Abstract
We have isolated and characterized AtREM1, the Arabidopsis ortholog of the cauliflower (Brassica oleracea) BoREM1. AtREM1 belongs to a large gene family of more than 20 members in Arabidopsis. The deduced AtREM1 protein contains several repeats of a B3-related domain, and it could represent a new class of regulatory proteins only found in plants. Expression of AtREM1 is developmentally regulated, being first localized in a few central cells of vegetative apical meristems, and later expanding to the whole inflorescence meristem, as well as primordia and organs of third and fourth floral whorls. This specific expression pattern suggests a role in the organization of reproductive meristems, as well as during flower organ development.
Plant morphogenesis depends upon the mitotic activity of cells in the shoot apical meristem (SAM). During the development of the plant, the SAM goes through different developmental phases, characterized by the type and features of the organs produced: leaves during the vegetative stage and flowers during the reproductive phase (Poethig, 1990). Transition from vegetative to reproductive development depends, in most annual plant species, on the acquisition of competence by the SAM and the presence of specific environmental conditions for temperature and photoperiod (for review, see Bernier et al., 1993).
The use of experimental genetic systems like Arabidopsis and snapdragon (Antirrhinum majus) has provided a tool to identify many of the genes involved in the regulation of flowering time and flower meristem and organ identity in those species. More than 80 genes whose mutations alter the time to flower have currently been identified in Arabidopsis, and many of them have been cloned and characterized at the molecular level (for review, see Koornneef et al., 1998; Piñeiro and Coupland, 1998; Simpson et al., 1999). In a similar way, mutations that alter the identity of flower meristems in Arabidopsis have allowed the identification of a set of genes required for the acquisition of flower meristem identity such as LEAFY and APETALA1 (Mandel et al., 1992; Weigel et al., 1992), as well as other genes like APETALA2 (AP2), CAULIFLOWER, and UNUSUAL FLORAL ORGANS, whose mutations enhance the phenotype of lfy and ap1 mutants (Jofuku et al., 1994; Ingram et al., 1995; Kempin et al., 1995).
In spite of the important role played by the genetic analysis in the identification of key genes in different biological processes, the results of current Arabidopsis genome sequencing projects show that only 8% of the gene sequences identified have been experimentally characterized to date (Lin et al., 1999; Mayer et al., 1999). This is likely the result of the absence of detectable phenotypic alterations for mutations at many loci due to genetic or functional redundancy. Molecular approaches designed to identify genes with particular expression patterns have proved to be useful complementary approaches for the identification of additional genes involved in specific biological processes (Sablowski and Meyerowitz, 1998).
To identify new genes involved in reproductive development, we constructed a cauliflower (Brassica oleracea) curd meristem cDNA library highly enriched in clones corresponding to transcripts specific to reproductive meristems (Franco-Zorrilla et al., 1999). Screening of this library allowed the characterization of one gene, Brassica oleracea reproductive meristem gene 1 (BoREM1), whose expression was specific to cauliflower curd meristems arrested in an early-inflorescence stage of development. BoREM1 belongs to a gene family in cauliflower and encodes a protein with features of transcriptional activators, but does not show homology to any protein of known function (Franco-Zorrilla et al., 1999). To understand the function of this new gene family in plants, we have isolated the corresponding Arabidopsis gene. Here, we report the molecular characterization of AtREM1, the ortholog of BoREM1 in Arabidopsis. AtREM1 belongs to a large gene family in Arabidopsis. Many REM family members are present in clusters of tandemly repeated genes. The deduced proteins of this family are characterized by the presence of a specific domain that we have named the REM domain. This domain shares some sequence similarity with the B3 domain found in the superfamily of B3-containing transcriptional regulators such as VIVIPAROUS1/ABSCISIC ACID INSENSITIVE 3 (VP1/ABI3; McCarty et al., 1991; Giraudat et al., 1992), FUSCA3 (FUS3; Luerssen et al., 1998), AUXIN RESPONSE FACTORS (ARFs; Ulmasov et al., 1997, 1999), and RAV1 (Kagaya et al., 1999). In situ hybridization analysis shows that AtREM1 expression is restricted to a few cells in the vegetative SAM and expands to the whole meristem in the inflorescence. AtREM1 is also expressed in the central zone of floral meristems and becomes progressively restricted to specific carpel cell types during the development of the flower. Finally, plants carrying a T-DNA insertion in AtREM1 coding sequence do not show any obvious mutant phenotype, likely as a consequence of gene duplication.
RESULTS
AtREM1 Belongs to a New Arabidopsis Gene Family
One Arabidopsis gene similar to BoREM1 was initially detected in DNA-blot hybridizations at moderate stringency conditions using different fragments corresponding to BoREM1 as probes (data not shown). As BoREM1 is expressed in the meristematic domes of the cauliflower curd (Franco-Zorrilla et al., 1999), we screened a cDNA library prepared from young inflorescences of Arabidopsis (Weigel et al., 1992) and we isolated a 1,775-bp cDNA. This cDNA, named AtREM1, contained a long open reading frame (ORF) starting at position 8, in agreement with the translation initiation consensus sequence defined for plants (Lütcke et al., 1987), and ending at position 1,561 (GenBank accession no. AF336344). Sequence comparison between the AtREM1 cDNA clone and the available genomic sequence of Arabidopsis revealed the existence of six introns in the AtREM1 genomic sequence and allowed its localization in bacterial artificial chromosome (BAC) clone F28 M20 (accession no. AL031004) from chromosome 4. Additional information on the 5′-untranslated region of AtREM1 was obtained from a genomic clone, and the transcription start site was mapped 51 bp upstream of the deduced initiation codon by primer extension (data not shown).
