirus (NeabNPV) is 84,264 bp in size, with a G+C content of 33.5%, and contains 93 predicted open reading frames (ORFs). Eleven predicted ORFs are unique to this baculovirus, 10 ORFs have a putative sequence homologue in the NeleNPV genome but not the NeseNPV genome, and 1 ORF (neab53) has a putative sequence homologue in the NeseNPV genome but not the NeleNPV genome. Specific repeat sequences are coincident with major genome rearrangements that distinguish NeabNPV and NeleNPV. Genes associated with these repeat regions encode a common amino acid motif, suggesting that they are a family of repeated contiguous gene clusters. Lepidopteran baculoviruses, similarly, have a family of repeated genes called the bro gene family. However, there is no significant sequence similarity between the NeabNPV and bro genes. Homologues of early-expressed genes such as ie-1 and lef-3 were absent in NeabNPV, as they are in the previously sequenced hymenopteran baculoviruses. Analyses of ORF upstream sequences identified potential temporally distinct genes on the basis of putative promoter elements.
The balsam fir sawfly (Neodiprion abietis, Hymenoptera, Symphyta, Diprionidae) is a native sawfly species that occurs throughout Canada and the United States. The balsam fir sawfly is a defoliating species that feeds primarily on the balsam fir Abies balsamea (L.) Mill. and spruce Picea spp. (96). An outbreak of the balsam fir sawfly in Newfoundland has persisted since 1991. This 15-year outbreak is in contrast to the typically observed outbreaks of 4- to 5-year durations that occur every 5 to 15 years. The current infestation, spanning more than 40,000 ha of balsam fir in western Newfoundland, threatens the significant silvicultural investment in that region, as severe defoliation greatly reduces tree growth and may cause tree mortality (81). The application of a virus naturally pathogenic to the balsam fir sawfly, the N. abietis nucleopolyhedrovirus (NeabNPV), may offer great potential in controlling sawfly populations (76). Recently, aerial application of NeabNPV has successfully initiated collapse of growing and peaking balsam fir sawfly populations (71).
NeabNPV is a member of the family Baculoviridae, a well-studied family of invertebrate viruses that have large, double-stranded, circular DNA genomes within rod-shaped nucleocapsids that are enveloped and occluded. The two genera of baculoviruses, Nucleopolyhedrovirus (NPV) and Granulovirus (GV), are typically distinguished by the size and localization of their occlusion bodies (OBs). NPVs have larger nucleus-localized OBs, ranging in diameter from 1 to 5 μm, with singly or multiply enveloped virions. GVs have smaller OBs with diameters of 0.3 to 0.6 μm in which single enveloped virions are normally occluded, and these OBs are distributed throughout the cell following nuclear disintegration. GVs have only been isolated from lepidopteran hosts, while NPVs have been isolated from lepidopterans, hymenopterans, and dipterans. NeabNPV is a singly enveloped NPV that infects the balsam fir sawfly.
Thirty baculovirus genomes are available in GenBank. Of these, the lepidopteran baculoviruses are the most highly represented (27 sequenced genomes). Two NPVs that infect the diprionid sawflies Neodiprion sertifer (NeseNPV) (26) and Neodiprion lecontei (NeleNPV) (62) and one that infects the mosquito Culex nigripalpus (Diptera, Culicidae), CuniNPV (2), have been completely sequenced. Baculovirus genomes range in size from 81.8 kb to 161 kb and have G+C contents ranging from 32% to 56%. The sawfly NPVs have both the smallest genomes and the lowest G+C contents of any baculoviruses sequenced to date (26, 62).
Baculovirus phylogeny was first resolved with the conserved polyhedrin gene sequence. This analysis identified two distinct taxa within the lepidopteran NPVs, termed groups I and II (103). More recently, full-genome phylogenetic analyses suggested that single-gene phylogenies might not accurately resolve evolutionary relationships among baculoviruses (44). Instead, seven genes are considered to share the topology of whole-genome phylogenies (44). Complete-genome phylogeny, along with gene content and gene order phylogenies, suggests that the lepidopteran-infecting baculoviruses form three distinct clades, i.e., GV, group I NPV, and group II NPV (43, 44). The dipteran-infecting baculovirus CuniNPV is taxonomically distinct from the lepidopteran-infecting baculoviruses (44). Recent genome sequencing of the hymenopteran-infecting baculoviruses NeseNPV and NeleNPV has indicated that they represent a separate clade that is distinct from both lepidopteran-infecting and dipteran-infecting baculoviruses (26, 44, 62).
Here we report the genome sequence of NeabNPV. Our data provide further insight into the evolution of sawfly baculoviruses. Furthermore, a detailed analysis of the upstream sequences of predicted NeabNPV open reading frames (ORFs) identified promoter motifs that potentially discriminate temporally distinct genes. Twenty-nine genes are believed to be conserved throughout all of the fully sequenced baculovirus species. This set of genes, however, does not include a number of essential early-expressed genes (ie-1, lef-3, pp31). Because of the essential role that these genes play in the lepidopteran baculovirus life cycle, we investigated promoter characteristics that distinguish early- and late-expressed genes. A subset of predicted early-expressed genes may include functional analogues for these genes.
MATERIALS AND METHODS
Source of NeabNPV.
NeabNPV OBs were produced and purified as described by Moreau et al. (71) This procedure involved thawing frozen (−20°C) larvae in 5 volumes of 0.3% sodium dodecyl sulfate (SDS) solution, followed by homogenization. The sample was twice treated with a process of filtration through a 1-mm2 plastic mesh, followed by resuspension of solid matter in 0.3% SDS and rehomogenization and refiltration until the filtrate was clear. Filtrates were then pooled, filtered twice through eight layers of cheesecloth, and centrifuged for 15 min at 9,000 × g. The supernatant was discarded, and the NeabNPV OB pellet was resuspended in 0.3% SDS. Centrifugation and resuspension were repeated until a clear supernatant was obtained. The NeabNPV OB pellet was then resuspended in water and concentrated to 109 OBs/ml.
Virus DNA purification.
NeabNPV OB suspensions were washed with sterile double-distilled H2O and centrifuged (15,000 × g, 5 min at 20°C) three times and then treated with SDS (0.4%) under agitation for 1 h. The resultant suspension was washed and centrifuged (15,000 × g, 10 min at 20°C) three more times and incubated with dissociation buffer (0.1 M Na2CO3, 0.04 M sodium thioglycolate) for 20 min. Debris was pelleted twice by centrifugation (2,000 × g, 5 min at 4°C), and the supernatant was collected. The supernatant was washed, pelleted (15,000 × g, 30 min at 4°C), and resuspended in Tris-HCl. This was then incubated with 0.01 mg/ml proteinase K (Invitrogen) and 2% sarcosyl for 16 h at 50°C. The solution was then treated with DNAzol (Invitrogen) following the manufacturer's standard protocol, and purified NeabNPV DNA was resolved by 0.8% agarose pulse-field gel electrophoresis in 0.5× Tris-borate-EDTA buffer at 200 V with a switch time of 60 s for 15 h, followed by a switch time of 90 s for 8 h, at 14°C. NeabNPV DNA was stained with 0.5 μg/ml ethidium bromide, viewed under long-wavelength UV illumination (365 nm), and excised. Excised DNA was purified with the Fermentas DNA extraction kit (Fermentas) according to the manufacturer's protocol.
DNA cloning and sequencing.
NeabNPV DNA was fragmented by restriction enzyme digestion and by hydrodynamic shearing (HydroShear; GenMachine, San Carlos, CA). Partial restriction libraries were generated by digestion of NeabNPV DNA with HindIII (New England Biolabs) followed by ligation into pBluescriptII KS+ (Stratagene) or by digestion with EcoRI (New England Biolabs) followed by ligation into pT7/T3a18. Random libraries fragmented by hydrodynamic shearing were generated and ligated into pSMART (Lucigen Corporation). PCR was used to generate gap-spanning fragments. The result was an average of 15-fold sequence redundancy. DNA sequencing was conducted with an ABI3700 automated DNA sequencer (Applied Biosystems, Inc.), and the reactions were carried out with an ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer Applied Biosystems).
Genome sequence analysis.
The complete NeabNPV genome was automatically assembled by PHRED/PHRAP/CONSED with a PHRAP repeat stringency of 0.8 (22, 23). The contiguous sequence was manually edited and confirmed. ORFs were identified with the NCBI ORF Finder program (http://ncbi.nih.nlm.gov/gorf/gorf.html), and those encoding more than 50 amino acids with minimal overlap were considered putative genes. Characterization of the ORFs involved BLAST analysis with BLASTP and PSI-BLAST against the NCBI nonredundant protein database (3), as well as RPS-BLAST against the CDD database (69). Baculovirus putative homologues were accepted on the basis of a BLASTP (default parameters) E value of ≤0.001. Predicted ORFs were then analyzed with InterProScan (104), which integrates queries against the PROSITE (45), PRINTS (5), Pfam (6), ProDom (19), SMART (88), TIGRFAMs (35), PIR superfamily (101), SUPERFAMILY (29), Gene3D (13), and PANTHER (70) databases.
Genome parity and repeat analysis.
The repeat regions were identified by REPuter (57). The 5′ and 3′ limits were defined as the outermost repeat with no other repeats within 1 kb on the external side. The minimal repeat element was queried against the NeleNPV and NeseNPV genomes with BLASTN 2.2.6. Repeat regions were also illustrated by generating parity plots of the NeabNPV genome sequence against itself with PipMaker (89). Additionally, genome parity between NeseNPV and NeabNPV and between NeleNPV and NeabNPV was illustrated with PipMaker (89).
