Abstract
Noroviruses (Caliciviridae) are RNA viruses with a single-stranded, positive-oriented polyadenylated genome. To date, little is known about the replication strategy of norovirus, a so-far noncultivable virus. We have examined the initiation of replication of the norovirus genome in vitro, using the active norovirus RNA-dependent RNA polymerase (3Dpol), homopolymeric templates, and synthetic subgenomic or antisubgenomic RNA. Initiation of RNA synthesis on homopolymeric templates as well as replication of subgenomic polyadenylated RNA was strictly primer dependent. In this context and as observed for other enteric RNA viruses, i.e., poliovirus, a protein-primed initiation of RNA synthesis after elongation of the VPg by norovirus 3Dpol was postulated. To address this question, norovirus VPg was expressed in Escherichia coli and purified. Incubation of VPg with norovirus 3Dpol generated VPg-poly(U), which primed the replication of subgenomic polyadenylated RNA. In contrast, replication of antisubgenomic RNA was not primer dependent, nor did it depend on a leader sequence, as evidenced by deletion analysis of the 3′ termini of subgenomic and antisubgenomic RNA. On nonpolyadenylated RNA, i.e., antisubgenomic RNA, norovirus 3Dpol initiated RNA synthesis de novo and terminated RNA synthesis by a poly(C) stretch. Interestingly, on poly(C) RNA templates, norovirus 3Dpol initiated RNA synthesis de novo in the presence of high concentrations of GTP. We propose a novel model for initiation of replication of the norovirus genome by 3Dpol, with a VPg-protein-primed initiation of replication of polyadenylated genomic RNA and a de novo initiation of replication of antigenomic RNA.
Norovirus (NV) is a major pathogen of acute gastroenteritis, infecting infants and adults worldwide. The NV genome consists of a positive-sense, single-stranded, poly(A)-tailed RNA of approximately 7.5 kb, encapsidated in a protein core. It is subdivided in three open reading frames (ORF) encoding nonstructural and structural viral proteins. ORF1 encodes a polyprotein that is further processed by autocatalytic cleavage leading to at least five viral enzymes (21). ORF2 encodes the norovirus capsid and ORF3 a small basic protein predicted to play an essential role in infectivity (24). ORF2 and ORF3 genes constitute the viral subgenomic RNA, with a length of about 2.5 kb and a poly(A) tail. Furthermore, in norovirus, the genomic and subgenomic RNAs display identical sequences of about 35 nucleotides (nt) at their 5′ termini.
Norovirus RNA-dependent RNA polymerase (3Dpol; about 57 kDa) is located at the 3′ end of ORF1. It bears in its active site highly conserved motives among RNA viruses (GDD) as well as the canonical GLPSG motives characteristic of the Caliciviridae (1). Activity of the recombinant NV polymerase has already been demonstrated in vitro (2, 4, 19). However, discordant data on initiation of RNA synthesis by norovirus 3Dpol was reported. According to our observations (19) as well as those of Fukushi et al. (4), norovirus 3Dpol initiates RNA synthesis de novo on nonpolyadenylated templates, whereas initiation on polyadenylated templates is strictly primer dependent. In contrast, other authors have postulated a norovirus 3Dpol RNA initiation by a back-priming mechanism (2).
Initiation of RNA synthesis by viral RNA-dependent RNA polymerases has been classified into three major mechanisms so far: a de novo initiation, a primer-dependent initiation, and a back-priming-based initiation (7). However, in members of the Picornaviridae, e.g., poliovirus and foot-and-mouth disease virus, and also in animal caliciviruses, such as the rabbit hemorrhagic disease virus, an additional mechanism of initiation of RNA synthesis termed “protein priming” has been previously described (8, 10, 12-17, 22). In those viruses, initiation of RNA synthesis relies upon uridylylation and subsequent elongation of a viral protein, designated VPg (virion protein, genome-linked), in the presence of the polyadenylated genomic RNA. This “protein-primed” initiation occurs after the annealing of the elongated VPg-poly(U) to the poly(A) tail of the viral genome. Interestingly, the norovirus genome encodes a VPg. This viral protein is predicted to play an important role in the replication of the norovirus genome, similar to the role of picornavirus VPg. However, in picornavirus, the VPg is 22 amino acids in length. In norovirus, the VPg has a predicted length of 133 amino acids and a molecular mass of 15.8 kDa. It remains unclear so far whether norovirus 3Dpol is able to uridylylate and elongate the VPg, leading subsequently to protein-primed initiation of replication of the viral genome. Furthermore, initiation of replication of antigenomic viral RNA remains unclear. Norovirus antigenomic RNA is predicted to play an important role in the replication of the norovirus genome, being the template used for synthesis of genomic RNA.
In this study, we have investigated the replication of the norovirus genome in vitro using recombinant active norovirus 3Dpol and the subgenomic RNA as a surrogate for the norovirus genome. Our results suggest that norovirus 3Dpol initiates replication of the viral genome in a protein-primed manner, after elongation of VPg. In contrast, norovirus 3Dpol initiates replication of antigenomic RNA de novo on a poly(C) stretch, added at the 3′ terminus by terminal transferase activity. Those differential mechanisms of initiation of RNA synthesis reflect a flexibility of norovirus 3Dpol modulated by its RNA templates. They also suggest that, in comparison to other single-stranded RNA viruses, norovirus uses a different strategy for replication of the viral genome.
MATERIALS AND METHODS
Generation of RNA templates.
Synthetic RNA templates were generated by in vitro transcription of cDNA as previously described (19). Poly(A) tailing of synthesized RNA was performed with a poly(A)-tailing kit (Ambion) according to manufacturer's instructions. Purification of the poly(A)-tailed RNA was performed with a MEGAClear kit (Ambion) according to manufacturer's instructions and the products visualized on agarose gels after ethidium bromide staining.
Heterologous expression and purification of recombinant NV proteins.