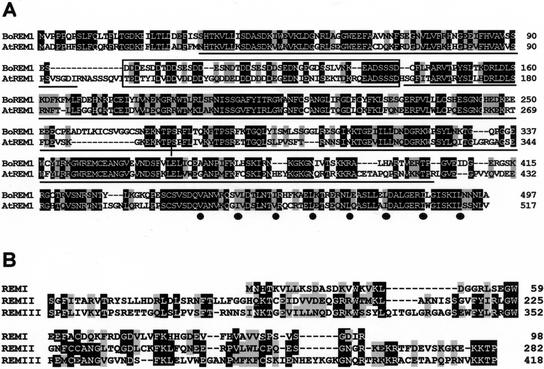
The deduced AtREM1 protein consists of 517 amino acids with a calculated molecular mass of 58.4 kD (Fig. 1A). It is rich in Lys, Asp, and Glu residues, which confer to the protein a pI of 5.24. Sequence analysis revealed the presence of two long (positions 160–282 and 283–418) and one short (position 30–98) semiconserved characteristic repeats in the AtREM1 deduced protein, referred to as the REM domains. Approximately 50% of the residues are similar within these repeats (Fig. 1B). AtREM1 also contains an acidic domain near the amino terminus, where 75% of the residues are negatively charged, two stretches of basic residues, resembling bipartite NLSs (Raikhel, 1992), as well as a putative coiled-coil domain at the carboxy terminus formed by eight hydrophobic repeats (Fig. 1A). The alignment between AtREM1 and BoREM1 from cauliflower (Franco-Zorrilla et al., 1999) showed 61% identity and 73% similarity (Fig. 1A). Proteins of known functions similar to AtREM1 could not be identified in the databases.
Figure 1.
The deduced AtREM1 protein. A, Alignment of the deduced AtREM1 protein and its ortholog from cauliflower (accession no. AF051772). Identical residues are in black, and conservative substitutions are in gray. Characteristic domains of both proteins are as follows: REM domains, including putative nuclear localization signals (NLSs), are underlined. The acidic domain is enclosed by a box and the putative coiled-coil domain is labeled with black dots under hydrophobic residues. B, Amino acid alignment of the three REM domains found in AtREM1.
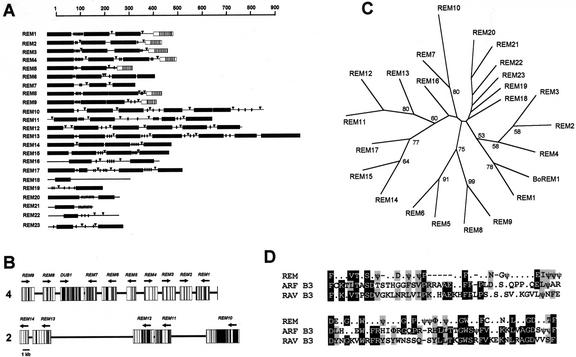
As previously observed for BoREM1 in cauliflower (Franco-Zorrilla et al., 1999), AtREM1 (REM1) also belongs to a gene family in Arabidopsis. Genomic DNA sequences from the Arabidopsis Genome Initiative have revealed the existence of at least 23 sequences characterized by the presence of duplicated REM domains (Table I; Fig. 2). Many of them also have acidic domains and/or the heptad hydrophobic repeats at their C-terminal regions. Fourteen of those sequences are organized as arrays of tandemly repeated sequences. In fact, REM1 is part of a tandem array of nine related genes in chromosome 4 (Fig. 2B), and there is one additional cluster formed by five putative gene sequences in chromosome 2 (Fig. 2B). The rest of the related sequences were found in independent chromosomal positions. REM1 showed highest similarity to the corresponding BoREM1 from cauliflower, suggesting that AtREM1 is the ortholog gene of BoREM1 (Fig. 2C). Similarity searches in databases using the REM repeats as a query did not reveal any similar protein in yeast or animal systems. However, a consensus sequence within the REM domain showed similarity to conserved residues within the B3 domain of the superfamily of B3-containing transcriptional regulators, composed by VP1/ABI3, FUSCA3, ARFs, and RAVs (Fig. 2D; McCarty et al., 1991; Giraudat et al., 1992; Ulmasov et al., 1997, 1999; Luerssen et al., 1998; Kagaya et al., 1999).
Table I.
Predicted REM genes found in Arabidopsis databases
| BAC Accession | Chr. | Gene | |
|---|---|---|---|
| REM1 | AL031004 | 4 | F28M20.200 |
| REM2 | AL031004 | 4 | F28M20.190 |
| REM3 | AL031004 | 4 | F28M20.190 |
| REM4 | AL031004 | 4 | F28M20.180 |
| REM5 | AL031004 | 4 | F28M20.170 |
| REM6 | AL031004 | 4 | F28M20.160 |
| REM7 | AL031004 | 4 | F28M20.150 |
| REM8 | AL031004 | 4 | F28M20.130 |
| REM9 | AL031004 | 4 | F28M20.120 |
| REM10 | AC006954 | 2 | F27A10.1 |
| REM11 | AC006954 | 2 | F25P17.1 |
| REM12 | AC006954 | 2 | F25P17.2 |
| REM13 | AC006954 | 2 | F25P17.5 |
| REM14 | AC006954 | 2 | F25P17.5 |
| REM15 | AF013293 | 4 | IG5I10.15 |
| REM16 | AL035678 | 4 | F17M5.40 |
| REM17 | AC006535 | 1 | T24P13.6 |
| REM18 | AC005310 | 2 | F19D11.1 |
| REM19 | AC007504 | 1 | F13F21.8 |
| REM20 | AL132966 | 3 | F4P12.10 |
| REM21 | AC018907 | 3 | F28L1.16 |
| REM22 | AL096859 | 3 | T6H20.200 |
| REM23 | AL353994 | 5 | F17I14.30 |
Figure 2.