Promoter sequence analysis.
Sequences 160 bp upstream of the predicted ORF start codons were analyzed for potential promoter motifs. These sequences were compared to experimentally verified promoter elements with SIGNAL SCAN v4.05 (82) to query the TRANSFAC database (99). Potential promoter elements were identified by aligning all upstream sequences with AlignACE v3.0 (48). The sequences were also analyzed for common known baculovirus promoter elements, including the TATA sequence (TATA, TATAW), CAKT, and DTAAG elements. These were also sought in the −40-bp to −20-bp region of the upstream sequences. Only putative predicted promoter elements with a P value of <0.05 were reported, except for TATA elements that were overrepresented in the −160-bp or −40-bp to −20-bp upstream sequences and a putative CAKT element, because of its established function as a baculovirus promoter element (84).
Calculation of P values.
To generate a P value for a sequence of a given length, 1,000 random loci of equal length (160 or 20 bp) were sampled. The expected frequency of occurrence was calculated for each motif in this data set. The P value for observing x motif-containing putative promoter elements out of N ORFs given an expected frequency Fe and a cumulative binomial distribution was calculated as follows: P(occ ≤ x) = [N!/x! × (N − x)!] × (Fe)x × (1 − Fe)N − x.
Nucleotide sequence accession number.
The NeabNPV genome sequence has been deposited in GenBank under accession number DQ317692.
RESULTS AND DISCUSSION
Nucleotide sequence analysis.
The NeabNPV genome was 84,264 bp in size, making it among the smallest baculovirus genomes, along with NeleNPV at 81,755 bp (62), NeseNPV at 86,462 bp (26), and Adoxophyes orana GV at 99,657 bp (100). The G+C content was equivalent to that of NeleNPV and NeseNPV at 33.5%. By comparison, the G+C contents of lepidopteran baculovirus genomes range from 32.4% for Cryptophlebia leucotreta GV (60) to 57.5% for Xestia c-nigrum GV (40).
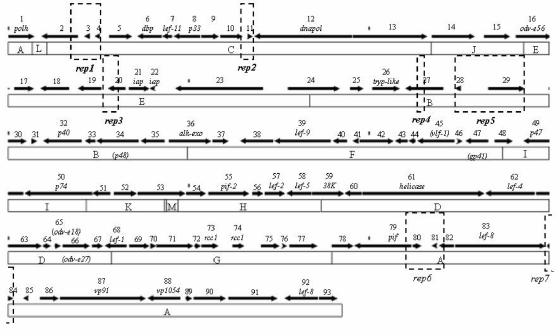
The NeabNPV genome contained 93 potential methionine-initiated ORFs (Fig. 1; Table 1). This is greater than the number of ORFs predicted to be encoded by the NeleNPV (89 ORFs) or NeseNPV (90 ORFs) genome (26, 62). Similar to the genome of NeseNPV, the NeabNPV ORFs are biased in orientation such that 60.9% are oriented clockwise and 39.1% are oriented in the opposite direction.
FIG. 1.
Linear sequence map and HindIII physical map of the circular NeabNPV genome. The transcriptional direction of each ORF is represented by an arrow labeled with the ORF number. Known baculovirus predicted homologues are presented below the ORF numbers. Repeat regions (rep) are shown as triangles. Repeat regions depicted in Fig. 2 are outlined with dotted lines.
TABLE 1.
NeabNPVORFs
| ORF | Name | Position | Length (amino acids) | Predicted sequence homologuea
|
Feature of coding sequenced | Putative promoter elementsc | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| NeleNPV | NeseNPV | AcMNPV | PxGVb | CuniNPV | ||||||
| 1 | polyhedrin (polh) | 0 > 741 | 247 | 1 (99) 247 | 1 (93) 246 | 8 (46) 245 | 4 (36) 248 | TATA box, DTAAG, M7, M8, M9 | ||
| 2 | 906 < 1953 | 349 | 5 (84) 52 | 3 (14) 388 | Cyclin-like F-box domain, RNA-binding motif | M1, M2 | ||||
| rep1 | 1881 > 2755 | |||||||||
| 3 | 2093 < 2273 | 60 | M1, M2 | |||||||
| 4 | 2380 < 2551 | 57 | TATA box, M1, M2, M4 | |||||||
| 5 | 2797 > 3469 | 224 | 13 (62) 247 | 20 (21) 270 | Similar to AcMNPV P33, signal peptide | TATA box, Zeste, DTAAG, M1, M6, M7, M8, M9 | ||||
| 6 | dbp | 3557 < 4268 | 237 | 14 (88) 236 | 22 (45) 251 | 25 (19) 316 | TATA box, M2, M5 | |||
| 7 | lef-11 | 4248 < 4557 | 103 | 15 (84) 101 | 23 (43) 100 | TATA box, CAKT, M9 | ||||
| 8 | p33 | 4570 < 5341 | 257 | 16 (87) 256 | 24 (51) 252 | 92 (19) 259 | 76 (18) 250 | 14 (14) 370 | DTAAG, M3, M6 | |
| 9 | 5342 > 5855 | 171 | 17 (91) 170 | 25 (47) 170 | TATA box, DTAAG | |||||
| 10 | 5872 > 6523 | 217 | 18 (77) 216 | 26 (44) 218 | 1 predicted TM domain, signal peptide | TATA box, DTAAG, M7 | ||||
| rep2 | 6570 > 6954 | |||||||||
| 11 | 6635 > 6809 | 58 | 2 predicted TM domains | TATA box, M3 | ||||||
| 12 | dnapol | 6795 < 9555 | 920 | 20 (84) 923 | 28 (64) 913 | 65 (23) 984 | 94 (23) 979 | 91 (17) 1138 | DNA polymerase type B family | Zeste, CREB/ATF-47, CAKT, M6 |
| 13 | 9553 > 11638 | 695 | 21 (66) 728 | 29 (28) 792 | 93 (9) 651 | 67 (13) 1357 | Putative ATPase, putative IMP dehydrogenase domain; prefoldin structure | CREB/ATF-47, M3, M6, M7 | ||
| 14 | 11742 > 12963 | 407 | 22 (63) 380 | 3 (21) 388 | ||||||
| 15 | 13175 > 13937 | 254 | 25 (35) 261 | Leucine-rich repeat | TATA box, DTAAG, M3 | |||||
| 16 | odv-c56 | 14060 > 15071 | 337 | 23 (92) 336 | 38 (71) 339 | 148 (34) 376 | 16 (38) 351 | 101 (20) 361 | 1 predicted TM domain, IMP dehydrogenase/GMP reductase domain | TATA box, DTAAG, M5, M7, M9 |
| 17 | 15159 > 15738 | 193 | 24 (92) 192 | 39 (50) 192 | TATA box, DTAAG, M3 | |||||
| 18 | 15870 < 16698 | 276 | 25 (52) 261 | M3, M4, M9 | ||||||
| 19 | 16912 < 17596 | 228 | 26 (67) 226 | TATA box, Zeste, M1, M6, M9 | ||||||
| 20 | 17757 < 18267 | 170 | 2 (34) 108 | TATA box | ||||||
| rep3 | 17804 > 18262 | |||||||||
| 21 | iap | 18332 > 18941 | 203 | 11 (75) 260 | 17 (24) 181 | 27 (23) 286 | 98 (21) 281 | Baculovirus IAP repeat domain | TATA box, Zeste, M1, M7 | |
| 22 | 18912 < 19074 | 54 | CAKT, M7 | |||||||
| 23 | 19610 < 22079 | 823 | 9 (60) 794 | 16 (27) 637 | TATA box, M3 | |||||
| 24 | 22749 > 24213 | 488 | 8 (50) 526 | 11 (16) 461 | Ring finger domain, zinc finger, LMP repeat | TATA box, M3 | ||||
| 25 | 24466 > 24868 | 134 | 7 (71) 131 | TATA box, M3, M6, M7 | ||||||
| 26 | trypsin-like protein | 25091 > 25871 | 260 | 6 (96) 259 | 7 (73) 258 | Trypsin-like serine protease | DTAAG, M3, M6 | |||
| 27 | 25989 < 27069 | 360 | 4 (73) 332 | 9 (20) 416 | TATA box, Zeste, M4 | |||||
| rep4 | 26312 > 26527 | |||||||||
| 28 | 27373 < 27559 | 62 | Signal peptide | Zeste, CAKT, M1, M6, M9 | ||||||
| rep5 | 27427 > 29532 | |||||||||
| 29 | 28291 > 29335 | 348 | 5 (84) 52 | 3 (14) 388 | F-box domain | TATA box, CREB/ATF-47 | ||||
| 30 | 29972 > 30539 | 189 | 27 (51) 190 | 37 (29) 133 | TATA box, M1, M2 | |||||
| 31 | 30612 < 30869 | 86 | TATA box, CAKT, M5 | |||||||
| 32 | p40 | 30950 < 32051 | 367 | 29 (91) 366 | 35 (64) 368 | TATA box, Zeste | ||||
| 33 | 32105 < 32423 | 106 | 30 (75) 116 | 34 (47) 117 | TATA box, DTAAG, M7 | |||||
| 34 | p48 | 32419 < 33628 | 403 | 31 (85) 390 | 33 (54) 402 | 103 (10) 387 | IMP dehydrogenase/GMP reductase domain | TATA box, M6 | ||
| 35 | 33646 < 34348 | 234 | 32 (88) 238 | 32 (63) 235 | 40 (17) 206 | Zeste, M6 | ||||
| 36 | alk-exo | 34451 > 35663 | 404 | 33 (82) 402 | 31 (48) 399 | 133 (22) 419 | 106 (25) 378 | 54 (15) 367 | IAP repeat restriction endonuclease like | |
| 37 | 35659 > 36133 | 158 | 34 (84) 157 | 152 (14) 408 | 1 predicted TM domain, signal peptide | TATA box, CAKT, DTAAG, M6, M9 | ||||
| 38 | 36417 < 37371 | 318 | 36 (60) 319 | 1 predicted TM domain, signal peptide | TATA box, M6 | |||||