Expression and purification of recombinant norovirus 3Dpol as well as mutated 3Dpol (YGD343GD344G) were performed as previously described (19). For expression and purification of recombinant VPg, cDNA was generated by PCR from NV clone pUS-NorII (GenBank accession number AY741811) using primers 172-Nor-VPg-NheI-for (5′-CGCTAGCGGCAAGAAAGGGAGGAACA AGACTG-3′) and 173-Nor-VPg-HindIII-rev (5′-CATAAGCTTCTCAAAACTGAGTTTCTCATTGTAGTC-3′) and under the same conditions as those previously described (19), with slight modifications. Briefly, cDNA was cloned into the pET-28b(+) vector (Novagen), the expression vector sequenced and used to transform Escherichia coli BL21(DE3)pLysS cells. After induction of cells grown at 37°C, cultures were incubated at 25°C overnight. Cell pellets obtained from 250-ml cultures were washed once in 4 ml phosphate-buffered saline and 1% Triton X-100 (Sigma). Cells were treated with DNase (10 U/ml) for 15 min at 37°C and then sonicated on ice and resuspended in 40 ml of binding buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, 5 mM imidazole). After centrifugation, the cleared lysate was obtained. The His6-tagged VPg was bound on a Ni-nitrilotriacetic acid-Sepharose resin (Novagen) preequilibrated with the binding buffer. The bound protein was washed with the binding buffer containing 60 mM imidazole and eluted with the binding buffer containing 100 mM imidazole. The eluted protein was then dialyzed against buffer A (25 mM Tris-HCl [pH 8.0], 1 mM β-mercaptoethanol, 50 mM NaCl, 5 mM MgCl2, 10% glycerol, and 0.1% Triton X-100). Protein concentration was determined with a bicinchoninic acid protein assay kit (Pierce) based on the biuret reaction. The purified protein was resuspended in glycerol to a final volume of 50% and stored at −20°C.
Western blot analysis of the purified recombinant proteins.
Western blot analysis was performed using Penta-His antibody mouse monoclonal immunoglobulin G1 (QIAGEN) as previously described (19).
RNA-dependent RNA polymerase assays.
The RNA-dependent RNA polymerase activity of NV 3Dpol was assessed in vitro as previously described (19). For primer-dependent initiation of RNA synthesis by norovirus 3Dpol on a homopolymeric RNA template, 1 μg homopolymeric poly(U), poly(G), poly(C), or poly(A) was used as a template, together with 150 pmol of oligo(A)20, oligo(C)20, oligo(G)20, or oligo(U)20 RNA primer per reaction mixture under the conditions described above. Similarly, primer-dependent initiation of RNA synthesis by norovirus 3Dpol on poly(A)-tailed subgenomic RNA (1 μg) was investigated in the presence of 150 pmol of oligo(U)20 RNA primer or a sequence-specific RNA oligonucleotide primer (5′-AAAAGACACUAAAGAAAG-3′) per reaction mixture. In parallel, initiation of RNA synthesis by norovirus 3Dpol was investigated under the same conditions as those described above, except that antisubgenomic RNA (1 μg/reaction mixture) was used in the absence (primer independent) and in the presence of 150 pmol of an RNA (5′-AUGAAGAUGGCGUCGAAUGACGCCAACCCAUCU-3′) or DNA (5′-ATGAAGATGGCGTCGAATGACGCCAACCCATCT-3′) oligonucleotide primer per reaction mixture.
VPg uridylylation assay.
Recombinant norovirus VPg (1 μg) was incubated with norovirus 3Dpol under the same conditions as those used for the assessment of its RNA-dependent RNA polymerase activity, except that reaction buffer (50 mM HEPES [pH 7.5], 1 mM MnCl2), UTP (10 μM), and 1 μg poly(A) RNA were added to the reaction mixture, whereas ATP, GTP, and CTP were omitted from the reaction mixture. The reaction mixture was incubated at 30°C for 2 h. The reaction products were visualized on 12% sodium dodecyl sulfate (SDS)-polyacrylamide gels by autoradiography. To further assess the specificity of the uridylylation reaction, the same conditions were used but in the presence of [γ-32P]UTP (800 Ci/mmol).
VPg-poly(U) elongation assay.
Recombinant norovirus VPg was incubated with norovirus 3Dpol under the same conditions as those used for uridylylation experiments, except that 100 μM UTP was added to the reaction mixture. The reaction was performed at 30°C for 2 h. The reaction products were visualized on 12% SDS-polyacrylamide gels or formaldehyde-agarose gels by autoradiography as described above. To further assess the specificity of the elongation reaction, the elongated VPg-poly(U) was submitted to digestion with RNases A and V1 (Ambion) at final concentrations of 1 μg/μl and 10 U/μl, respectively, for 2 h. The digested VPg-poly(U) was then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
5′ mapping of the NV 3Dpol replication product.
The 5′ end of the 3Dpol replication product was mapped using an RNA ligase-mediated rapid amplification of cDNA ends approach. Subgenomic RNA was first replicated in the 3Dpol assay as previously described (19). The reaction was followed by ligation of a GeneRacer RNA oligonucleotide (5′-CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA-3′; Invitrogen) to the 5′ end of the replicated RNA using T4 RNA ligase (Invitrogen). The RNA ligation reaction mixture with a total volume of 12 μl consisted of 8 μl replicated RNA, 0.25 μg RNA oligonucleotide, 1 μl 10× ligase buffer (330 mM Tris-acetate [pH 7.8], 660 mM potassium acetate, 100 mM magnesium acetate, 5 mM dichlorodiphenyltrichloroethane), 10 mM ATP, 40 U RNasin (Promega), and 5U T4 RNA ligase (Invitrogen). After incubation at 37°C for 1 h, the RNA was reverse transcribed. The reaction mixture (20 μl) consisted of 10 μl of ligated RNA, 2.5 μM of 138-NV-ORF3 primer (5′-ATGACAAAGGCTCTGGACTGGA-3′), and 0.5 mM of dATP, dGTP, dCTP, or dTTP. After incubation at 65°C for 5 min followed by chilling on ice, 200 U Superscript III reverse transcriptase (RT) (Invitrogen), 5× Superscript III RT buffer, 40 U RNasin, and 0.1 M dichlorodiphenyltrichloroethane were added, followed by incubation at 50°C for 1 h. After inactivation of the RT reaction mixture at 70°C for 15 min, 2 U of RNase H (Invitrogen) was added and the reaction mixture incubated at 37°C for 20 min. Two microliters of the reaction mixture was then used for amplification of cDNA by PCR. The PCR mixture (50 μl) consisted of 2 μl cDNA template; 5 μl of 10× Herculase polymerase reaction buffer (Stratagene); 0.2 mM each of dATP, dCTP, dGTP, and dTTP; 5% dimethyl sulfoxide; 1 μM of GeneRacer 5′ primer (5′-CGACUGGAGCACGAGGACACUGA-3′; Invitrogen) and 139-NV-ORF3 primer (5′-GCATTGCCAGGAAGAACTAATC-3′); and 5 U Herculase Hotstart polymerase (Stratagene). Initial denaturation was carried out for 5 min at 94°C. This was followed by 35 cycles of denaturation for 30 s at 94°C, annealing for 1 min at 55°C, and extension for 1 min at 72°C. A final extension step was carried out for 7 min. PCR products were separated by 1% agarose gel electrophoresis in the presence of ethidium bromide, and the gel was visualized under UV light. For second-round amplification, master mix composition and cycling conditions were identical to those used for first-round PCR, except that primers GeneRacer 5′-nested (5′-GGACACUGACAUGGACUGAAGGAGUA-3′) and 140-NV-ORF3 (5′-GCTCTCAACACATCATTTGTCACC-3′) were used. Amplicons were sequenced as previously described (18, 20). Sequences were generated in both directions for each amplified viral nucleic acid.