The REM gene family in Arabidopsis. A, Schematic representation of the deduced REM proteins from predicted genes found in databases. The REM domains are shown as black boxes. Short stretches of acidic residues are represented as oval chains, and putative NLSs are represented as triangles. Only seven of the deduced proteins related to REM1 have heptad hydrophobic repeats at their C terminus (vertical bars in an open box). B, Physical map of a 29-kb genomic region including REM1 and related sequences in tandem in chromosome 4 and another cluster of related genes in chromosome 2. ORFs are represented by white boxes and introns are shown with black bars. The direction of the deduced ORFs are indicated with arrows. DUB-1 corresponds to a gene sequence with similarity with a deubiquitinating enzyme. C, Unrooted phylogenetic tree showing the relationships among the Arabidopsis REM proteins and BoREM1. Branches with a support of 50 or more are indicated. D, Alignment of consensus sequences from REM domain and B3 domains from ARFs and RAVs proteins. A consensus at 60% for REM domain was obtained by alignment of one deduced REM domain from each predicted gene. A consensus for B3 domain from ARFs was obtained from alignment of ARF1 to ARF10 as shown in Ulmasov et al. (1999), and that corresponding to RAV from alignment of RAV1 and RAV2 as shown in Kagaya et al. (1999). Identical residues are in black, and conservative substitutions are in gray. Hydrophobic residues (L, V, I, W, F, and Y) are represented as ψ and aromatic amino acids (W, F, and Y) are represented as Φ. Hyphens are introduced for optimization of the alignment.
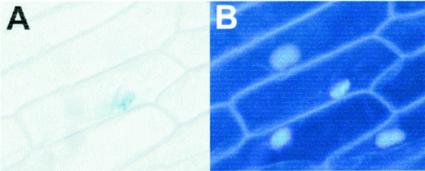
As the characteristic REM domain resembles a DNA-binding domain and the deduced REM1 protein contains two stretches of basic residues that could act as NLSs, we studied the subcellular localization of the REM1 protein. For this purpose, we used a translational fusion between REM1 and the Escherichia coli β-glucuronidase (GUS) in transient expression assays (Varagona et al., 1991). REM1 targeted most GUS activity into the nucleus of onion (Allium cepa) epidermal cells (Fig. 3). This nuclear localization of REM1 supports the functionality of the bipartite NLSs and a nuclear function for REM1.
Figure 3.
Subcellular localization of REM1 protein. A, Histochemical GUS detection in a transient expression analysis on epidermal onion cells of a REM1-GUS fusion protein. B, 4,6-Diamidino-2-phenylindole staining of the same group of cells.
AtREM1 Is Preferentially Expressed in Inflorescence Apices
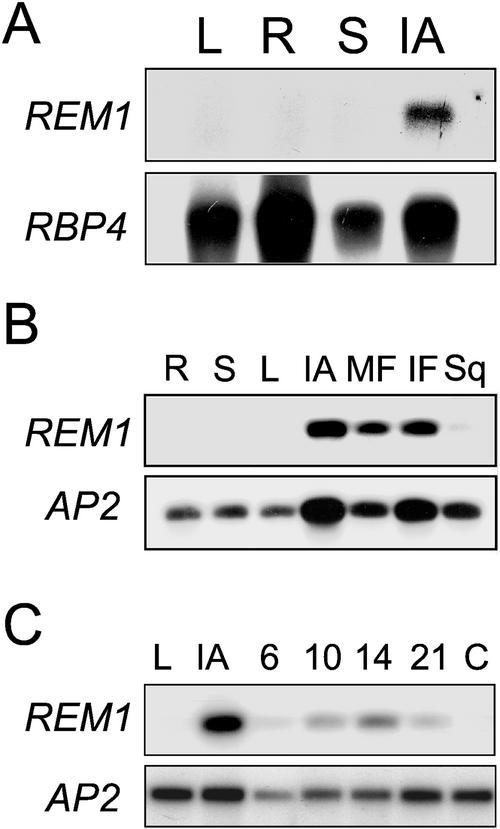
The expression of REM1 was analyzed by RNA-blot hybridization using poly(A)+ RNA from different organs of adult plants. As shown in Figure 4A, a transcript of approximately 2 kb was detected in inflorescence apices, being apparently absent in the rest of the organs analyzed. A similar result was also obtained in RT-PCR experiments (Fig. 4B), which showed that REM1 is also expressed in immature and mature flowers, and in siliques at lower level. To determine the time course of expression of REM1 in the shoot apex, we performed RT-PCR experiments using total RNA isolated from the apex at different times after germination. The results showed that REM1 transcripts could already be detected in apices excised from 6-d-old seedlings grown in petri dishes under long-day photoperiods, and that their levels slightly increased as development progressed (Fig. 4C).
Figure 4.
Expression analysis of REM1. A, An RNA blot containing 1.5 μg of poly(A)+ RNA from different organs of Arabidopsis was hybridized with the cDNA REM1 and a probe corresponding to ribosomal protein L3 as a control for gel loading (RBP4; Kim et al., 1990). B, Expression analysis of AtREM1 and AP2 by reverse transcriptase (RT)-PCR in different organs from adult plants. RT-PCR products were blotted onto membranes and hybridized with the corresponding probes. C, RT-PCR analysis of expression of REM1 in apices from young seedlings at different stages of development. C, Cotyledon and hypocotyl from young seedlings; IA, inflorescence apex; IF, immature flower, before anthesis; L, leaf; MF, flower at anthesis; R, root; S, stem; Sq, silique.
