Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is the etiologic agent for Kaposi's sarcoma (KS) and primary effusion lymphoma (PEL). We previously reported that hypoxia activates KSHV lytic replication and that the promoter for open reading frame 34 (ORF34) contains a functional hypoxia-responsive element (HRE). ORF34 is part of a cluster of lytic genes (ORF34-37) that includes ORF36, a phosphotransferase, and ORF37, a shutoff exonuclease. Rapid amplification of cDNA ends analysis revealed that they share a common polyadenylation signal but have two start sites. Two transcripts were identified, one 3.4 kb encoding ORF35-37, and the other 4.2 kb encoding ORF34 and also having coding potential for ORF35-37. Exposure of PEL cell lines to hypoxia induced messages of lengths consistent with those of these transcripts. Reporter assays with Hep3B cells showed activation of both transcripts by hypoxia. The ORF34-37 promoter region has six consensus HREs. Sequential deletion, site-directed mutagenesis experiments, and Northern blot analysis of RNA produced by constructs indicated that the second HRE (HRE-2) plays a critical role in the hypoxic activation of both RNA transcripts. The ORF35-37 transcript was upregulated by cotransfected hypoxia-inducible factor (HIF). Electrophoretic mobility shift assays demonstrated that HRE-2 and ancillary sequences bind and compete for HIF with hypoxic Hep3B nuclear extract. The activation of this gene cluster by hypoxia may have implications for the pathogenesis of PEL and KS. Moreover, the activation of ORF36 by hypoxia might be exploited to develop targeted therapy for PEL, which arises in a hypoxic environment (pleural effusions).
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV-8), is a gamma-2 herpesvirus originally identified in the biopsy specimen of a Kaposi's sarcoma lesion (8). It is the etiologic agent for Kaposi's sarcoma and two other tumors, primary effusion lymphoma (PEL) and multicentric Castleman's disease (6, 57, 62). KSHV shares significant sequence homology with herpesvirus saimiri and Epstein-Barr virus (EBV) (49, 54). Like other herpesviruses, KSHV can establish latent or lytic infections. Lytic gene expression can be induced by treatment of latently infected cells with chemical agents such as 12-0-tetradecanoyl 13-acetate (TPA) and sodium butyrate (1, 47, 72). However, relatively little is known about the factors responsible for activating KSHV under physiologic conditions.
We previously reported that KSHV could be induced to lytic replication by hypoxia and provided evidence that hypoxia may be a physiologic activator for this virus (11, 24). Cells exposed to hypoxic conditions accumulate hypoxia-inducible factor 1 (HIF-1), which plays an important role in regulating the cellular response (69). HIF-1 is a heterodimeric transcription factor consisting of HIF-1α and HIF-1β subunits, the latter also known as aryl hydrocarbon receptor nuclear translocator (33, 34, 68). HIF-1α is constitutively expressed but rapidly degraded in normoxic cells (29, 35, 55). This degradation involves the hydroxylation of proline-402 and proline-564 residues in an oxygen-dependent degradation domain of HIF-1α (30, 31, 71). The hydroxylated HIF-1α binds to the von Hippel-Lindau tumor suppressor protein, which leads to polyubiquination and degradation via the proteasome (10, 36, 46, 51). In the presence of oxygen, activity of HIF-1α is also regulated by hydroxylation of asparagine-803 within the C-terminal transactivation domain (15, 28, 38, 39). Under hypoxic conditions, these modifications of HIF-1α do not occur, leading to accumulation of the protein and increased ability to recruit coactivators (3, 15, 28, 39, 58). These enzymatic modifications can also be inhibited by iron chelation and cobalt ions (28).
Under conditions of hypoxia, HIF-1α accumulates and associates with HIF-1β, which is constitutively expressed, to form a heterodimer in the nucleus. This heterodimer then binds to hypoxia-responsive elements (HREs) within the promoter or enhancer of various hypoxia-responsive target genes and upregulates expression of these genes (3, 15, 28, 45). Several dozen HIF-1 target genes have been identified, including erythropoietin (EPO), vascular endothelial growth factor, and glucose transporter 1 (17, 25, 58, 59, 61). The core consensus sequence that HIF-1 heterodimers bind to in the promoter region of these genes has been identified as 5′-RCGTG-3′, although variants of this sequence have also been reported previously (28, 60).
Another hypoxia-responsive protein, called endothelial PAS domain protein 1 (EPAS1), or HIF-2α, that is structurally and functionally related to HIF-1α has also been identified (14, 16, 67). Like HIF-1α, HIF-2α also forms a functional heterodimer with HIF-1β (called HIF-2) and activates gene transcription from HRE-containing target genes (14, 16, 67). Certain cellular genes are preferentially responsive to HIF-1 or to HIF-2 (27, 70). A third protein involved in the response to hypoxia, designated HIF-3α, has also been identified (21).
We recently reported that the promoter region of open reading frame 34 (ORF34), a lytic KSHV gene of unknown function, is strongly activated by hypoxia (24). ORF34 along with ORF35-37 is part of a cluster of genes all oriented in the same direction within the KSHV genome. The function of ORF34 and of ORF35 is not known. ORF36 is a phosphotransferase that can phosphorylate ganciclovir, and ORF37 has recently been identified as a shutoff exonuclease (SOX) (5, 19). In the present study, we show that each of the genes of this cluster is upregulated by hypoxia in KSHV-infected PEL cell lines. We further show that these genes are transcribed from two different promoters that form two mRNA species sharing a common poly(A) at the 3′ end. The longer mRNA moiety encodes ORF34-37, while the shorter mRNA is more abundant and encodes ORF35-37. Finally, we show that the promoter of the transcript for ORF35 to ORF37 can be activated by hypoxia, utilizing the same HRE sequence that we previously reported was able to activate ORF34 (24). Thus, each of these genes, including phosphotransferase and SOX, can be upregulated by hypoxia, a finding that may be important in the pathogenesis of KSHV-associated diseases and possibly as a therapeutic target.
MATERIALS AND METHODS
Cell lines and culture conditions.
The PEL cell lines BC-3 and BCBL-1, harboring HHV-8 only (1, 53), and JSC-1, dually infected with EBV and HHV-8 (4), were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum at 37°C in an atmosphere of 95% air and 5% CO2 (normoxia). Where noted, the cells were exposed to hypoxia by culture in an incubator with 1% O2 and 5% CO2 as described elsewhere (11). In some experiments, 100 μM cobalt chloride (CoCl2) was utilized as a chemical mimic of hypoxia. Lytic activation of KSHV in BC-3 and BCBL-1 cells was also induced by treatment with TPA (25 ng/ml), and lytic activation of JSC-1 cells was induced with 0.3 mM sodium butyrate (0.3 mM/ml). In some experiments, the cells were exposed to TPA (25 ng/ml) plus the DNA synthesis inhibitor phosphonoacetic acid (PAA) (300 μg/ml). The cells were harvested at various time intervals after exposure to these conditions, as noted below. Hep3B, a human hepatoma cell line, was obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. This line was used for transient transfection and cotransfection experiments, as described below.
RNA isolation and Northern blot analysis.
Total cellular RNA was isolated from PEL cells that were cultured for 24 h under various conditions by using TRIzol reagent (Invitrogen, California) following the manufacturer's protocol. Hep3B cells transfected with pSEAP reporters were harvested by trypsinization at the end of 48 h of transfection and washed once with phosphate-buffered saline. Total RNA was isolated from the Hep3B cells by using TRIzol and treated with RQ1 RNase-free DNase (Promega) before Northern blot analysis was performed. Northern blot hybridization analysis was performed using a nonisotopic digoxigenin (DIG)-labeled probe generated by PCR following the supplier's protocol (Roche Applied Science, Indianapolis, IN). Briefly, 5 μg of total RNA was fractionated on a 0.9% agarose gel containing 2% formaldehyde and subsequently transferred to Hybond N nylon membranes (Amersham, Piscataway, NJ) in the presence of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) overnight by capillary transfer. After the RNA was fixed to the membrane by UV cross-linking, the membranes were prehybridized for 30 min and hybridized with DIG Easy Hyb buffer (Roche Applied Science) overnight at 50°C with full-length DIG-labeled gene-specific DNA probes incorporated by PCR (specific probe sequences are available on request). The membranes were washed twice for 5 min each with low stringency buffer (2× SSC, 0.1% sodium dodecyl sulfate) at room temperature and then with high stringency buffer (0.1× SSC, 0.1% sodium dodecyl sulfate) at 50°C for 15 min each. After the washing and blocking steps, the membranes were incubated for 30 min with an appropriate dilution of anti-DIG antibody conjugated with alkaline phosphatase. The membranes were then washed twice in washing buffer for 15 min each, equilibrated in equilibration buffer, incubated with CDP-Star solution for 5 min, and exposed to film with an intensifying screen. The membranes were stripped with stripping buffer (50% formamide, 2× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}]) at 65°C for 1 h and then rehybridized with a β-actin probe generated from HHV-8 cDNA as a loading control.
5′ and 3′ RACE.
Rapid amplification of cDNA ends (RACE) was performed using a SMART RACE cDNA amplification kit (BD Biosciences Clontech, California) according to the manufacturer's instructions. One microgram of total RNA prepared from TPA-treated BCBL-1 cells after 48 h of induction was used for first-strand cDNA synthesis. Briefly, for 5′ RACE, first-strand cDNA was primed using a 5′ CDS primer at the 3′ end of the RNA. A SMART 11A oligonucleotide was then added to the 5′ end. After the templates were switched, the cDNA was amplified by PCR with a forward primer, 10× universal primer mix,which is included with the kit, and a gene-specific reverse primer for each of the ORFs: 34, 35, 36, and 37. For 3′ RACE, first-strand cDNA was synthesized with 3′ CDS primer A, also provided with the kit. After the templates were switched, the cDNA was PCR amplified using 10× universal primer mix and gene-specific forward primers for ORF34, -35, -36, and -37. The PCR products were gel purified (Bio-Rad, Hercules, CA), cloned into pGEM-T Easy vector (Promega, Wisconsin), and sequenced to identify the 5′ and 3′ ends of each transcript.