| 39 | lef-9 | 37363 < 38953 | 530 | 37 (87) 503 | 40 (62) 507 | 62 (29) 516 | 99 (31) 494 | 59 (15) 590 | TATA box, M3, M7, M9 | |
| 40 | 38981 < 39404 | 141 | 38 (87) 141 | 41 (48) 147 | 68 (20) 192 | 96 (15) 128 | 58 (18) 137 | 1 predicted TM domain | TATA box, M7 | |
| 41 | 39533 < 39722 | 73 | TATA box, DTAAG, M7, M8, M9 | |||||||
| 42 | 39720 > 40707 | 329 | 39 (70) 353 | 42 (24) 444 | Zeste, M3, M7 | |||||
| 43 | 40703 < 41081 | 126 | 40 (87) 125 | 43 (39) 125 | TATA box, DTAAG, M7 | |||||
| 44 | 41077 < 41308 | 74 | 41 (90) 76 | 44 (63) 82 | 1 predicted TM domain, signal peptide | CREB/ATF-47, M6 | ||||
| 45 | vlf-1 | 41310 < 42375 | 355 | 42 (86) 354 | 45 (65) 353 | 77 (24) 379 | 89 (29) 346 | 18 (16) 358 | DNA breaking-rejoining enzyme, catalytic core, IMP dehydrogenase/GMP reductase domain | TATA box, CAKT, DTAAG |
| 46 | 42458 > 42617 | 53 | 1 predicted TM domain, signal peptide | TATA box | ||||||
| 47 | gp41 | 42649 < 43315 | 222 | 44 (94) 270 | 47 (60) 312 | 80 (30) 409 | 87 (19) 283 | TATA box, DTAAG, M4, M8 | ||
| 48 | 43484 > 44012 | 176 | 45 (90) 175 | 48 (75) 176 | 81 (36) 233 | 86 (34) 191 | 106 (14) 186 | 2 predicted TM domains | Zeste, DTAAG, M3 | |
| 49 | p47 | 44255 < 45434 | 393 | 46 (87) 389 | 49 (61) 386 | 40 (22) 401 | 51 (20) 386 | 73 (10) 439 | Zeste, CAKT, M7 | |
| 50 | p74 | 45435 < 47340 | 635 | 47 (86) 633 | 50 (63) 634 | 138 (38) 645 | 49 (31) 578 | 74 (32) 681 | 3 predicted TM domains, IMP dehydrogenase/GMP reductase domain | TATA box, M3 |
| 51 | 47317 < 47854 | 179 | 48 (69) 178 | 51 (28) 142 | Double-stranded RNA-binding motif | TATA box, M8, M9 | ||||
| 52 | 47916 > 48585 | 223 | 49 (76) 218 | 52 (40) 220 | Zinc finger | TATA box, Zeste | ||||
| 53 | 48577 > 49933 | 452 | 53 (48) 386 | 1 predicted TM domain IMP dehydrogenase/GMP reductase domain | TATA box, M4, M5, M6 | |||||
| 54 | 49929 > 50508 | 193 | 51 (83) 192 | 54 (47) 184 | TATA box, CREB/ATF-47, DTAAG, M6, M9 | |||||
| 55 | pif-2 | 50537 > 51692 | 385 | 52 (93) 384 | 55 (73) 384 | 22 (43) 382 | 37 (46) 368 | 38 (43) 403 | 1 predicted TM domain, signal peptide | TATA box, DTAAG, M3, M6, M7 |
| 56 | 51761 > 52094 | 111 | 53 (88) 104 | 56 (57) 115 | TATA box, DTAAG, M7 | |||||
| 57 | lef-2 | 52095 > 52683 | 196 | 54 (80) 195 | 57 (37) 200 | 6 (16) 210 | 25 (11) 225 | TATA box, M4 | ||
| 58 | lef-5 | 52709 < 53417 | 236 | 55 (88) 235 | 58 (57) 230 | 99 (27) 265 | 69 (31) 247 | Zinc finger | TATA box, M9 | |
| 59 | 38K | 53407 > 54325 | 306 | 56 (88) 305 | 59 (59) 304 | 98 (31) 320 | 70 (31) 340 | 87 (25) 303 | IMP dehydrogenase/GMP reductase domain, putative phosphatase | Zeste, DTAAG |
| 60 | 54310 < 54817 | 169 | 57 (92) 168 | 60 (63) 170 | 96 (25) 173 | 71 (24) 161 | 90 (25) 202 | 1 predicted TM domain, signal peptide | TATA box, M5 | |
| 61 | helicase | 54803 > 58214 | 1,137 | 58 (89) 1134 | 61 (59) 1143 | 95 (18) 1221 | 72 (18) 1124 | 89 (12) 1332 | IMP dehydrogenase/GMP reductase domain, P-loop containing nucleoside triphosphate hydrolases | M7 |
| 62 | lef-4 | 58210 < 59617 | 469 | 59 (85) 468 | 62 (52) 477 | 90 (22) 464 | 78 (25) 432 | 96 (16) 497 | TATA box, M4, M6 | |
| 63 | 59633 > 60962 | 443 | 60 (89) 442 | 63 (52) 441 | 142 (20) 477 | TATA box, DTAAG, M3, M6 | ||||
| 64 | 60970 > 61210 | 80 | 61 (89) 78 | 64 (43) 78 | 1 predicted TM domain | TATA box, DTAAG, M4, M5 | ||||
| 65 | odv-e18 | 61285 > 61489 | 68 | 62 (95) 85 | 65 (83) 83 | 1 predicted TM domain, signal peptide, aspartate and glutamate racemases signature 1 | TATA box, DTAAG | |||
| 66 | odv-e27 | 61503 > 62292 | 263 | 63 (94) 262 | 66 (71) 260 | 144 (21) 290 | 80 (12) 287 | CAKT, DTAAG | ||
| 67 | 62319 > 62652 | 111 | 64 (87) 110 | 67 (70) 108 | 145 (20) 77 | 12 (27) 98 | 1 predicted TM domain, signal peptide, putative chitin-binding motif | TATA box, DTAAG | ||
| 68 | lef-1 | 62659 < 63295 | 212 | 65 (85) 211 | 68 (54) 190 | 14 (20) 266 | 55 (27) 251 | 45 (26) 235 | DNA primase | TATA box, CREB/ATF-47, M5 |
| 69 | 63352 > 63940 | 196 | 66 (89) 193 | 69 (63) 190 | 115 (24) 204 | 29 (28) 181 | 46 (27) 203 | 1 predicted TM domain, signal peptide | TATA box, Zeste, M9 | |
| 70 | 63939 > 64119 | 60 | 1 predicted TM domain, signal peptide | TATA box, Zeste, DTAAG, M6, M7 | ||||||
| 71 | 64093 > 65158 | 355 | 67 (86) 354 | 70 (57) 357 | 109 (21) 390 | 43 (20) 414 | 69 (17) 409 | TATA box, M3, M7, M9 | ||
| 72 | 65157 > 65367 | 70 | 68 (89) 69 | 71 (53) 71 | 1 predicted TM domain | TATA box, DTAAG | ||||
| 73 | rcc1 | 65353 > 65851 | 166 | 69 (80) 136 | 72 (60) 118 | RCC1 domain, 1 predicted TM domain | TATA box, M3, M5, M6 | |||
| 74 | rcc1 | 66214 > 66565 | 117 | 71 (71) 99 | 72 (7) 118 | RCC1 domain, 1 predicted TM domain, signal peptide | TATA box, M7 | |||
| 75 | 66996 > 67545 | 183 | 72 (64) 283 | 75 (39) 273 | TATA box, M7 | |||||
| 76 | 67599 > 67755 | 52 | TATA box | |||||||
| 77 | 67787 > 68585 | 266 | 74 (81) 265 | 77 (36) 284 | TATA box, M4, M7 | |||||
| 78 | 68962 > 69592 | 210 | 75 (79) 209 | 78 (27) 143 | TATA box, CREB/ATF-47, M9 | |||||
| 79 | pif | 69573 < 71187 | 538 | 76 (91) 530 | 79 (63) 519 | 119 (29) 530 | 7 (28) 536 | 29 (30) 523 | 1 predicted TM domain, signal peptide, zinc finger | Zeste, CREB/ATF-47, DTAAG, M1, M6, M9 |
| rep6 | 71014 > 72092 | |||||||||
| 80 | 71218 > 71485 | 89 | 35 (25) 97 | TATA box, CAKT, M7 | ||||||
| 81 | 71728 < 71899 | 57 | 2 (51) 108 | TATA box, M3, M6 | ||||||
| 82 | 71923 < 72379 | 152 | 77 (70) 149 | 80 (45) 156 | 28 (13) 284 | TATA box, CREB/ATF-47 | ||||
| 83 | lef-8 | 72377 > 74909 | 844 | 78 (87) 843 | 81 (69) 846 | 50 (30) 876 | 109 (32) 838 | 26 (20) 922 | Subunits of DNA dependent RNA polymerase | TATA box, Zeste, CAKT, M5 |
| rep7 | 74887 > 75140 | |||||||||
| 84 | 74926 > 75151 | 75 | 2 (28) 108 | TATA box | ||||||
| 85 | 75391 < 75556 | 55 | TATA box, DTAAG, M4, M8 | |||||||
| 86 | 75878 > 76430 | 184 | 81 (89) 148 | 83 (66) 145 | Capsid structural protein | TATA box, M1, M5, M8 | ||||
| 87 | vp91 | 76438 > 78844 | 802 | 82 (82) 803 | 84 (51) 824 | 83 (23) 847 | 84 (21) 533 | 35 (22) 741 | 1 predicted TM domain, signal peptide, IMP dehydrogenase/GMP reductase domain, chitin-binding peritrophin A domain | TATA box, Zeste, DTAAG, M4, M7, M9 |
| 88 | vp1054 | 78845 < 79787 | 314 | 83 (93) 313 | 85 (60) 310 | 54 (20) 365 | 115 (16) 311 | 8 (13) 329 | IMP dehydrogenase/GMP reductase domain, DNA polymerase beta, N terminals like | |
| 89 | 79912 > 80125 | 71 | 84 (81) 69 | 86 (58) 73 | ||||||
| 90 | 80132 > 81056 | 308 | 85 (83) 305 | 87 (38) 311 | TATA box, DTAAG, M6 | |||||
| 91 | 81100 > 82480 | 460 | 87 (54) 476 | 18 (11) 301 | TATA box, M7 | |||||
| 92 | vp39 | 82640 < 83588 | 316 | 88 (98) 315 | 89 (71) 312 | 89 (18) 347 | 79 (14) 320 | 24 (10) 289 | Putative RNA 2′-phosphotransferase | CAKT, M2, M5, M6 |
| 93 | 83597 > 84140 | 181 | 89 (80) 183 | 90 (43) 192 | Putative RNA 2′-phosphotransferase | TATA box, Zeste, DTAAG | ||||
The ORF number of a putative homologue is shown with the percent amino acid identity in parentheses, followed by the length of the homologue.