3′ mapping of the NV 3Dpol replication product.
The 3′ terminus of the 3Dpol replication product was mapped using a CapFinder approach as described by others (3). Briefly, the product of the 3Dpol reaction was reverse transcribed with a Superscript one-step RT-PCR with Platinium Taq kit (Invitrogen). The RT-PCR mixture (50 μl) consisted of 13 μl RNA template; 23 μl of 2× reaction mixture containing 0.4 mM each of dATP, dCTP, dGTP, and dTTP and 2.4 mM MgSO4; 1 μM each of primer CapFinder-dGTP (5′-GAGAGAACGCGTGACGAGAGACTGACAGGGGGGGGH-3′) or CapFinder-dCTP (5′-GAGAGAACGCGTGACGAGAGACTGACACCCCCCCCH-3′) and 197-Nor-ORF2 (5′-AGCCACCTGCATAACCATTG-3′); and 2 μl RT/Platinium Taq enzyme mixture. Reverse transcription was carried out for 45 min at 50°C. This was followed by denaturation for 2 min at 94°C and 40 cycles of denaturation for 30 s at 94°C, annealing for 1 min at 55°C, and extension for 1 min at 72°C. A final extension step was carried out for 7 min. Amplified fragments were visualized on a 1% agarose gel containing 0.25% ethidium bromide. Amplicons were sequenced as described above. Sequences were generated in both directions for each amplified viral nucleic acid.
Deletion analysis of the 3′ termini of subgenomic and antisubgenomic RNA.
Deletion analysis was performed by generating cDNA fragments harboring deletions at the 3′ termini of the corresponding subgenomic and antisubgenomic RNA. Generation of synthetic RNA transcripts was performed as described above.
RESULTS
Generation of norovirus full-length subgenomic RNA transcripts.
In vitro transcription of norovirus full-length subgenomic cDNA (2,473 nt) yielded a single-stranded RNA transcript that was visualized on formaldehyde-agarose gel. Tailing of the synthetic RNA by poly(A) polymerase resulted in the addition of about 150 nt at its 3′ terminus, yielding a full-length subgenomic poly(A) RNA.
Heterologous expression and purification of recombinant norovirus proteins.
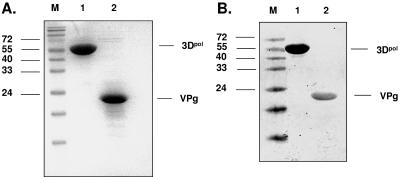
Expression and purification of recombinant norovirus 3Dpol as well as mutated 3Dpol (YGD343GD343G) were performed as previously described (19), yielding a recombinant protein (Fig. 1A). Similarly, expression of recombinant norovirus VPg was performed with E. coli BL21(DE3)pLysS cells (Novagen). A fusion protein containing a His6 tag at its N and C termini was overexpressed at 25°C and purified by Ni-nitrilotriacetic acid affinity chromatography as a soluble protein of 19.8 kDa (169 amino acids in length) (Fig. 1A). Western blot analysis of the elution fraction with anti-His antibodies confirmed the reactivity at an apparent molecular mass of about 20 kDa (Fig. 1B). The fraction containing norovirus VPg was dialyzed and used for the uridylylation assays. In addition, the same fraction containing the bacterial cell lysates after transformation with the expression vector alone without the gene of interest was used as a negative control in the uridylylation experiments.
FIG. 1.
Expression and purification of norovirus 3Dpol and VPg in E. coli. (A) SDS-PAGE analysis of the expression products. Lane 1, wild-type norovirus 3Dpol; lane 2, norovirus VPg. M, molecular mass marker (kDa). (B) Western blot analysis of the expression products. Lane 1, wild-type norovirus 3Dpol; lane 2, norovirus VPg.
Norovirus 3Dpol initiates RNA synthesis on homopolymeric RNA templates.
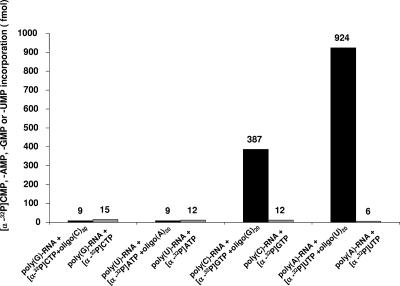
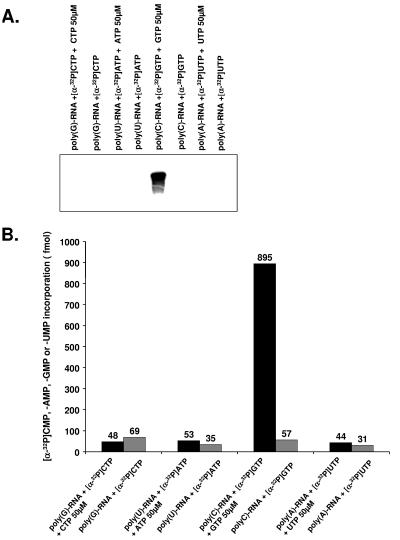
In the first step, purified norovirus 3Dpol was treated with micrococcal nuclease, eliminating a possible contamination with DNA- or RNA-priming fragments that may launch replication of norovirus RNA. In the second step, initiation of RNA synthesis by norovirus 3Dpol was examined on homopolymeric RNA templates. In the absence of an oligonucleotide primer, no incorporation of [α-32P]AMP, [α-32P]CMP, [α-32P]GMP, or [α-32P]UMP was seen (Fig. 2). In contrast, the addition of a complementary oligonucleotide RNA primer to the reaction mixture led to the incorporation of [α-32P]GMP and [α-32P]UMP on homopolymeric poly(C) and poly(A) templates, respectively. Interestingly, the addition of cold GTP to the reaction mixture (50 μM) allowed the incorporation of [α-32P]GMP even in the absence of an oligonucleotide RNA primer (Fig. 3A and B). This was observed on poly(C) templates, indicating that synthesis of RNA from poly(C) homopolymeric templates can take place in a primer-independent manner. For poly(G), poly(U), and particularly poly(A) templates, no incorporation of [α-32P]CMP, [α-32P]AMP, or [α-32P]UMP was seen in the absence of a primer, even after the addition of cold CTP, ATP, or UTP, respectively.
FIG. 2.
Primer-dependent initiation of RNA synthesis on homopolymeric templates. RNA synthesis was performed in the presence (black bars) or in the absence (gray bars) of a cRNA oligonucleotide primer. Incorporation of [α-32P]CMP, [α-32P]AMP, [α-32P]GMP, or [α-32P]UMP was measured after trichloroacetic acid precipitation and collection on G/C glass fiber filters. Incorporation values are indicated.