Reporter and expression plasmids.
The full-length reporter plasmid for ORF35-37 (35-37P1/1891 or 35-37P1) contains 2,300 nucleotides (nt) spanning the region from nt −1891 to nt +409, where nt +1 indicates the transcription start site of the mRNA for ORF 35-37 as determined by 5′ RACE. This plasmid includes the methionine initiation codon of ORF35 at nt +73 and extends to before the ATG codon of ORF36 at nt +409. The sequences and locations of the primers used to amplify this fragment were as follows: 35-37P-F1, 5′-CTAGCTAGCCTGGGTCCTCTTACGAAT-3′, nt 53676 to 53693; and 35-37P-R1, 5′-ATAGATCTCTAGGCGCACGGCCACCTC-3′, nt 55976 to 55957 (underlined sequences represent NheI and BglII restriction linkers, respectively). Another promoter, 35-37P2, containing 1,940 nucleotides spanning the region from nt −1891 to nt +49, which is 360 nucleotides short at the 3′ end of the full-length promoter, was also constructed with the same forward primer at the 5′ end and the following reverse primer at the 3′ end: 35-37P-R2, 5′-ATAGATCTGCCGGTATGTCATACGATGG-3′, nt 55615 to 55596. These numbers and sequences are according to those described by Russo et al. (54). These promoter regions were PCR amplified from BCBL-1 cellular DNA and were inserted into pGL3-Basic, a promoterless reporter vector without an enhancer (Promega, Wisconsin), in a sense orientation relative to a luciferase coding sequence at the NheI and BglII sites to generate the reporter plasmids GL3B-35/37P1 and GL3B-35/37P2, respectively. For certain experiments, 35-37P1 and 35-37P2 were excised from pGL3 vector and cloned into the NheI and HindIII sites of pSEAP2-Basic vector (Clontech, California) in a sense orientation relative to the SEAP coding sequence to generate pSEAP-35/37P1 and pSEAP-35/37P2 reporters. SEAP is a secreted form of human placental alkaline phosphatase and is used as a reporter (12). In addition to being used for reporter gene analysis, cells transfected with the SEAP reporter can be studied for RNA and protein analyses.
A series of 5′ deletions of the reporter plasmid 35-37P1/1891 were constructed by the use of different primers at the 5′ end and a common primer, 35-37P-R1, at the 3′ end. The sequences and locations of the forward primers were as follows: 35-37P D1, 5′-CTAGCTAGCGTAACAGCCGTTCAGAAA-3′, nt 54206 to 54223; 35-37P D2, 5′-CTAGCTAGCTGCAAGTATGTTGATAGGG-3′, nt 54366 to 54384; and 35-37P D3, 5′-CTAGCTAGCCTACTTCCAGGTCACATGTA-3′, nt 55236 to 55255 (underlined sequences represent NheI restriction linkers). These primers were used for the amplification of deletions 35-37P1/1361, 35-37P1/1201, and 35-37P1/331, respectively. The amplified products were cloned into the same sites of the pGL3-Basic vector, as described above. To clone 102-bp surrounding sequences of HRE-2 in a simian virus 40 (SV40) promoter, the following PCR primers were used: 35-37P-HRE+ ancillary F, 5′-ATAGGTACCTTCTGGCCGCCTGGTA-3′, nt 54295 to 54310; and 35-37P-HRE+ancillary R, 5′-AGCAGATCTGCCTATTGGAGTCCCTA-3′, nt 54396 to 54380 (underlined sequences represent KpnI and BglII restriction linkers). Using 35-37P1/1891 as a template, these primers were cloned into the KpnI and BglII sites, respectively, of pGL3P.
The K5 promoter contains the region spanning nt −690 to nt +309 upstream of the ATG initiation of K5, where nt +1 indicates the transcription start site of the gene as described previously (23). The sequences and locations of the primers used to clone this region were as follows: K5P-F1, 5′-CCCAAGCTTGAAACCCCAAATAGCCTT-3′, nt 27482 to 27465; and K5P-R1 5′-AATAAGCTTCTCTGCAGCTGGGGTGGA-3′,nt 26484 to 26501 (underlined sequences denote HindIII restriction linkers). This region was cloned into the HindIII sites of pGL3-Basic vector. Expression plasmids encoding human HIF-1α (pHAHIF1α-pcDNA3) and HIF-2α (hEPAS1-pcDNA3), gifts of Eric Huang (NCI, NIH) and Steven L. McKnight (University of Texas), respectively, have been described elsewhere (29, 67). Expression plasmid pHAHIF1αm (P402A, P564A), which produces a mutant, HIF-1α (HIF-1αm), with alanine substitutions at residues 402 and 564 that are functional but resistant to degradation in normoxic cells, was also a gift from Eric Huang (NCI, NIH).
Site-directed mutagenesis.
Plasmids expressing mutagenized or deleted HRE were constructed using the PCR-based QuikChange site-directed mutagenesis kit (Stratagene, California) according to the manufacturer's protocol. Briefly, the mutation or deletion was carried out on the full-length 35-37P1/1891 construct with synthetic oligonucleotide primers containing the mutated or deleted HRE sequence. In these experiments, the 35-37P1/1891 construct is labeled 35-37PW to indicate that it is the wild-type promoter. The primer sequence used to generate mutation 1 in the HRE-2 sequence has been shown elsewhere (24). The primer sequences used to generate mutation 2 in HRE-2 were 35-37P HRE mut2F (5′-TTGATCGGCCGTGGATTTGTGCGCGTCCTC-3′) and 35-37P HRE mut2R (5′-GAGGACGCGCACAAATCCACGGCCGATCAA-3′). This mutant plasmid contains a 3-nt substitution in the HRE-2 element, changing the HRE-2 sequence from GACGTG to TTTGTG as underlined (the position of the putative HRE sequence is shown in bold). The following primer pairs were used to generate the HRE-2-deleted plasmids 35-37P HRE dF (5′-TTGATCGGCCGTGGA*CGCGTCCTCGCG-3′) and 35-37P HRE dR (5′-CGCGAGGACGCG*TCCACGGCCGATCAA-3′) (* indicates the deleted HRE-2 sequence). These sequences are complementary to each other. The above primers were also used to generate the SV40 promoter HRE-2 mutation and deletion. To generate the ancillary mutant plasmid, the following primer pairs were used: 35-37P HASmF (5′-CGTCCTCGCGCATTTACCGCATCTGCAAG-3′) and 35-37P HASmR (5′-CTTGCAGATGCGGTAAATGCGCGAGGACG-3′). These sequences are complementary to each other. The mutant plasmid contains a 3-nt substitution in the ancillary sequence, changing the ancillary sequence from CACAC to TTTAC as underlined (the position of the putative ancillary sequence is shown in bold). The reaction was performed for 18 cycles at 95°C for 30 s, 55°C for 1 min, and 68°C for 7 min. The PCR product was digested with DpnI at 37°C for 60 min to digest the parental vector. The treated DNA was used to transform XL1-Blue supercompetent cells. All deletion and mutation constructs were confirmed by sequencing.
Transfection and reporter assays.
All reporter experiments were performed with Hep3B cells, using 12-well plates, transfected with Fugene transfection reagent (Roche Applied Science) according to the supplier's protocol. On the day before transfection, 1.3 × 105 cells were plated per well in a 12-well plate and were transfected with 550 ng of various reporter plasmids and 50 ng of an internal control plasmid, pSV-β-gal (Promega, Wisconsin), which was used to normalize transfection efficiency. After transfection, cells were allowed to grow under normoxic conditions for 30 h and then exposed to either 21% or 1% oxygen for another 18 h. At the end of 48 h of incubation, cells were washed twice with phosphate-buffered saline, lysed with 250 μl of 1× reporter lysis buffer (Promega), and freeze-thawed once. Samples were centrifuged at 13,000 × g for 5 min, and 20 μl and 50 μl of cell lysates were used for luciferase and β-galactosidase (β-gal) activities, respectively. The luciferase value was normalized to the β-gal value to correct for transfection efficiency. The normalized value for the reporters incubated at 21% oxygen was set at unity, compared to the results obtained under conditions of hypoxia, and expressed as induction (n-fold). For SEAP reporter assays, after 18 h of hypoxic incubation, 15 μl of cell culture supernatant was assayed for SEAP activity by using a chemiluminescence detection kit (Clontech). Cell lysates (50 μl) were assayed for β-gal activity, which was normalized as described above.
For Northern analysis, transfection of SEAP reporters containing pSEAP-35/37P1, pSEAP-35/37P2 wild, and HRE2 mutants was performed in a 10-cm dish. On the day before transfection, 1.5 × 106 cells were plated in a 10-cm dish and allowed to grow overnight. Cells were transfected with 6 μg of DNA using Fugene transfection reagent, allowed to grow under normoxic conditions for 30 h, and then exposed to either 21% or 1% oxygen for another 18 h. For cotransfection experiments with HIF plasmids, cells were transfected with 300 ng of wild or mutant reporter of 35/37P1-1891 and with 250 ng of HIF-1α, HIF-1αm, or HIF-2α expression plasmid. As a control, cells were transfected with pcDNA3.1 empty vector. Cells also received 50 ng of an internal control plasmid, pSV-β-gal, for normalization of transfection efficiency. After transfection, cells were incubated under normoxic conditions for 48 h and cell lysates were obtained and assayed for luciferase activity as described above. The results of the mutant reporter transfected with empty vector were set at unity, compared to the results of the wild-type promoter, and expressed as induction (n-fold). All results are reported as the means and standard deviations (SD) of three experiments, and samples in each experiment were performed in triplicate.
Electrophoretic mobility shift assay.