PxGV: Platella xylostella GV.
Promoter elements and abbreviations are defined in Table 2. A TATA box was defined as a TATA sequence within the −40 to −20-bp upstream region of the ORF or a TATAW sequence within 160 bp upstream of the ORF. CAKT was reported only when it occurred within the −40 to −20-bp upstream region of the ORF. All other elements were reported if they occurred within 160 bp upstream of the ORF.
Summary of BLAST, PSI-BLAST, RPS-BLAST, and INTERPRO SignalScan analyses.
NeabNPV gene content.
Twenty-nine genes are shared among all of the baculovirus genomes sequenced to date (26, 62). Putative homologues of all 29 of these conserved genes are found in NeabNPV. Of the 93 predicted NeabNPV ORFs, 72 have corresponding homologues in NeseNPV and 81 have corresponding homologues in NeleNPV. Eleven NeabNPV predicted ORFs are unique to NeabNPV (neab3, neab4, neab11, neab22, neab28, neab31, neab41, neab46, neab70, neab76, and neab85). Ten NeabNPV predicted ORFs (neab15, neab18, neab19, neab20, neab25, neab37, neab38, neab80, neab81, and neab84) have a putative sequence homologue in the NeleNPV genome but not in the NeseNPV genome, while one NeabNPV predicted ORF (neab53) has a putative sequence homologue in the NeseNPV genome but not in the NeleNPV genome.
Potential envelope fusion proteins in sawfly baculoviruses.
In Autographa californica multiple NPV (AcMNPV), the GP64 protein appears to be a critical protein for cell-to-cell transmission of the baculovirus budded virus, as gp64 deletion mutants were localized to insect midguts (10). In the Lymantria dispar NPV genome, where a gp64 homologue was not identified (58), the gene encoding a potential gp64 analog, ld130, was proposed and its Spodoptera exigua NPV sequence homologue was later shown to mediate pH-dependent membrane fusion between cells (50). An orthologue for gp64 was not identified in NeabNPV, NeleNPV, or NeseNPV (26, 62) by BLAST analysis. Sequence homology searches, however, may not be the most effective means of identifying analogs for envelope fusion proteins, as L. dispar NPV Ld130 does not have significant sequence similarity to AcMNPV GP64. According to the original criteria on the basis of which Ld130 was proposed as a potential envelope fusion protein (58), candidate protein analogs should possess both an N-terminal signal and a transmembrane (TM) domain.
Two NeabNPV ORFs, neab10 and neab44, possess these characteristics and may be candidate envelope fusion genes. InterProScan analysis (see Materials and Methods) of the predicted gene products indicated the presence of both an N-terminal signal and a TM domain. The putative sequence homologues of these two gene products in NeleNPV and NeseNPV also encode a potential N-terminal signal and a potential TM domain (26, 62). Alternatively, the sawfly baculoviruses may not encode an envelope fusion protein or produce a budded virus since for almost all known nonlepidopteran NPVs virus replication occurs only in the midgut epithelium (24).
NeabNPV repeats.
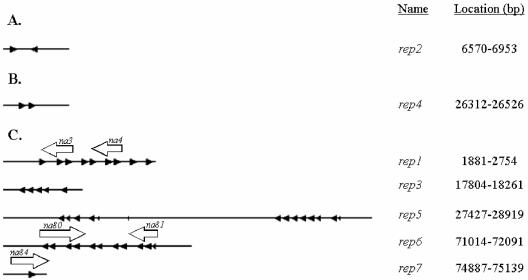
Two repeat regions (rep2 and rep4) each contain a cluster of localized repeat elements that are not interspersed throughout the genome (Fig. 2A and B). The remaining repeat clusters (rep1, rep3, rep5, rep6, and rep7), however, have a repetitive element in common with the consensus sequence 5′-CAACTTGTCAAATGTGTTGGACCTCGAGCCCAACAAACGCGACATCT-3′ that is interspersed at multiple loci throughout the genome (Fig. 2C and 3A). This core sequence is present in NeleNPV direct repeat 5 but does not occur in the NeseNPV genome. The loci known as homologous regions, hrs, of the lepidopteran-infecting NPVs have been found to function as enhancers of early gene transcription (30, 33) and origins of DNA replication (56, 80). The hrs of the lepidopteran-infecting NPVs have also been implicated as sites of DNA recombination (49). In NeabNPV, the location of this repeat element appears to coincide with major rearrangement events between the NeleNPV and NeabNPV genomes (Fig. 3B).
FIG. 2.
Loci of the NeabNPV genome with repeating elements. The triangles represent the repeated core element in the direction indicated by the point. The name and genome location of the repeat region are indicated on the right, and the repeat element consensus sequences are as follows: A, ATAAAAAACAGTAAATATT(C/T)CAATACGATGC AAACGCACGTGATTAATGT; B, TCAG(A/C)ATTGTCGTTGTTGTTTT(G/C)AG TGTTTTCTGT(G/A)TTATTTC; C, AGATGTCGCGTTTGTTGGGCTCGAGGT CCAACACATTTGACAAGTTG. White arrows indicate the orientation and location of NeabNPV ORFs depicted in Fig. 4.
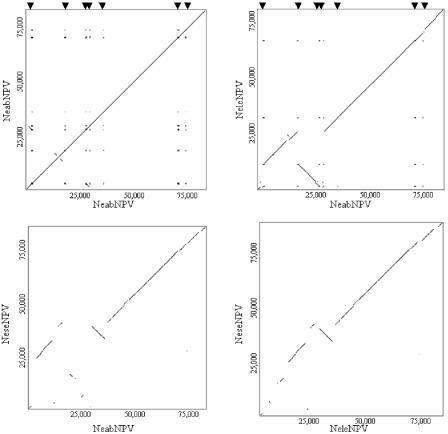
FIG. 3.
Genome parity plots. Genome plots were derived by comparing (A) NeabNPV against itself, (B) NeabNPV against NeleNPV, (C) NeabNPV against NeseNPV, and (D) NeleNPV against NeseNPV. The axes are labeled with the genome name and the distance (in base pairs). The interspersed repeat sites of NeabNPV are labeled on plots A and B. Triangles above plots A and B represent the locations of interspersed repeats 1 to 7.
Repeated genes.
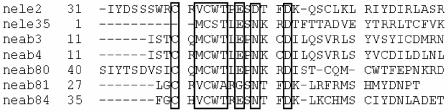
Lepidopteran baculovirus genomes, except Spodoptera exigua NPV (51), Plutella xylostella GV (39), and A. orana GV (100), commonly encode members of a family of repeated genes known as bro genes. These genes are typically repeated elements of the baculovirus genomes, and it has been proposed that bro gene products are involved in DNA replication and transcription (105), as well as in maximizing polyhedron formation in specific host species (8). Outside of the family Baculoviridae, bro and bro-like genes have been found in ascoviruses, insect iridoviruses, entomopoxviruses (ALI gene family), the phycodnavirus ectocarpus siliculosus virus, bacteriophages, and prophages integrated into bacterial genomes (2, 7, 8, 53). Despite the widespread occurrence of bro genes, no homologues of these genes were found in NeabNPV or the other sawfly baculoviruses. Five ORFs unique to NeabNPV and NeleNPV (neab3, neab4, neab80, neab81, and neab84), however, have in common an amino acid motif duplicated in two NeleNPV ORFs (Fig. 4) and are proximal to interspersed genome repeats (rep1 and rep7). These repeated amino acid motifs are shared in two genes in NeleNPV but were not previously reported (62). Duplicated genes flanked by repeated regions were reported in NeseNPV (26), but they do not share the amino acid motifs of the NeabNPV and NeleNPV repeated genes.