FIG. 3.
De novo initiation of RNA synthesis on homopolymeric templates. (A) Reaction products were analyzed on formaldehyde-agarose gel and visualized by autoradiography. RNA synthesis was performed in the presence or absence of 50 μM cold CTP, ATP, GTP, or UTP for poly(G) RNA, poly(U) RNA, poly(C) RNA, or poly(A) RNA templates, respectively. (B) RNA synthesis was performed in the presence (black bars) or in the absence (gray bars) of 50 μM cold CTP, ATP, GTP, or UTP for poly(G) RNA, poly(U) RNA, poly(C) RNA, or poly(A) RNA templates, respectively. Incorporation of [α-32P]CMP, [α-32P]AMP, [α-32P]GMP, or [α-32P]UMP was measured after trichloroacetic acid precipitation and collection on G/C glass fiber filters. Incorporation values are indicated.
Our results indicate that norovirus 3Dpol initiates RNA synthesis on or poly(C) or poly(A) homopolymeric templates in a primer-dependent manner. In addition, de novo initiation is possible on poly(C) templates in the presence of high concentrations of purine nucleotides.
Norovirus 3Dpol replicates subgenomic poly(A)-tailed RNA in a primer-dependent manner.
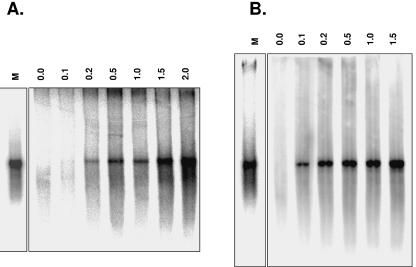
The activity of norovirus 3Dpol was assessed in vitro, with full-length subgenomic poly(A)-tailed RNA used as a template. Norovirus 3Dpol was able to replicate full-length subgenomic RNA (2,473 nt) in a primer-dependent manner (Fig. 4A). Interestingly, when using an RNA primer (5′-AAAAGACACUAAAGAAAG-3′) specific for the sequence adjacent to the poly(A) tail, replication also occurred (Fig. 4B). No replication was observed in the absence of an oligo(U)20 RNA primer on poly(A)-tailed RNA templates.
FIG. 4.
Primer-dependent replication of full-length subgenomic polyadenylated RNA by norovirus 3Dpol. In all reactions, synthetic subgenomic polyadenylated RNA was used as a template. Reaction products were analyzed on formaldehyde-agarose gels and visualized by autoradiography. (A) The reaction was performed in the presence of an oligo(U)20 RNA primer as indicated (mM). M, marker (in vitro transcribed subgenomic polyadenylated RNA). (B) The reaction was performed in the presence of an RNA oligonucleotide complementary to the sequence contiguous to the poly(A) tail. RNA oligonucleotide primer concentrations (mM) are indicated.
These results correlate with the precedent observation on homopolymeric templates, indicating that norovirus 3Dpol initiates RNA synthesis and replication of subgenomic poly(A)-tailed RNA in a primer-dependent manner.
Uridylylation and elongation of norovirus VPg by norovirus 3Dpol.
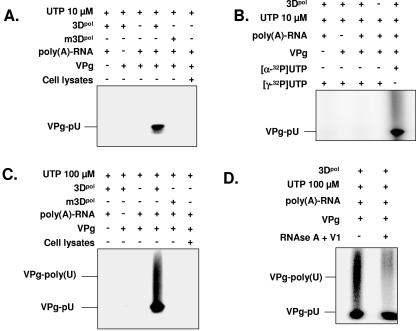
We next examined whether norovirus 3Dpol was able to uridylylate VPg, yielding VPg-pU. SDS-polyacrylamide gel electrophoresis of the reaction products showed that norovirus 3Dpol uridylylates VPg (Fig. 5A). No uridylylation was seen when mutated norovirus 3Dpol was used or in the absence of VPg, in the absence of poly(A) RNA, or in the presence of bacterial cell lysates, demonstrating the specificity of the reaction to VPg, poly(A) RNA, and norovirus 3Dpol, respectively. To determine the specificity of the uridylylation reaction, [γ-32P]UTP was used instead of [α-32P]UTP in the reaction. In this case and as expected, no signal was detected (Fig. 5B), indicating that the signal observed in the presence of [α-32P]UTP is specific and does not result from radiolabeling of VPg.
FIG. 5.
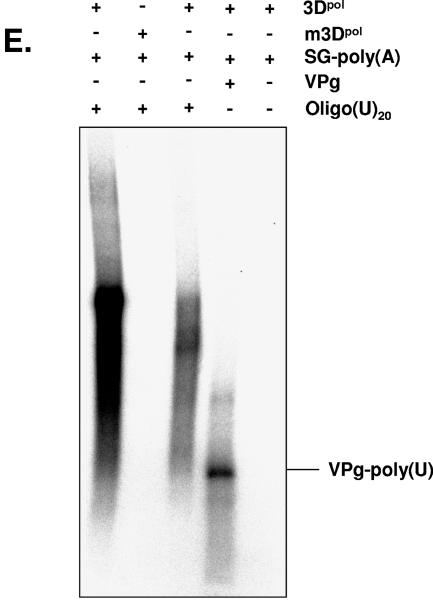
Uridylylation and elongation of VPg by norovirus 3Dpol. Uridylylation and elongation reactions were performed in the presence of poly(A) RNA. Products were analyzed on 12% SDS-polyacrylamide gel or formaldehyde-agarose gel and visualized by autoradiography. (A) Uridylylation of VPg by norovirus 3Dpol and analysis of the uridylylation reaction by SDS-PAGE. Uridylylation of VPg (1 μg) was performed in the presence of UTP (10 μM) or in the presence or absence of poly(A) RNA (1 μg), 3Dpol, mutated 3Dpol (m3Dpol), or a bacterial cell lysate, as indicated. (B) Analysis of the specificity of uridylylation of VPg by norovirus 3Dpol and analysis of the reaction by SDS-PAGE. Uridylylation of VPg (1 μg) was performed in the presence of UTP (10 μM), in the presence or absence of poly(A) RNA (1 μg) or 3Dpol, and in the presence of [γ-32P]UTP, as indicated. As a control, the uridylylation reaction was performed in the presence of [α-32P]UTP, as described above. (C) Elongation of VPg by norovirus 3Dpol and analysis of the elongation reaction by SDS-PAGE. Elongation of VPg (1 μg) was performed in the presence of UTP (100 μM) or in the presence or absence of poly(A) RNA (1 μg), 3Dpol, m3Dpol, or a bacterial cell lysate, as indicated. (D) Analysis of the specificity of the elongation of VPg by norovirus 3Dpol and analysis of the reaction by SDS-PAGE. Elongation of VPg (1 μg) was performed in the presence of UTP (100 μM), poly(A) RNA (1 μg), and 3Dpol, as indicated. The elongated VPg-poly(U) was then treated with a mixture of RNase A and RNase V1 for 2 h at 37°C, as indicated. The reaction products were visualized on SDS-PAGE gels. (E) Protein-dependent initiation of replication by VPg. The subgenomic polyadenylated RNA was incubated in the presence of an oligo(U)20 RNA primer and wild-type 3Dpol and 0.4 mM of ATP, GTP, CTP, and UTP, yielding a replicated antisubgenomic RNA. In contrast, incubation of the subgenomic polyadenylated RNA [SG-poly(A)] with oligo(U)20 RNA in the presence of m3Dpol or with 3Dpol but without an oligo(U)20 RNA primer did not yield a replication product. Incubation of the subgenomic RNA with VPg (1 μg) and 3Dpol as well as UTP (100 μM), but without ATP, GTP, or CTP, allowed initiation of RNA synthesis.