Nuclear extracts were prepared from Hep3B cells, as described previously (24), cultured at normoxia, and exposed to hypoxia for 24 h. An electrophoretic mobility shift assay was performed using a DIG gel shift kit (Roche Applied Science) according to the manufacturer's protocol. Synthetic oligonucleotides were annealed with their corresponding reverse complementary strands and labeled by DIG at their 3′ end. The sequence of each sense strand probe is given in Results. Labeled DNA probes were incubated with 7.5 μg of nuclear extract for 20 min at 4°C. poly(dI-dC) was included per reaction to reduce nonspecific binding. For competition assays, labeled probes were incubated with increasing amounts (50× and 250×) of cold unlabeled double-stranded oligonucleotides, either wild type or mutated, with the nuclear extract. The protein-DNA complexes were separated on a 6% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA buffer, transferred to a nylon membrane, and probed with the appropriate dilution of antidigoxigenin antibody conjugated to alkaline phosphatase, and bands were visualized by nitroblue tetrazolium and BCIP (5-bromo-4-chloro-3-indolylphosphate).
RESULTS
Genomic organization of ORF34-37 gene cluster.
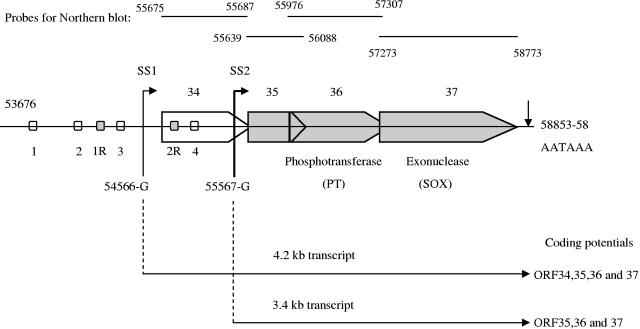
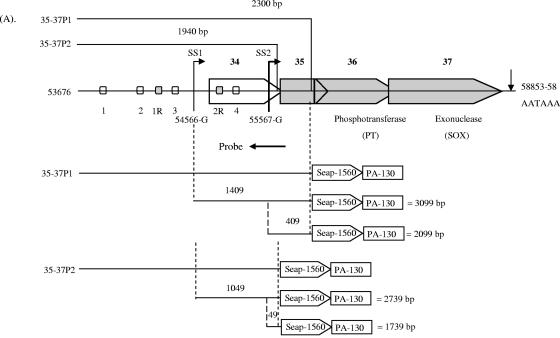
We previously reported that the ORF34 promoter contained a functional hypoxia response element (24). Examination of the genomic organization of KSHV reveals that ORF34 is the furthest upstream gene of a cluster of several overlapping genes, ORF34 to ORF37, all oriented in the same direction (54). This genomic organization and previously reported data (5) suggested that transcription of these genes may be regulated by a complex mechanism. Before we could analyze the possible activation of ORF35, ORF36, and ORF37, it was necessary to investigate the organizational structure of this gene cluster. To this end, we performed RACE analysis for each of these genes in this cluster. This analysis (see the supplemental material for details) revealed two separate transcription start sites at the 5′ end and a single common poly(A) signal at the 3′ end (Fig. 1). Furthermore, it showed that ORF34 transcription starts at position 54566(G), indicated as start site 1 (SS1), while the transcription of ORF35, ORF36, and ORF37 starts at position 55567(G), indicated as SS2 (Fig. 1). All the genes are polyadenylated through a common poly(A) signal at the 3′ end (AATAAA at positions 58853 to 58858). Sequence analysis of the RACE results of ORF34, ORF35, ORF36, or ORF37 showed no evidence of splicing of the mRNA in this region. Thus, two transcripts were identified in this region: a longer 4.2-kb transcript with coding potential for ORF34-37 and a shorter 3.4-kb transcript with a downstream start site and coding potential for ORF35-37 (Fig. 1).
FIG. 1.
Overview of the genomic organization of the KSHV ORF34-37 lytic gene cluster. Genomic organization reveals that the genes in this cluster overlap with each other. ORF34 and ORF35 are of unknown function, whereas ORF36 and ORF37 encode a viral phosphotransferase and a shutoff exonuclease, respectively. 5′ RACE identified the putative transcription start of the mRNA encoding ORF34 at position 54566-G (SS1) and that of the predominant mRNA encoding ORF35, ORF36, and ORF37 at position 55567-G (SS2); these are indicated by thin and thick arrows, respectively. 3′ RACE identified that all the genes use one poly(A) signal at positions 58853 to 58858 for transcription termination. For clarity, ORF34 is left white and ORF35, ORF36, and ORF37 are filled with gray. All numbers are according to those described by Russo et al. (54). Shown above are the full-length gene probes used for Northern blot analyses. Shown below are the two mRNA transcripts identified for this region, their sizes in kilobases (kb), and their coding potentials.
Hypoxic activation of the ORF34-37 gene cluster in PEL cells.
We previously demonstrated by reverse transcription- PCR analysis that mRNA of ORF34 can be induced by hypoxia from KSHV-infected BCBL-1 cells (24). Extending these observations, we sought to determine whether the message for ORF35, ORF36, and ORF37 could also be upregulated by hypoxia or hypoxia-mimicking chemicals. To investigate this, we performed Northern blot analysis, a more quantitative analysis than that of standard reverse transcription-PCR, on each of the genes in the ORF34-37 cluster. We chose two different PEL cell lines for this study: JSC-1, a cell line dually infected with EBV and KSHV that has a high KSHV viral copy number (4), and BC-3, a cell line infected with KSHV only that has a lower viral copy number (1). These lines were cultured under various conditions, including exposure to hypoxia for 24 h, treatment with 100 μM CoCl2, induction with 0.3 mM/ml sodium butyrate, induction with 25 ng/ml TPA, and also induction with 25 ng/ml TPA in the presence of PAA, as described in Materials and Methods. After 24 h of induction, total RNA was isolated and hybridized to each gene-specific probe.
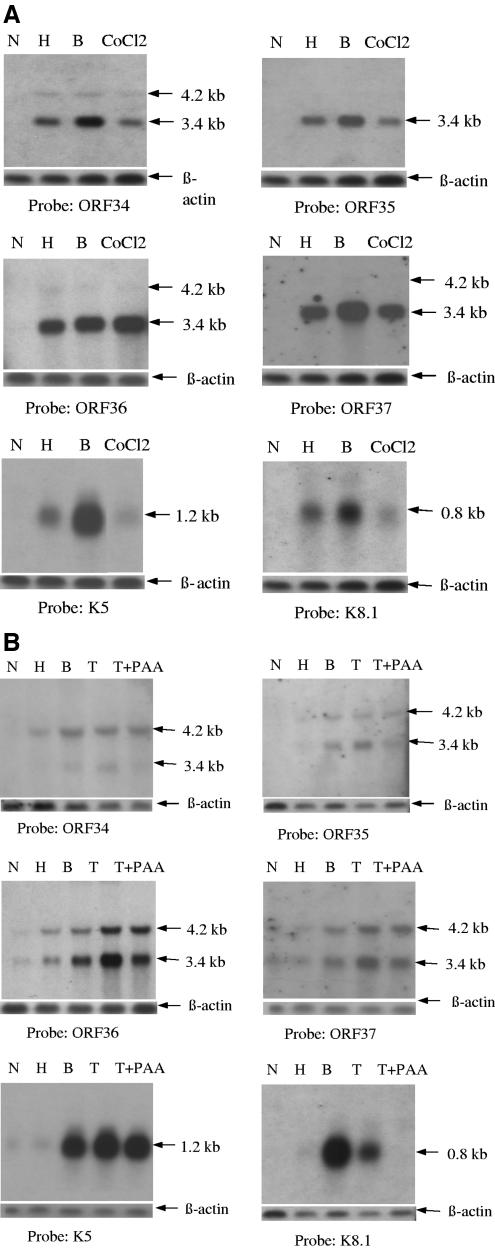
The results indicate that JSC-1 cells exposed to hypoxia have increased levels of mRNA expression when probed with full-length ORF34, ORF35, ORF36, and ORF37 probes (Fig. 2A). Bands of 3.4 kb and 4.2 kb were observed, which are consistent with the lengths of the transcripts originating from the two start sites. It should be noted that the 3.4-kb transcript includes part of the distal sequence of ORF34 (Fig. 1), and this shorter transcript can thus be detected with the full-length ORF34 probe utilized here. The 3.4-kb band was substantially more abundant, and using certain probes (especially ORF35), it was hard to visualize the 4.2-kb transcript. This is consistent with that observed with the RACE analysis described above, which provided evidence that ORF34 is transcribed from the 4.2-kb transcript while ORF35, ORF36, and ORF37 are transcribed from the more abundant 3.4-kb transcript.
FIG. 2.
Northern blot analysis results of the effect of hypoxia, CoCl2, and other inducers of different KSHV genes in PEL cells. (A) JSC-1 cells were cultured under conditions of normoxia (N), exposed to hypoxia (H), induced with 0.3 mM sodium butyrate (B), or exposed to 100 μm CoCl2 (CoCl2) per ml of culture medium. After 24 h, 5 μg of total cellular RNA was loaded per lane of a 0.9% agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized to DIG-labeled full-length probes for the indicated genes (Fig. 1, top). (B) BC-3 cells, which are more tightly latent than JSC-1 cells, were also cultured for 24 h, and the RNA was then hybridized with the same probes that were used for panel A. In this case, cells were also treated with 25 ng/ml of TPA alone (T) or TPA plus 300 μg/ml of phosphonoacetic acid (T+PAA), a viral DNA synthesis inhibitor. The cellular housekeeping gene, the β-actin gene, was used to calibrate the loading control. Arrowheads indicate the sizes of the bands in kilobases (kb).