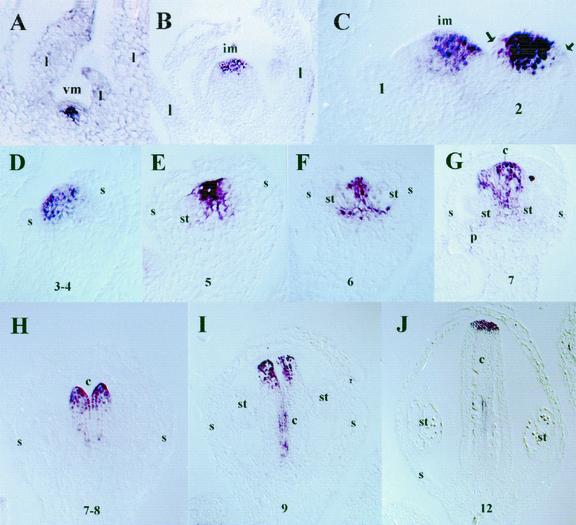
To analyze the distribution of AtREM1 transcripts in the plant apex, we performed in situ hybridization experiments on Arabidopsis apices collected at different times during the development of the plant. Plants were grown under short-day photoperiods to extend the period of vegetative development. Sections of plant tissue were hybridized with REM1 digoxygenin-labeled riboprobes. Under those experimental conditions, REM1 mRNA was not detectable in young seedlings (data not shown). Later, during vegetative development, 12 to 17 d after sowing when the SAM was actively producing leaf primordia, REM1 was detected at low levels in the inner cell layers of the SAM (Fig. 5A). At flowering, the SAM shape changes and starts producing flower meristems at its periphery. At this time, REM1 was also detected in the SAM, but its expression differed quantitatively and qualitatively from the vegetative expression. The signal was higher and spread throughout the meristem cell layers (Fig. 5, B and C). During reproductive development, REM1 was expressed in the shoot apex and during flower development. Although REM1 expression was absent at stage 1 of flower meristems, it appeared at stage 2 in the central area of the flower meristem (Fig. 5C). In later stages 3 and 4, when sepal primordia initiate and grow to enclose the bud, the AtREM1 hybridization signal became excluded from the developing sepals (Fig. 5, C and D). During stages 5 and 6, when petal and stamen primordia first became visible, the hybridization signal was further restricted to the central area of the floral meristem that gives rise to the gynoecium and to small groups of cells at the tip and at the base of stamens (Fig. 5, E and F). This latter group of cells could correspond to the presumptive nectaries (data not shown). After stage 6, REM1 expression was restricted to the developing gynoecium being further restricted, during late stage 7, to the medial ridge of cells (Fig. 5, G and H). At stage 9, different regions of the gynoecium became morphologically distinct from one another and REM1 mRNA was detected in the septum, style, and stigma (Fig. 5I). This expression was maintained until the end of flower development (Fig. 5J). REM1 was also detected in lateral shoot meristems with a similar pattern to that shown in the primary SAM (data not shown). Thus, in agreement with the results of RNA-blot hybridizations and RT-PCR experiments, REM1 was expressed in SAMs throughout development, although quantitative and qualitative differences were observed between vegetative and reproductive expression patterns. Furthermore, REM1 was also expressed in floral meristems from stage 2, with an expression pattern that became progressively restricted to the gynoecium area that gives rise to the style, stigma, and septum. Transgenic plants expressing the GUS reporter gene (Jefferson et al., 1987) fused to the 5′-flanking region of REM1 (from −1,212 to +62, relative to the transcriptional start site) reproducibly showed a GUS expression pattern consistent with the results of RNA-blot hybridization, RT-PCR, and in situ hybridization (data not shown).
Figure 5.
Expression patterns of REM1 during Arabidopsis development. RNA in situ hybridization of Arabidopsis (ecotype Col) probed with REM1. All sections are longitudinal. A, 17-short-day rosette. The SAM, vm, produces leaves in its periphery. REM1 signal is absent from L1 and is not detected in leaf primordia. B, Inflorescence SAM, im, and young floral meristems. Signal is detected in all three layers of the im. In stage 1 flower, REM1 mRNA is not detectable. In stage 2 flowers, REM1 is expressed in the central area of the floral dome. Arrows indicate the sepal anlagen. C, Stage 2 and 3 flowers showing REM1 expression excluded from the sepal anlagen. D, Stage 3 and 4 flower. E, Stage 5 flower. Stamen primordia are now visible. REM1 signal becomes restricted to the carpel anlagen. F, Stage 6 flower. G, Stage 7 flower. H, Stage 7 and 8 flower, REM1 mRNA accumulates in the medial ridge. I, Stage 9 flower. REM1 is expressed in the septum, style, and stigma. J, Stage 12 flower shortly before anthesis. REM1 expression is detected in the stigma and septum. Controls using REM1 sense probes gave no signal (not shown). vm, Vegetative SAM; im, inflorescence SAM; l, leaf primordia; s, sepal; p, petal; st, stamen; c, carpel. The numbers indicate stages according to Smyth et al. (1990).