In order to further characterize the role of VPg in initiating RNA synthesis, elongation experiments were performed in vitro under the same conditions as those used for uridylylation of VPg, except that UTP at a final concentration of 100 μM was used. Similar to the uridylylation reaction, norovirus 3Dpol was able to elongate VPg (Fig. 5C) only in the presence of poly(A) RNA but not when mutated norovirus 3Dpol was used or in the absence of VPg or in the presence of bacterial cell lysates. Furthermore, the sensitivity of the elongation product to RNase digestion was examined by treating the reaction mixture with RNases A and V1. As shown in Fig. 5D, the elongation product was completely digested by RNases A and V1 in the reaction. In contrast, VPg-pU was resistant to both RNase A and RNase V1. These results show that VPg-poly(U) consists of a poly(A)-poly(U) dimer covalently linked to VPg, whereas the VPg-pU results from uridylylation of VPg.
Priming of norovirus subgenomic poly(A) RNA replication by uridylylated VPg.
To further elucidate the priming mechanism of norovirus 3Dpol, purified recombinant VPg was incubated with full-length subgenomic poly(A)-tailed RNA as well as 100 μM UTP under the same conditions as those used for replication, except that ATP, CTP, and GTP were omitted from the reaction mixture. In the absence of the oligo(U)20 primer, no replication was observed. After the addition of 1 μg of VPg, incorporation of [α-32P]UMP in the nascent strand was evidenced, indicating that elongation of VPg-poly(U) and subsequent priming of RNA synthesis had occurred (Fig. 5E). These data suggest that initiation of replication of polyadenylated subgenomic RNA depends on elongation of VPg.
Norovirus 3Dpol initiates replication at the template's 3′ end.
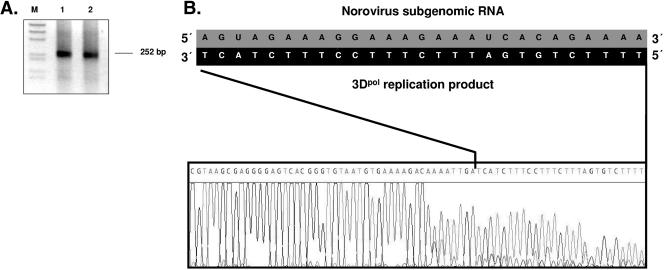
In order to determine the initiation site of norovirus 3Dpol, subgenomic nonpolyadenylated RNA was used as a template in the primer extension approach. Amplification of the 5′ end of the norovirus 3Dpol replication product yielded a 252-bp amplicon (Fig. 6A). Sequencing of the amplicon showed that its 5′ terminus is identical to the 3′ end of the synthetic RNA template, indicating that NV 3Dpol initiates replication at the template's 3′ end (Fig. 6B).
FIG. 6.
Primer extension analysis at the 5′ terminus of the replication product of norovirus 3Dpol. In all reactions, synthetic subgenomic RNA was used as a template for the replication reaction, yielding a replicated RNA corresponding to the antisubgenomic RNA. (A) Lanes 1 and 2, reverse transcription and amplification of the 5′ terminus of the replicated RNA by RNA ligase-mediated rapid amplification of cDNA ends. Amplified cDNA was visualized on 2% agarose gel by UV transillumination after ethidium bromide staining. (B) Sequence analysis of the amplification product. The sequences of the template (3′ terminus of norovirus subgenomic RNA) and the 3Dpol replication product (5′ terminus) are shown.
Norovirus 3Dpol terminates replication at the template's 5′ end.
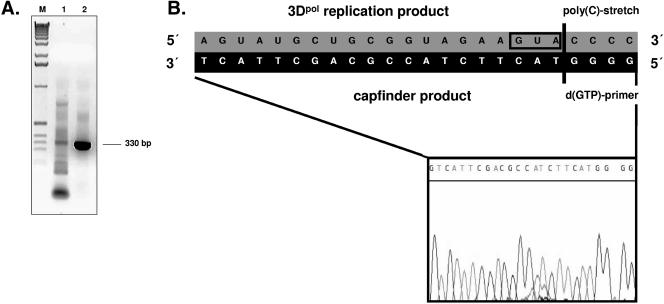
In order to determine the termination site of norovirus 3Dpol replication, subgenomic nonpolyadenylated RNA was used as a template in the CapFinder approach. Amplification of the 3′ end of the norovirus 3Dpol replication product yielded an amplicon of about 330 bp, but only with the CapFinder-d(GTP) primer and not the CapFinder-d(CTP) primer, indicating an annealing of the CapFinder-d(GTP) primer to a poly(C) stretch present on the 3′ terminus of the template (Fig. 7A). Sequencing of the amplicon showed that it displays a poly(G) stretch at its 5′ terminus, corresponding to the CapFinder-d(GTP) primer, thus reflecting the poly(C) stretch at the 3′ terminus of the replication product (Fig. 7B).
FIG. 7.
CapFinder analysis of the 3′ terminus of the replication product of norovirus 3Dpol. In all reactions, synthetic subgenomic RNA was used as a template for the replication reaction, yielding a replicated RNA corresponding to the antisubgenomic RNA. (A) Amplification of the 3′ terminus of replicated RNA by CapFinder analysis. Amplified cDNA was visualized on 2% agarose gel by UV transillumination after ethidium bromide staining. Lane 1, reverse transcription and amplification of the 3′ terminus, with the CapFinder-poly(dCTP) oligonucleotide used as the reverse primer; lane 2, reverse transcription and amplification of the 3′ terminus, with the CapFinder-poly(dGTP) oligonucleotide used as the reverse primer. (B) Sequence analysis of the amplification product. The sequences of the template (3′ terminus of the 3Dpol replication product) and the CapFinder product (5′ terminus) are shown. The 3′ end of the CapFinder-d(GTP) oligonucleotide, the corresponding poly(C) stretch, and the predicted start codon of the subgenomic RNA (located at the 3′ end of the replicated RNA) are highlighted.