Exposure of these lines to CoCl2 also induced approximately equal expression levels of the transcripts compared to exposure of the lines to hypoxia, with the exception of the ORF36 probe, which showed somewhat higher levels of mRNA expression after exposure to CoCl2. Expression of the transcripts after induction by hypoxia was slightly less than that after induction by butyrate. As a control, we also probed the same blots of RNA with a probe for K5, which has previously been shown to be an early gene, and with a probe for K8.1, which is a true late gene (7, 23, 43, 65). Expression of K5 and K8.1 in JSC-1 cells exposed to hypoxia was substantially less than that induced by exposure to butyrate, and that induced by exposure to CoCl2 was even less than that induced by exposure to hypoxia (Fig. 2A). None of the probes could detect messages in cells exposed to normoxia alone.
The expression pattern of these genes in BC-3 cells was similar to that in JSC-1 cells, although the intensities were in general weaker and there was relatively greater expression of the larger (4.2 kb) transcript (Fig. 2B). In BC-3 cells, the messages detected by probes for ORF34, ORF35, ORF36, and ORF37 were all induced by hypoxia, although their intensity was somewhat less than that in cells exposed to butyrate or TPA. In contrast to the genes of the ORF34-37 cluster, there was minimal induction of either K5 or K8.1 in BC-3 cells exposed to hypoxia compared to cells exposed to butyrate or TPA (Fig. 2B). These results suggested that as in JSC-1 cells, there was selectively greater expression of ORF34-37 in BC-3 cells by hypoxia than that of either K5 or K8.1, in each case using butyrate or TPA induction as a reference. We then sought to define whether ORF34 to ORF37 were late lytic genes. To this end, we treated the BC-3 cells with 300 μg/ml of PAA, an antiviral drug, to inhibit the late genes whose expression is dependent on viral DNA replication. Transcription of the ORF34-37 gene cluster was minimally inhibited by PAA, suggesting that these are not late genes (Fig. 2B). As a control, we analyzed K5 and K8.1, which were previously identified as early and late genes, respectively (7, 23, 43, 65). Expression of K5 was essentially unaffected in the presence of PAA, while expression of K8.1 was inhibited. Taken together, these results indicate that hypoxia and hypoxia mimics can induce the specific messages for the ORF34-37 cluster of genes and that they do not appear to be late genes.
Functional analysis of the ORF35-37 promoter.
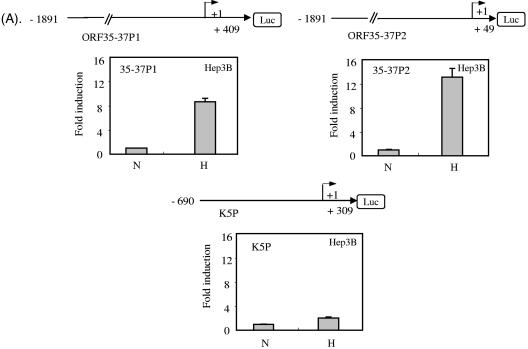
Since hypoxia strongly upregulated ORF35, ORF36, and ORF37 mRNA in PEL cell lines, we were interested in analyzing the promoter region of these genes in a reporter assay system. We also analyzed the K5 promoter for comparison. As discussed earlier, the start site (SS2) of the 3.4-kb mRNA, which encodes ORF35-37, is downstream of the start site (SS1) of the 4.2-kb mRNA. As shown in Fig. 3A, a stretch of the promoter regions containing different lengths upstream of the ORF35-37 and K5 genes was cloned into the pGL3-Basic luciferase reporter vector. For ORF35-37, we constructed two different promoters: the longer promoter, ORF35-37P1, extends to just before the ATG translation initiation codon of ORF36, and the shorter one, ORF35-37P2, had the same start site but was 360 nt shorter at the 3′ end and extends to just before the coding sequence of ORF35. These constructs were transiently transfected into Hep3B cells, and the luciferase activities were determined under various conditions. As shown in Fig. 3A, ORF35-37P1 and ORF35-37P2 were upregulated 8.66-fold and 13.07-fold, respectively, by hypoxia compared to their upregulation by normoxia, whereas the K5 promoter was activated only 1.99-fold by hypoxia. This result is consistent with our Northern blot analysis showing relatively strong upregulation of the 3.4-kb transcript that encodes ORF35-37 mRNA by hypoxia compared to upregulation by butyrate.
FIG.3.
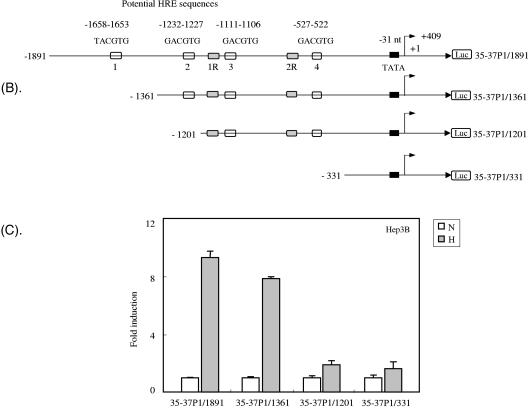
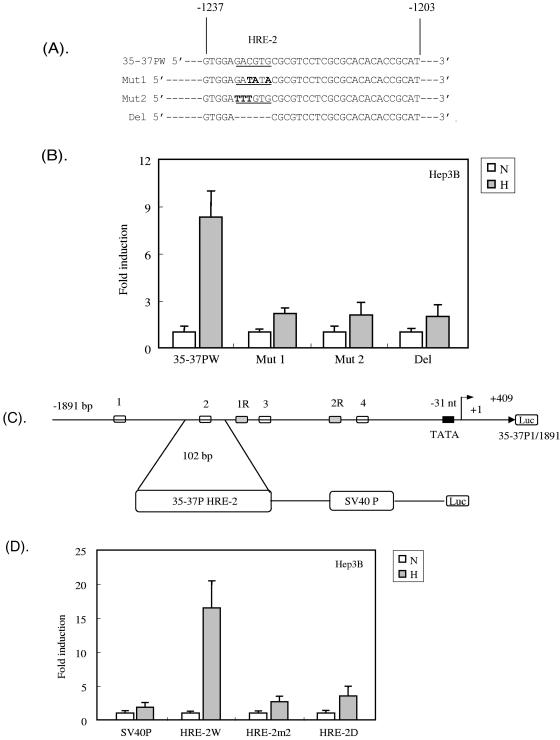
Activation of the ORF35-37 promoter by hypoxia. (A) Hep3B cells were transiently transfected with 550 ng of reporter plasmid encoding the promoter region ORF35-37P1, ORF35-37P2, or K5. The promoter region ORF35-37P1 included +409 bp downstream of the ORF35-37 mRNA transcription start site (SS2) at position 55567; this sequence included the methionine initiation codon of ORF35 at +73 bp and extended to +409 bp, which is just before the methionine initiation codon of ORF36. The promoter region ORF35-37P2 included +49 bp downstream of the SS2 mRNA transcription start site, which is 360 bp shorter at the 3′ end than promoter region ORF35-37P1. The construction of the K5 promoter has been described elsewhere (23). Fifty nanograms of an internal control plasmid, pSV-β-gal, was cotransfected for normalization of transfection efficiency. Cells were allowed to grow under normoxia conditions for about 30 h and then exposed to either normoxia (N) or hypoxia (H) for another 18 h. After normalization of the luciferase (Luc) value to the β-gal value, the induction (n-fold) was calculated compared to the mean normalized value for normoxia alone, which was set at unity. The results depicted are the means from three independent experiments, each done in triplicate, except for ORF35-37P2, which was done in duplicate. Error bars represent one SD, except for ORF35-37P2, for which they represent the range of results. (B) Deletion and reporter constructs of ORF35-37P1. The 5′ deletions were made starting with the ORF35-37P1/1891 construct and were named according to the 5′ end relative to the transcription start site (SS2) for ORF35-37 mRNA at position 55567 as determined by RACE. Shown are the upstream potential HRE sequences in this promoter region. The potential HRE sequences 1, 2, and 3 were previously studied for activity in the ORF34 promoter (24). Putative HRE number 4 is within the coding region of ORF34. The SS2 RNA transcription start site for ORF35-37 at position 55567, which was determined by RACE, is designated nt +1 and indicated by an arrowhead. A potential TATATA box −31 nt upstream of the RNA cap is indicated by a black box. (C) Activity of the full-length ORF35-37P1/1891 and deletion reporters under normoxic and hypoxic conditions. Cells were cotransfected with the full-length and deletion reporter constructs, as described for panel A. After normalization, the induction (n-fold) was calculated as described for panel A. The results depicted are the means from three independent experiments, each done in triplicate, with error bars representing one SD.
To understand this hypoxia responsiveness, we undertook an analysis of the promoter for the 3.4-kb mRNA that encodes ORF35-37 (GL3B-35/37P1). Sequence analysis of the 2,300-bp region of this promoter revealed four potential positively oriented hypoxia response elements with the sequence T/GACGTG, three of them in the previously described ORF34 promoter region and one within the coding region of ORF34 (Fig. 3B). In addition, this region had two reverse-oriented HRE sequences, one between HRE-2 and HRE-3 and one within the coding region of ORF34. In order to identify the region that principally mediated the response to hypoxia, we constructed a series of 5′ progressive deletion mutants of the promoter for ORF35-37 as shown in Fig. 3B and inserted them into the pGL3-Basic reporter vector. Reporter analysis of Hep3B cells indicated that the cells transfected with constructs 35/37P1-1891 and 35/37P1-1361 were activated 9.3-fold and 7.84-fold, respectively, by hypoxia, whereas cells transfected with constructs 35/37P1-1201 and 35/37P1-331 were activated only 1.89-fold and 1.62-fold, respectively (Fig. 3C). These results indicate the presence of cis-acting sequences that are responsive to hypoxia located between positions −1361 and −1201 in the 5′ flanking region of the promoter for ORF35-37. This region contains the HRE-2 consensus sequence.
HRE-2 mediates most of the hypoxia responsiveness of the ORF35-37 promoter.