AtREM1 Knockout Plants Do Not Show Conspicuous Abnormalities
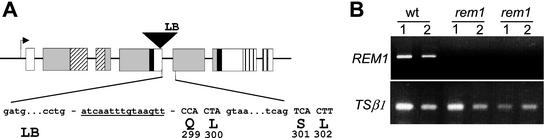
An Arabidopsis line carrying a T-DNA insertion in REM1 was identified by PCR screening of the T-DNA tagged lines obtained by Feldmann (1991). Segregation analysis for resistance to kanamycin revealed the presence of a unique T-DNA insertion in this line. Plants homozygous for the insertion were identified by PCR genotyping, which allowed the amplification of a single 1-kb PCR product with primers corresponding to the left border of the T-DNA and the 3′ end of REM1 sequence, and failed to amplify the 2-kb fragment with 5′ and 3′ primers corresponding to the wild-type genomic sequence (Fig. 6). Sequence determination of the LB/3′-PCR product indicated that the T-DNA was inserted in the coding region of REM1 just before the Gln codon in position 299 and, therefore, it might be a null mutation. In fact, RNA-blot hybridization and RT-PCR analysis with RNA obtained from inflorescence apices of this mutant (rem1-1) failed to detect the accumulation of REM1 transcripts, as well as other mRNA species that could be derived from alternate splicing of the disrupted REM1 gene (Fig. 6B). Mutant plants were grown and analyzed under short- and long-day photoperiodic conditions and no phenotypic differences were detected regarding flowering time or reproductive development compared with sibling wild-type plants.
Figure 6.
A, Isolation of a mutant line carrying a T-DNA insertion in REM1. Map of the insertion of the T-DNA in the coding sequence of REM1. Exons are represented with boxes and characteristic domains of REM1 are as in Figure 1. Between the left border of the T-DNA (LB) and REM1, a 16-bp fragment of unknown derivation was found (underlined sequence). B, REM1 expression in wild-type and in two rem1 homozygous lines. 1 and 2 denote different concentrations of cDNA employed in PCR reactions (see “Materials and Methods”). PCR corresponding to Trp synthase β-subunit was used as a control.
DISCUSSION
The identification of AtREM1 demonstrates the validity of using cauliflower meristems to isolate Arabidopsis genes specifically expressed in its reproductive apex. Using a strategy based on the search for specific expression patterns in pools of enhancer- or gene-trap lines, Campisi et al. (1999) reported the isolation of promoter trap lines exhibiting GUS staining patterns in the inflorescence. In fact, one of them carries one insertion between REM3 and REM4 showing specific inflorescence apex expression (Campisi et al., 1999; T. Jack, personal communication). Thus, both strategies may contribute to the identification of new genes expressed in a small number of cells. Consistent with this, REM genes are underrepresented in expressed sequence tag (EST) databases. Only two ESTs from Arabidopsis corresponding to REM-containing genes have been identified. One of them (GenBank accession no. AV566479), isolated from a green siliques cDNA library, corresponds to REM2, whereas the other one (accession no. H76710) does not correspond to any gene listed in Table I. In addition, a rice (Oryza sativa) EST (accession no. AU30260) from immature leaves including apical meristems, as well as two tomato (Lycopersicon esculentum) ESTs from fruits and carpels (accession nos. AW441198 and AI489095, respectively) can be identified.
AtREM1 Contains Three Repetitions of a B3-Related DNA-Binding Domain
The REM1 deduced protein has features characteristic of regulatory proteins (Figs. 1 and 2). It contains a distinctive repetition of a basic domain that we have named the REM domain. This domain is related to the B3 domain of the VP1/ABI3-ARF family of transcriptional regulators (McCarty et al., 1991; Giraudat et al., 1992; Ulmasov et al., 1997), which has been shown to bind DNA in vitro and in vivo (Suzuki et al., 1997; Ulmasov et al., 1997). In addition to VP1/ABI3 and ARFs, RAV1 and RAV2 constitute another group of B3-containing proteins (Okamuro et al., 1997; Kagaya et al., 1999). This group shows an additional AP2-DNA-binding domain and it has been shown that AP2 and B3 domains maintain autonomous DNA-binding activities to different DNA motifs (Kagaya et al., 1999). Consensus sequences alignment in Figure 2D shows that the REM domain shares 28% to 30% similarity to both types of B3 domains, which are more related between them (45% similarity). Therefore, the REM domain is more distantly related to the B3 domain found in ARFs and RAVs families of transcriptional regulators. The presence of a putative DNA-binding domain, a functional NLS, and the acidic domain, which has been proposed to function as an activation domain (Mitchell and Tjian, 1989), suggests that REM1 can function as a transcriptional regulator. In addition, the presence of a putative coiled-coil domain in its carboxy terminus suggests that it could specifically interact with other proteins (O'Shea et al., 1989).
AtREM1 Expression Pattern Suggests a Role in Reproductive Development
Different B3-containing proteins have been involved in different aspects of plant development from seed maturation (ABI3) and embryo development (MON/ARF5) to reproductive development (ETT/ARF3) (Giraudat et al., 1992; Sessions et al., 1997; Hardtke and Berleth, 1998). REM1 has a very specific pattern of transcript accumulation in the SAM, starting in a few cells in the L3 layer of the vegetative SAM and expanding to the three meristematic layers in the inflorescence meristem (Fig. 4, A and B). Some genes are involved in flower transition, such as AGL20 in Arabidopsis, SaMADS A, the putative ortholog of AGL20 in white mustard (Sinapis alba), and AGL8/FRUITFUL are also expressed in the SAM (Mandel and Yanofsky, 1995; Menzel et al., 1996; Gu et al., 1998; Lee et al., 2000). Furthermore, the expression pattern of REM1 during flower development from stage 2 until stage 7 is coincident to that observed for AGL8/FUL (Fig. 4). Expression of REM1 and AGL8/FUL differs from that stage on, when both genes become expressed in complementary regions. Another B3-containing gene, ETT/ARF3, is also expressed in inflorescence meristems and in developing flowers, although flower expression pattern does not coincide with that of REM1 (Sessions et al., 1997). Thus, the pattern of expression of REM1 in the SAM, flower meristems, and in carpels suggests a role for this gene in different aspects of reproductive development. The lack of an obvious phenotypic alteration in the T-DNA insertion mutant of REM1, likely due to genetic redundancy, precludes having a clue of the role of this gene in plant development. In this way, at least one homologous gene in the REM1 cluster, REM3, could also have a redundant function because its promoter-driven GUS expression suggests that it is expressed with a similar pattern (T. Jack, personal communication).