Primer-independent initiation of replication on norovirus antisubgenomic RNA.
In order to assess the ability of norovirus 3Dpol to initiate RNA synthesis on heteropolymeric antisubgenomic RNA, norovirus antisubgenomic RNA was synthesized by in vitro transcription. Norovirus 3Dpol was able to replicate antisubgenomic RNA in the absence of an RNA primer or in the presence of an RNA primer or a DNA primer complementary to the first 21 nt at the 3′ terminus of the template (Fig. 8A). These results indicate that norovirus 3Dpol does not depend on the first 21 nt of antisubgenomic RNA to initiate replication, as the blocking of the sequence with an RNA (5′-AUGAAGAUGGCGUCGAAUGACGCCAACCCAUCU-3′) or DNA (5′-ATGAAGATGGCGTCGAATGACGCCAACCCATCT-3′) primer did not abolish replication. Strand separation analysis of the norovirus 3Dpol replication product allowed complete resolution of the double-stranded RNA on 1.25 M formaldehyde-agarose gels, indicating that the replication product did not consist of two RNA strands covalently linked, as would had been expected in the case of a back-priming initiation of norovirus 3Dpol RNA synthesis (Fig. 8B).
FIG. 8.
De novo initiation of RNA synthesis on norovirus antisubgenomic RNA. In all reactions, synthetic antisubgenomic RNA was used as a template for the replication reaction. RNA reaction products were analyzed on native agarose gels and visualized by UV transillumination after ethidium bromide staining. (A) Lanes 1 to 3, RNA synthesis in the presence of wild-type 3Dpol, in the presence of mutated 3Dpol, and in the absence of 3Dpol, respectively. Lanes 4 to 6, RNA synthesis in the presence of wild-type 3Dpol and a 3′ terminus cRNA oligonucleotide, in the presence of mutated 3Dpol and a 3′ terminus cRNA oligonucleotide, and with a 3′ terminus cRNA oligonucleotide but without NV 3Dpol, respectively. Lanes 7 to 9, RNA synthesis in the presence of wild-type 3Dpol and a 3′ terminus cDNA-oligonucleotide, in the presence of mutated 3Dpol and a 3′ terminus cDNA-oligonucleotide, and with a 3′ terminus cDNA-oligonucleotide but without NV 3Dpol, respectively. T, template RNA. R, replication product. M, RNA molecular size marker (kb). (B) Strand separation analysis of the replication products. Reaction products were analyzed on formaldehyde-agarose gels and visualized by UV transillumination after ethidium bromide staining. Lanes 1 and 2, template (antisubgenomic RNA) and norovirus 3Dpol replication products, respectively.
Tentative characterization of the initiation site of replication of subgenomic RNA and antisubgenomic RNA by norovirus 3Dpol.
To further exclude the possibility of a sequence-specific initiation of RNA synthesis by norovirus 3Dpol, deletion analysis was performed on norovirus subgenomic and antisubgenomic RNA. Therefore, nine subgenomic and antisubgenomic RNA truncated at their 3′ termini were synthesized and used in vitro for RNA replication. As shown in Fig. 9 and 10, RNA replication is not dependent on a specific sequence located at the 3′ termini of either subgenomic or antisubgenomic norovirus RNA. The truncated RNAs did not exclusively bear a pyrimidine as the ultimate nucleotide at their 3′ termini, indicating that in vitro, de novo initiation on heteropolymeric templates is not strictly dependent on the presence of a pyrimidine as the ultimate nucleotide at the template's 3′ termini. These data also indicate that RNA synthesis by norovirus 3Dpol is not sequence dependent.
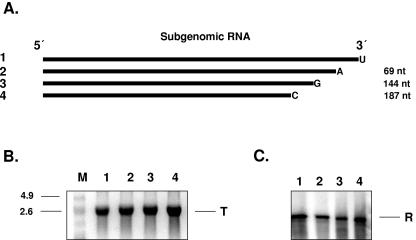
FIG. 9.
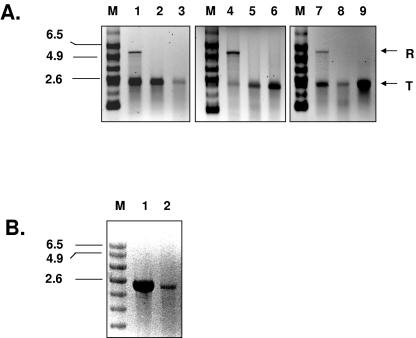
Deletion analysis of the 3′ terminus of the subgenomic RNA template. (A) Schematic representation of deleted subgenomic RNA used as a template. Sizes of deleted fragments are indicated (nt). The ultimate nucleotides at the template's 3′ ends are indicated. (B) In vitro transcription of subgenomic RNA. Reaction products were analyzed on formaldehyde-agarose gel and visualized by UV transillumination after ethidium bromide staining. Lane 1, undeleted subgenomic RNA; lanes 2 to 4, subgenomic RNAs with successive deletions at their 3′ termini. M, RNA molecular size marker (kb). (C) Replication of 3′ terminus-deleted subgenomic RNAs. Reaction products were analyzed on formaldehyde-agarose gel and visualized by autoradiography. Lanes 2 to 4, replication of synthetic subgenomic RNAs with successive deletions at their 3′ termini. T, template RNA; R, replication product.
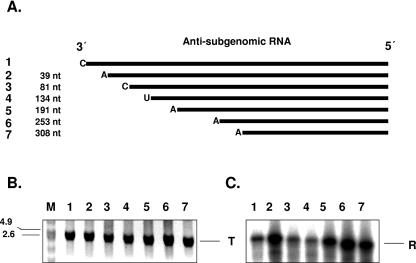
FIG. 10.
Deletion analysis of the 3′ terminus of antisubgenomic RNA. (A) Schematic representation of deleted antisubgenomic RNA used as a template. Sizes of deleted fragments are indicated (nt). The ultimate nucleotides at the template's 3′ ends are indicated. (B) In vitro transcription of antisubgenomic RNA. Reaction products were analyzed on formaldehyde-agarose gel and visualized by UV transillumination after ethidium bromide staining. Lane 1, undeleted antisubgenomic RNA; lanes 2 to 7, antisubgenomic RNA with successive deletions at their 3′ termini. M, RNA molecular size marker (kb). (C) Replication of 3′ terminus-deleted antisubgenomic RNA. Reaction products were analyzed on formaldehyde-agarose gel and visualized by autoradiography. Lanes 2 to 7, replication of antisubgenomic RNAs with successive deletions at their 3′ termini. T, template RNA; R, replication product.