A mutagenesis strategy was then utilized to assess whether HRE-2 was indeed a hypoxia-responsive region for ORF35 to ORF37. Using site-directed mutagenesis, we made two constructs with mutations in the core HRE-2 sequence and then made a third construct in which the whole HRE-2 sequence was deleted. All the mutations and deletions were made in the context of the full-length ORF35-to-ORF37 promoter region, as shown in Fig. 4A. Reporter assays with Hep3B cells indicated that while the wild-type promoter for ORF35-37 is activated 8.32-fold by hypoxia, the activities of both HRE-2-mutated and HRE-2-deleted reporter plasmids were activated only about 2-fold (Fig. 4B). These results indicate that HRE-2 plays a critical role in the hypoxic activation of this promoter. To further evaluate the hypoxia responsiveness of HRE-2, we tested its activity in a heterologous promoter. A 102-bp sequence surrounding and including HRE-2 was subcloned upstream of the SV40 promoter and used to drive luciferase in the pGL3 promoter vector, as shown in Fig. 4C. We also mutated the HRE-2 sequence in this system and deleted the whole HRE-2 sequence. Transient transfection in Hep3B cells indicated that the SV40 promoter itself was activated about 1.88-fold by hypoxia but that the promoter with the HRE-2 fragment linked to it was activated 16.47-fold by hypoxia. Similar to results reported earlier, the activities were reduced to only 2.69- and 3.50-fold by the mutated and HRE-2-deleted plasmids, respectively (Fig. 4D). These results also confirmed that HRE-2 is important for hypoxic activation.
FIG. 4.
Effects of site-directed mutagenesis and deletion of HRE-2 on 35-37P1/1891 in response to hypoxia. (A) Relevant sequences of the wild-type 35-37P1/1891 (referred to here as 35-37PW) and mutant HRE-2 reporter plasmids. Two different HRE-2 mutant reporters were constructed (Mut1 and Mut2), each containing a 3-base substitution (shown in bold) within the core HRE-2 sequence (shown underlined). In another construct (Del), the entire HRE sequence was deleted. All mutations and the deletion were made in the context of the full-length promoter (ORF35-37P1/1891). (B) Hep3B cells were cotransfected with wild-type promoter (35-37PW) reporter plasmid, HRE-2 mutant reporter plasmid, or an HRE-2-deleted reporter plasmid, assayed as described for Fig. 3A. The results depicted are the means from three independent experiments, each done in triplicate, with error bars representing one SD. (C) Constructs used to assess hypoxic activation of the HRE-2-containing fragment in a heterologous promoter. A 102-bp fragment of the ORF35-37P1/1891 promoter (−1,272 bp to −1,170 bp relative to the SS2 start site) containing wild-type or mutant HRE-2 sequence was cloned upstream of the SV40 promoter driving luciferase in the pGL3 vector as shown. (D) Hep3B cells were cotransfected with the constructs described above containing the 102-bp wild-type fragment (HRE-2W) or with the fragment containing the Mut2 (HRE-2m2) or HRE Del (HRE2D) sequence as described for panel A. SV40P was utilized as a control. The cells were cultured under normoxic (N) or hypoxic (H) conditions and assayed as described for Fig. 3A. The results depicted are the means from three independent experiments, each done in triplicate, with error bars representing one SD.
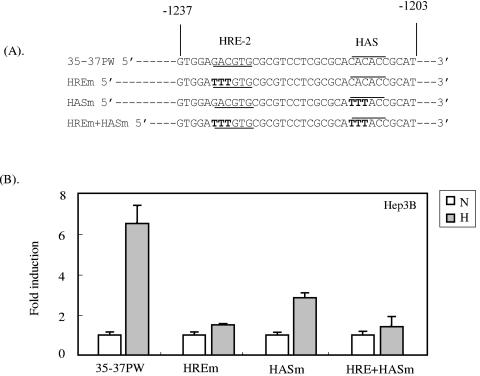
As more has been learned about cellular HREs, it has become apparent that most consist of both a sequence that binds HIF, which is termed the hypoxia binding sequence (HBS), and an additional element, termed the HIF ancillary sequence (HAS) (37, 41, 45, 48, 50). The HBS is the short sequence that was first identified as the HRE, and these two terms are sometimes used interchangeably. The HAS is just downstream of the HBS and is required for full hypoxic activation. The hypoxia-responsive promoters of human EPO, human transferring receptor, human DEC1, and human ceruloplasmin have HASs that contain CACAG/C downstream of their HRE sequences (41, 45, 48, 50). Examination of the promoter sequence for ORF35-37 revealed that it has also a CACAC sequence downstream of HRE-2 (Fig. 5A). To investigate whether this CACAC sequence of the ORF35-37 promoter plays a role in the activation by hypoxia, we introduced a mutation in the HAS alone, in the HRE-2 sequence alone, and in both sequences in the context of the full-length promoter as shown in Fig. 5A. Reporter assays with Hep3B cells showed that mutation in the HAS alone reduced the reporter activity to about half of that observed with the wild-type HRE promoter. When the reporter contained both the HRE mutation and the HAS mutation, its activity was reduced to near control levels. These results suggest that the promoter region for ORF35-37 requires an HAS for full hypoxic activation.
FIG. 5.
ORF35-37 promoter requires an HIF ancillary sequence for full activation. (A). Relevant sequences of the wild-type HRE-2 region (35-37PW), the HRE-2 HBS Mut 2 mutant (MREm), a mutant of the putative HRE-2 HAS (HASm), and combined HRE-2 and HAS mutants. HRE-2 HBSs and their mutations are underlined, while putative HASs and their mutations are overlined. Mutant substitutions of the HRE-2 HBS and HAS are shown in bold. All the mutants were constructed in the context of the full-length promoter (35-37P1/1891). (B) Hep3B cells were cotransfected with the wild-type 35-37/PW plasmid or the mutants described above and exposed to normoxia or hypoxia, and the luciferase activity was determined and normalized as described for Fig. 3A. The results depicted are the means from three independent experiments, each done in triplicate, with error bars representing one SD.
In cells transfected with the ORF35-37 promoter region, the majority of mRNA on exposure to hypoxia is produced by the downstream start site (SS2) mRNA via HRE-2.
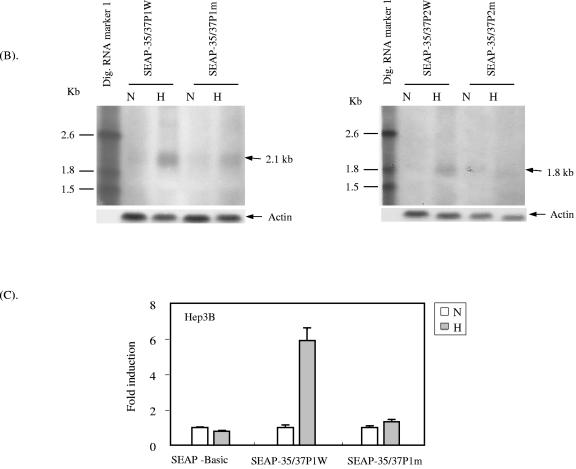
Since the ORF35-37 promoter has two different transcription start sites (SS1 at position 54566, initiating the 4.2-kb message encoding ORF34-37, and SS2 at position 55567, initiating the 3.4-kb message encoding ORF35-37) and because the 35-37P1 constructs described above also included the upstream start site, it remained possible that the induction by hypoxia of these constructs involved initiation from SS1. To clarify whether hypoxia upregulated production of mRNA from the downstream start site, we attempted to analyze the mRNA produced by the luciferase reporter constructs described above by Northern blot analysis but encountered technical difficulties in working with this system. As an alternative approach, we utilized the pSEAP2-Basic vector. We cloned both ORF35-37P1 and ORF35-37P2 wild and HRE-2 mutant reporters into this vector, as described in Materials and Methods. Figure 6A shows the coding potentials from the two start sites when 35-37P1 or 35-37P2 is inserted into SEAP vectors.
FIG.6.
Upregulation by hypoxia of the mRNA transcript starting at SS2 (position 55567) in cells transfected with ORF35-37P. (A) Diagram showing the lengths of potential mRNA transcripts originating from the two identified ORF35-37P start sites (positions 54566 and 55567) in constructs made by inserting either of two ORF35-37P sequences (35-37P1 or 35-37P2) into the SEAP reporter vector. In the pSEAP2-Basic vector, the SEAP gene coding sequence is 1,560 bp long and is followed by a 130-bp SV40 late polyadenylation signal as shown. For 35-37P1, if transcription is initiated from the upstream (SS1, position 54566) transcription start site, the encoded transcript size would be 3,099 bp (1,409 + 1,560 + 130). Similarly, from the downstream (SS2, position 55567) transcription start site, the transcript size would be 2,099 bp (409 + 1,560 + 130). For 35-37P2, the respective transcript sizes would be 2,739 bp and 1,739 bp, respectively. (B) Northern analysis results showing the upregulation by hypoxia of the second transcript (initiating at SS2, position 55567) in ORF35-37P. Hep3B cells were transiently transfected with SEAP-35/37P1W, SEAP-35/37P2W wild-type reporters, or reporters containing the Mut2 HRE-2 sequence (SEAP-35/37P1m and SEAP-35/37P2m). Total RNA was isolated after 48 h of transfection including 18 h of normoxic or hypoxic incubation. Five micrograms of total RNA was obtained from each culture, Northern blotted through a 0.9% agarose-formaldehyde gel, and probed with an antisense single-stranded DNA probe labeled with DIG (probe shown by an arrow in panel A). N, normoxia; H, hypoxia. The cellular housekeeping gene, the actin gene, was used to calibrate the loading control. DIG-labeled RNA molecular weight markers are shown on the left side of each blot. Arrowheads indicate the sizes of the predominant bands in kilobases (kb). (C) Reporter assay results showing the activation of SEAP-35/37 wild-type promoter by hypoxia. Hep3B cells were transfected with SEAP-Basic, SEAP35-37P1W (wild type), and SEAP-35/37P1m (HRE-2 Mut2) reporters and cultured under conditions of normoxia for 30 h. The cells were then cultured under conditions of hypoxia (H) or normoxia (N) for an additional 18 h. Cell culture supernatant (15 μl) was assayed for SEAP activity using a chemiluminescence detection kit (Clontech). The SEAP value was normalized to the β-gal value to correct for transfection efficiency. Induction (n-fold) was calculated as described for Fig. 3A. The results depicted are the means from three independent experiments, each done in triplicate, with error bars representing one SD.