FIG. 4.
Alignment of conserved motifs repeated in multiple predicted ORFs of the NeabNPV and NeleNPV genomes. Gaps in the alignment are indicated by dashes, and conserved amino acids are outlined.
The G+C composition of the putative NeabNPV ORFs within the repeat regions contrasts significantly with the genome average. While the predicted coding sequences of the NeabNPV genome had a mean G+C content of 34% ± 4%, eight putative ORFs had a G+C content of greater than 40%, including four ORFs within the repeated segments, neab3 (45% G+C), neab4 (47% G+C), neab80 (44% G+C), and neab81 (46% G+C). In bacteria, the G+C content is homogeneous throughout genomes and heterogeneous between genomes (73). Furthermore, genome G+C content is related to phylogeny (78) and has been used to predict genes that have been acquired by horizontal gene transfer (27, 28, 74). As a result, G+C content is often used as a genome signature and can be used to compare baculovirus genomes. Given that the base composition of the repeat region sequences contrasts so greatly with the average of the sawfly baculovirus genomes and that the repeat regions are found in NeabNPV and NeleNPV but not in NeseNPV, it is possible that a common ancestor of NeabNPV and NeleNPV acquired those elements by horizontal transfer following its divergence from NeseNPV.
NeabNPV evolution.
Sawfly baculoviruses represent a distinct taxon of the virus family Baculoviridae (26, 62). Nucleotide sequence plots revealed greater extended colinearity between NeabNPV and NeleNPV than between NeabNPV and NeseNPV (Fig. 3B and C). Furthermore, NeabNPV and NeleNPV share the same conserved loci compared to NeseNPV (Fig. 3C and D). Although all three host species originally inhabited North America, N. sertifer is believed to have migrated via the Bering Land Bridge to Eurasia. Following this migration, N. sertifer became extinct in the New World only to be reintroduced in ca. 1925 (67). This geographic isolation of host species is supported by the viral DNA sequences and is consistent with the divergence of NeseNPV from NeabNPV and NeleNPV.
The parity plots (Fig. 3) illustrate that the region of the genome from 37 to 84 kb (neab41 to neab93), relative to the polyhedrin gene, holds more colinearity than the preceding region. Among the first 40 ORFs in NeabNPV, only 67.5% have predicted sequence homologues in NeseNPV. Among ORFs 41 to 93, however, 87% have predicted sequence homologues. Similarly, among the first 40 ORFs, only 6 (0.15%) are conserved in all of the sequenced baculovirus genomes, whereas among ORFs 41 to 93, 23 (43.4%) are conserved in all of the sequenced baculovirus genomes.
Genome recombination is a mechanism for host range expansion in baculoviruses (55). Arends and Jehle (4) noted that inversions of genome loci are often flanked by hrs in the lepidopteran baculoviruses. The repeat element in the NeabNPV and NeleNPV genomes appears to be analogous to the hrs in that its expansion and interspersal may promote recombination between repeats. Given the role of genome recombination in host range expansion, the repeat sequences may have contributed significantly to the evolution of NeabNPV and NeleNPV following their ancestral divergence from NeseNPV.
Promoter analysis.
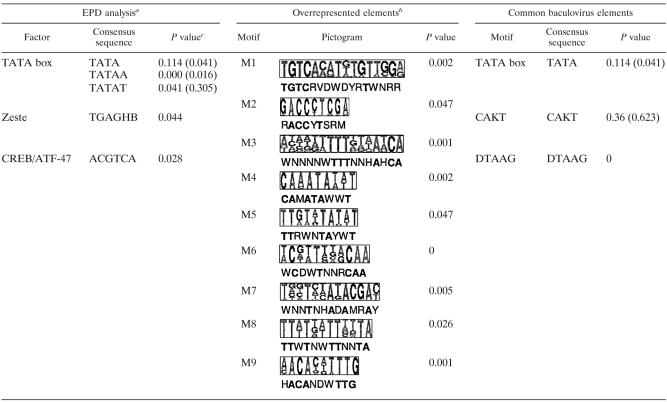
Gene expression of lepidopteran-infecting baculoviruses is temporally regulated, with four distinct stages, immediate-early, early, late, and very late. Immediate-early and delayed-early genes are expressed before DNA replication and are transcribed by insect host RNA polymerase II (25, 46). These early genes may encode promoter elements such as the TATA element that acts as an assembly site for the host RNA polymerase II transcription complex (32, 83) and the CAKT sequence element that enhances gene expression and serves as a transcription start site in early-expressed baculovirus genes (9, 83).
Putative TATA elements were identified upstream of 69 NeabNPV ORFs, and CAKT elements were identified in the −40- to −20-bp region upstream of 12 ORFs. On the basis of the lepidopteran baculovirus model, ORFs with both elements should represent early-expressed genes. Seven ORFs with putative TATA box motifs also have putative CAKT motifs. Among these are suspected homologues of the viral RNA polymerase subunit gene lef-8 (31), lef-11, and vlf-1 and four uncharacterized ORFs. CAKT was also identified in six predicted TATA-less promoters, including the early-expressed dnapol (16, 47, 65, 93) and p47 (15, 61) genes, as well as the late-expressed odv-e27 (12) and vp39 genes (11, 41, 92), and two uncharacterized ORFs. The occurrence of TATA elements was significantly more frequent in the regions upstream of the predicted ORFs than in the whole genome, suggesting that they are general transcription motifs. The CAKT sequence was not significantly overrepresented either 160 bp upstream or in the −40- to −20-bp upstream regions, relative to the predicted translation start sites of NeabNPV ORFs. This may be due to the fact that the early genes would represent a small subset of viral genes and this element would not be a general transcription element. Alternatively, the absence of lepidopteran baculovirus immediate-early gene homologues in NeabNPV might suggest that sawfly baculoviruses utilize an alternative mechanism for early gene regulation.
Late gene expression in lepidopteran baculoviruses is associated with a DTAAG sequence motif (72). The DTAAG promoter element acts as the initiation site for transcription mediated by the virus-encoded RNA polymerase, and the strength of expression from this site depends on the context of the sequence between it and the translational initiation site (68, 77, 82, 83, 97). The DTAAG motif was a common element in NeabNPV and was overrepresented in the regions 160 bp upstream of the predicted ORF translation initiation sites.
Among the predicted NeabNPV ORFs that have well-characterized putative sequence homologues in other baculoviruses, there is little evidence that the presence of these three promoter elements is indicative of the temporal expression of the genes. From a genome analysis perspective, the DTAAG sequence occurred in NeabNPV early genes as often as in late genes. However, it is likely that this site acts as an alternate start site for later transcripts. Whether the viral RNA polymerase of sawfly baculoviruses utilizes a DTAAG motif sequence as a transcription start site remains to be experimentally verified.
In order to explore alternative approaches to baculovirus promoter analysis, we examined two features. The first approach involved determining whether there was a general overrepresentation of host transcription factor binding sites upstream of NeabNPV ORFs and whether the occurrence of these binding sites is biased toward genes within a similar temporal group. We hypothesized that early baculovirus promoters would rely on host transcription factors to a greater degree than later genes, as they utilize the host transcription mechanism.
The second involved using AlignACE (48) to perform multiple alignments to identify conserved motifs. This approach successfully identified core promoter elements in Drosophila (75), and we hypothesized that it would identify overrepresented conserved promoter motifs in the NeabNPV genome. In particular, we were interested in whether the multiple-alignment method would identify core promoter elements likely to be employed by the insect host cell transcriptional apparatus or common baculovirus elements.
To address the first question regarding the utilization of known host transcription factor binding sites, the upstream sequences were queried against the TRANSFAC database. Three experimentally confirmed insect promoter elements were overrepresented in the upstream sequences of predicted NeabNPV ORFs. One was the TATA box, a core promoter element common in insect and baculovirus genomes. Another was the motif TGAGHB, which corresponds to the DNA binding motif for the Zeste protein.
Zeste is a transcription factor that facilitates transvection, the mechanism by which a gene can trans regulate the expression of a homologue (21, 54, 102). It has been observed that AcMNPV hr1 can act as an enhancer in trans in mammalian cells (95). Although the hr1 region did not permit transvection of genes encoded by host insect genomes, it is possible that hymenopteran baculovirus genes could utilize Zeste-mediated trans regulation. The zeste promoter motif has been identified upstream of seven genes with a bias toward early-expressed genes, including dnapol (16, 47, 65, 93), iap-3 (14, 52), p40 (66), p47 (15, 61), and 38K(pp31) (91). Two genes with this sequence upstream, pif and vp91, are expressed late in lepidopteran NPVs (34, 87, 90). zeste promoter motifs were identified in the upstream sequences of a further 13 predicted NeabNPV ORFs.
A CREB/ATF47 binding site (known as the cis-acting replication element) mediates transcription in response to members of the CREB/ATF protein family. This is a large family of leucine zipper transcription factors (38) that, despite their diverse activities, have in common the abilities to respond to environmental signals and maintain cellular homeostasis. Some ATF proteins (ATF2, ATF3, and ATF6) play a role in mediating stress response (37), CREB and ATF1 regulate transcription in response to intracellular cyclic AMP concentrations (36), and ATF4 acts as a negative regulator of cis-acting replication element-dependent transcription (20). ATF proteins are important elements in regulating early transcription of members of the family Adenoviridae. The adenovirus E1A protein interacts with host-encoded ATF-2 to activate transcription of several adenovirus early genes (63). In the NeabNPV genome, the CREB/ATF binding sites were observed upstream of both the putative early-expressed genes dnapol (16, 47, 65, 93) and lef-1 (79), as well as the putatively late-expressed gene pif (34, 90). The remaining six ORFs with upstream CREB/ATF binding sites had no characterized homologues.