DISCUSSION
In this study, initiation of replication of the norovirus genome by norovirus RNA-dependent RNA polymerase (3Dpol) was examined in vitro, using a recombinant active norovirus RNA-dependent RNA polymerase as well as subgenomic RNA as a surrogate for genomic RNA. Genomic and subgenomic RNA are both polyadenylated at their 3′ termini and bear at their 5′ termini identical 33-nt sequences (5′-AUGAAGAUGGCGUCGAAUGACGCCAACCCAUCU-3′ in NV/Dresden174/1997/GE [GenBank accession number AY741811]), making it possible to address the initiation of replication of the norovirus genome using as a surrogate the subgenomic RNA. We found that norovirus 3Dpol initiates RNA synthesis in a primer-dependent manner on polyadenylated templates, whereas initiation of RNA synthesis on heteropolymeric templates occurs de novo. Our results also suggest that initiation of replication of the norovirus genome is VPg-protein primed, whereas replication of antigenomic RNA occurs de novo.
Norovirus RNA-dependent RNA polymerase is predicted to play an essential role in the replication of the norovirus genome. To date, many aspects relative to norovirus replication remain unclear, mainly because of the lack of a cell culture system for isolation of human norovirus. Indeed, the mechanisms by which norovirus 3Dpol initiates replication as well as the nature of its replication products remain so far uncharacterized. Understanding the molecular mechanisms of norovirus replication in vitro may shed light on the replication strategy used by human norovirus, a major agent of acute gastroenteritis. It is also an important prerequisite for the development of antiviral drugs directed against this essential viral enzyme.
In this study and in the first step, initiation of RNA synthesis by norovirus 3Dpol was examined on homopolymeric templates and found to be strictly primer dependent. As expected, this primer dependency was also observed when using norovirus subgenomic polyadenylated RNA as a template, strictly requiring the addition of an oligo(U)20 RNA primer for initiation of RNA synthesis. However, primer dependency was not limited to homopolymeric templates, as a heteropolymeric sequence-specific RNA primer was also able to initiate replication of polyadenylated subgenomic RNA in a concentration-dependent manner (Fig. 4B). This strict primer dependency was overcome only on homopolymeric poly(C) RNA templates in the presence of GTP, indicating a de novo initiation mechanism on this template.
In the next step, the role of norovirus VPg in initiating RNA synthesis was addressed. The VPg is postulated to display features essential for infectivity. However, its role in initiating replication of norovirus RNA is still not understood. In this study, we have postulated that initiation of replication on norovirus genomic RNA is protein primed, hence depending on uridylylation and subsequent elongation of norovirus VPg by 3Dpol, similar to protein-primed initiation of replication of poliovirus genomic RNA (13-16). Here, full-length polyadenylated subgenomic RNA was used, as both genomic and subgenomic RNA bear at their 3′ termini identical poly(A) tails, making it possible to address this question on full-length subgenomic RNA. We have expressed recombinant VPg in E. coli and purified it to a milligram concentration. Incubation of norovirus VPg with 3Dpol in the presence of a poly(A) template led to uridylylation of VPg and formation of VPg-pU, as evidenced by SDS-PAGE analysis. Uridylylation was strictly dependent on the presence of VPg, 3Dpol, and poly(A) RNA in the reaction mixture, and incubation of [γ-32P]UTP did not lead to labeling of VPg, sustaining the specificity of the uridylylation reaction. Incubation of VPg with 3Dpol in the presence of a poly(A) template and high concentrations of cold UTP led to elongation of VPg and generation of VPg-poly(U). This was not observed in the absence of poly(A) RNA template or VPg or when mutated 3Dpol was used, indicating the strict dependency of the uridylylation reaction on this template, the VPg, and 3Dpol. Furthermore, the elongated VPg-poly(U) was sensitive to RNase treatment, yielding VPg-pU that is, in turn, RNase resistant. We then examined whether norovirus 3Dpol was able to initiate RNA synthesis on polyadenylated subgenomic RNA in the presence of VPg but in the absence of an oligo(U)20 primer. Incubation of norovirus polyadenylated subgenomic RNA with VPg and 3Dpol allowed initiation of RNA synthesis. These observations strongly suggest a protein-primed initiation of replication of the norovirus genome by 3Dpol, as already reported for animal caliciviruses, i.e., feline calicivirus and rabbit hemorrhagic disease virus (10, 23, 25).
We then examined whether initiation of replication on antigenomic RNA is template dependent or occurs de novo. To address this question, antisubgenomic RNA was synthesized in vitro. Our results indicate that norovirus polymerase carries out replication of antisubgenomic RNA in a primer- and sequence-independent manner. Indeed, norovirus 3Dpol yielded an RNA product in the absence of an exogenous primer but not when 3Dpol was omitted or the inactive 3Dpol mutant was used. This primer-independent replication of nonpolyadenylated subgenomic RNA templates raised the question as to whether RNA sequences located at the 3′ terminus of NV genomic RNA might play an important role in initiating replication. In this context, the 42- to 78-nt-long untranslated region located at the 3′ termini of the genomic and subgenomic RNA is of interest. This sequence is predicted to form a stable hairpin structure (5). Stable RNA secondary structures located at the 3′ termini of single-stranded RNA viruses have been reported to play an important role in the initiation of genomic replication (6, 9, 11, 15). In norovirus, this hairpin at the 3′ end of the genome is conserved all over the genogroups (5). In our study, deletion of this hairpin structure as well as contiguous sequences did not influence replication of subgenomic RNA in vitro. Similarly, deletion of the 3′ terminus of antisubgenomic RNA as well as contiguous sequences did not inhibit RNA replication. Accordingly, initiation of replication in NV does not seem to strictly depend on a stable RNA secondary structure at the 3′ terminus of the genomic RNA or on the 3′ terminus of the antigenomic RNA. Interestingly, a poly(C) stretch was evidenced by CapFinder analysis of the 3′ terminus of antisubgenomic RNA. Norovirus 3Dpol has been shown to display a terminal transferase activity on the 3′ terminus of single-stranded RNA, with a strong preference for CTP (19). Therefore, the addition of a poly(C) stretch at the 3′ terminus of the genome may result from the terminal transferase activity of norovirus 3Dpol. Interestingly, norovirus 3Dpol is capable of de novo initiation on antisubgenomic RNA templates, as shown in Fig. 8. On the basis of these data, we postulate that initiation of replication of antisubgenomic RNA by norovirus 3Dpol occurs de novo on a poly(C) stretch, added at the 3′ terminus of the antisubgenomic RNA by norovirus 3Dpol terminal transferase activity. This is strongly supported by the observation that de novo initiation on RNA genomes preferentially occurs on pyrimidines (here a cytosine), being the ultimate T+1 nucleotide, after the addition of an N+1 nucleotide (7). Strikingly, comparison of the 5′ termini of norovirus genomes sustains our hypothesis, as all NV genomes bear a guanidine 5′ monophosphate at the 5′ termini of their genomic and subgenomic RNAs (5), presumably resulting from transcription of the ultimate cytidine at the 3′ terminus of antigenomic RNA.