Hep3B cells were transfected with these reporter constructs and cultured under normoxic or hypoxic conditions. Total RNA was isolated and treated with RQ1 RNase-free DNase to eliminate any traces of DNA contamination. Five micrograms of total RNA was subjected to electrophoresis by Northern blot analysis and hybridized to an antisense digoxigenin-labeled single-strand DNA probe. The results revealed that cells exposed to hypoxia produced predominant bands of 2.1 kb and 1.8 kb when cells were transfected with the wild-type 35-37P1 and 35-37P2, respectively (Fig. 6B). These bands are consistent with the expected mRNA produced from the downstream (SS2, position 55567) start site for ORF35-37. The bands were barely detectable in cells cultured under conditions of normoxia and were also fainter in cells transfected with HRE-2 mutant reporters (Fig. 6B). Longer exposure of these blots also revealed weak bands of 3 kb and 2.7 kb, which are the expected sizes if the transcription is initiated from the upstream (SS1, position 54566) start site (data not shown). In parallel experiments, we also assessed the reporter activity of SEAP-35/37P1 in Hep3B transfected cells. As shown in Fig. 6C, hypoxia upregulated SEAP induction in cells transfected with wild-type ORF35/37P1 but not in those transfected with mutant HRE-2. These results demonstrate that hypoxia upregulates transcription of mRNA preferentially from the downstream start site (SS2) that encodes ORF35-37 and that this effect is mediated largely by HRE-2. Thus, hypoxia can upregulate production of mRNA from both the SS1 and SS2 start sites, encoding ORF34-37 and ORF35-37, respectively, and in each case, this effect is mediated largely through HRE-2.
Response of ORF35-37 promoter to HIF-1 or HIF-2, as assessed by cotransfection experiments.
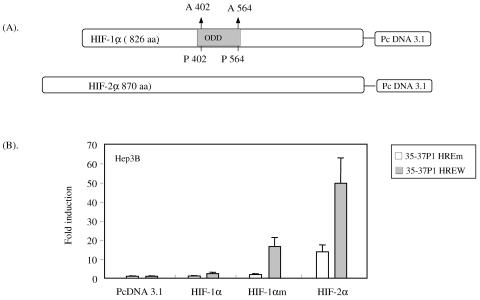
The question still remained as to whether the response of the promoter region for ORF35 to ORF37 to hypoxia was mediated through HIF and, if so, whether the promoter could respond to HIF-1, HIF-2, or both. A difficulty in assessing these effects is that HIF-1α protein is unstable under conditions of normoxia because it is hydroxylated at two specific proline residues (P402 and P564), leading to the binding of the von Hippel-Lindau tumor suppressor protein and degradation. To address this issue, we utilized a mutant HIF-1α plasmid (HIF-1αm) in which the proline residues at positions P402 and P564 were mutated to alanine (P402A and P564A) as shown in Fig. 7A. We have previously found that HIF-2α is not as rapidly degraded under conditions of normoxia, and a mutated form was not needed to perform cotransfection experiments (70). Hep3B cells were transfected with 300 ng of ORF35-37 wild-type or HRE mutant reporter plasmid (Fig. 4A). The cells were then cotransfected with 250 ng of plasmid encoding HIF-1α, HIF-1αm, or HIF-2α (Fig. 7A). Cells also received 50 ng of internal control plasmid to normalize the transfection efficiency. After 48 h of incubation at 21% oxygen, the cells were harvested and assayed for luciferase expression. Cotransfection with the wild-type HIF-1α plasmid increased the ORF35-37 wild-type reporter activity only 2.4-fold (Fig. 7B). However, when the same promoter was cotransfected with the degradation-resistant HIF-1αm plasmid, the promoter was activated 16.5-fold (Fig. 7B). This extent of activation is similar to that found with a number of cellular genes activated by hypoxia (17, 25, 27, 58, 59, 61, 70). Cotransfection with the HIF-2 plasmid resulted in an even greater increase in activity of 49.6-fold.
FIG. 7.
ORF35-37 wild-type promoter (35-37PW) is strongly activated by a degradation-resistant HIF-1 plasmid in normoxic cells. (A) Diagram showing the constructs of the HIF-1α and HIF-2α expression plasmids that were used to cotransfect Hep3B cells with 35-37P1 HRE wild-type and HRE-2 Mut-2 mutant reporter plasmids. Degradation-resistant HIF-1α (HIF-1αm) plasmid has two proline-to-alanine mutations in the oxygen-dependent domain (ODD) at positions 402 (P402A) and 564 (P564A) that allows the protein to escape from degradation and be functional at normal oxygen tension. (B) Hep3B cells were cotransfected with 300 ng of either wild-type ORF35-37P1 (HREW) or ORF35-37P1 containing the Mut2 HRE-2 (HREm) reporter plasmid along with 250 ng of wild-type HIF-1α, mutant HIF-1αm, or wild-type HIF-2α plasmid. As a control, cells were cotransfected with pcDNA3.1 empty vector. Cells also received 50 ng of an internal control plasmid, and the total amount of DNA was adjusted by the addition of the appropriate amount of filler plasmid. Cells were incubated under normoxic conditions for 48 h, and the luciferase activity was determined and normalized as described for Fig. 3A. The normalized value of the mutant reporter (HREm) cotransfected with empty vector was set at unity, compared to the results obtained with the wild-type reporter (HREW), and expressed as induction (n-fold). Bars depict the means from three independent experiments, each done in triplicate, with error bars representing one SD.
HIF binds to the HRE-2 sequence of the ORF35-37 promoter.
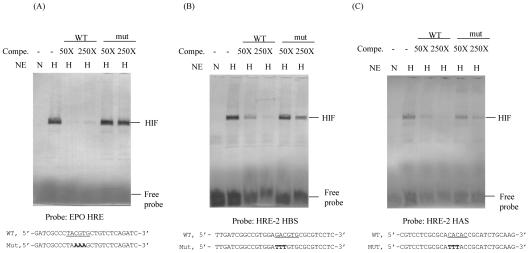
To determine whether the HRE-2 sequence identified in the ORF35-37 promoter region binds to HIF, electrophoretic mobility shift assays were performed using nuclear extract from normoxic and hypoxic Hep3B cells. For a control in these experiments, we used the HRE from EPO, a prototype hypoxia-inducible gene that has a well-characterized HRE (45). As shown in Fig. 8, EPO HRE, the putative HRE-2 HBS, and the putative HRE-2 HAS each formed a complex in nuclear extracts from hypoxic Hep3B cells but not in extracts from normoxic Hep3B cells. This complex is specific, since an excess of unlabeled wild-type probe (50× and 250×) is competed with the DIG-labeled wild-type probe in a dose-dependent manner, whereas unlabeled oligonucleotides with mutations in the HRE or ancillary sequence are largely ineffective, even at a 250-fold excess. This result is consistent with our previous finding (24) that HRE-2 can be competed with EPO HRE in an HIF-1 enzyme-linked immunosorbent assay. Taken together, these data demonstrate that HIF binds to the consensus HRE-2 sequence present within the ORF35-37 promoter.
FIG. 8.
Results of electrophoretic mobility shift assays showing the specificity of DNA-protein interaction using digoxigenin-labeled double-stranded probes. Synthetic oligonucleotides were labeled by DIG at the 3′ end. Each labeled probe was incubated with nuclear extract (NE) produced from normoxic (N) or hypoxic (H) Hep3B cells. To demonstrate the specificity of the DNA-protein interaction, increased quantities (50× and 250×) of unlabeled wild-type (WT) (same sequence as that of probe) or mutated (mut) oligonucleotides were added to the binding reaction. The protein-DNA complexes were separated on a 6% nondenaturing polyacrylamide gel, blotted to a nylon membrane, and probed with antidigoxigenin antibody conjugated to alkaline phosphatase. The sequence of each sense strand probe or competing oligonucleotide (Compe.) used is shown at the bottom of each blot. The wild-type HRE sequence is underlined, and the mutant sequence is shown in bold. The positions of the HIF complex and the free probe are shown. The blots shown in panels A, B, and C are probed with EPO HRE, ORF35-37P HRE-2 HBS, and ORF35-37 HRE-2 HAS, respectively.
DISCUSSION
In this study, we show that genes in the ORF34-37 cluster of KSHV are members of the growing family of genes that are transcriptionally activated by hypoxia through HIFs. Although genes in the cluster are transcribed from two different promoters, both RNA transcripts respond to HIF primarily through the same HRE sequence (HRE-2).
ORF34, along with ORF35-37, comprises a cluster of genes all oriented in the same direction. By RACE analysis, we found that the RNA species for these genes are transcribed from two different promoters but share a common poly(A) signal at positions 58853 to 58858. ORF34 is transcribed from the upstream start site (SS1) at position 54566 (SS1), while RNAs for ORF35, ORF36, and ORF37 are transcribed predominantly from a downstream start site (SS2) at position 55567, which is within the ORF34 coding region. It is not uncommon for two or more genes within KSHV to share a common promoter or poly(A) signal. For example, it has been reported that KSHV RTA, K8, and K8.1 transcripts are initiated from alternative promoters but share a common poly(A) signal (40, 44, 63, 66, 73). Also, ORF71(v-FLIP), ORF72 (v-Cyclin), and ORF73 (LANA) are transcribed from a common promoter and share a common poly(A) signal (13, 32, 56, 64, 73). Interestingly, ORFK13 (OX-2) and ORF74 (v-GPCR), which are transcribed in the opposite direction of ORF71-73, also share a single promoter and use a common poly(A) signal at the 3′ end (64). It is unclear at this time exactly how the gene products of ORF36 and ORF37 are translated. ORF35, ORF36, and ORF37 overlap, and we did not find any evidence of gene splicing. It is quite possible that translation of ORF36 and ORF37 involves translation reinitiation. Alternatively, it is possible that it involves leaky scanning of the upstream start sites or internal ribosome entry sites, as has been described for other KSHV genes (2, 20, 42). While the results here suggest that there is greater production of the 3.4-kb transcript, this does not exclude the possibility that ORF36 and/or ORF37 is in fact translated from the 4.2-kb transcript. Additional research will be needed to understand these issues. In either case, the results of this paper show that these genes, in addition to ORF34 and ORF35, are upregulated by hypoxia through HRE-2.