The REM Gene Family
REM1 belongs to a large gene family characterized by the presence of one to six REM repeats. Most of them also show predicted acidic regions and potential NLSs, but the predicted coiled-coil region is a feature characteristic of REM1 and other deduced proteins in the cluster on chromosome 4 (Fig. 2A). Thus, the presence of the B3-related domain or REM domain identifies a new gene family formed by more than 20 putative genes in Arabidopsis. As with other groups within the B3 superfamily of transcription factors, the REM family is present only in plants; no related proteins have been found in any bacterial, fungal, or animal sequence present in databases.
Successive genome duplication events have likely generated the repeated intragenic structure, as well as the tandem duplicated arrays of REM genes found in different chromosomes. The low sequence similarity observed between REM sequences in the cluster of chromosome 2 and those in the cluster on chromosome 4 indicates that they do not derive from a recent duplication event. In fact, BAC F28 m20 containing REM1 is part of a 664-kb region in chromosome 4 that is duplicated in chromosome 2 (Blanc et al., 2000). In general, REM genes are more related within each cluster than between clusters. In this way, most of the deduced proteins encoded by gene sequences in the chromosome 4 cluster contain heptad hydrophobic repeats at their C terminus (Fig. 2A) that are absent in other REM family members. Thus, REM genes in the cluster on chromosome 4, with the exception of REM7, could constitute a subgroup within the REM family. It is likely that duplications and divergence within clusters were originated after the chromosome duplication.
Among all the REM sequences found in Arabidopsis, AtREM1 is the most related at nucleotide and amino acid sequence level to BoREM1 from cauliflower (Figs. 1A and 2C). Furthermore, AtREM1 is more related to BoREM1 than to any other REM-like sequence in the Arabidopsis genome. This would indicate that the chromosomal duplication that gave rise to segments of chromosomes 2 and 4, as well as former events of gene duplication within the AtREM1 cluster, predate the divergence of the Brassica and Arabidopsis genus. This conclusion is consistent with previous observations of extensive duplication and colinearity between Brassica and Arabidopsis genomes (Schmidt, 2000).
In conclusion, we have identified a new family of putative regulatory proteins in Arabidopsis. The specific tissue expression patterns of at least two of the members (Campisi et al., 1999) suggest a role for some of the encoded proteins in the regulation of the vegetative to reproductive transition, as well as in carpel development. Unfortunately, genetic redundancy precludes the generation of mutant phenotypes, and the tandem array of the genes requires strategies that allow the generation of knockouts for several genes simultaneously. We hope that deletion of several or all the REM related genes in BAC F28 m20 on chromosome 4 would provide information about the role of this set of genes in Arabidopsis reproductive development.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis plants used in this work belonged to Landsberg erecta, Columbia (Col), and Wassilewskija-2 ecotypes. Plants were grown at 22°C under long-day (16 h of light) or short-day (8 h of light) conditions, at an intensity of 140 μE m−2 s−1 in a mixture of soil:vermiculite (3:1). For some experiments, plants were grown under sterile conditions in petri dishes containing Murashige and Skoog salts supplemented with 1% (w/v) Suc and solidified with 0.8% (w/v) agar.
Isolation of cDNA and Genomic Clones Corresponding to AtREM1
A cDNA library prepared from young inflorescences of Arabidopsis (ecotype Landsberg erecta; Weigel et al., 1992) was screened (5 × 105 pfu) with the CM6.2 fragment (287 bp) corresponding to the 3′ end of BoREM1 (Franco-Zorrilla et al., 1999). Hybridization was performed at 55°C using standard procedures (Sambrook et al., 1989). Eight phages giving positive hybridization were identified, and plasmids were excised in vivo and converted into EcoRI-XhoI inserts in pBS SK (Stratagene, La Jolla, CA). A genomic clone was isolated from an Arabidopsis (ecotype Col) genomic DNA library in a λGEM11 (Promega, Madison, WI) after screening with the AtREM1 cDNA using standard procedures (Sambrook et al., 1989). A 4-kb EcoRI restriction fragment was cloned in pBS KS, and different restriction fragments were subcloned for sequence determination. The transcription start site for AtREM1 was identified by primer extension analysis (Sambrook et al., 1989) using the primer 5′-AATGGTTTGTCTCCGGTACG-3′ for reverse transcription and 1 μg of poly(A)+ RNA from inflorescence apices as template.
DNA Sequencing and Analysis
Sequencing was carried out using Sequenase Ver 2.0 (U.S. Biochemical Corp., Cleveland) and synthetic primers. All sequences were analyzed for similarity to databases using the internet site of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and The Arabidopsis Information Resource (http://www.Arabidopsis.org), running the BLAST programs (Altschul et al., 1990). Other sequence analysis and manipulations were carried out using the PC-Gene (IntelliGenetics, Mountain View, CA) and GCG (Genetics Computer Group, Madison, WI) packages. To construct the phylogenetic tree, predicted proteins were aligned with CLUSTALW, 100 bootstrapped data sets were obtained with SEQBOOT, distance matrices were calculated with PROTDIST-Dayhoff PAM matrix algorithm, trees were constructed with NEIGHBOR, and a consensus tree was obtained with CONSENSE. SEQBOOT, PROTDIST, NEIGHBOR, and CONSENSE are from the PHYLIP package (Felsenstein, 1989).