By the second approach, alignment of upstream sequences identified nine sequences that we designated M1 to M9 (Table 1 and Table 2). Contrary to our prediction, promoter alignment did not report the core Drosophila promoter elements or the known baculovirus promoter elements. Within these overrepresented conserved motifs, however, certain spatial and temporal patterns were observed.
TABLE 2.
Predicted putative promoter elements for NeabNPV
Transcription factors derived from SignalScan analysis of the eukaryote promoter database (see Materials and Methods). Only those that occur significantly more often in the upstream sequence than in the whole genome were reported.
AlignAce analysis of upstream sequences identified a number of putative common elements. Only those that occur significantly more often in the upstream sequence than in the whole genome were reported.
The P value represents a statistical test that the element occurs more often −160 bp upstream of predicted ORFs than in the whole NeabNPV genome. The value in parentheses represents a statistical test that the element occurs more often in the −40 to −20-bp region upstream of predicted ORFs than in the whole NeabNPV genome.
The M1 motif was not overrepresented in genes known to be temporally distinct but rather was clustered spatially. Of the 10 occurrences of this motif, 8 are within a 30-kb region (747 to 29,973 bp) within the more variable half of the circular NeabNPV genome. Similarly, five of six ORFs with the upstream M2 motif were within an approximately 5-kb region (bp 82,641 to 84,264 and bp 0 to 3,558). The M8 motif was upstream of the late-expressed polh (17, 18, 86, 94) and gp41 (64, 98) genes, as well as five other putative ORFs. Five of the seven ORFs with an upstream M8 motif also possess putative TATA and DTAAG motifs. Motifs M2 and M8 occurred in only six and seven upstream sequences, respectively, but these still occur there significantly more often than in the whole genome. Motif M3 occurred upstream of 10 predicted ORFs, including p33, trypsin-like, lef-9, p74, pif-2, and rcc1. There is no available temporal characterization for all of these genes except lef-9 and p74, which are both expressed late (1, 31, 59, 85). The other motifs (M4, M5, M6, and M7) did not appear to have characteristic clustering or temporal properties.
Our hypothesis that host transcription factor binding sites might be enriched in the promoters of early-expressed baculovirus genes appears to hold for the Zeste protein binding site but not for the CREB/ATF47 binding site. Conversely, motifs characterized by promoter alignment with AlignACE did not correspond to temporally grouped genes; however, the spatial clustering of genes with these motifs is intriguing. These patterns and hypotheses require experimental confirmation and emphasize the need for more sophisticated analyses of baculovirus promoters.
Concluding remarks.
Although this study represents the third sawfly baculovirus genome to be completely sequenced, our analysis conforms to previous observations that the sawfly baculoviruses represent a distinct taxon (26, 62); our study suggests a possible mechanism for sawfly baculovirus evolution through genome rearrangement events between interspersed genome repeats. Rearrangement between these repeats may be similar to the genome arrangements described between the repeated lepidopteran hrs. Although not as well conserved as the lepidopteran baculovirus bro genes, predicted genes encoded within the NeabNPV repeats have an amino acid motif in common.
Our comparison of gene content and genome rearrangement agrees with the hypothesis proposed by Herniou et al. (42) that baculoviruses coevolve with their host. We observed that the geographically isolated Old World baculovirus NeseNPV has diverged more from the New World sawfly baculoviruses NeabNPV and NeleNPV.
The intriguing absence of immediate-early gene homologues and membrane fusion proteins in NeabNPV is consistent with previous studies of nonlepidopteran baculoviruses (2, 26, 62) and raises interesting questions regarding how early gene expression is mediated and how the sawfly baculovirus infection process differs from that of the lepidopteran baculoviruses. These questions, along with the renewed interest in sawfly baculoviruses in pest management, provide us with an opportunity to examine the unique and exciting characteristics of the baculoviruses outside the order Lepidoptera.
Acknowledgments
This work was supported by grants from the Natural Sciences and Engineering Research Council (NSERC) through an Industrial Research Partnership with Forest Protection Limited (FPL) in Fredericton, New Brunswick, Canada, the NSERC Biocontrol Network, and through a grant from Natural Resources Canada and the Canadian Forest Service.
We thank John Taylor for comments on and corrections to the manuscript.
REFERENCES
- 1.Acharya, A., and K. P. Gopinathan. 2002. Characterization of late gene expression factors lef-9 and lef-8 from Bombyx mori nucleopolyhedrovirus. J. Gen. Virol. 83:2015-2023. [DOI] [PubMed] [Google Scholar]
- 2.Afonso, C. L., E. R. Tulman, Z. Lu, C. A. Balinsky, B. A. Moser, J. J. Becnel, D. L. Rock, and G. F. Kutish. 2001. Genome sequence of a baculovirus pathogenic for Culex nigripalpus. J. Virol. 75:11157-11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Arends, H. M., and J. A. Jehle. 2002. Homologous recombination between the inverted terminal repeats of defective transposon TCp3.2 causes an inversion in the genome of Cydia pomonella granulovirus. J. Gen. Virol. 83:1573-1578. [DOI] [PubMed] [Google Scholar]
- 5.Attwood, T. K., M. D. Croning, D. R. Flower, A. P. Lewis, J. E. Mabey, P. Scordis, J. N. Selley, and W. Wright. 2000. PRINTS-S: the database formerly known as PRINTS. Nucleic Acids Res. 28:225-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bawden, A. L., K. J. Glassberg, J. Diggans, R. Shaw, W. Farmerie, and R. W. Moyer. 2000. Complete genomic sequence of the Amsacta moorei entomopoxvirus: analysis and comparison with other poxviruses. Virology 274:120-139. [DOI] [PubMed] [Google Scholar]
- 8.Bideshi, D. K., S. Renault, K. Stasiak, B. A. Federici, and Y. Bigot. 2003. Phylogenetic analysis and possible function of bro-like genes, a multigene family widespread among large double-stranded DNA viruses of invertebrates and bacteria. J. Gen. Virol. 84:2531-2544. [DOI] [PubMed] [Google Scholar]
- 9.Blissard, G. W., P. H. Kogan, R. Wei, and G. F. Rohrmann. 1992. A synthetic early promoter from a baculovirus: roles of the TATA box and conserved start site CAGT sequence in basal levels of transcription. Virology 190:783-793. [DOI] [PubMed] [Google Scholar]
- 10.Blissard, G. W., and J. R. Wenz. 1992. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 66:6829-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blissard, G. W., R. L. Quant-Russell, G. F. Rohrmann, and G. S. Beaudreau. 1989. Nucleotide sequence, transcriptional mapping, and temporal expression of the gene encoding p39, a major structural protein of the multicapsid nuclear polyhedrosis virus of Orgyia pseudotsugata. Virology 168:354-362. [DOI] [PubMed] [Google Scholar]
- 12.Braunagel, S. C., H. He, P. Ramamurthy, and M. D. Summers. 1996. Transcription, translation, and cellular localization of three Autographa californica nuclear polyhedrosis virus structural proteins: ODV-E18, ODV-E35, and ODV-EC27. Virology 222:100-114. [DOI] [PubMed] [Google Scholar]
- 13.Buchan, D. W., A. J. Shepherd, D. Lee, F. M. Pearl, S. C. Rison, J. M. Thornton, and C. A. Orengo. 2002. Gene3D: structural assignment for whole genes and genomes using the CATH domain structure database. Genome Res. 12:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carpes, M. P., M. E. de Castro, E. F. Soares, A. G. Villela, F. J. Pinedo, and B. M. Ribeiro. 2005. The inhibitor of apoptosis gene (iap-3) of Anticarsia gemmatalis multicapsid nucleopolyhedrovirus (AgMNPV) encodes a functional IAP. Arch. Virol. 150:1549-1562. [DOI] [PubMed] [Google Scholar]
- 15.Carstens, E. B., A. L. Lu, and H. L. Chan. 1993. Sequence, transcriptional mapping, and overexpression of p47, a baculovirus gene regulating late gene expression. J. Virol. 67:2513-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaeychomsri, S., M. Ikeda, and M. Kobayashi. 1995. Nucleotide sequence and transcriptional analysis of the DNA polymerase gene of Bombyx mori nuclear polyhedrosis virus. Virology 206:435-447. [DOI] [PubMed] [Google Scholar]
- 17.Choi, J. Y., Y. L. Kim, and J. M. Yang. 1998. Molecular cloning and analysis of transcription initiation in the Anagrapha falcifera multiple nucleocapsid polyhedrosis virus polyhedrin gene. Mol. Cells 8:537-543. [PubMed] [Google Scholar]
- 18.Chou, C. M., C. J. Huang, C. F. Lo, G. H. Kou, and C. H. Wang. 1996. Characterization of Perina nuda nucleopolyhedrovirus (PenuNPV) polyhedrin gene. J. Invertebr. Pathol. 67:259-266. [DOI] [PubMed] [Google Scholar]