Using two different PEL cell lines exposed to normoxia or hypoxia, Northern blot analysis generally revealed two bands of 3.4 kb and 4.2 kb in length when cells were probed with each of the genes in the ORF34-37 cluster. Similar results have been reported by Cannon et al. using Northern blot analysis to study JSC-1 cells stimulated by TPA and butyrate (5). The 4.2-kb transcript corresponds in size to that predicted from the first start site (from which ORF34 is transcribed), and the more abundant 3.4-kb transcript corresponds in size to that predicted from the second start site (from which ORF35, ORF36, and ORF37 are predominantly transcribed, as determined by RACE). It is not surprising that the 4.2-kb transcript was in most cases also detected with the probes for ORF35, ORF36, and ORF37, as this transcript also has coding potential for those genes. Also, the upstream region of the 3.4-kb transcript for ORF35 to ORF37 includes part of the ORF34 gene sequence, and this accounts for the detection of the 3.4-kb message when the transcripts were probed with the full-length ORF34 used in these experiments. While the 3.4-kb transcript was more abundant than the 4.2-kb transcript in both cell lines, there seemed to be relatively greater expression of the 4.2-kb transcript in the BC-3 line than in the JSC-1 cell line. The reasons for this are not clear at this time but might be related to the fact that JSC-1 has higher lytic KSHV replication or is coinfected with EBV.
We have previously shown that HIF-induced activation of ORF34 was mediated largely by HRE-2, which spans the region from positions −231 to −226 from the ORF34 transcription start site (24). The promoter for ORF35 to ORF37 was found to be strongly activated by hypoxia as well, and we were interested in determining whether this was mediated by the same HRE sequence or whether a different HRE or mix of HREs was responsible. This question was of particular interest because of the identification of an additional consensus HRE sequence (HRE-4) within the ORF34 coding region. A series of deletion and mutation experiments revealed that as with ORF34, only HRE-2 played a major role in the activation of the promoter for ORF35 to ORF37 by hypoxia. Also, as with ORF34, the activation of ORF35-37 was more responsive to HIF-2 than to HIF-1.
Previously, we have also shown that this HRE-2 sequence can bind to HIF, as assessed by an HIF-1 binding and competition enzyme-linked immunosorbent assay utilizing nuclear extracts of Hep3B and BCBL-1 cells (24). We now show further the binding of HRE-2 to HIF in an electrophoretic mobility shift assay. Most of the known functional HREs of cellular genes contain a sequence, termed the HAS, that is downstream and adjacent to the HBS and that is required for full hypoxic activation (37, 41, 45, 48, 50). Analysis of the promoter region around HRE-2 revealed the presence of a downstream ancillary-like sequence (CACAC). Using site-directed mutagenesis studies of reporter plasmids containing the ancillary sequence of this HRE, we have shown that mutation of this putative HAS markedly reduces the hypoxia responsiveness of the promoter for ORF35 to ORF37 (Fig. 5A and B). Thus, we show that a viral HRE is organizationally similar to cellular HREs in that it has both an HBS and an HAS and both are required for full function.
Activation of herpesviruses from the resting to the lytic state generally involves induction of one or more switch genes which in turn activate a cascade of viral genes in an orderly transcriptional program. In the case of KSHV, the lytic switch gene is the Rta gene, and we have previously shown that the promoter region of the Rta gene can be activated to a limited extent by hypoxia and to a greater extent by transfection with HIF-1 or HIF-2 (24). The results of the present study indicate that genes of the ORF34-37 cluster can be substantially and directly activated by hypoxia through the binding of HIF-1 or HIF-2 to an HRE. Northern blot analyses, for which results are shown in Fig. 2, provide evidence that the ORF34-37 cluster is activated to a greater extent under conditions of hypoxia than two other viral genes (K5 and K8.1) that are not known to have a functional HRE in their promoter region. This genetic structure would enable KSHV to have a greater, or perhaps an earlier, activation of ORF34-37 in a hypoxic environment than under other conditions of viral activation. This specific activation is similar to the activation of KSHV viral interleukin-6 by alpha interferon through binding to an interferon-stimulated response element in the promoter region of viral interleukin-6 (9). ORF34 and ORF35 are proteins of unknown function that have sequence homology with the EBV genes BGLF3 and BGLF4, respectively (54). ORF36 is a phosphotransferase or serine kinase that has been hypothesized to phosphorylate regulatory proteins and in this way functions to regulate expression of key cellular or viral genes (5, 52). Recently, it has also been shown that ORF36 can activate the c-Jun N-terminal protein kinase signaling pathway (22). ORF37, a homolog of DNA exonucleases in other herpesviruses, and its product (SOX) have recently been shown to inhibit host gene expression (19). It is unclear at this time what evolutionary advantage the direct activation of these genes would provide for KSHV. Activation of ORF37, for example, may provide a mechanism for KSHV to rapidly shut off host genes in latently infected cells that are exposed to hypoxia. An understanding of these issues may provide insights into the strategies for KSHV infection and replication under various conditions.
Finally, the results of this study could lead to therapeutic considerations for the treatment of KSHV-associated malignancies. ORF36 has been shown to phosphorylate the antiherpes drug ganciclovir, and the phosphorylated form of this drug can be toxic to human cells (5, 52). Thus, under conditions of hypoxia or hypoxia mimics, KSHV-infected cells may substantially activate ganciclovir or related drugs. PEL, for example, arises in body cavities and forms effusions, such as pleural effusions, and these generally are hypoxic (18, 26). Our group is currently exploring possible treatment strategies for PEL based on this approach.
Supplementary Material
Acknowledgments
We thank Steven L. McKnight (University of Texas) for the expression plasmid encoding HIF-2α and Eric Huang (National Cancer Institute) for HIF-1α wild and mutant plasmids. We also thank Hiroaki Mitsuya of the HIV and AIDS Malignancy Branch, NCI, for advice and for critical reading of the manuscript.
This research was supported by the Intramural Research Program of the National Cancer Institute, NIH.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Arvanitakis, L., E. A. Mesri, R. G. Nador, J. W. Said, A. S. Asch, D. M. Knowles, and E. Cesarman. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648-2654. [PubMed] [Google Scholar]
- 2.Bieleski, L., and S. J. Talbot. 2001. Kaposi's sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J. Virol. 75:1864-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruick, R. K., and S. L. McKnight. 2002. Transcription. Oxygen sensing gets a second wind. Science 295:807-808. [DOI] [PubMed] [Google Scholar]
- 4.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon, J. S., F. Hamzeh, S. Moore, J. Nicholas, and R. F. Ambinder. 1999. Human herpesvirus 8-encoded thymidine kinase and phosphotransferase homologues confer sensitivity to ganciclovir. J. Virol. 73:4786-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 7.Chandran, B., C. Bloomer, S. R. Chan, L. Zhu, E. Goldstein, and R. Horvat. 1998. Human herpesvirus-8 ORF K8.1 gene encodes immunogenic glycoproteins generated by spliced transcripts. Virology 249:140-149. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee, M., J. Osborne, G. Bestetti, Y. Chang, and P. S. Moore. 2002. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science 298:1432-1435. [DOI] [PubMed] [Google Scholar]
- 10.Cockman, M. E., N. Masson, D. R. Mole, P. Jaakkola, G. W. Chang, S. C. Clifford, E. R. Maher, C. W. Pugh, P. J. Ratcliffe, and P. H. Maxwell. 2000. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 275:25733-25741. [DOI] [PubMed] [Google Scholar]
- 11.Davis, D. A., A. S. Rinderknecht, J. P. Zoeteweij, Y. Aoki, E. L. Read-Connole, G. Tosato, A. Blauvelt, and R. Yarchoan. 2001. Hypoxia induces lytic replication of Kaposi sarcoma-associated herpesvirus. Blood 97:3244-3250. [DOI] [PubMed] [Google Scholar]
- 12.Deng, H., J. T. Chu, M. B. Rettig, O. Martinez-Maza, and R. Sun. 2002. Rta of the human herpesvirus 8/Kaposi sarcoma-associated herpesvirus up-regulates human interleukin-6 gene expression. Blood 100:1919-1921. [DOI] [PubMed] [Google Scholar]
- 13.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ema, M., S. Taya, N. Yokotani, K. Sogawa, Y. Matsuda, and Y. Fujii-Kuriyama. 1997. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. USA 94:4273-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fedele, A. O., M. L. Whitelaw, and D. J. Peet. 2002. Regulation of gene expression by the hypoxia-inducible factors. Mol. Interv. 2:229-243. [DOI] [PubMed] [Google Scholar]
- 16.Flamme, I., T. Frohlich, M. von Reutern, A. Kappel, A. Damert, and W. Risau. 1997. HRF, a putative basic helix-loop-helix-PAS-domain transcription factor is closely related to hypoxia-inducible factor-1 alpha and developmentally expressed in blood vessels. Mech. Dev. 63:51-60. [DOI] [PubMed] [Google Scholar]