RNA Isolation and Analysis
Isolation of total and poly(A)+ RNA and northern hybridizations were previously described (Franco-Zorrilla et al., 1999). Minipreparations of plant RNA were performed using the FastRNA Green Kit (Bio 101, Vista, CA) followed by a treatment with RNase free DNaseI (Roche Molecular Biochemicals, Summerville, NJ). RNAs were quantified and their integrity was tested in ethidium bromide-stained agarose gels. For RT-PCR reactions, 250 ng (for 50-μL reactions) of RNA was used as template in one-step RT-PCR reactions with GeneAmp Thermostable rTth Reverse Transcriptase RNA PCR kit (PerkinElmer, Norwalk, CT). First-strand synthesis was accomplished from downstream primer 5′-CCGTCTTCTCCGATATGTTC-3′. PCR with the same primer and the oligonucleotide 5′-TACCGGAGACAAACCATTTC-3′ subsequently allowed the amplification of a 397-bp cDNA fragment. As a control for RT-PCR reactions, we used the AP2 gene as in Putterill et al. (1995). In all cases, reverse transcription was carried out at 63°C for 30 min, followed by a PCR reaction at 94°C for 45 s and 63°C for 45 s for 30 cycles. PCR reactions were loaded in 1.7% (w/v) agarose gels, transferred onto Hybond N+ membranes, and hybridized with the corresponding DNA probes. RT-PCR experiments were performed at least twice with different RNA samples, and a representative result is presented.
In Situ Hybridization
Digoxygenin labeling of RNA probes, tissue preparation, and hybridization were done as described by Coen et al. (1990). The REM1 probe used was designed to prevent cross-hybridization with other REM genes. To obtain a 3′-1,250-bp insert as a template for REM1 riboprobes synthesis, the REM1 cDNA was digested with XbaI and religated in pBS SK. The hybridized sections were visualized with Nomarski optics.
Transient Transformation of Onion (Allium cepa) Epidermis Cells
The construct for transient expression was generated by fusing a PCR-amplified full-length REM1 cDNA in frame to the GUS reporter gene into the vector pBI221 from CLONTECH (Palo Alto, CA). Transformation of onion epidermal cells using a PDS 1000 helium particle gun (Bio-Rad, Hercules, CA) and histochemical staining were performed as described in Varagona et al. (1991).
Isolation of the Insertional T-DNA Line rem1
A total of 6,000 Arabidopsis (ecotype Wassilewskija-2) T-DNA-tagged lines (Feldmann, 1991) in six pools of 1,000 lines each was PCR-screened for an insertion in REM1 with oligonucleotides ARA6 5′-AAATTTCGTACCGGAGACAAACCATTTCTC-3′ and ARA7 5′-TCTTTGTTTCTTTCCCCATCTTCAGTCTCAC-3′ corresponding to 5′ and 3′ ends of REM1, respectively, and LB-RB primers as in Krysan et al. (1996) for T-DNA borders. After identification of a positive pool, a bidimensional secondary screening (Azpiroz-Leehan and Feldmann, 1997) was performed with oligonucleotides LB and ARA7 on 20 DNA pools from 100 lines each, which allowed the identification of a 10-line subpool. Seeds corresponding to 10 individual lines were grown to isolate the rem1 mutant. PCR performed with ARA6 and RB primers failed to amplify any fragment, probably due to a partial deletion of the region of the right border homologous to RB primer. PCR reactions were performed in a 2400 thermocycler (PerkinElmer) at 94°C for 30 s, at 68°C for 1 min, and at 72°C for 1 min 30 s, 35 cycles. For RT-PCR expression studies in rem1 homozygous lines, 5 μg of total RNA from inflorescence apices was treated with DNase and was employed in first-strand cDNA synthesis using SuperScript II (Gibco-BRL, Grand Island, NY) and an oligo T17 as primer. cDNAs were 250- and 500-fold diluted in subsequent PCR reactions. Primers used in these experiments were ARA6 and ARA7 for REM1 expression and CTCATGGCCGCCGGATCTGA and CTTGTCTCTCCATATCTTGAGCA corresponding to TSβ1 as a control (Berlyn et al., 1989).
ACKNOWLEDGMENTS
We thank the Arabidopsis Biological Resource Center, Ohio State University (Columbus), for providing the DNA pools corresponding to the Feldmann collection and the cDNA library, and Dr. Nigel Crawford for supplying the genomic DNA library. We thank Dr. Thomas Jack for sharing unpublished results and for his critical reading of the manuscript.
Footnotes
This work was supported by Comisión Interministerial de Ciencia y Tecnologia (Spain; grant no. AGF98-0206). Support to research activity at Centro Nacional de Biotecnología is provided through specific agreement between Consejo Superior de Investigaciones Cientificas and Instituto Nacional de Investigacion y Tecnologia Agraria y Alimentaria. J.M.F.-Z. was funded by a predoctoral fellowship from Dirección General de Investigación Científica y Tecnológica (Spain). B.F.-C. was a recipient of a postdoctoral fellowship from Instituto Nacional de Investigacion y Tecnologia Agraria y Alimentaria (Spain), and P.C. and J.A.J. are recipients of postdoctoral Ministerio de Educación y Ciencia (Spain) contracts.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010323.