- 19.Corpet, F., J. Gouzy, and D. Kahn. 1999. Recent improvements of the ProDom database of protein domain families. Nucleic Acids Res. 27:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Cesare, D., and P. Sassone-Corsi. 2000. Transcriptional regulation by cyclic AMP-responsive factors. Prog. Nucleic Acid Res. Mol. Biol. 64:343-369. [DOI] [PubMed] [Google Scholar]
- 21.Duncan, I. W. 2002. Transvection effects in Drosophila. Annu. Rev. Genet. 36:521-556. [DOI] [PubMed] [Google Scholar]
- 22.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 23.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 24.Federici, B. A. 1993. Viral pathobiology in relation to insect control, p. 81-101. In N. Beckage, S. N. Thompson, and B. A. Federici (ed.), Pathogens, vol. 2. Academic Press, San Diego, Calif. [Google Scholar]
- 25.Fuchs, L. Y., M. S. Woods, and R. F. Weaver. 1983. Viral transcription during Autographa californica nuclear polyhedrosis virus infection: a novel RNA polymerase induced in infected Spodoptera frugiperda cells. J. Virol. 48:641-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Maruniak, A., J. E. Maruniak, P. M. Zanotto, A. E. Doumbouya, J. C. Liu, T. M. Merritt, and J. S. Lanoie. 2004. Sequence analysis of the genome of the Neodiprion sertifer nucleopolyhedrovirus. J. Virol. 78:7036-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Vallve, S., A. Romeu, and J. Palau. 2000. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 10:1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Vallve, S., A. Romeu, and J. Palau. 2000. Horizontal gene transfer of glycosyl hydrolases of the rumen fungi. Mol. Biol. Evol. 17:352-361. [DOI] [PubMed] [Google Scholar]
- 29.Gough, J., K. Karplus, R. Hughey, and C. Chothia. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313:903-919. [DOI] [PubMed] [Google Scholar]
- 30.Guarino, L. A., and M. D. Summers. 1986. Interspersed homologous DNA of Autographa californica nuclear polyhedrosis virus enhances delayed-early gene expression. J. Virol. 60:215-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guarino, L. A., B. Xu, J. Jin, and W. Dong. 1998. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J. Virol. 72:7985-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guarino, L. A., and M. Smith. 1992. Regulation of delayed-early gene transcription by dual TATA boxes. J. Virol. 66:3733-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guarino, L. A., M. A. Gonzalez, and M. D. Summers. 1986. Complete sequence and enhancer function of the homologous DNA regions of Autographa californica nuclear polyhedrosis virus. J. Virol. 60:224-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gutierrez, S., I. Kikhno, and M. Lopez Ferber. 2004. Transcription and promoter analysis of pif, an essential but low-expressed baculovirus gene. J. Gen. Virol. 85:331-341. [DOI] [PubMed] [Google Scholar]
- 35.Haft, D. H., B. J. Loftus, D. L. Richardson, F. Yang, J. A. Eisen, I. T. Paulsen, and O. White. 2001. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res. 29:41-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hai, T., and M. G. Hartman. 2001. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 273:1-11. [DOI] [PubMed] [Google Scholar]
- 37.Hai, T., C. D. Wolfgang, D. K. Marsee, A. E. Allen, and U. Sivaprasad. 1999. ATF3 and stress responses. Gene Expr. 7:321-335. [PMC free article] [PubMed] [Google Scholar]
- 38.Hai, T. W., F. Liu, W. J. Coukos, and M. R. Green. 1989. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 3:2083-2090. [DOI] [PubMed] [Google Scholar]
- 39.Hashimoto, Y., T. Hayakawa, Y. Ueno, T. Fujita, Y. Sano, and T. Matsumoto. 2000. Sequence analysis of the Plutella xylostella granulovirus genome. Virology 275:358-372. [DOI] [PubMed] [Google Scholar]
- 40.Hayakawa, T., R. Ko, K. Okano, S. I. Seong, C. Goto, and S. Maeda. 1999. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology 262:277-297. [DOI] [PubMed] [Google Scholar]
- 41.Heldens, J. G., Y. Liu, D. Zuidema, R. W. Goldbach, and J. M. Vlak. 1998. A highly conserved genomic region in baculoviruses: sequence analysis of an 11.3 kbp DNA fragment (46.5-55.1 m.u.) of the Spodoptera exigua multicapsid nucleopolyhedrovirus. Virus Res. 55:187-198. [DOI] [PubMed] [Google Scholar]
- 42.Herniou, E. A., J. A. Olszewski, D. R. O'Reilly, and J. S. Cory. 2004. Ancient coevolution of baculoviruses and their insect hosts. J. Virol. 78:3244-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herniou, E. A., J. A. Olszewski, J. S. Cory, and D. R. O'Reilly. 2003. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 48:211-234. [DOI] [PubMed] [Google Scholar]
- 44.Herniou, E. A., T. Luque, X. Chen, J. M. Vlak, D. Winstanley, J. S. Cory, and D. R. O'Reilly. 2001. Use of whole genome sequence data to infer baculovirus phylogeny. J. Virol. 75:8117-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofmann, K., P. Bucher, L. Falquet, and A. Bairoch. 1999. The PROSITE database, its status in 1999. Nucleic Acids Res. 27:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoopes, R. R., and G. F. Rohrmann. 1991. In vitro transcription of baculovirus immediate early genes: accurate mRNA initiation by nuclear extracts from both insect and human cells. Proc. Natl. Acad. Sci. USA 88:4513-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang, J., and D. B. Levin. 2001. Identification, transcription and sequence analysis of the Spodoptera littoralis nucleopolyhedrovirus (SpliNPV) DNA polymerase gene. Arch. Virol. 146:303-326. [DOI] [PubMed] [Google Scholar]
- 48.Hughes, J. D., P. W. Estep, S. Tavazoie, and G. M. Church. 2000. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 296:1205-1214. [DOI] [PubMed] [Google Scholar]
- 49.Hyink, O., R. A. Dellow, M. J. Olsen, K. M. Caradoc-Davies, K. Drake, E. A. Herniou, J. S. Cory, D. R. O'Reilly, and V. K. Ward. 2002. Whole genome analysis of the Epiphyas postvittana nucleopolyhedrovirus. J. Gen. Virol. 83:957-971. [DOI] [PubMed] [Google Scholar]
- 50.IJkel, W. F., M. Westenberg, R. W. Goldbach, G. W. Blissard, J. M. Vlak, and D. Zuidema. 2000. A novel baculovirus envelope fusion protein with a proprotein convertase cleavage site. Virology 275:30-41. [DOI] [PubMed] [Google Scholar]
- 51.IJkel, W. F., E. A. van Strien, J. G. Heldens, R. Broer, D. Zuidema, R. W. Goldbach, and J. M. Vlak. 1999. Sequence and organization of the Spodoptera exigua multicapsid nucleopolyhedrovirus genome. J. Gen. Virol. 80(Pt. 12):3289-3304. [DOI] [PubMed] [Google Scholar]
- 52.Ikeda, M., K. Yanagimoto, and M. Kobayashi. 2004. Identification and functional analysis of Hyphantria cunea nucleopolyhedrovirus iap genes. Virology 321:359-371. [DOI] [PubMed] [Google Scholar]
- 53.Jakob, N. J., K. Muller, U. Bahr, and G. Darai. 2001. Analysis of the first complete DNA sequence of an invertebrate iridovirus: coding strategy of the genome of Chilo iridescent virus. Virology 286:182-196. [DOI] [PubMed] [Google Scholar]
- 54.Kennison, J. A., and J. W. Southworth. 2002. Transvection in Drosophila. Adv. Genet. 46:399-420. [DOI] [PubMed] [Google Scholar]
- 55.Kondo, A., and S. Maeda. 1991. Host range expansion by recombination of the baculoviruses Bombyx mori nuclear polyhedrosis virus and Autographa californica nuclear polyhedrosis virus. J. Virol. 65:3625-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kool, M., J. T. Voeten, R. W. Goldbach, J. Tramper, and J. M. Vlak. 1993. Identification of seven putative origins of Autographa californica multiple nucleocapsid nuclear polyhedrosis virus DNA replication. J. Gen. Virol. 74:2661-2668. [DOI] [PubMed] [Google Scholar]
- 57.Kurtz, S., J. V. Choudhuri, E. Ohlebusch, C. Schleiermacher, J. Stoye, and R. Giegerich. 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29:4633-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuzio, J., M. N. Pearson, S. H. Harwood, C. J. Funk, J. T. Evans, J. M. Slavicek, and G. F. Rohrmann. 1999. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology 253:17-34. [DOI] [PubMed] [Google Scholar]
- 59.Kuzio, J., R. Jaques, and P. Faulkner. 1989. Identification of p74, a gene essential for virulence of baculovirus occlusion bodies. Virology 173:759-763. [DOI] [PubMed] [Google Scholar]
- 60.Lange, M., and J. A. Jehle. 2003. The genome of the Cryptophlebia leucotreta granulovirus. Virology 317:220-236. [DOI] [PubMed] [Google Scholar]
- 61.Lapointe, R., D. W. Back, Q. Ding, and E. B. Carstens. 2000. Identification and molecular characterization of the Choristoneura fumiferana multicapsid nucleopolyhedrovirus genomic region encoding the regulatory genes pkip, p47, lef-12, and gta. Virology 271:109-121. [DOI] [PubMed] [Google Scholar]
- 62.Lauzon, H. A., C. J. Lucarotti, P. J. Krell, Q. Feng, A. Retnakaran, and B. M. Arif. 2004. Sequence and organization of the Neodiprion lecontei nucleopolyhedrovirus genome. J. Virol. 78:7023-7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu, F., and M. R. Green. 1990. A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1a protein. Cell 61:1217-1224. [DOI] [PubMed] [Google Scholar]