- 17.Forsythe, J. A., B. H. Jiang, N. V. Iyer, F. Agani, S. W. Leung, R. D. Koos, and G. L. Semenza. 1996. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 16:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funahashi, A., T. K. Sarkar, and R. C. Kory. 1971. PO 2, PCO 2, and pH in pleural effusion. J. Lab. Clin. Med. 78:1006. [PubMed] [Google Scholar]
- 19.Glaunsinger, B., and D. Ganem. 2004. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell 13:713-723. [DOI] [PubMed] [Google Scholar]
- 20.Grundhoff, A., and D. Ganem. 2001. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:1857-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu, Y. Z., S. M. Moran, J. B. Hogenesch, L. Wartman, and C. A. Bradfield. 1998. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3alpha. Gene Expr. 7:205-213. [PMC free article] [PubMed] [Google Scholar]
- 22.Hamza, M. S., R. A. Reyes, Y. Izumiya, R. Wisdom, H. J. Kung, and P. A. Luciw. 2004. ORF36 protein kinase of Kaposi's sarcoma herpesvirus activates the c-Jun N-terminal kinase signaling pathway. J. Biol. Chem. 279:38325-38330. [DOI] [PubMed] [Google Scholar]
- 23.Haque, M., J. Chen, K. Ueda, Y. Mori, K. Nakano, Y. Hirata, S. Kanamori, Y. Uchiyama, R. Inagi, T. Okuno, and K. Yamanishi. 2000. Identification and analysis of the K5 gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 74:2867-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haque, M., D. A. Davis, V. Wang, I. Widmer, and R. Yarchoan. 2003. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains hypoxia response elements: relevance to lytic induction by hypoxia. J. Virol. 77:6761-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris, A. L. 2002. Hypoxia—a key regulatory factor in tumour growth. Nat. Rev. Cancer 2:38-47. [DOI] [PubMed] [Google Scholar]
- 26.Houston, M. C. 1981. Pleural effusion: diagnostic value of measurements of PO2, PCO2, and pH. South. Med. J. 74:585-589. [PubMed] [Google Scholar]
- 27.Hu, C.-J., L.-Y. Wang, L. A. Chodosh, B. Keith, and M. C. Simon. 2003. Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol. Cell. Biol. 23:9361-9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, L. E., and H. F. Bunn. 2003. Hypoxia-inducible factor and its biomedical relevance. J. Biol. Chem. 278:19575-19578. [DOI] [PubMed] [Google Scholar]
- 29.Huang, L. E., J. Gu, M. Schau, and H. F. Bunn. 1998. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 95:7987-7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 31.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 32.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang, B. H., E. Rue, G. L. Wang, R. Roe, and G. L. Semenza. 1996. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem. 271:17771-17778. [DOI] [PubMed] [Google Scholar]
- 34.Jiang, B. H., G. L. Semenza, C. Bauer, and H. H. Marti. 1996. Hypoxia-inducible factor 1 levels vary exponentially over a physiologically relevant range of O2 tension. Am. J. Physiol. 271:C1172-C1180. [DOI] [PubMed] [Google Scholar]
- 35.Kallio, P. J., W. J. Wilson, S. O'Brien, Y. Makino, and L. Poellinger. 1999. Regulation of the hypoxia-inducible transcription factor 1alpha by the ubiquitin-proteasome pathway. J. Biol. Chem. 274:6519-6525. [DOI] [PubMed] [Google Scholar]
- 36.Kamura, T., S. Sato, K. Iwai, M. Czyzyk-Krzeska, R. C. Conaway, and J. W. Conaway. 2000. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl. Acad. Sci. USA 97:10430-10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura, H., A. Weisz, T. Ogura, Y. Hitomi, Y. Kurashima, K. Hashimoto, F. D'Acquisto, M. Makuuchi, and H. Esumi. 2001. Identification of hypoxia-inducible factor 1 ancillary sequence and its function in vascular endothelial growth factor gene induction by hypoxia and nitric oxide. J. Biol. Chem. 276:2292-2298. [DOI] [PubMed] [Google Scholar]
- 38.Lando, D., D. J. Peet, J. J. Gorman, D. A. Whelan, M. L. Whitelaw, and R. K. Bruick. 2002. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lando, D., D. J. Peet, D. A. Whelan, J. J. Gorman, and M. L. Whitelaw. 2002. Asparagine hydroxylation of the HIF transactivation domain: a hypoxic switch. Science 295:858-861. [DOI] [PubMed] [Google Scholar]
- 40.Lin, S. F., D. R. Robinson, G. Miller, and H. J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol. 73:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lok, C. N., and P. Ponka. 1999. Identification of a hypoxia response element in the transferrin receptor gene. J. Biol. Chem. 274:24147-24152. [DOI] [PubMed] [Google Scholar]
- 42.Low, W., M. Harries, H. Ye, M. Q. Du, C. Boshoff, and M. Collins. 2001. Internal ribosome entry site regulates translation of Kaposi's sarcoma-associated herpesvirus FLICE inhibitory protein. J. Virol. 75:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu, M., J. Suen, C. Frias, R. Pfeiffer, M. H. Tsai, E. Chuang, and S. L. Zeichner. 2004. Dissection of the Kaposi's sarcoma-associated herpesvirus gene expression program by using the viral DNA replication inhibitor cidofovir. J. Virol. 78:13637-13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madan, A., and P. T. Curtin. 1993. A 24-base-pair sequence 3′ to the human erythropoietin gene contains a hypoxia-responsive transcriptional enhancer. Proc. Natl. Acad. Sci. USA 90:3928-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 47.Miller, G., L. Heston, E. Grogan, L. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyazaki, K., T. Kawamoto, K. Tanimoto, M. Nishiyama, H. Honda, and Y. Kato. 2002. Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J. Biol. Chem. 277:47014-47021. [DOI] [PubMed] [Google Scholar]
- 49.Moore, P. S., S. J. Gao, G. Dominguez, E. Cesarman, O. Lungu, D. M. Knowles, R. Garber, P. E. Pellett, D. J. McGeoch, and Y. Chang. 1996. Primary characterization of a herpesvirus agent associated with Kaposi's sarcomae. J. Virol. 70:549-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukhopadhyay, C. K., B. Mazumder, and P. L. Fox. 2000. Role of hypoxia-inducible factor-1 in transcriptional activation of ceruloplasmin by iron deficiency. J. Biol. Chem. 275:21048-21054. [DOI] [PubMed] [Google Scholar]
- 51.Ohh, M., C. W. Park, M. Ivan, M. A. Hoffman, T. Y. Kim, L. E. Huang, N. Pavletich, V. Chau, and W. G. Kaelin. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2:423-427. [DOI] [PubMed] [Google Scholar]
- 52.Park, J., D. Lee, T. Seo, J. Chung, and J. Choe. 2000. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) open reading frame 36 protein is a serine protein kinase. J. Gen. Virol. 81:1067-1071. [DOI] [PubMed] [Google Scholar]
- 53.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 54.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salceda, S., and J. Caro. 1997. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J. Biol. Chem. 272:22642-22647. [DOI] [PubMed] [Google Scholar]
- 56.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 73:1438-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schalling, M., M. Ekman, E. E. Kaaya, A. Linde, and P. Biberfeld. 1995. A role for a new herpes virus (KSHV) in different forms of Kaposi's sarcoma. Nat. Med. 1:707-708. [DOI] [PubMed] [Google Scholar]
- 58.Semenza, G. L. 2004. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology (Bethesda) 19:176-182. [DOI] [PubMed] [Google Scholar]
- 59.Semenza, G. L. 1999. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell Dev. Biol. 15:551-578. [DOI] [PubMed] [Google Scholar]
- 60.Semenza, G. L., B. H. Jiang, S. W. Leung, R. Passantino, J. P. Concordet, P. Maire, and A. Giallongo. 1996. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271:32529-32537. [DOI] [PubMed] [Google Scholar]
- 61.Semenza, G. L., and G. L. Wang. 1992. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12:5447-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 63.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Talbot, S. J., R. A. Weiss, P. Kellam, and C. Boshoff. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84-94. [DOI] [PubMed] [Google Scholar]
- 65.Tang, S., K. Yamanegi, and Z.-M. Zheng. 2004. Requirement of a 12-base-pair TATT-containing sequence and viral lytic DNA replication in activation of the Kaposi's sarcoma-associated herpesvirus K8.1 late promoter. J. Virol. 78:2609-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang, S., and Z. M. Zheng. 2002. Kaposi's sarcoma-associated herpesvirus K8 exon 3 contains three 5′-splice sites and harbors a K8.1 transcription start site. J. Biol. Chem. 277:14547-14556. [DOI] [PubMed] [Google Scholar]
- 67.Tian, H., S. L. McKnight, and D. W. Russell. 1997. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11:72-82. [DOI] [PubMed] [Google Scholar]
- 68.Wang, G. L., B. H. Jiang, E. A. Rue, and G. L. Semenza. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, G. L., and G. L. Semenza. 1993. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J. Biol. Chem. 268:21513-21518. [PubMed] [Google Scholar]
- 70.Wang, V., D. A. Davis, M. Haque, L. E. Huang, and R. Yarchoan. 2005. Differential gene up-regulation by hypoxia-inducible factor-1alpha and hypoxia-inducible factor-2alpha in HEK293T cells. Cancer Res. 65:3299-3306. [DOI] [PubMed] [Google Scholar]
- 71.Yu, F., S. B. White, Q. Zhao, and F. S. Lee. 2001. HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. USA 98:9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu, Y., J. B. Black, C. S. Goldsmith, P. J. Browning, K. Bhalla, and M. K. Offermann. 1999. Induction of human herpesvirus-8 DNA replication and transcription by butyrate and TPA in BCBL-1 cells. J. Gen. Virol. 80:83-90. [DOI] [PubMed] [Google Scholar]
- 73.Zheng, Z. M. 2003. Split genes and their expression in Kaposi's sarcoma-associated herpesvirus. Rev. Med. Virol. 13:173-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.