LITERATURE CITED
- Altschul F, Gish F, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Azpiroz-Leehan R, Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet. 1997;13:152–156. doi: 10.1016/s0168-9525(97)01094-9. [DOI] [PubMed] [Google Scholar]
- Berlyn MB, Last RL, Fink GR. A gene encoding the tryptophan synthase beta subunit of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1989;86:4604–4608. doi: 10.1073/pnas.86.12.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier G, Havelange A, Houssa C, Petitjean A, Leujeune P. Physiological signals that induce flowering. Plant Cell. 1993;5:1147–1155. doi: 10.1105/tpc.5.10.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M. Extensive duplication and reshuffling in the Arabidopsisgenome. Plant Cell. 2000;12:1093–1101. doi: 10.1105/tpc.12.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi L, Yang Y, Yi Y, Heilig E, Herman B, Cassista AJ, Allen DW, Xiang H, Jack T. Generation of enhancer trap lines in Arabidopsisand characterization of expression patterns in the inflorescence. Plant J. 1999;17:699–707. doi: 10.1046/j.1365-313x.1999.00409.x. [DOI] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R. floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum. Plant J. 1991;1:71–82. [Google Scholar]
- Felsenstein J. PHYLIP: Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Franco-Zorrilla JM, Fernández-Calvín B, Madueño F, Cruz-Alvarez M, Salinas J, Martínez-Zapater JM. Identification of genes specifically expressed in cauliflower reproductive meristems: molecular characterization of BoREM1. Plant Mol Biol. 1999;39:427–436. doi: 10.1023/a:1006130629100. [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the Arabidopsis ABI3gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q, Ferrándiz C, Yanofsky MF, Martienssen R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsisfruit development. Development. 1998;125:1509–1517. doi: 10.1242/dev.125.8.1509. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROSencodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram GC, Goodrich J, Wilkinson MD, Simon RD, Haughn GW, Coen ES. Parallels between unusual floral organs and fimbriata, genes controlling flower development in Arabidopsis and Antirrhinum. Plant Cell. 1995;7:1501–1510. doi: 10.1105/tpc.7.9.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BGW, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Ohmiya K, Hattori T. RAV1, a novel DNA binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 1999;27:470–478. doi: 10.1093/nar/27.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempin SA, Savidge B, Yanofsky MF. Molecular basis of the cauliflower phenotype in Arabidopsis. Science. 1995;267:522–525. doi: 10.1126/science.7824951. [DOI] [PubMed] [Google Scholar]
- Kim Y, Zhang H, Scholl RL. Two evolutionary divergent genes encode a cytoplasmic ribosomal protein in Arabidopsis thaliana. Gene. 1990;93:177–182. doi: 10.1016/0378-1119(90)90222-d. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Peeters AJM, Soppe W. Genetic control of flowering time in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:43–48. doi: 10.1146/annurev.arplant.49.1.345. [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Tax F, Sussman MR. Identification of transferred DNA insertions within Arabidopsisgenes involved in signal transduction and ion transport. Proc Natl Acad Sci USA. 1996;93:8145–8150. doi: 10.1073/pnas.93.15.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 2000;14:2366–2376. doi: 10.1101/gad.813600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito MI, Town CD, Fujii CY, Mason T, Bowman C, Barnstead M et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Luerssen K, Kirik V, Herrmann P, Misera S. FUSCA3 encodes a protein with a conserved VP1/ABI3-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 1998;15:755–764. doi: 10.1046/j.1365-313x.1998.00259.x. [DOI] [PubMed] [Google Scholar]
- Lütcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA. Selection of AUG initiation codons differs in plants and animals. EMBO J. 1987;6:43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF. The Arabidopsis AGL8 MADS box gene is expressed in the inflorescence meristems and is negatively regulated by APETALA1. Plant Cell. 1995;7:1763–1771. doi: 10.1105/tpc.7.11.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K, Schuller C, Wambutt R, Murphy G, Volckaert G, Pohl T, Dusterhoft A, Stiekema W, Entinn KD, Terryn N et al. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature. 1999;402:769–777. doi: 10.1038/47134. [DOI] [PubMed] [Google Scholar]
- McCarty DR, Hatori T, Carson CB, Vasil V, Lazar M, Vasil IK. The Viviparous-1developmental gene of maize encodes a novel transcriptional activator. Cell. 1991;66:895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- Menzel G, Apel K, Melzer S. Identification of two MADS box genes that are expressed in the apical meristem of the long-day plant Sinapis albain transition to flowering. Plant J. 1996;9:399–408. doi: 10.1046/j.1365-313x.1996.09030399.x. [DOI] [PubMed] [Google Scholar]
- Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA. 1997;94:7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea EK, Rutkowski R, Kim PS. Evidence that the leucine zipper is a coiled coil. Science. 1989;243:538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- Piñeiro M, Coupland G. The control of flowering time and floral identity in Arabidopsis. Plant Physiol. 1998;117:1–8. doi: 10.1104/pp.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS. Phase change and the regulation of shoot morphogenesis in plants. Science. 1990;250:923–930. doi: 10.1126/science.250.4983.923. [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsispromotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Raikhel N. Nuclear targeting in plants. Plant Physiol. 1992;100:1627–1632. doi: 10.1104/pp.100.4.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RWM, Meyerowitz EM. A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell. 1998;92:93–103. doi: 10.1016/s0092-8674(00)80902-2. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmidt R. Synteny: recent advances and future prospects. Curr Opin Plant Biol. 2000;3:97–102. doi: 10.1016/s1369-5266(99)00048-5. [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. ETTIN patterns the Arabidopsisfloral meristem and reproductive organs. Development. 1997;124:4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Gendall AR, Dean C. When to switch to flowering. Annu Rev Cell Dev Biol. 1999;15:519–550. doi: 10.1146/annurev.cellbio.15.1.519. [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kao CK, McCarty D. The conserved domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell. 1997;9:799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- Varagona MJ, Schmidt RJ, Raikhel NV. Monocot regulatory protein Opaque-2 is localized in the nucleus of maize endosperm and transformed tobacco plants. Plant Cell. 1991;3:105–113. doi: 10.1105/tpc.3.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;68:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]