- 64.Liu, J. C., and J. E. Maruniak. 1995. Nucleotide sequence and transcriptional analysis of the gp41 gene of Spodoptera frugiperda nuclear polyhedrosis virus. J. Gen. Virol. 76:1443-1450. [DOI] [PubMed] [Google Scholar]
- 65.Liu, J. J., and E. B. Carstens. 1995. Identification, localization, transcription, and sequence analysis of the Choristoneura fumiferana nuclear polyhedrosis virus DNA polymerase gene. Virology 209:538-549. [DOI] [PubMed] [Google Scholar]
- 66.Lu, A., A. Craig, R. Casselman, and E. B. Carstens. 1996. Nucleotide sequence, insertional mutagenesis, and transcriptional mapping of a conserved region of the baculovirus Autographa californica nuclear polyhedrosis virus (map unit 64.8-66.9). Can. J. Microbiol. 42:1267-1273. [DOI] [PubMed] [Google Scholar]
- 67.Lyons, L. A. 1964. The European pine sawfly, Neodiprion sertifer (Geoffr.) (Hymenoptera: Diprionidae). A review with emphasis on studies in Ontario. Proc. Entomol. Soc. Ont. 94:5-37. [Google Scholar]
- 68.Mans, R. M., and D. Knebel-Morsdorf. 1998. In vitro transcription of pe38/polyhedrin hybrid promoters reveals sequences essential for recognition by the baculovirus-induced RNA polymerase and for the strength of very late viral promoters. J. Virol. 72:2991-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mi, H., B. Lazareva-Ulitsky, R. Loo, A. Kejariwal, J. Vandergriff, S. Rabkin, N. Guo, A. Muruganujan, O. Doremieux, M. J. Campbell, H. Kitano, and P. D. Thomas. 2005. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 33:D284-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moreau, G., C. J. Lucarotti, E. G. Kettela, G. S. Thurston, S. Holmes, C. Weaver, D. B. Levin, and B. Morin. 2005. Aerial application of nucleopolyhedrovirus induces decline in increasing and peaking populations of Neodiprion abietis. Biol. Control 33:65-73. [Google Scholar]
- 72.Morris, T. D., and L. K. Miller. 1994. Mutational analysis of a baculovirus major late promoter. Gene 140:147-153. [DOI] [PubMed] [Google Scholar]
- 73.Muto, A., and S. Osawa. 1987. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl. Acad. Sci. USA 84:166-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ochman, H., E. Lerat, and V. Daubin. 2005. Examining bacterial species under the specter of gene transfer and exchange. Proc. Natl. Acad. Sci. USA 102:6595-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ohler, U., G. C. Liao, H. Niemann, and G. M. Rubin. 2002. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3. [Online.] http://genomebiology.com/2002/3/12/RESEARCH/0087. [DOI] [PMC free article] [PubMed]
- 76.Olofsson, E. 1973. Evaluation of a nuclear polyhedrosis virus as an agent for the control of the balsam fir sawfly, Neodiprion abietis Harr. Information report IP-X-2. Insect Pathology Research Institute, Department of the Environment, Canadian Forestry Service, Sault Ste. Marie, Ontario, Canada.
- 77.Ooi, B. G., C. Rankin, and L. K. Miller. 1989. Downstream sequences augment transcription from the essential initiation site of a baculovirus polyhedrin gene. J. Mol. Biol. 210:721-736. [DOI] [PubMed] [Google Scholar]
- 78.Osawa, S., T. H. Jukes, K. Watanabe, and A. Muto. 1992. Recent evidence for evolution of the genetic code. Microbiol. Rev. 56:229-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Passarelli, A. L., and L. K. Miller. 1993. Identification and characterization of lef-1, a baculovirus gene involved in late and very late gene expression. J. Virol. 67:3481-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pearson, M., R. Bjornson, G. Pearson, and G. Rohrmann. 1992. The Autographa californica baculovirus genome: evidence for multiple replication origins. Science 257:1382-1384. [DOI] [PubMed] [Google Scholar]
- 81.Piene, H., D. Ostaff, and E. Eveleigh. 2001. Growth loss and recovery following defoliation by the balsam fir sawfly in young, spaced balsam fir stands. Can. Entomol. 133:675-686. [Google Scholar]
- 82.Possee, R. D., and S. C. Howard. 1987. Analysis of the polyhedrin gene promoter of the Autographa californica nuclear polyhedrosis virus. Nucleic Acids Res. 15:10233-10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pullen, S. S., and P. D. Friesen. 1995. Early transcription of the ie-1 transregulator gene of Autographa californica nuclear polyhedrosis virus is regulated by DNA sequences within its 5′ noncoding leader region. J. Virol. 69:156-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pullen, S. S., and P. D. Friesen. 1995. The CAGT motif functions as an initiator element during early transcription of the baculovirus transregulator ie-1. J. Virol. 69:3575-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rashidan, K. K., N. Nassoury, P. N. Giannopoulos, and C. Guertin. 2005. Transcription, translation, and immunolocalization of ODVP-6E/ODV-E56 and p74 proteins: two highly conserved ODV-associated envelope proteins of Choristoneura fumiferana granulovirus. J. Biochem. Mol. Biol. 38:65-70. [DOI] [PubMed] [Google Scholar]
- 86.Rohel, D. Z., and P. Faulkner. 1984. Time course analysis and mapping of Autographa californica nuclear polyhedrosis virus transcripts. J. Virol. 50:739-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Russell, R. L., and G. F. Rohrmann. 1997. Characterization of P91, a protein associated with virions of an Orgyia pseudotsugata baculovirus. Virology 233:210-223. [DOI] [PubMed] [Google Scholar]
- 88.Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schwartz, S., Z. Zhang, K. A. Frazer, A. Smit, C. Riemer, J. Bouck, R. Gibbs, R. Hardison, and W. Miller. 2000. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 10:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Simon, O., S. Gutierrez, T. Williams, P. Caballero, and M. Lopez-Ferber. 2005. Nucleotide sequence and transcriptional analysis of the pif gene of Spodoptera frugiperda nucleopolyhedrovirus (SfMNPV). Virus Res. 108:213-220. [DOI] [PubMed] [Google Scholar]
- 91.Smith, G. E., J. M. Vlak, and M. D. Summers. 1982. In vitro translation of Autographa californica nuclear polyhedrosis virus early and late mRNAs. J. Virol. 44:199-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thiem, S. M., and L. K. Miller. 1989. Identification, sequence, and transcriptional mapping of the major capsid protein gene of the baculovirus Autographa californica nuclear polyhedrosis virus. J. Virol. 63:2008-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tomalski, M. D., J. G. Wu, and L. K. Miller. 1988. The location, sequence, transcription, and regulation of a baculovirus DNA polymerase gene. Virology 167:591-600. [PubMed] [Google Scholar]
- 94.van Strien, E. A., D. Zuidema, R. W. Goldbach, and J. M. Vlak. 1992. Nucleotide sequence and transcriptional analysis of the polyhedrin gene of Spodoptera exigua nuclear polyhedrosis virus. J. Gen. Virol. 73:2813-2821. [DOI] [PubMed] [Google Scholar]
- 95.Viswanathan, P., B. Venkaiah, M. S. Kumar, S. Rasheedi, S. Vrati, M. D. Bashyam, and S. E. Hasnain. 2003. The homologous region sequence (hr1) of Autographa californica multinucleocapsid polyhedrosis virus can enhance transcription from non-baculoviral promoters in mammalian cells. J. Biol. Chem. 278:52564-52571. [DOI] [PubMed] [Google Scholar]
- 96.Wallace, D. R., and J. C. Cunningham. 1995. Diprionid sawflies, p. 193-232. In J. A. Armstrong and W. G. H. Ives (ed.), Forest insect pests in Canada, vol. 1. Natural Resources Canada, Ottawa, Ontario, Canada. [Google Scholar]
- 97.Weyer, U., and R. D. Possee. 1988. Functional analysis of the p10 gene 5′ leader sequence of the Autographa californica nuclear polyhedrosis virus. Nucleic Acids Res. 16:3635-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Whitford, M., and P. Faulkner. 1992. Nucleotide sequence and transcriptional analysis of a gene encoding gp41, a structural glycoprotein of the baculovirus Autographa californica nuclear polyhedrosis virus. J. Virol. 66:4763-4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wingender, E. 1988. Compilation of transcription regulating proteins. Nucleic Acids Res. 16:1879-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wormleaton, S., J. Kuzio, and D. Winstanley. 2003. The complete sequence of the Adoxophyes orana granulovirus genome. Virology 311:350-365. [DOI] [PubMed] [Google Scholar]
- 101.Wu, C. H., H. Huang, L. S. Yeh, and W. C. Barker. 2003. Protein family classification and functional annotation. Comput. Biol. Chem. 27:37-47. [DOI] [PubMed] [Google Scholar]
- 102.Wu, C. T., and J. R. Morris. 1999. Transvection and other homology effects. Curr. Opin. Genet. Dev. 9:237-246. [DOI] [PubMed] [Google Scholar]
- 103.Zanotto, P. M., B. D. Kessing, and J. E. Maruniak. 1993. Phylogenetic interrelationships among baculoviruses: evolutionary rates and host associations. J. Invertebr. Pathol. 62:147-164. [DOI] [PubMed] [Google Scholar]
- 104.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847-848. [DOI] [PubMed] [Google Scholar]
- 105.Zemskov, E. A., W. Kang, and S. Maeda. 2000. Evidence for nucleic acid binding ability and nucleosome association of Bombyx mori nucleopolyhedrovirus BRO proteins. J. Virol. 74:6784-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]