According to our results, recombinant norovirus 3Dpol displays a relative flexibility as to initiation of replication that is modulated by the 3′-terminal sequence of its templates. Indeed, norovirus 3Dpol (i) initiates RNA synthesis on homopolymeric poly(A) and poly(C) templates in a primer-dependent and de novo manner, respectively, (ii) replicates norovirus polyadenylated subgenomic RNA only in the presence of an oligo(U)20 primer, (iii) uridylylates and elongates VPg in the presence of poly(A) RNA, (iv) initiates replication on polyadenylated subgenomic RNA in a VPg-protein-primed manner, (v) replicates norovirus antisubgenomic RNA in a primer-independent manner, starting at the very beginning of the 3′ terminus of template RNA, and finally (vi) adds a poly(C) stretch at the 3′ terminus of the replication product through terminal transferase activity, allowing de novo initiation on this template.
In summary, our results suggest a differential initiation of replication of genomic and antigenomic RNA by norovirus 3Dpol. The major determinants of this variability seem to be the poly(A) tail at the 3′ termini of genomic and subgenomic norovirus RNA as well as a poly(C) stretch at the 3′ terminus of the antigenomic RNA. In this context, the roles of other norovirus proteins, like the 2CNTPase, the N-terminal protein, and even the ORF3-encoded protein, remain to be determined.
Acknowledgments
We are grateful to Enno Jacobs for his continuous support.
This study was supported by a start-up grant from the Medical Faculty of Dresden (MeDDrive 2005) as well as the European project VIZIER (Comparative Structural Genomics of Viral Enzymes Involved in Replication), funded by the 6th Framework Programme of the European Commission under reference LSHG-CT-2004-511960.
REFERENCES
- 1.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181(Suppl. 2):S336-S348. [DOI] [PubMed] [Google Scholar]
- 2.Belliot, G., S. V. Sosnovtsev, K. O. Chang, V. Babu, U. Uche, J. J. Arnold, C. E. Cameron, and K. Y. Green. 2005. Norovirus proteinase-polymerase and polymerase are both active forms of RNA-dependent RNA polymerase. J. Virol. 79:2393-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franz, O., I. Bruchhaus, and T. Roeder. 1999. Verification of differential gene transcription using virtual northern blotting. Nucleic Acids Res. 27:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukushi, S., S. Kojima, R. Takai, F. B. Hoshino, T. Oka, N. Takeda, K. Katayama, and T. Kageyama. 2004. Poly(A)- and primer-independent RNA polymerase of Norovirus. J. Virol. 78:3889-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green, K. Y., R. M. Chanock, and A. Z. Kapikian. 2001. Human caliciviruses. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 6.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 7.Kao, C. C., P. Singh, and D. J. Ecker. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 8.Kuhn, R. J., H. Tada, M. F. Ypma-Wong, B. L. Semler, and E. Wimmer. 1988. Mutational analysis of the genome-linked protein VPg of poliovirus. J. Virol. 62:4207-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsen, G. R., B. L. Semler, and E. Wimmer. 1981. Stable hairpin structure within the 5′-terminal 85 nucleotides of poliovirus RNA. J. Virol. 37:328-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Machin, A., J. M. Martin Alonso, and F. Parra. 2001. Identification of the amino acid residue involved in rabbit hemorrhagic disease virus VPg uridylylation. J. Biol. Chem. 276:27787-27792. [DOI] [PubMed] [Google Scholar]
- 11.Nagy, P. D., and A. E. Simon. 1998. In vitro characterization of late steps of RNA recombination in turnip crinkle virus. I. Role of motif1-hairpin structure. Virology 249:379-392. [DOI] [PubMed] [Google Scholar]
- 12.Nayak, A., I. G. Goodfellow, and G. J. Belsham. 2005. Factors required for the uridylylation of the foot-and-mouth disease virus 3B1, 3B2, and 3B3 peptides by the RNA-dependent RNA polymerase (3Dpol) in vitro. J. Virol. 79:7698-7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul, A. V., X. Cao, K. S. Harris, J. Lama, and E. Wimmer. 1994. Studies with poliovirus polymerase 3Dpol. Stimulation of poly(U) synthesis in vitro by purified poliovirus protein 3AB. J. Biol. Chem. 269:29173-29181. [PubMed] [Google Scholar]
- 14.Paul, A. V., J. Peters, J. Mugavero, J. Yin, J. H. van Boom, and E. Wimmer. 2003. Biochemical and genetic studies of the VPg uridylylation reaction catalyzed by the RNA polymerase of poliovirus. J. Virol. 77:891-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 17.Rieder, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohayem, J., S. Berger, T. Juretzek, O. Herchenroeder, M. Mogel, M. Poppe, J. Henker, and A. Rethwilm. 2004. A simple and rapid single-step multiplex RT-PCR to detect Norovirus, Astrovirus and Adenovirus in clinical stool samples. J. Virol. Methods 118:49-59. [DOI] [PubMed] [Google Scholar]
- 19.Rohayem, J., K. Jager, I. Robel, U. Scheffler, A. Temme, and W. Rudolph. J. Gen. Virol., in press. [DOI] [PubMed]
- 20.Rohayem, J., J. Munch, and A. Rethwilm. 2005. Evidence of recombination in the norovirus capsid gene. J. Virol. 79:4977-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seah, E. L., J. A. Marshall, and P. J. Wright. 1999. Open reading frame 1 of the Norwalk-like virus Camberwell: completion of sequence and expression in mammalian cells. J. Virol. 73:10531-10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semler, B. L., C. W. Anderson, R. Hanecak, L. F. Dorner, and E. Wimmer. 1982. A membrane-associated precursor to poliovirus VPg identified by immunoprecipitation with antibodies directed against a synthetic heptapeptide. Cell 28:405-412. [DOI] [PubMed] [Google Scholar]
- 23.Sosnovtsev, S., and K. Y. Green. 1995. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology 210:383-390. [DOI] [PubMed] [Google Scholar]
- 24.Sosnovtsev, S. V., G. Belliot, K. O. Chang, O. Onwudiwe, and K. Y. Green. 2005. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 79:4012-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sosnovtsev, S. V., and K. Y. Green. 2000. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology 277:193-203. [DOI] [PubMed] [Google Scholar]